Abstract
Current therapy for cystic fibrosis (CF) focuses on minimizing the microbial community and the host’s immune response through the aggressive use of airway clearance techniques, broad-spectrum antibiotics, and treatments that break down the pervasive endobronchial biofilm. Antibiotic selection is typically based on the susceptibility of individual microbial strains to specific antibiotics in vitro. Often this approach cannot accurately predict medical outcomes because of factors both technical and biological. Recent culture-independent assessments of the airway microbial and viral communities demonstrated that the CF airway infection is considerably more complex and dynamic than previously appreciated. Understanding the ecological and evolutionary pressures that shape these communities is critically important for the optimal use of current therapies (in both the choice of therapy and timing of administration) and the development of newer strategies. The climax–attack model (CAM) presented here, grounded in basic ecological principles, postulates the existence of two major functional communities. The attack community consists of transient viral and microbial populations that induce strong innate immune responses. The resultant intense immune response creates microenvironments that facilitate the establishment of a climax community that is slower-growing and inherently resistant to antibiotic therapy. Newer methodologies, including sequence-based metagenomic analysis, can track not only the taxonomic composition but also the metabolic capabilities of these changing viral and microbial communities over time. Collecting this information for CF airways will enable the mathematical modeling of microbial community dynamics during disease progression. The resultant understanding of airway communities and their effects on lung physiology will facilitate the optimization of CF therapies.
Keywords: cystic fibrosis, airway ecology, metagenomics
Clinical Relevance
Newer technologies have provided a clearer picture regarding the complexity of airway infection in Cystic fibrosis (CF), demonstrating dynamic interactions between bacterial, viral, and fungal communities. Applying established ecological principles to the study of CF airway infection has produced important biological and pathophysiological insights into CF airway infection, and will facilitate the optimal use of current and future therapies.
Our view of cystic fibrosis (CF) airway infection is shifting from one based on the analysis of single microbial strains under laboratory conditions to one grounded in an ecological, systems-based assessment of entire communities interacting in situ with the host immune response. Laboratory studies of single pathogens provide valuable information about their potential roles in CF. However, colonization of the CF airway is not achieved by a single strain or species acting in isolation. The CF airway is a complex ecosystem hosting a heterogeneous community including bacteria, viruses, and fungi (1–3). Immigration (colonization) events, the order of arrival, and the ensuing interactions between microbial strains further complicate the work of the physician treating a patient with CF. Future breakthroughs in the control of infection in the CF lung will most likely come as a result of our increased understanding of the ecological and evolutionary processes shaping these microbial communities (4, 5).
Here we advocate for an ecological perspective that recognizes the chronic CF airway infection as a polymicrobial community, and that informs new treatments consistent with established ecological principles. In this light, CF infection is no longer viewed as the invasion of a single pathogen, such as Pseudomonas aeruginosa, that could be countered by the administration of antibiotics targeting that organism. Instead, disease is viewed as the result of dynamic changes in airway microbial populations, with the concomitant alteration of community physiology. Such a perspective focuses our attention on the factors that regulate microbial community stability and function, the nature of interactions between bacterial and phage populations, and the influences that control microbial populations. The well-studied responses of host innate and adaptive immune systems are now seen to be acting within this dynamic ecological theater and, in turn, to be eliciting evolutionary responses from the microbial community, all of which affects the health of the host.
This ecological perspective has important therapeutic implications, offering the prospect of novel approaches to treatment. For example, instead of antibiotics directly targeting a specific pathogen, the control of chronic airway infection may be achieved by disturbing some factor within the lung that is essential for the growth and persistence of pathogenic organisms, such as the presence of another community member, a nutrient, or another environmental attribute. Alternatively, the patient’s immune responses may be manipulated to favor a shift in the composition and function of the microbial community toward a less pathogenic state. Successful implementation requires the ability to predict and manipulate the outcomes of potential clinical treatments. As a step toward that goal, we propose a model, grounded in ecosystem concepts (6, 7), of the microbial communities within the lungs of patients with CF. Such a model can optimize the use of current therapies and also indicate potential novel therapeutic approaches. It also provides a framework for interpreting the results of past research and clinical experience, and can set directions for productive future investigations.
The Microbiota Of The Cf Lung Ecosystem
During CF infection, the lungs house a complex ecosystem consisting of dynamic communities of bacteria, fungi, and viruses, many (possibly most) of which remain to be characterized. The accumulation of mucus in the bronchial tree creates an environment conducive to colonization by various opportunistic pathogens. Initially the community is dominated by aerobes, succeeded later by facultative and strictly anaerobic microbial populations (8). Thus, in younger patients with CF, Staphylococcus aureus, Haemophilus influenzae, and P. aeruginosa are common, whereas in older patients, P. aeruginosa dominates the ecosystem, often accompanied by other pathogens such as S. aureus, Burkholderia cepacia complex, Stenotrophomonas maltophilia, and Achromobacter spp. Recently, large numbers of anaerobic bacteria were identified in the sputum of patients with CF, although their clinical significance remains uncertain (9). Moreover, nontuberculous Mycobacterium species have been isolated in patients with CF. Their frequency, ranging from less than 10% to greater than 20% in different regions of the United States, may reflect the varying environmental prevalence of these organisms (10–13). Among the fungi present, Aspergillus spp. and Candida spp. are most frequently observed, but more drug-resistant species, such as Scedosporium azoospermia, are becoming common (14). The viruses include both those that potentially infect humans as well as bacteriophages (phages) that prey upon the bacterial community (15).
