Abstract
Taste buds are found in a distributed array on the tongue surface, and are innervated by cranial nerves that convey taste information to the brain. For nearly a century, taste buds were thought to be induced by nerves late in embryonic development. However, this view has shifted dramatically. A host of studies now indicate that taste bud development is initiated and proceeds via processes that are nerve-independent, occur long before birth, and governed by cellular and molecular mechanisms intrinsic to the developing tongue. Here we review the state of our understanding of the molecular and cellular regulation of taste bud development, incorporating important new data obtained through the use of two powerful genetic systems, mouse and zebrafish.
Keywords: Mouse, zebrafish, molecular genetics, Sox2, Wnt, Shh, Notch, Fgf, miR-200
1. Introduction
Taste is a primary sense of all vertebrates, reliably conveying important chemical information from the mouth to the brain to regulate ingestion. For most animals, sweet, moderately salty or certain amino acid stimuli trigger appetitive behaviour, while bitter or sour substances typically are rejected. The ability of animals to distinguish between nutritional, versus potentially lethal, fermented or unripe food items may mean the difference between survival and death. Human beings also rely on the taste system to select delicious foods and beverages based, in part, on taste preferences for the 5 basic taste stimuli; sweet, sour, salt, bitter, and umami (the taste of glutamate). Because taste is involved in ingestive behaviour, this sensory system is inherently linked to human diseases that result from obesity, such as stroke, heart disease and diabetes.
Taste buds are multicellular receptor organs that transduce taste stimuli in the oral cavity. Multiple taste cells within buds are innervated by cranial nerve fibers, which in turn convey taste information to the brain. For nearly a century, taste buds were thought to be induced by nerve contact late in embryonic development. Over the past decade, however, this view has shifted dramatically. A host of studies now indicate that taste bud development is initiated and proceeds via processes that are nerve-independent, occur long before birth, and are governed by cellular and molecular mechanisms intrinsic to the developing tongue. Thus our primary focus here is a review of recent advances in our understanding of the molecular and cellular mechanisms of taste bud development that occur in the embryonic oropharynx.
Another aspect of our review is to emphasize the progress made in detailed mapping of the fates of embryonic cell populations that contribute to the developing taste periphery. In large part, these studies have entailed use of cutting edge molecular genetic mouse models, and each has provided new insights into the cellular mechanisms governing specification and differentiation of taste epithelium. These types of genetic tools have also been instrumental in revealing fundamental molecular machinery involved in these early processes.
Finally, while the focus of the field has for decades been on rodent model systems, more recently amphibian embryos, and now the genetic zebrafish and medaka systems are providing new insights and accelerating the pace of discovery. Here we highlight the newest molecular genetic data concerning taste bud development in fish.
2. The anatomy of the taste system of vertebrates
The sense of taste or gustation is mediated by multicellular taste buds, which reside primarily within the oral and pharyngeal cavities. Taste receptor cells transduce taste stimuli, i.e., sweet, bitter, sour, salt and umami, into electrochemical signals and transmit these signals to afferent nerve fibers of the VIIth, IXth and Xth cranial nerve ganglia, which in turn convey taste information to the first central taste relay located in the hindbrain, the nucleus of the solitary tract . To date, all vertebrate groups have been found to have the same organization of these taste system elements, albeit with some modification over evolutionary time. For example, zebrafish and catfish, in addition to oral buds, have taste buds embedded in the epidermis of the head or over the entire body surface, respectively [1–3]. Nonetheless, these external taste receptor organs comprise a heterogeneous population of fusiform cells, which are innervated by branches of the VIIth cranial nerve, and the central projections of these fibers likewise terminate in the hindbrain in regions associated with taste function [4].
Another evolutionarily variable aspect of the taste periphery is the degree to which taste buds reside in specialized taste papillae versus directly in the epithelium. For example, taste buds in the mouse tongue are housed in 3 types of taste papillae [5]; anteriorly a single taste bud is situated in each of a dispersed array of fungiform papillae, while posteriorly, several hundred taste buds are housed in a single midline circumvallate papilla (CVP), as well as in bilateral foliate papillae. All 3 types of papillae possess a mesenchymal core surrounded by invaginated epithelium, in which taste buds are located. Taste buds in mammals are also found in the soft palate, the epiglottis and the larynx, however these lack papillae. Likewise, in fish and amphibians, taste buds are found embedded directly in the oral and pharyngeal epithelia or in specialized taste papillae, depending both on species as well as location [6–8].
3. Taste bud cellular composition in mice and fish
Each taste bud is composed of a heterogeneous population of fusiform cells, which parse into 3 morphological types (I, II and III), but physiologically likely represent at least 5 distinct functional classes (sweet, sour, salt, bitter and umami). Enormous advances have been made in our understanding of taste receptor cell function in the past decade, and these are reviewed elsewhere. Here we focus briefly on the conserved cellular and molecular properties of taste buds across vertebrate taxa. Historically, taste cells were classified into dark, intermediate and light cell types. Subsequently, with the advent of our ability to identify gene products expressed in taste bud cells, in mammals this dichotomy was superseded by Type I, II and III taste cell nomenclature (for a full review see [9].
In mammals, Type I cells are considered glial-like, express both GLAST and NTPdase2, and are thought to be the most numerous cell type within each bud [10,11]. Type II cells express a host of proteins (including Plcβ2 and Trpm5) which are common to the transduction of sweet, bitter and umami stimuli [12–15] as well as other molecules that are likely required for their function including calcium-binding proteins [16]. However, each of these 3 taste stimuli is transduced by a separate subtype of Type II cells, which is identified by the taste receptor protein each expresses. For example, sweet-detecting Type II cells express the heterodimeric G-protein-coupled receptors T1R1 and T1R3, whereas umami/glutamate-responsive cells express T1R2 with T1R3 [17]. Bitter detector cells express T2R proteins, of which there are ~30 in mice [18] . Altogether, Type II taste cells make up 20% of the taste bud cell population [19]. Finally, the Type III cells, which are least common, respond to acidic or sour stimuli, and express PKD2L1 and carbonic anhydrase 4 [20–22]. These cells also selectively take up Serotonin, and are readily identified with antisera against this neuromodulator [23–25]. Type III cells are also the only taste cell type in rodents which forms conventional presynaptic specializations onto sensory afferents [26], and are thought therefore to be more “neural” than Types I and II [27]. In fact, based on these innervation criteria, another taste cell type nomenclature has been suggested: type II cells are termed “receptor cells”, while type III cells are “presynaptic cells”.
The degree to which the details of mammalian cell type categorization extend to other vertebrates is generally not known, however, work in zebrafish and several other teleost species is revealing strong similarities with mammals. As discussed above, subsets of fish taste cells co-express elements of the common transduction cascade for bitter/sweet/umami, including Plcβ2, Trpm5, and specific G proteins [14,28,29], as well as T1R and T2R receptor proteins [30] and calcium-binding proteins (Fig. 2B, MK- unpublished data). As in mammals, fish T1R1 and T1R2 receptor are each co-expressed with T1R3, but in mutually exclusive subsets of cells, as are T2R receptors [31]. Importantly, functional tests of zebrafish taste receptors in heterologous cells indicate that T1R heterodimers respond to amino acids, while T2Rs are bitter sensitive [32]. In sum, these data infer a strong similarity, at the molecular and functional levels between teleost and mammalian Type II/receptor cells. Fish (and amphibia) also have a Serotonergic taste cell type, although the morphology of these cells is distinct from mammalian Type III/presynaptic taste cells. Rather than a fusiform shape as in mammals, Serotonergic taste cells are located basally within anamniote taste buds and have extensive radial processes that appear to contact both afferent sensory nerve fibers and fusiform cells within taste buds [33–36]. While the precise evolutionary relationship between Serotonin-immunoreactive taste cells of mammals versus those of fish and amphibians remains unresolved, it is clear in all species examined to date that the Serotonin/Type III/ presynaptic cells and sweet-bitter-umami/Type II/receptor cells comprise non-overlapping cell populations within vertebrate taste buds.
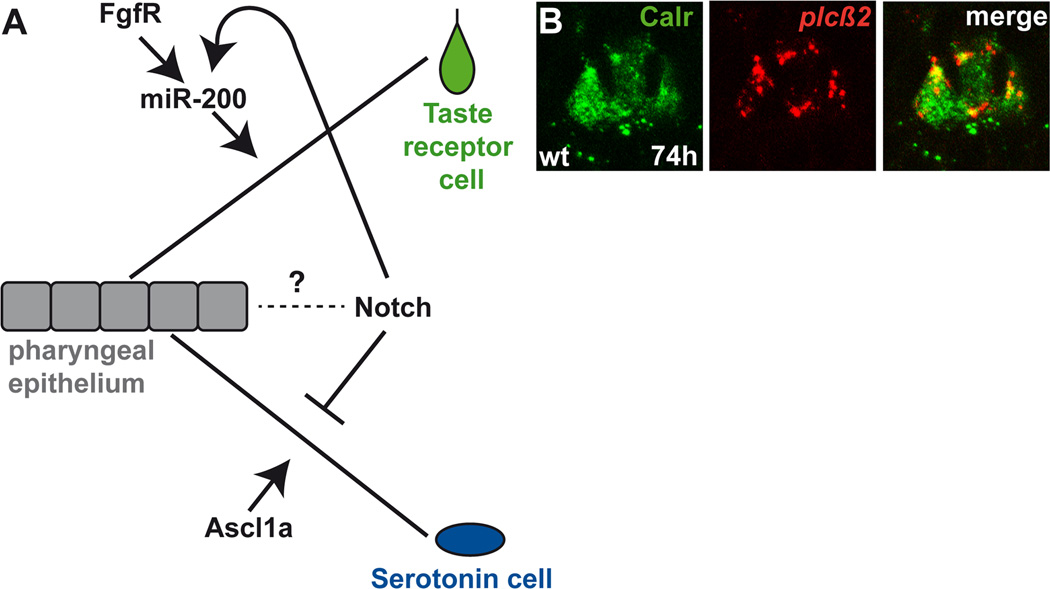
Figure 2.
A. Schematic representation of the key signals that are required for the late steps of development of taste receptor and Serotonin-expressing cells of the zebrafish taste buds. Fgf receptor (FgfR), miR-200 and Ascl1a activity are required within pharyngeal epithelial cells to drive taste receptor and Serotonin cell formation, respectively. Activation of Notch signaling is necessary for taste receptor cell formation whereas inhibition of this signal is required for Serotonin cell formation. Notch likely has additional functions in taste bud development within the pharyngeal epithelium. B. A 2µm optical section through a wild-type zebrafish taste bud reveals taste receptor cells co-expressing Calretinin (Calr) and plcβ2.
4. Defining the embryonic origin of vertebrate taste buds
Taste buds are found in the epithelia lining the oral and pharyngeal cavity, and like epidermis of the skin, taste cells are continually renewed throughout adult life [37,38]. Based on their location and this regenerative feature, taste bud cells have long been considered to be modified epithelial cells which therefore were presumed to arise directly from the local epithelium [39,40]. Subsequently, this hypothesis was addressed experimentally; in both axolotl (a type of salamander) [41] embryos, taste buds were shown to develop from labelled epithelial precursor cells. Importantly in the axolotls, a potential contribution to taste buds from the subepithelial mesenchyme, which is derived from the cranial neural crest [42] was explicitly ruled out. These findings have since been reinforced by the elegant demonstration that taste bud cells in adult mice derive directly from proliferative basal keratinocytes in the tongue epithelium [43,44].
More recently, to assay for a direct contribution of neural crest to developing taste buds in mice, we employed a tissue specific Cre-lox approach to label and then follow the fate of neural crest cells in mouse embryos. A well-characterized Wnt1Cre allele drives reporter allele expression in the neural crest beginning prior to its migration from the dorsal neural tube [45] and use of this allele combination revealed further that neural crest cells contribute extensively to the subepithelial mesenchyme of the tongue, but not to the lingual epithelium [46]. Likewise, we found robust labeling in the mesenchyme of embryonic and adult tongues, but not in the embryonic lingual epithelium; nor did this embryonic cell population contribute cells to taste buds in adult mice [47]. However, a recent study employing additional Cre-lox tools now suggests that in fact, the neural crest contributes an apparent majority of cells to taste buds in mice [48]. What could account for these very different findings?
Liu and colleagues [48], also using Wnt1Cre crossed with reporter mice to map the neural crest, identified exceedingly rare instances (1–2 cells per mouse) of a neural crest cell closely associated with immature taste buds at birth. However, for the majority of their studies, they employed a less specific transgenic line where Cre recombinase is driven by a 1.1kb promoter fragment of the myelin protein P0 [49]. This promoter fragment does not recapitulate the native expression of P0 [50], and in fact, drives Cre activity and thus ectopic reporter expression in both oral endoderm and ectoderm, as well as the notochord, early in development [49] (their figure 3) [51] (their figure 4a). Finally, it is not known to what extent the 1.1kb P0 promoter fragment drives Cre expression in postnatal epithelium, which could also account for the discrepancies in cell labeling. Thus it is not surprising that P0Cre yields a far different pattern of reporter gene expression at birth from that due to Wnt1Cre activity: although lingual subepithelial mesenchyme is robustly labelled as expected, fully 90% of immature taste buds also express the reporter, as do large patches of lingual epithelium, a finding inconsistent with results from all published studies of cranial neural crest cell fate, as well as the contribution of the embryonic ectoderm to the epithelium lining the oral cavity [41,52–55]. Thus, while these new data are intriguing, they also illustrate the need to systematically define the expression of P0Cre throughout both developmental and postnatal stages.