In the past 5 years, culture-independent assessments of the microbial and viral communities in CF airway secretions (sputum and bronchial alveolar lavage fluid) have revealed a more varied community than had been evident using standard microbial culture techniques. Based on the 16S ribosomal RA (rRNA) sequences present, this approach has shown not only that the microbial communities are more diverse than previously appreciated, but that this diversity may peak during adolescence, before significant structural remodeling of the lung (16, 17). Recent metagenomic studies characterizing the microbial and viral communities within lung tissue dissected from distinct lung regions have added the spatial dimension, reporting that different lobes housed distinctly different communities, thus further complicating clinical treatment (15, 18).
Control Of Microbial Populations
Effective, ecologically based CF treatment could minimize lung damage and loss of function by reducing or altering microbial communities in the lung. Current clinical therapies, such as airway clearance therapy and the administration of antibiotics, directly (but often indiscriminately) decrease microbial populations. The human immune system also exerts powerful top-down control of microbes in the lung, but its actions have detrimental long-term repercussions. Although it protects the lungs in the short term by killing microbes, the immune response maintains ongoing inflammation during chronic CF airway infections. This, in turn, leads to airway remodeling and the creation of a spatial structure with ensuing effects on microbial diversity and community stability that are likely to make microbial populations more difficult to eradicate (19).
Lung bacterial populations are also subject to top-down control by phages. Phages comprise the most abundant entities in all characterized ecosystems, with typically about 10 phages for every bacterial cell. Because most phages quickly kill their bacterial hosts, they are responsible for significant bacterial mortality. Unlike other agents of top-down control, phages are selective predators, each type infecting only related strains or species of bacteria. Thus, their top-down control also affects the relative abundances of bacterial populations (20). Phages have been isolated from various sites within the human body, including the airway secretions (20–25) and lung tissue (15) of patients with CF. Significantly, phages were absent from late stage biofilms in CF lungs, suggesting that biofilm formation may help bacteria escape this top-down control. Further characterization of the phage community and its impact on bacteria present in the CF lung can be expected to provide new insights into the top-down control of the infection, including the potential use of phages for novel therapeutic options (26).
Other controls, termed “bottom-up,” limit the growth and reproduction of microbial populations by limiting or partitioning essential resources. The CF airway provides a wide range of nutrients including amino acids, phospholipids, DNA, and a range of sugars (27), and any of these resources may fluctuate across space and time. This chemical complexity, along with spatial heterogeneity, affords ecological opportunity to bottom-up controls. Such resident controls are likely critical to the emergence and maintenance of microbial diversity in the CF lung (19, 28, 29). The potential therapeutic use of bottom-up controls may arise in the form of a low concentration of a limiting micronutrient or the inhibition of a critical nutrient pathway in a specific environment. For example, the inhibition of nitric oxide synthase in the CF airway may create a shortage of nitrite and nitrate, both of which are critical for the growth of denitrifying microbial populations (30).
Landscape Ecology
Landscape ecology considers the impacts of a heterogeneous environment on the organisms that reside therein. In the CF lung, this environmental heterogeneity is evident in the patchy distribution of resources, the spatial structuring of the lung over distances both small and large, and regional differences in microbial communities (18). A heterogeneous environment affords ecological opportunity. Provided that resources are limited, ecologically diverse populations will evolve (30, 31). A heterogeneous environment also supports the maintenance of this diversity (19, 32). In contrast, a homogeneous environment limits both the opportunity for diversification and the level of diversity that can be maintained.
Interactions among component organisms are central to the functioning of any microbial community (33–36). For example, excreted metabolites provide opportunities for both cooperative (syntrophic) and antagonistic (allelopathic) interactions. Small signaling molecules, such as the homoserine lactones or 4-hydroxy-2-heptylquinoline-N-oxide, allow populations to coordinate activities in a variety of antagonistic or mutually beneficial ways (37, 38). In the context of the CF lung, a full range of interactions is likely. The extent and nature of these interactions will reflect the degree of environmental heterogeneity as well as the level of connectivity that allows for the movement of organisms or resources from place to place.
How heterogeneous and independent are macroenvironments within the CF lung? Although each lobe constitutes a spatially distinct region, and the bronchioles are clearly separate spatially and physiologically from alveoli, the question remains, to what extent do microbes experience these as distinct niches? In other words, does the lung provide a well-mixed environment with resources and organisms dispersed uniformly throughout, or is it a cluster of isolated niches? Understanding the extent of connectivity between populations within a single lung will influence how we think about the emergence of antibiotic resistant strains, the mucoid phenotype, and mutator strains within lung populations. Of these three factors, the evolution of antibiotic resistance is currently the most pressing issue. If connectivity between lobes is high, then physicians would be right to be concerned about the detection of an antibiotic-resistant bacterial strain that could rapidly spread throughout the lung, supplanting susceptible types when under selection pressure from antibiotic treatment. If, instead, the lung is spatially structured with little connectivity between niches, then an antibiotic-resistant bacterial strain evolving, for example, in the right lower lobe would be unlikely to have an opportunity to colonize the right upper lobe. Because phages in the CF lung harbor a reservoir of antibiotic resistance genes (18), the phage-mediated spread of antibiotic resistance from lobe to lobe may also be important.