In mammals, lingual taste buds reside in epithelial-mesenchymal specializations, termed taste papillae. In mouse embryos, the first indication of taste development occurs at midgestation with the appearance of taste placodes, which are foci of columnar epithelial cells in an otherwise cuboidal lingual epithelium. These placodes undergo epithelial morphogenesis to surround a mesenchymal core, and taste buds then differentiate within the epithelial compartment at or within 1 week of birth. It has been assumed that taste placodes are papilla precursors, which morph into taste papillae, which then in turn produce taste buds from a subset of papilla epithelial cells [56]. However, it is equally plausible that specification of taste bud precursor cells represents the primary patterning event, and these taste precursors instead induce the papillae to form around them.
In addition to morphological changes, taste placodes express a number of gene products as they form. Importantly among these, taste placode cells turn on focal expression of Shh [57], a diffusible protein known to regulate cell fate and survival in a wide array of tissues during development [58]; and Shh expression persists in a subset of taste papilla epithelial cells through the remainder of embryogenesis. To map the fate of the Shh+ taste placode cells in embryonic mice, we used an allele where a tamoxifen-sensitive form of Cre recombinase is “knocked in” under the control of the native Shh locus [59]. Treating pregnant females with tamoxifen at precise times in development allowed us to determine if and when Shh-expressing placode cells contribute to taste buds, taste papillae or to both. In fact, we found that taste placodes are precursors of taste bud cells only, and do not contribute to taste papillae [60]. Moreover, these findings support a model in which taste bud precursor cell induction is the primary event in taste bud development, while taste papillae form in response to signals emitted from newly induced precursor cells.
5. Tissue interactions in taste bud development
5.1. Nerve-dependent induction
The prevailing model of taste bud development has been neural induction, where late in embryogenesis taste nerves invade the lingual epithelium and induce taste bud precursors from an otherwise homogeneously competent epithelium [47,61,62]. However, over the past 20 years, experimental tests have firmly rejected this hypothesis. Using both grafting and culture approaches combined with molecular marker expression, we showed that amphibian taste buds differentiate without innervation [57], consistent with early results [63,64]. Studies in rodent embryos using explant culture (mouse, rat) and engineered genetic tools (mouse) have confirmed that embryonic development of taste bud precursors is likewise nerve-independent in mammals [65–71].
However, in contrast to amphibians, differentiation of mammalian taste bud cells appears to require innervation; this may be because the majority of taste buds in rodents differentiate postnatally. Specifically, at birth, taste buds are detectable morphologically as onion-shaped clusters of cells within the regularly arranged papilla epithelium. These cells express markers of embryonic taste precursor cells (Shh, Sox2, cytokeratin K8) – and do so in the absence of innervation in the case of Shh but they lack expression of differentiated taste cell markers. It is not until a few days after birth that taste bud cells in rodents first differentiate in most of the taste fields, including the anterior fungiform and posterior circumvallate papillae ([47] for review). In a series of benchmark studies, Oakley and colleagues showed that differentiation of taste cells required an intact innervation during the first postnatal week, and that even if the injured nerve was allowed to reinnervate the circumvallate papilla, the number of differentiated taste buds that eventually resided within the epithelium was permanently diminished ([72] for review)[57,73]. These data thus argue for a critical postnatal period during which nerves are required for differentiation of embryonic taste buds in rodents.
5.2. Epithelial-mesenchymal interactions
Taste buds embedded in taste papillae are often categorized as ectodermal appendages, which like teeth, feathers, and hair follicles, have epithelial and mesenchymal components [74,75]. Interactions between subepithelial mesenchyme and epithelium are absolutely essential for development of these appendages, and have been proposed to underlie taste papilla morphogenesis [76]. All connective tissues of the tongue, and specifically the subepithelial mesenchyme, is derived from the neural crest [56] as discussed above, while the tongue musculature arises from hypoglossal somitic mesoderm [46,77]. Although the neural crest-derived mesenchyme almost certainly does not contribute directly to taste buds (see discussion above), signals from this compartment to the overlying lingual epithelium are important for discrete aspects of taste bud and papilla development.
In mouse embryos, signals from the lingual mesenchyme clearly play an important role in development of the taste periphery. Specifically, lingual mesenchyme defines the regions of the lingual epithelium that produce taste organs [78]. Likewise, identified signals secreted from the mesenchyme, e.g. follistatin, Fgfs, affect development of both anterior fungiform and posterior circumvallate taste papillae, respectively ([79,80] and see below). In sum, these findings support an important role for mesenchymal signals in development of taste epithelium in rodents. However, in amphibians, cranial mesenchyme is not required for taste bud development; taste buds reliably develop and differentiate in oral explants devoid of both neural crest- and mesoderm-derived cell populations [81,82]. One way to reconcile the apparent mesenchyme-independent development of amphibian taste buds from the clear function of mesenchyme in mammals is to consider species differences in taste organ morphology. Axolotl taste buds are embedded in the oral epithelium and are not housed in papillae, while studies in mammalian embryos to date have focused primarily on lingual taste buds that reside in taste papillae. Thus, in mammals, development of taste buds and papillae are intertwined such that mesenchyme likely influences papilla formation, which in turn would feed back on taste bud development. In this model then, taste bud precursors would be induced via epithelium-intrinsic mechanisms in both rodents and amphibians, while papillae formation and refinement of taste organ pattern in mammals would require further interaction with lingual mesenchyme. However, equally plausible, in amphibians the mesenchyme may be dispensable for taste bud induction, but may supply signals that play a role in patterning taste bud precursors, an option that remains to be tested.
5.3. The taste bud differences along the A-P axis
Taste buds in mammals and fish occur primarily within the oral and pharyngeal cavities, and in broad terms, taste buds found anteriorly do not differ from those found posteriorly. Increasingly, however, researchers are finding evidence of many molecular differences in rodent taste buds located in anterior versus posterior papillae. For example, in mice, the coordinated expression of sweet versus umami receptor heterodimers with specific G proteins by type II/receptor cells varies between the fungiform (anterior) and circumvallate (posterior) taste buds [83,84]. Likewise, expression of a BMP4 reporter allele differs between fungiform papillae, where labelled cells are detected immediately adjacent (perigemmal) to taste buds, and circumvallate papillae, where labelled cells are found both perigemmally and within taste buds [85].