Cf Disease States As Alternate Stable States
In any ecosystem, many combinations of biological and environmental conditions can yield the same recognizable community, referred to as a stable state (or an attractor). When a resilient ecosystem is perturbed, it recovers and returns to its original “stable” state. However, a stronger perturbation may shift an ecosystem into an alternative (and also stable) state that maintains a different community of organisms. Restoration ecology seeks to identify the perturbations that cause ecosystems to shift from one state to another, and then use appropriate perturbations to restore ecosystems to their more desirable state.
This same restoration approach can be applied to the CF lung. When a CF airway infection is viewed as an ecosystem, two alternative states, in the simplest configuration, are evident: a quiescent, clinically stable state, and an exacerbated state. The quiescent state resembles an ecosystem that is dominated by stable populations that are well-adapted to their particular biological niche. Perturbations in the local environment that alter top-down or bottom-up population controls can alter this stable community, possibly shifting it to an alternative state.
The Climax–Attack Model Of The Cf Lung Ecosystem
The ecological principles already discussed suggest numerous potential therapeutic strategies that may prove efficacious in shifting the CF community to a less pathogenic state. However, each tactic needs to be evaluated in advance to determine its optimal mode of application, and to predict possible adverse effects. Mathematical modeling of the interactions between the CF microbial community and environmental factors can be used to test potential therapies. For instance, modeling may suggest that a particular therapy could be more effective in reducing overall microbial abundance when applied at a particular stage in the disease cycle, or it may identify therapies that can shift the CF airway ecosystem from a sick, exacerbated state to a healthier quiescent state. In this manner, the coupling of ecological modeling with methods from microbial ecology can lead us to better control of CF airway infections.
The Climax And Attack Communities
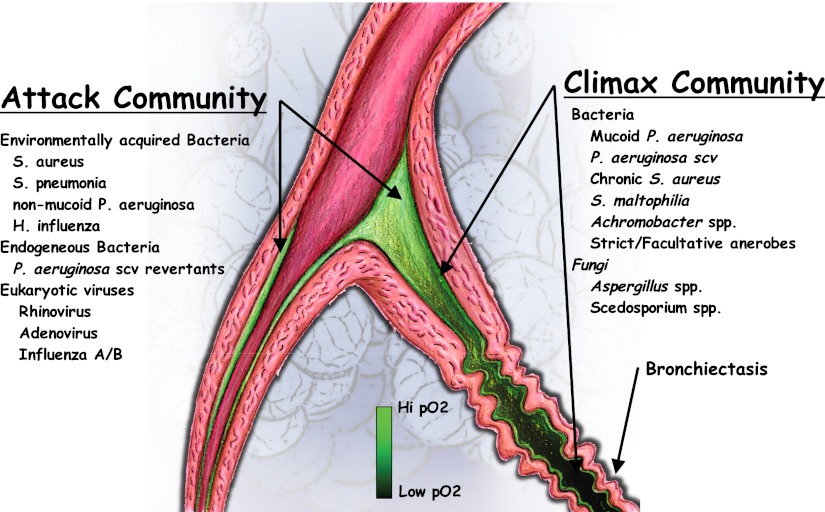
Based on current ecological models and knowledge of the human airway microbial community, we posit the existence of two broad functional classes of CF microbial communities: a virulent, transient attack community that is associated with pulmonary exacerbations, and a climax community that dominates during relatively stable periods (Figure 1). Our proposed ecological climax–attack model (CAM) is based on the abundant, persistent “keystone” populations that characterize the microbial communities present in the quiescent and the exacerbated states (3, 39–42). Both attack and climax communities can coexist in a given patient; sequential attack communities dominate the airway microbiota before significant airway remodeling, whereas climax communities prevail in heavily remodeled airways.
Figure 1.
Proposed climax–attack model for cystic fibrosis. The attack communities consist of pathogens such as Streptococcus spp., Staphylococcus spp., and eukaryotic viruses. These communities elicit strong immune responses and scarring. The scar tissue is then colonized by the climax community, which consists of the facultative anaerobe Pseudomonas aeruginosa (P. aeruginosa) at the periphery and strict anaerobes in the central region.
In the climax–attack community model, a taxonomic unit such as P. aeruginosa, as identified in the microbiology laboratory or by culture-independent means (e.g., 16S rRNA sequencing), can belong to both communities. For this reason, our model focuses not only on taxonomic characterizations, but importantly on functional differences. These two communities are not the same as the statistically defined “core” and “satellite” communities previously proposed (3, 43), but are defined by their metabolic capabilities, as determined by metagenomic, metatranscriptomic, and functional microbiological studies. By sampling an attack or climax community and then sequencing the DNA from the microbes present, one generates a metagenome, that is, a set of gene sequences for that community, some of which can be assigned a known function. On this basis, it is appropriate to define an attack metagenome as a microbial metagenome that is enriched in the virulence genes that elicit intense innate immune responses and subsequently initiate airway remodeling. Similarly, a climax metagenome is enriched in genes that promote the chronic colonization of CF airways, elicit the antibody-dependent adaptive immune response, and decrease antibiotic susceptibility. Metagenomic analysis is particularly advantageous here, because it does not limit investigation to identified cultured microbes. Instead, it enables the characterization of metabolic capabilities of the entire microbial community within an individual patient, a community that is likely to differ in detail over time and between patients. A critical task facing CF-related metagenomic studies involves characterizing the attack and climax metagenomes, identifying the changes that occur over time, and correlating these changes with their clinical context, including the therapies administered.