One reason that may account for these differences is that anterior taste buds have been proposed to derive from ectoderm, while posterior taste buds arise from endoderm. The degree to which the germ layer of origin affects molecular characteristics of taste buds is not known, although in axolotls, taste buds only form in the oral ectoderm if this tissue is paired with early endoderm suggesting that signals from the endoderm are key to taste bud development. Most recently, using molecular genetic lineage tracing, the anterior limit of the endoderm has been identified in mice [86]. Using embryos carrying Sox17Cre expressed in the endoderm and the R26RLacZ reporter allele, Tucker and colleagues [53] showed that the CV and foliate taste epithelia derive from the endoderm, whereas the anterior tongue, comprising the fungiform domain, is devoid of label, and is therefore ectodermal. Likewise, in zebrafish, fate mapping experiments using the photoconvertible protein Kaede and endoderm transplants showed that endoderm generates only pharyngeal arch (posterior) Calretinin-expressing taste bud cells, whereas unlabeled taste buds on the lips are presumed ectodermal [87]. This difference in embryonic origin may underlie reports where the same developmental signaling pathway has different effects on anterior versus posterior taste development (see below).
6. Molecular mechanisms of taste bud development
The focus of studies of taste bud development has now shifted from a neural induction model to processes intrinsic to the tongue, and in particular to investigations of the molecular mechanisms involved. To date, regulation of taste bud development involves a number of pathways known to play critical roles in a variety of tissues throughout development, as well as in adult homeostasis and disease. Foremost among these are the Shh and Wnt/β-catenin pathways. Here we review new data on the role of distinct signaling pathways, as well as their interactions, in taste bud development in mice and zebrafish.
6.1. Wnt/β-catenin: the primary inductive signal for taste placodes
Wnts are short range, diffusible factors comprising 19 genes. Wnt ligands bind Frizzled receptors (of which there are 9 in mammals), and then signal via at least 3 intracellular pathways, although the canonical or β-catenin pathway is the best studied [88]. In the canonical pathway, the destruction complex (Axin/Gsk3-β/APC) keeps cytoplasmic levels of β-catenin low in the absence of ligand. Wnt binding of the Frizzled receptor dissociates the destruction complex, allowing β-catenin levels to rise, which results in transit of β-catenin to the nucleus where it functions to activate Wnt target gene transcription.
Studies of the role of Wnts in the developing taste system have focused exclusively on the anterior fungiform taste field in mice. Several Wnt ligands are expressed in overlapping patterns in the developing tongue epithelium around the time of taste placode formation, suggesting a role for this pathway in taste bud induction. For example, Wnt10b is expressed in taste placodes. Wnt10a is excluded from placodes, yet is expressed broadly in the tongue epithelium. In addition, examination of mouse embryos carrying the engineered Wnt/β-catenin reporter allele, TOPGAL, revealed that taste placode cells and lingual epithelium are responding to these early Wnt signals [89–91]. Consistent with the expression data, Wnt/β-catenin function is required and sufficient for taste placode formation (Figure 1A). When β-catenin is genetically deleted from the lingual epithelium, taste placodes fail to develop, whereas when β-catenin is stabilized and thus activated, again using tissue-specific genetic tools, the entire tongue surface is transformed into a continuous array of taste bud precursors surrounded by enlarged papillae. Loss of Wnt10b, whose expression is restricted to taste placodes, however, does not phenocopy the loss of β-catenin, as taste placodes still form, inferring that other Wnts are involved [90].
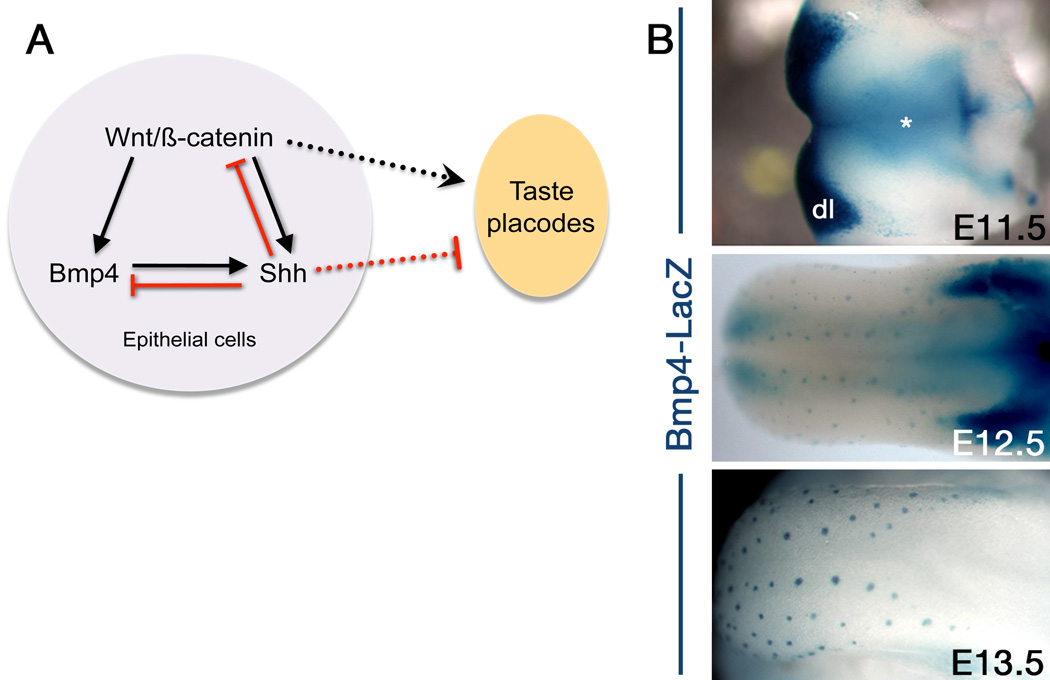
Figure 1.
Development of taste placodes is regulated by interactions between the Wnt, Shh and Bmp pathways. A. Wnt/β-catenin signaling is required upstream of Shh and Bmp4 for taste placode specification. Shh represses Wnt and Bmp4, as well as its own expression, while Bmp4, at least early on, is a positive regulator of Shh. See text for details. Black arrows = positive regulation ; red lines = negative regulation. Dashed lines indicate that the Wnt pathway promotes, while Shh inhibits taste placode formation. B. Embryonic tongues taken from Bmp4-LacZ mice [122] were reacted with Xgal, and reveal that Bmp4 is expressed initially in the lingual mesenchyme (E11.5 *) as well as in the dental lamina (dl) of the lower jaw. By E12.5, Bmp4-LacZ reports in the developing fungiform placodes, and this expression deepens by E13.5. Anterior is left in all 3 panels.