Recent studies demonstrated that the sputum during many pulmonary exacerbations is dominated by bacterial taxa in the airways that are similar to those taxa in a clinically stable state (42). The climax–attack model is consistent with these findings, because changes in the communities are functional, and are therefore not necessarily identified by standard microbiological assessments or 16S rRNA sequence taxonomic assessments. For example, acute pulmonary exacerbations can result from aggressively growing revertants derived from slowly growing, endogenous microbial populations, or from a change in bacterial gene expression eliciting stronger immune responses (44, 45) (Floyd Romesberg, personal communication). In either instance, the signs and symptoms of a pulmonary exacerbation could result without major changes in the dominating taxa during a more stable period. In other cases, the attack communities associated with exacerbations are predicted to be dynamic, and are frequently acquired through environmental exposure or aspiration, and include populations such as Streptococcus spp., Staphylococcus spp., nonmucoid Pseudomonas spp., and eukaryotic viruses (e.g., influenza and rhinoviruses). These environmentally acquired strains are more obvious in early disease, before established lung remodeling, but are certainly known to be associated with succession events in patients with more advanced airway remodeling. In all cases, the attack community may express a significant complement of wild-type virulence factors, elicit strong responses from the innate immune system, and produce sudden illnesses and abrupt decline in lung health. The resultant clinically observed physiological changes are generally limited in time and space, and can be reversed by successful therapy. These changes result from both the immune response and the growth of biofilm in the airways, and include dyspnea, the production of sputum, coughing, and systemic signs of inflammation. Remodeling is limited by the transient nature of the community and by successful exacerbation therapies. These dynamic attack communities dominate the conducting airways of patients with early-stage CF before significant airway remodeling. However, their fast growth and immunogenic nature also enable them to initiate and propagate permanent airway remodeling, thereby creating the microenvironments that allow for the establishment of a chronic climax community (46).
In contrast, the model predicts that climax communities are composed of persistent, slow-growing microbial and fungal “keystone” populations whose metabolisms are well adapted to the CF airway microenvironment (31, 39, 44, 47). These organisms include commonly cultured microbial populations such as mucoid Pseudomonas spp. (46), chronic Staphylococcus spp., S. maltophilia, Achromobacter spp., and other strict anaerobes. Island biogeography theory (48), as supported by recent experiments (49), shows that community membership, function, and evolution also depend on the immigration history, including the sequence and timing of arrival of each organism.
The climax community thrives in remodeled airways, ensconced within biofilms of their own making that generally provide protection from the patient’s immune response. Although this community does not cause sudden, transient drops in lung function, together with the attack community it is responsible for the steady progressive loss of lung function over time. In most cases, the pace of airway remodeling by the climax community drives the long-term prognosis because of its prolonged stimulation of innate and adaptive immune responses, particularly evident when dominated by the mucoid P. aeruginosa or B. cepacia complex. For example, the destructive antibody response to the Pseudomonas-produced alginate is a major determinant of lung function in patients with CF (44). The climax community is generally resistant to standard antibiotic therapy, and is best controlled by airway clearance therapies (e.g., inhaled hypertonic saline or inhaled recombinant human DNase) and, to a certain extent, by very high concentrations of inhaled antibiotics. Its rapid expansion is facilitated by the frequent exacerbations caused by attack communities, by noncompliance with chronic therapies, or conceivably by the loss of other top-down controls (e.g., reduced lytic phage populations, and impaired immune system function). Although attack communities are usually associated with pulmonary exacerbations, a slow, unimpeded expansion of the climax community because of medical nonadherence is also associated with signs and symptoms of pulmonary exacerbations.
Cam And Clinical Therapies
The CAM can inform the development of effective clinical protocols for different stages of disease. For example, early in a patient’s life, the climax community is small or nonexistent. The microbial, viral, and fungal populations are primarily those characteristic of attack communities. Eradicating these early infections is important to minimize their impact and to delay the development of a climax community. At some point, however, the cumulative damage allows for the establishment of a climax community. Based on the CAM, one can predict that treatments will exert different effects on the two communities. For example, the attack community would be expected to be more susceptible to antibiotics. Thus, the broad-spectrum antibiotics used to kill susceptible members of an attack community will select and favor the growth of the nonsusceptible microbes present.
Based on the CAM, we would expect various possible etiologies for CF pulmonary exacerbations. This model enables us to relate exacerbations to various factors such as a patient’s clinical history, physiology, and airway community function, as revealed by metagenomic analysis. For example, expansion of the climax community would be predicted to occur during periods of therapeutic noncompliance or inefficacious therapy in patients with significant preexisting airway remodeling. A sharp decline in lung function because of the acquisition of virulent environmental strains would be predicted to create a metagenomic functional profile distinct from that during exacerbations attributable to secondary viral infections, the inhalation of toxic substances (e.g., smoke or dust), or other causes.
Mathematical Models Of Microbial Communities In The Cf Lung
The ecological Climax-Attack Model (CAM) provides a framework for modeling specific aspects of the CF airway disease. Such models assist in the interpretation and synthesis of prior observations and form a coherent basis for the development and evaluation of new therapies. Any useful model must address both the cyclical nature of the disease progression and the dynamic spatial heterogeneity of disease states throughout the lung. We discuss three approaches to modeling CF lung disease: statistical models that extract significant correlations from the CF Foundation Patient Registry; physiological models that correlate the degree of airway remodeling with decreasing lung function and increasing age; and community dynamics (ecological) models that express the spatially and temporally complex interactions between the host, the bacteria, and the phage.