6.2. Sonic hedgehog (Shh): a negative regulator of taste placode formation
Shh is a well studied diffusible protein that is a key regulator of proliferation and cell survival in multiple developing tissues, and thus Shh was among the first developmental regulatory genes found to be expressed in taste placodes [92]. While Shh expression is initially broad throughout the developing tongue epithelium, expression consolidates in taste placodes as they form [58,93]. Ptch1, a Shh receptor and target, and Gli1, a downstream target gene, are likewise expressed early on broadly in both epithelial and mesenchymal compartments of the tongue. As Shh expression focalizes, so does that of Ptch1 and Gli1, so that expression of the latter 2 resolves to cups of cells comprising both epithelium and mesenchyme surrounding each Shh-expressing epithelial cell cluster.
Functional tests of Shh to date have been performed solely via short-term culture of embryonic tongues. When tongue explants are taken prior to formation of taste placodes, and cultured in the presence of a function-blocking antibody against Shh [67] or with a Shh pathway inhibitor [66], fungiform taste placodes increase in both size and number. Moreover, when Shh signaling is inhibited, taste placodes form in a posterior domain of the tongue surface, which, in control tissue and intact animals, is always devoid of taste buds. Consistent with this repressive function of Shh, when tongue explants are cultured with excess Shh ligand, taste placodes are dramatically reduced (Figure 1A) [90]. Interestingly, when Shh function is inhibited, Shh expression is expanded in the more numerous and enlarged taste placodes [67], indicating that Shh’s impact on its own expression is indirect. One candidate for this intermediary is the Wnt/β-catenin pathway. Specifically, β-catenin is required for Shh expression by taste placode cells, placing Shh downstream of Wnt [91]. However, Wnt signaling in taste placodes is also negatively regulated by Shh; in vitro inhibition of Shh results in expanded placode-specific Wnt activity, in an increased number of taste placodes, whereas excess of Shh activity has the opposite effect (Figure 1A) [90].
Interestingly, Shh has been shown to promote taste bud development in cave forms of the teleost fish, Astyanax mexicanum. Numerous evolutionary changes have occurred that distinguish the cave form, which is blind, from the sighted form, which lives in surface waters. While cavefish have degenerated eyes, they have significantly more taste buds than surface fish. This increase in taste bud number, however, appears to be indirect, and instead is downstream from a Shh-driven expansion of the entire oral-pharyngeal apparatus, a process that commences well before taste bud development [94,95].
6.3. BMPs : a role in placode induction and patterning
Bone morphogenetic proteins (BMPs) comprise a subfamily of the enormous TGFβ family of secreted factors. Of these, BMPs 2, 4, and 7 are expressed in the developing tongue epithelium in a spatiotemporal pattern consistent with a role in taste placode induction and patterning (Figure 1B) [79,93,96,97]. In fact, when tongue explants are cultured in excess of BMP 2, 4 or 7, both the number and size of taste placodes are increased, as is their expression of Shh (Figure 1A) [97]. A pro-placodal function is reinforced for BMP7 by elegant genetic studies from the Calof lab [79]. They likewise show that BMP7 functions upstream of Shh, and further show BMP7’s role is downstream of Wnt/β-catenin during taste placode patterning. However, when tongue explants are treated with excess of BMP 2, 4 or 7 after placode induction, both placodes and Shh expression are diminished [97]. Interestingly, genetic loss of follistatin, a BMP antagonist expressed exclusively within the lingual mesenchyme, is permissive for taste placode formation, as follistatin restricts BMP function in patterning the overlying epithelium [79]. By contrast, follistatin administered to cultured tongue explants does not repress taste placode development [97]. However, this latter result was achieved in cultured embryonic tongues in which taste placodes had already formed, and thus the critical window for follistatin repressive function likely was missed.
6.4. Sox2: a critical transcription factor for taste bud cell lineage
Sox2, a member of the metazoan conserved Sry-related High Mobility Group box transcription factors, regulates the transcription of a variety of genes during stem cell differentiation in the nervous system [98,99]. Addressing its functions in vivo has been possible only in hypomorphic mutants since Sox2-null embryos die before gastrulation [100]. Genetic mapping analysis using a tamoxifen inducible Keratin-14 Cre recombinase shows that bipotential progenitors located adjacent to the taste buds express Sox2 and generate both taste bud and keratinized epithelial cells. Within this progenitor pool, cells of the keratinocyte lineage lose Sox2 expression whereas cells with taste bud fate upregulate Sox2 [44]. Consistent with this, in the hypomorph Sox2EGFP/LP embryos, filiform papillae are normal but the number of fungiform and palatal taste buds is gradually reduced and then completely absent at birth, indicating that Sox2 activity is necessary for taste bud formation [101]. Interestingly, Sox2 overexpression in lingual epithelium converts filiform papillae to dome-like structures, reminiscent of fungiform taste papillae, but they are devoid of differentiated taste bud cells. Therefore, Sox2 activity is not sufficient for the differentiation of taste bud cells.
Although, Sox2 loss and gain of function data are not available in fish, a difference in the expression of this transcription factor has been reported [87]. Sox2 is expressed throughout the oropharyngeal epithelium and in immature taste bud cells, as in mice, but it is absent from differentiating Serotonin- and Calretinin-expressing taste cells. A prediction from this observation is that Sox2 may be required for the progenitor and immature phase of taste bud cells but as differentiation proceeds, Sox2 expression is downregulated to allow the expression of late differentiation genes. This would be consistent with the conversion of filiform epithelium to structures reminiscent of taste papillae but devoid of taste cells after overexpression of Sox2 in mouse lingual epithelium [101]. As for the epistatic relations of Sox2 with other signaling molecules, activation of Wnt/β-catenin signaling enhances Sox2 expression in taste buds as well as promotes ectopic Sox2 expression in the lingual epithelium, indicating that Sox2 acts downstream of Wnt/β-catenin. In contrast, the expression of neurotrophic factors Bdnf and NT3 is downregulated in the Sox2 hypomorph embryos suggesting that Sox2 lies upstream of these factors during taste bud formation [101].
6.5. Fgf signaling: regulator of the progenitor field size and taste bud cell type differentiation
Fgf ligands bind to tyrosine kinase receptors (RTK) and activate multiple intracellular cascades including Ras/Raf/Erk1/2, PLCgamma/PKC and PI3K/AKT. Appropriate levels of activation of RTK signaling is ensured by four Sprouty proteins that are induced by Fgf and other growth factors and act mainly as RTK inhibitors [102]. The Fgf pathway is a primary example of a signaling pathway that regulates anterior versus posterior taste bud development in a differential manner. Specifically, in Spry2−/− mice, the number of fungiform (anterior) taste placodes is significantly reduced but in striking contrast the CV (posterior) placode is initially enlarged and CV papillae ultimately duplicated. While the CVP of Spry1−/− mice is comparable to controls, the combined genetic deletion of Spry2 and Spry1 results in multiple small CVPs, revealing a synergistic effect of Spry1 and Spry2 in repressing CV taste placode formation. Sprouty activity is regulated by Fgf10, which is expressed by the mesenchyme directly beneath the developing CV placode. The CVP is absent in Fgf10−/− mice and as expected, when Fgf10 gene dosage is reduced in Spry2−/− mutants, taste bud development is rescued, i.e, only one CVP forms. Therefore, tightly regulated Fgf signaling is critical for development of a single CVP [80].