The Statistical Model
The CF Foundation Patient Registry and individual case data provide excellent sources for longitudinal data that include repeated observations of the same individuals. For example, registry data have been used to examine the effects of age and gender as predictors of future forced expiratory volumes in 1 second (FEV1) (50). As a first step toward developing this model, data mining and exploratory analyses of registry data would be used to classify cases based on the severity of disease, and to identify key factors driving CF lung disease within each class. As one example, this approach can identify the variables that best predict changes in FEV1 over time. A similar study of pediatric patients with CF using European patient data examined many variables. However, derived variables such as regression coefficients were not considered. The process of building a linear mixed model such as this for longitudinal data is iterative and requires a series of model-fitting steps and repeated searching for the most significant correlations among data (51). This strategy balances statistical and biomedical considerations with the goal of identifying disease classes and the most physiologically significant factors in each class.
The Physiological Model
A physiological model tracks the persistent effects of an attack community (e.g., scarring attributable to the host inflammatory response) and the growth and spatial distribution of a climax community by modeling biofilm expansion, increased mucus plugging, and altered airway function over time. The model is based on the fractal structure of the lung (52–54). It assumes that the rate of mucus accretion in an infected bronchiole is a constant characteristic of the local attack community. When the accumulating mucus fills the bronchiole, the passage becomes blocked, thus making all distal lung volumes inaccessible and thereby altering the FEV1 value. In this model, the distal bronchioles also become infected because of the branching tree design of the airways (55–57). Formally, time delay differential equations are used (58), with switching at certain values of key variables such as mucus volume and scarring.
To provide the best match between the model and clinical observations, values are determined for several parameters: the probability of onset of infection at any site, the rate of mucus buildup, the scarring rate, and the scarring threshold for the onset of irreversible restructuring. In the simplest version of this model, treatments reset the mucus volume of the infected regions to zero unless scarring has exceeded the scarring threshold at time of treatment. Simulations can then be run, and their predicted outcomes can be compared with the data for the patient FEV1 responses available in lung function databases. The model is then adjusted and the cycle repeated for several iterations (59).
The Community Dynamics Model
This approach involves the challenging task of integrating several factors to predict clinically significant changes in microbial populations. These interacting factors include: (1) host immune responses and clinical therapies, (2) the spatial heterogeneity of CF airways, (3) the transitioning of aerobic microenvironments that support a rapidly growing planktonic attack community to anaerobic niches populated by slowly growing climax populations within biofilms, and (4) phage predation. Accurate modeling of advanced, chronically infected airways of patients with CF will require validated submodels that represent each of these four factors. Currently, we lack sufficient empirical data to develop and test a comprehensive model. However, we can approach the modeling of phage predation using models already developed for predator–prey interactions, and this submodel can serve as an example. The populations of predators (e.g., phages) and prey (e.g., bacteria) are often observed to vary cyclically. An increased prey population supports more predators; more predators reduce the number of prey; and fewer prey cause a crash in the predator population, allowing the prey population to rebound. These dynamic fluctuations have been simulated for phages and bacteria, using generalized Lotka–Volterra modeling based on a pair of linear differential equations. Applying this model to the CF lung is complicated by local variation in the concentrations of phage or bacteria because of microbial-scale spatial heterogeneity, as well as variations over time because of the effects of the immune system and clinical therapies.
Implications for Clinical Treatment
Each of our three modeling approaches quantitatively describes important aspects of disease progression in patients with CF. The models are interrelated insofar as one model predicts values for parameters used by another. Together, they provide a valuable adjunct to clinical treatment. It is neither possible nor desirable to perform all available treatments, in various combinations and each with varied timing of administration, on all patients. When a patient with CF presents, a physician must recommend a treatment protocol, perhaps choosing one antibiotic from among five possibilities, each with different treatment schedules. This is where the models become particularly valuable, because essentially all possibilities can be explored by computer simulations to predict which regimen would most likely increase the patient’s FEV1 most quickly.
However, the ability of these models to describe the CF lung ecosystem and to predict its behavior depends on the assignment of verified values to the parameters involved. To that end, we suggest that the acquisition of certain data for airways and lung tissues in CF is essential: (1) population dynamics for microbial communities; (2) the distribution of microbial taxonomic and metabolic diversity across space and time; (3) nutrient sources for microbial communities; (4) growth rates of specific microbes; (5) the types and numbers of viruses; and (6) the rates of phage-mediated mortality of microbes. With values for these variables, modeling the CF lung microbial–viral community will be possible, using a variety of standard and proposed ecological models (S. Zarei, unpublished results).
Microbial ecologists have already developed the methods needed to assign values for these parameters. The next step involves applying them to the CF lung. Initial studies using metagenomic analyses of lung microbial communities have been reported (1, 3, 5, 15, 17, 18). Continuing investigations can provide critical information on the population dynamics and community structure of microbial airway communities in health and disease. There is a reasonable expectation that such studies may identify an as-yet-unrecognized Achilles’ heel within the pathogenic microbial populations of CF lungs. Metagenomic studies also can provide information on the natural history of viral communities in the CF lung, an essentially unexplored topic. Of particular relevance are the complex relationships between bacteria and their phage predators, as well as the possibility that viruses pathogenic to humans may be associated with the progression of lung disease.
Conclusions
The recent methodological developments that spurred rapid advances in microbial ecology also provide new opportunities for understanding and controlling disease states in the CF lung. We propose an ecological model of the diseased lung that identifies two interrelated microbial communities: the aggressive attack community, and the more stable climax community. The ecological approach that we advocate considers the interplay of CF airway microbial and viral communities. Modeling these interactions offers the potential to predict the effects of therapeutic interventions, thus dispelling much of the current therapeutic empiricism and leading to a more effective use of CF therapies. Lastly, applying the principles of restoration ecology may lead to new therapeutic approaches designed to shift CF airway communities toward a less pathogenic state.
Supplementary Material
Acknowledgments
The authors thank Angela Wang and Mike Furlan for their critical reading and comments on the manuscript, and Sean Needham for the medical illustration.
APPENDIX: DEFINITIONS
Bottom-up population control: The control of the abundance of organisms by limited resources (e.g., nutrients) or environmental factors (e.g., oxygen concentration).
Landscape ecology: The study of the spatial heterogeneity of biotic and abiotic factors in an ecosystem, focusing on the accompanying interactions and their consequences for ecological processes.
Metagenomics: The culture-independent genomic analysis of an assemblage of microorganisms collected from an environment (e.g., the human gut and the CF lung). Phylogenetic typing discloses “who is there,” whereas functional metagenomics reveal the metabolic capabilities of the entire sampled community (i.e., “What are they doing?”).
Microbes: Bacteria and archaea. Although archaea play important roles in biogeochemical cycles, none are known to be pathogens.
Microbiota: The microscopic organisms inhabiting a particular environment, including bacteria, archaea, viruses, and fungi.
Phage (or bacteriophage): A virus that infects bacteria.
Top-down population control: The control of the abundance of organisms by predation. For microbial populations in CF airways, top-down control is effected by bacteriophages and the immune system.
Footnotes
This work was supported by grant 55676 from the Cystic Fibrosis Research, Inc., and by National Institutes of Health grant RO1 56586A (F.R.).
Originally Published in Press as DOI: 10.1165/rcmb.2012-0059PS on October 25, 2012
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Rogers GB, Carroll MP, Serisier DJ, Hockey PM, Jones G, Bruce KD. Characterization of bacterial community diversity in cystic fibrosis lung infections by use of 16S ribosomal DNA terminal restriction fragment length polymorphism profiling. J Clin Microbiol 2004;42:5176–5183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Willner D, Furlan M, Haynes M, Schmieder R, Angly FE, Silva J, Tammadoni S, Nosrat B, Conrad D, Rohwer F. Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PLoS ONE 2009;4:e7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van der Gast CJ, Walker AW, Stressmann FA, Rogers GB, Scott P, Daniels TW, Carroll MP, Parkhill J, Bruce KD. Partitioning core and satellite taxa from within cystic fibrosis lung bacterial communities. ISME J 2011;5:780–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harrison F. Microbial ecology of the cystic fibrosis lung. Microbiology 2007;153:917–923 [DOI] [PubMed] [Google Scholar]
- 5.Klepac-Ceraj V, Lemon KP, Martin TR, Allgaier M, Kembel SW, Knapp AA, Lory S, Brodie EL, Lynch SV, Bohannan BJ, et al. Relationship between cystic fibrosis respiratory tract bacterial communities and age, genotype, antibiotics and Pseudomonas aeruginosa. Environ Microbiol 2010;12:1293–1303 [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez-Brito B, Li L, Wegley L, Furlan M, Angly F, Breitbart M, Buchanan J, Desnues C, Dinsdale E, Edwards R, et al. Viral and microbial community dynamics in four aquatic environments. ISME J 2010;4:739–751 [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez-Valera F, Martin-Cuadrado AB, Rodriguez-Brito B, Pasic L, Thingstad TF, Rohwer F, Mira A. Explaining microbial population genomics through phage predation. Nat Rev Microbiol 2009;7:828–836 [DOI] [PubMed] [Google Scholar]
- 8.Lipuma JJ. The changing microbial epidemiology in cystic fibrosis. Clin Microbiol Rev 2010;23:299–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tunney MM, Field TR, Moriarty TF, Patrick S, Doering G, Muhlebach MS, Wolfgang MC, Boucher R, Gilpin DF, McDowell A, et al. Detection of anaerobic bacteria in high numbers in sputum from patients with cystic fibrosis. Am J Respir Crit Care Med 2008;177:995–1001 [DOI] [PubMed] [Google Scholar]
- 10.Ziedalski TM, Kao PN, Henig NR, Jacobs SS, Ruoss SJ. Prospective analysis of cystic fibrosis transmembrane regulator mutations in adults with bronchiectasis or pulmonary nontuberculous mycobacterial infection. Chest 2006;130:995–1002 [DOI] [PubMed] [Google Scholar]