In zebrafish, compromised Fgf receptor signaling also dramatically affects taste bud development, however, in fish, both anterior (mouth) and posterior (pharyngeal arch, palate) taste bud cells are similarly lost [87]. The number of taste bud cells is also reduced in the Fgf8a−/− zebrafish mutants although less significantly compared to larvae with compromised Fgf receptor activity. As Fgf signaling is required for the formation of other oropharyngeal components including epithelial and cartilagenous cells [103–105] an indirect effect on taste bud development cannot be excluded. Surprisingly, when Fgf receptor signaling is compromised specifically within the posterior pharyngeal epithelium, just prior to the appearance of differentiated taste bud cells, the Calretinin-expressing taste receptor cell population is severely reduced, but the number of Serotonin-expressing cells is unaffected (Figure 2A) [87]. These data implicate Fgf signalling in cell fate decisions within specified taste bud precursors, in addition to its early function in taste bud patterning. In sum, these new studies in mice and fish implicate a requirement for Fgf signaling in several steps of taste bud development.
6.6. Notch signaling: how many functions during taste bud formation?
The primary components of the Notch pathway are Notch receptors, which upon ligand (Delta/Jagged) binding, are cleaved by gamma-secretases. This cleavage event releases an intracellular domain of Notch which is able to enter the nucleus and convert the CBF1 complex from a repressor to an activator of transcription of Notch-specific target genes. Ligand-receptor interaction occurs only after appropriate processing of the ligand that includes ubiquitinilation by E3 ubiquitin-ligases such as Mindbomb. Among Notch targets, the Hes transcription factors antagonize proneural genes like Mash1 to block early, and thus inappropriate, neuronal gene expression in the nervous system [106,107].
Notch receptors, ligands, and transcriptional targets have a dynamic expression profile during posterior taste bud development in both fish and mice. Before taste bud appearance, Notch related molecules are widely expressed in the epithelium but are progressively restricted to differentiating taste cells or to epithelial cells adjacent to taste buds, suggesting time dependent, and changing functions of Notch signaling during taste bud formation [87,108] . Among Notch pathway members, Hes1 binds to promoters of both Plcβ2 and Ip3r3 (both expressed by type II taste receptor cells), while Hes1 protein is localized in the nucleus of undifferentiated taste bud cells. Therefore, Ota and colleagues [109] proposed that Hes1 may repress expression of taste cell specific molecules in precursor cells. Indeed, in Hes1-null mice, the number of type II taste receptor cells is increased 5-fold compared to controls suggesting that in the absence of Hes1, aberrant transcription of taste receptor-specific genes takes place in precursor cells.
When Notch activity is blocked in Mindbomb zebrafish mutants, excess Serotonin-expressing taste cells develop, the taste progenitor cell pool is reduced, and the number of Calretinin-expressing taste receptor cells is decreased (Figure 2A) [87]. In a complementary manner, Notch activation just prior to cell differentiation has the opposite effect; Serotonin-expressing cells are absent while the Calretinin-expressing taste receptor cell population is maintained but not increased. These results support the hypothesis that cells with activated Notch signaling maintain a progenitor state as predicted by the mouse model in which Hes1 and therefore its Notch-repressing activity is lost [109]. Consistent with the role of Notch in the formation/maintenance of a progenitor pool for taste bud cells, the severe decrease of taste receptor cells provoked by compromised Fgf signaling in fish is restored by Notch activation (Figure 2A) [87].
Mash1 (Ascl1a in zebrafish) expression is restricted to taste bud presynaptic/Serotonin-expressing cells. In Mash1−/− mice, taste bud organisation at birth appears normal and some presynaptic cells are present, but these fail to acquire Serotonin identity [110]. In the Ascl1a−/− zebrafish larvae, taste bud cells are devoid of Serotonin but whether only the neurotransmitter or the entire presynaptic cell population is lost remains unclear (Figure 2A). However, taste buds are rather disorganized and the number of Calretinin-expressing taste receptor cells is slightly increased in Ascl1a−/− zebrafish, which may reflect the absence of the presynaptic cells [87]. One interpretation of these differences between Mash1−/− and Ascl1a−/− taste buds in mice and fish, respectively, is that fish taste buds are not tightly confined to papillae and thus cell organization defects may be more obvious compared to mouse. Alternatively, one report [111] suggested that Mash1 may be expressed in mice in an intermediate immature cell type with the potential to become a taste receptor or presynaptic cell; this scenario is consistent with the slight increase of taste receptor cells in Ascl1a−/− fish.
Altogether these data show that Notch signaling is likely active in progenitor cells, and that Mash1/Ascl1a is necessary for Serotonin/presynaptic cell identity. However, Notch signaling may fulfil additional yet-to-be discovered functions in taste bud formation, as has been shown in other sensory organs (e.g.[112]). For instance, Notch signaling can promote cell identity through lateral inhibition. Skn-1a is a POU homeodomain protein specifically expressed in taste receptor and basal (progenitor) cells. In Skn1a-null mice, presynaptic cells are expanded at the expense of taste receptor cells suggesting a binary competence for progenitor cells to give rise to taste receptor or presynaptic cells [113]. It will be of interest to analyse if and how Notch signaling and Skn-1a interact to regulate taste bud cell fate.
6.7. miR-200: a crossroad integrating information from diverse signaling pathways to promote taste bud cell formation
microRNAs (miRNAs) are 20–22 nucleotide RNA molecules that bind to complementary sequences, usually located in the 3’UTR of target mRNAs, and block the expression of the corresponding proteins via either mRNA degradation or translational inhibition [114,115]. miRNAs regulate diverse processes during development and disease, including cell differentiation (e.g. [116–118]).
The miR-200 family is expressed in taste buds and in particular in taste receptor cells [87,119,120]. Knock-down of miR-200 results in severe reduction of the number of Calretinin-expressing taste receptor cells, showing that these miRNAs are required for the formation of this taste cell type (Figure 2A) [87]. The number of Serotonin cells is mildly reduced in miR-200 knockdown, suggesting that this is a secondary and/or indirect defect. miR-200 expression relies on intact Fgf and Notch signaling activity, as embryos with either compromised Fgf or Notch signaling have severely decreased miR-200 expression. In addition, when miR-200 activity is compromised, activation of Notch signaling fails to restore the number of taste receptor cells, further supporting that miR-200 acts downstream of Notch signaling. One of miR-200 direct targets is Sox2, whose expression is regulated by miR-200 during taste bud development [87,121]. But as miR-200 have many additional bioinformatically predicted targets, it is likely that many of them are also involved in taste bud cell formation or function. Therefore, miR-200 may reflect a molecular relay where information from an array of signaling pathways converges and is integrated to regulate the formation of functional taste buds.