- 11.Forslow U, Geborek A, Hjelte L, Petrini B, Heurlin N. Early chemotherapy for non-tuberculous mycobacterial infections in patients with cystic fibrosis. Acta Paediatr Scand 2003;92:910–915 [DOI] [PubMed] [Google Scholar]
- 12.Olivier KN, Weber DJ, Wallace RJ, Jr, Faiz AR, Lee JH, Zhang Y, Brown-Elliot BA, Handler A, Wilson RW, Schechter MS, et al. Nontuberculous mycobacteria: I. Multicenter prevalence study in cystic fibrosis. Am J Respir Crit Care Med 2003;167:828–834 [DOI] [PubMed] [Google Scholar]
- 13.Olivier KN. The natural history of nontuberculous mycobacteria in patients with cystic fibrosis. Paediatr Respir Rev 2004;5 (Suppl. A):S213–216 [DOI] [PubMed] [Google Scholar]
- 14.Marshall B, Hazel L. Cystic Fibrosis Foundation registry report: annual data report. Bethesda, MD: Cystic Fibrosis Foundation; 2010 [Google Scholar]
- 15.Willner D, Haynes MR, Furlan M, Hanson N, Kirby B, Lim YW, Rainey PB, Schmieder R, Youle M, Conrad D, et al. Case studies of the spatial heterogeneity of DNA viruses in the cystic fibrosis lung. Am J Respir Cell Mol Biol 2012;46:127–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cox MJ, Allgaier M, Taylor B, Baek MS, Huang YJ, Daly RA, Karaoz U, Andersen GL, Brown R, Fujimura KE, et al. Airway microbiota and pathogen abundance in age-stratified cystic fibrosis patients. PLoS ONE 2010;5:e11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris JK, De Groote MA, Sagel SD, Zemanick ET, Kapsner R, Penvari C, Kaess H, Deterding RR, Accurso FJ, Pace NR. Molecular identification of bacteria in bronchoalveolar lavage fluid from children with cystic fibrosis. Proc Natl Acad Sci USA 2007;104:20529–20533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Willner D, Haynes MR, Furlan M, Schmieder R, Lim YW, Rainey PB, Rohwer F, Conrad D. Spatial distribution of microbial communities in the cystic fibrosis lung. ISME J 2012;6:471–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rainey PB, Buckling A, Kassen R, Travisano M. The emergence and maintenance of diversity: insights from experimental bacterial populations. Trends Ecol Evol 2000;15:243–247 [DOI] [PubMed] [Google Scholar]
- 20.Lennon JT, Martiny JB. Rapid evolution buffers ecosystem impacts of viruses in a microbial food web. Ecol Lett 2008;11:1178–1188 [DOI] [PubMed] [Google Scholar]
- 21.Goerke C, Wolz C. Regulatory and genomic plasticity of Staphylococcus aureus during persistent colonization and infection. Int J Med Microbiol 2004;294:195–202 [DOI] [PubMed] [Google Scholar]
- 22.Hanlon GW, Denyer SP, Olliff CJ, Ibrahim LJ. Reduction in exopolysaccharide viscosity as an aid to bacteriophage penetration through Pseudomonas aeruginosa biofilms. Appl Environ Microbiol 2001;67:2746–2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iacocca VF, Sibinga M, Barbero GJ. Respiratory tract bacteriology in cystic fibrosis. Am J Dis Child 1963;106:315–324 [DOI] [PubMed] [Google Scholar]
- 24.Ojeniyi B. Bacteriophages in sputum of cystic fibrosis patients as a possible cause of in vivo changes in serotypes of Pseudomonas aeruginosa. APMIS 1988;96:294–298 [DOI] [PubMed] [Google Scholar]
- 25.Sekaninova G, Kolarova M, Pillich J, Semenka J, Slavikova H, Kubickova D, Zajicova V. Pseudomonas aeruginosa phage lysate as an immunobiological agent: 1. Selection of Pseudomonas aeruginosa clinical strains for phage lysate preparation. Folia Microbiol (Praha) 1999;44:93–97 [DOI] [PubMed] [Google Scholar]
- 26.Carmody LA, Gill JJ, Summer EJ, Sajjan US, Gonzalez CF, Young RF, LiPuma JJ. Efficacy of bacteriophage therapy in a model of Burkholderia cenocepacia pulmonary infection. J Infect Dis 2010;201:264–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Son MS, Matthews WJ, Jr, Kang Y, Nguyen DT, Hoang TT. In vivo evidence of Pseudomonas aeruginosa nutrient acquisition and pathogenesis in the lungs of cystic fibrosis patients. Infect Immun 2007;75:5313–5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacLean RC, Bell G. Experimental adaptive radiation in Pseudomonas. Am Nat 2002;160:569–581 [DOI] [PubMed] [Google Scholar]
- 29.Rainey PB, Travisano M. Adaptive radiation in a heterogeneous environment. Nature 1998;394:69–72 [DOI] [PubMed] [Google Scholar]
- 30.Marozkina NV, Gaston B. Nitrogen balance in the ecosystem of the cystic fibrosis lung. Am J Respir Crit Care Med 2011;183:1290–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schluter D. The ecology of adaptive radiation. Oxford: Oxford University Press; 2000 [Google Scholar]
- 32.Nguyen D, Singh PK. Evolving stealth: genetic adaptation of Pseudomonas aeruginosa during cystic fibrosis infections. Proc Natl Acad Sci USA 2006;103:8305–8306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sutherland IW, Hughes KA, Skillman LC, Tait K. The interaction of phage and biofilms. FEMS Microbiol Lett 2004;232:1–6 [DOI] [PubMed] [Google Scholar]
- 34.Torsvik V, Ovreas L, Thingstad TF. Prokaryotic diversity: magnitude, dynamics, and controlling factors. Science 2002;296:1064–1066 [DOI] [PubMed] [Google Scholar]
- 35.Wommack KE, Ravel J, Hill RT, Chun J, Colwell RR. Population dynamics of Chesapeake Bay virioplankton: total-community analysis by pulsed-field gel electrophoresis. Appl Environ Microbiol 1999;65:231–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hansen SK, Rainey PB, Haagensen JA, Molin S. Evolution of species interactions in a biofilm community. Nature 2007;445:533–536 [DOI] [PubMed] [Google Scholar]
- 37.Hoffman LR, Deziel E, D’Argenio DA, Lepine F, Emerson J, McNamara S, Gibson RL, Ramsey BW, Miller SI. Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proc Natl Acad Sci USA 2006;103:19890–19895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duerkop BA, Ulrich RL, Greenberg EP. Octanoyl-homoserine lactone is the cognate signal for Burkholderia mallei BmaR1–BmaI1 quorum sensing. J Bacteriol 2007;189:5034–5040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang L, Jelsbak L, Marvig RL, Damkiaer S, Workman CT, Rau MH, Hansen SK, Folkesson A, Johansen HK, Ciofu O, et al. Evolutionary dynamics of bacteria in a human host environment. Proc Natl Acad Sci USA 2011;108:7481–7486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Magurran AE. Species abundance distributions over time. Ecol Lett 2007;10:347–354 [DOI] [PubMed] [Google Scholar]
- 41.Magurran AE, Henderson PA. Explaining the excess of rare species in natural species abundance distributions. Nature 2003;422:714–716 [DOI] [PubMed] [Google Scholar]
- 42.Zhao J, Schloss PD, Kalikin LM, Carmody LA, Foster BK, Petrosino JF, Cavalcoli JD, VanDevanter DR, Murray S, Li JZ, et al. Decade-long bacterial community dynamics in cystic fibrosis airways. Proc Natl Acad Sci USA 2012;109:5809–5814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ulrich W, Zalewski M. Abundance and co-occurrence patterns of core and satellite species of ground beetles on small lake islands. Oikos 2006;114:338–348 [Google Scholar]
- 44.Bjarnsholt T, Jensen PO, Fiandaca MJ, Pedersen J, Hansen CR, Andersen CB, Pressler T, Givskov M, Hoiby N. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr Pulmonol 2009;44:547–558 [DOI] [PubMed] [Google Scholar]
- 45.Rogers GB, Carroll MP, Bruce KD. Enhancing the utility of existing antibiotics by targeting bacterial behaviour? Br J Pharmacol 2012;165:845–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Worlitzsch D, Tarran R, Ulrich M, Schwab U, Cekici A, Meyer KC, Birrer P, Bellon G, Berger J, Weiss T, et al. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J Clin Invest 2002;109:317–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D’Argenio DA, Miller SI, Ramsey BW, Speert DP, Moskowitz SM, et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci USA 2006;103:8487–8492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.MacArthur R, Wilson E. The theory of island biogeography. Princeton: Princeton University Press; 1967 [Google Scholar]
- 49.Fukami T, Beaumont HJ, Zhang XX, Rainey PB. Immigration history controls diversification in experimental adaptive radiation. Nature 2007;446:436–439 [DOI] [PubMed] [Google Scholar]
- 50.Edwards LJ. Modern statistical techniques for the analysis of longitudinal data in biomedical research. Pediatr Pulmonol 2000;30:330–344 [DOI] [PubMed] [Google Scholar]
- 51.Kraemer R, Blum A, Schibler A, Ammann RA, Gallati S. Ventilation inhomogeneities in relation to standard lung function in patients with cystic fibrosis. Am J Respir Crit Care Med 2005;171:371–378 [DOI] [PubMed] [Google Scholar]
- 52.Anderson JC, Babb AL, Hlastala MP. A fractal analysis of the radial distribution of bronchial capillaries around large airways. J Appl Physiol 2005;98:850–855 [DOI] [PubMed] [Google Scholar]
- 53.Glenny RW, Bernard SL, Robertson HT. Pulmonary blood flow remains fractal down to the level of gas exchange. J Appl Physiol 2000;89:742–748 [DOI] [PubMed] [Google Scholar]
- 54.West BJ. Physiology in fractal dimensions: error tolerance. Ann Biomed Eng 1990;18:135–149 [DOI] [PubMed] [Google Scholar]
- 55.Kitaoka H, Suki B. Branching design of the bronchial tree based on a diameter–flow relationship. J Appl Physiol 1997;82:968–976 [DOI] [PubMed] [Google Scholar]
- 56.Parker JC, Cave CB, Ardell JL, Hamm CR, Williams SG. Vascular tree structure affects lung blood flow heterogeneity simulated in three dimensions. J Appl Physiol 1997;83:1370–1382 [DOI] [PubMed] [Google Scholar]
- 57.Weibel E. Design of airways and blood vessels considered as branching trees. : Crystal RG, West JB, editors The lung: scientific foundations. New York: Raven Press; 1991. pp. 711–720 [Google Scholar]
- 58.Guckenheimer J, Holmes P. Nonlinear oscillations, dynamical systems, and bifurcations of vector fields. New York: Springer-Verlag; 1983 [Google Scholar]
- 59.Maki D, Thompson M. Mathematical modeling and computer simulation. Pacific Grove, CA: Thomson Brooks/Cole; 2006 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.