7. In Closing…
The sense of taste plays a key role in our ability to select palatable and nutritious foods, as well as to reject those substances that are toxic or decomposing. While our understanding of taste system function has expanded dramatically over the past 2 decades, how this system develops embryonically is only now receiving increased attention and study. Molecular genetic tools in mouse have become steadily more numerous and sophisticated, as well as more tractable for use in the taste system; these approaches have rapidly advanced understanding of key molecular and cellular mechanisms of taste development. In addition, zebrafish is a powerful model system, where genetic interactions among signalling pathways can be elegantly tested. Again, use of zebrafish has likewise significantly advanced understanding of molecular regulation of taste bud development. Most critically, use of both models already has provided key and complementary insights into the fundamental and evolutionary conserved mechanisms governing formation of this important sensory system.
Highlights.
Early induction and patterning of taste buds occur via processes inherent to the developing tongue.
Interactions of the Wnt, BMP, Shh and FGF pathways regulate early taste bud formation.
Signals from the mesenchyme to the overlying epithelium are important regulators of taste bud pattern.
Regulation of the Notch pathway and FGF signaling by miR-200 controls cell fate decisions within multicellular taste buds.
Molecular genetic approaches in both mouse and zebrafish underlie recent advances in our understanding of taste development.
Acknowledgements
We are grateful to Frederic Rosa and IBENS for support, and to the Rocky Mountain Taste and Smell Center (P30 DC004657) for . This work was funded by the ANR-09-BLAN-077 (MK) and NIH/NIDCD DC008373 and DC003947 (LAB).
Abbreviations
- CVP
circumvallate papilla
- RTK
tyrosine kinase receptor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Northcutt RG. Taste buds: development and evolution. Brain Behav Evol. 2004;64:198–206. doi: 10.1159/000079747. [DOI] [PubMed] [Google Scholar]
- 2.Landacre F. On the place of origin and method of distribution of taste buds in Ameirus melas. J Comp Neurol. 1907;17:1–66. [Google Scholar]
- 3.LeClair EE, Topczewski J. Development and regeneration of the zebrafish maxillary barbel: a novel study system for vertebrate tissue growth and repair. PloS One. 2010;5:e8737. doi: 10.1371/journal.pone.0008737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atema J. Structures and functions of the sense of taste in the catfish (Ictalurus natalis) Brain Behav Evol. 1971;4:273–294. doi: 10.1159/000125438. [DOI] [PubMed] [Google Scholar]
- 5.Kinnamon J. Organization and innervation of taste buds. In: Finger TE, Silver W, editors. The neurobiology of taste and smell. New York: Wiley; 1987. pp. 277–297. [Google Scholar]
- 6.Finger TE. The gustatory system of teleost fish. In: Northcutt RG, Davis RE, editors. Fish neurobiology. New York: Wiley; 1983. pp. 285–309. [Google Scholar]
- 7.Reutter K. Taste organ in the bullhead (Teleostei) Adv Anat Embryol Cell Biol. 1978;55:1–98. doi: 10.1007/978-3-642-67008-4. [DOI] [PubMed] [Google Scholar]
- 8.Reutter K, Breipohl W, Bijvank G. Taste bud types in fishes II. Scanning electron microscopical investigations on Xiphophorus helleri Heckel (Poeciliidae, Cyprinodontiformes, Teleostei) Cell Tissue Res. 1974;153:151–165. doi: 10.1007/BF00226604. [DOI] [PubMed] [Google Scholar]
- 9.Finger TE, Simon S. Cell biology of taste epithelium. In: Finger TE, Silver W, Restrepo D, editors. The neurobiology of taste and smell. New York: Wiley; 2000. pp. 287–314. [Google Scholar]
- 10.Bartel DL, Sullivan SL, Lavoie EG, Sévigny J, Finger TE. Nucleoside triphosphate diphosphohydrolase-2 is the ecto-ATPase of type I cells in taste buds. J Comp Neurol. 2006;497:1–12. doi: 10.1002/cne.20954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawton DM, Furness DN, Lindemann B, Hackney CM. Localization of the glutamate-aspartate transporter, GLAST, in rat taste buds. Eur J Neurosci. 2000;12:3163–3171. doi: 10.1046/j.1460-9568.2000.00207.x. [DOI] [PubMed] [Google Scholar]
- 12.Clapp TR, Medler KF, Damak S, Margolskee RF, Kinnamon SC. Mouse taste cells with G protein-coupled taste receptors lack voltage-gated calcium channels and SNAP-25. BMC Biol. 2006;4:7. doi: 10.1186/1741-7007-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miura H, Nakayama A, Shindo Y, Kusakabe Y, Tomonari H, Harada S. Expression of gustducin overlaps with that of type III IP3 receptor in taste buds of the rat soft palate. Chem Senses. 2007;32:689–696. doi: 10.1093/chemse/bjm036. [DOI] [PubMed] [Google Scholar]
- 14.Yoshida Y, Saitoh K, Aihara Y, Okada S, Misaka T, Abe K. Transient receptor potential channel M5 and phospholipase C-beta2 colocalizing in zebrafish taste receptor cells. Neuroreport. 2007;18:1517–1520. doi: 10.1097/WNR.0b013e3282ec6874. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Hoon MA, Chandrashekar J, Mueller KL, Cook B, Wu D, et al. Coding of sweet, bitter, and umami tastes: different receptor cells sharing similar signaling pathways. Cell. 2003;112:293–301. doi: 10.1016/s0092-8674(03)00071-0. [DOI] [PubMed] [Google Scholar]
- 16.Rebello MR, Aktas A, Medler KF. Expression of calcium binding proteins in mouse type II taste cells. J Histochem Cytochem. 2011;59:530–539. doi: 10.1369/0022155411402352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao GQ, Zhang Y, Hoon MA, Chandrashekar J, Erlenbach I, Ryba NJP, et al. The receptors for mammalian sweet and umami taste. Cell. 2003;115:255–266. doi: 10.1016/s0092-8674(03)00844-4. [DOI] [PubMed] [Google Scholar]
- 18.Chandrashekar J, Mueller KL, Hoon MA, Adler E, Feng L, Guo W, et al. T2Rs function as bitter taste receptors. Cell. 2000;100:703–711. doi: 10.1016/s0092-8674(00)80706-0. [DOI] [PubMed] [Google Scholar]
- 19.Ma H, Yang R, Thomas SM, Kinnamon JC. Qualitative and quantitative differences between taste buds of the rat and mouse. BMC Neurosci. 2007;8:5. doi: 10.1186/1471-2202-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang AL, Chen X, Hoon MA, Chandrashekar J, Guo W, Tränkner D, et al. The cells and logic for mammalian sour taste detection. Nature. 2006;442:934–938. doi: 10.1038/nature05084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chandrashekar J, Yarmolinsky D, von Buchholtz L, Oka Y, Sly W, Ryba NJP, et al. The taste of carbonation. Science. 2009;326:443–445. doi: 10.1126/science.1174601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kataoka S, Yang R, Ishimaru Y, Matsunami H, Kinnamon JC, Finger TE. The candidate sour taste receptor, PKD2L1, is expressed by type III taste cells in the mouse. Chem Senses. 2008;33:243–254. doi: 10.1093/chemse/bjm083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dvoryanchikov G, Tomchik SM, Chaudhari N. Biogenic amine synthesis and uptake in rodent taste buds. J Comp Neurol. 2007;313:302–313. doi: 10.1002/cne.21494. [DOI] [PubMed] [Google Scholar]
- 24.Kim D, Roper SD. Localization of serotonin in taste buds: a comparative study in four vertebrates. J Comp Neurol. 1995;353:364–370. doi: 10.1002/cne.903530304. [DOI] [PubMed] [Google Scholar]
- 25.Takeda M. Uptake of 5-hydroxytryptophan by gustatory cells in the mouse taste bud. Arch Histol Jpn. 1977;40:243–250. doi: 10.1679/aohc1950.40.243. [DOI] [PubMed] [Google Scholar]
- 26.Yang R, Stoick CL, Kinnamon JC. Synaptobrevin-2-like immunoreactivity is associated with vesicles at synapses in rat circumvallate taste buds. J Comp Neurol. 2004;471:59–71. doi: 10.1002/cne.20021. [DOI] [PubMed] [Google Scholar]
- 27.Chaudhari N, Roper SD. The cell biology of taste. J Cell Biol. 2010;190:285–296. doi: 10.1083/jcb.201003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aihara Y, Yasuoka A, Yoshida Y, Ohmoto M, Shimizu-Ibuka A, Misaka T, et al. Transgenic labeling of taste receptor cells in model fish under the control of the 5’-upstream region of medaka phospholipase C-beta 2 gene. Gene Exp Patterns. 2007;7:149–157. doi: 10.1016/j.modgep.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 29.Asano-Miyoshi M, Abe K, Emori Y. Co-expression of calcium signaling components in vertebrate taste bud cells. Neurosci Lett. 2000;283:61–64. doi: 10.1016/s0304-3940(00)00911-3. [DOI] [PubMed] [Google Scholar]
- 30.Ohmoto M, Okada S, Nakamura S, Abe K, Matsumoto I. Mutually exclusive expression of Gαia and Gα14 reveals diversification of taste receptor cells in zebrafish. J Comp Neurol. 2011;519:1616–1629. doi: 10.1002/cne.22589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishimaru Y, Okada S, Naito H, Nagai T, Yasuoka A, Matsumoto I, et al. Two families of candidate taste receptors in fishes. Mech Dev. 2005;122:1310–1321. doi: 10.1016/j.mod.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 32.Oike H, Nagai T, Furuyama A, Okada S, Aihara Y, Ishimaru Y, et al. Characterization of ligands for fish taste receptors. J Neurosci. 2007;27:5584–5592. doi: 10.1523/JNEUROSCI.0651-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delay R, Taylor R, Roper SD. Merkel-like basal cells in Necturus taste buds contain serotonin. J Comp Neurol. 1993;335:606–613. doi: 10.1002/cne.903350411. [DOI] [PubMed] [Google Scholar]
- 34.Toyoshima K, Shimamura A. Monoamine-containing basal cells in the taste buds of the newt Triturus pyrrhogaster. Arch Oral Biol. 1987;32:619–621. doi: 10.1016/0003-9969(87)90034-3. [DOI] [PubMed] [Google Scholar]
- 35.Toyoshima K, Nada O, Shimamura A. Fine structure of monoamine-containing basal cells in the taste buds on the barbels of three species of teleosts. Cell Tissue Res. 1984;235:479–484. doi: 10.1007/BF00226942. [DOI] [PubMed] [Google Scholar]
- 36.Zachar PC, Jonz MG. Confocal imaging of Merkel-like basal cells in the taste buds of zebrafish. Acta Histochem. 2012;114:101–115. doi: 10.1016/j.acthis.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 37.Beidler LM, Smallman RL. Renewal of cells within taste buds. J Cell Biol. 1965;27:263–272. doi: 10.1083/jcb.27.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farbman AI. Renewal of taste bud cells in rat circumvallate papillae. Cell Tissue Kinet. 1980;13:349–357. doi: 10.1111/j.1365-2184.1980.tb00474.x. [DOI] [PubMed] [Google Scholar]
- 39.Cook MH, Neal HV. Are the taste buds of elesmobranchs endodermal in origin? J Comp Neurol. 1921;33:45–63. [Google Scholar]
- 40.Johnston JB. The limit between the ectoderm and entoderm in the mouth, and the origin of taste buds. I. Amphibians. Am J Anat. 1910;10:41–67. [Google Scholar]
- 41.Barlow L, Northcutt RG. Embryonic origin of amphibian taste buds. Dev Biol. 1995;169:273–285. doi: 10.1006/dbio.1995.1143. [DOI] [PubMed] [Google Scholar]
- 42.Hörstadius S. The neural crest: its properties and derivatives in the light of experiemental research. Oxford: Oxford University Press; 1950. [Google Scholar]
- 43.Luo X, Okubo T, Randell S, Hogan BL. Culture of endodermal stem/progenitor cells of the mouse tongue. In Vitro Cell Dev Biol Anim. 2009;45:44–54. doi: 10.1007/s11626-008-9149-2. [DOI] [PubMed] [Google Scholar]
- 44.Okubo T, Clark C, Hogan BL. Cell lineage mapping of taste bud cells and keratinocytes in the mouse tongue and soft palate. Stem Cells. 2009;27:442–450. doi: 10.1634/stemcells.2008-0611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- 46.Chai Y, Jiang X, Ito Y, Bringas P, Han J, Rowitch DH, et al. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- 47.Thirumangalathu S, Harlow DE, Driskell AL, Krimm RF, Barlow LA. Fate mapping of mammalian embryonic taste bud progenitors. Development. 2009;136:1519–1528. doi: 10.1242/dev.029090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu HX, Komatsu Y, Mishina Y, Mistretta CM. Neural crest contribution to lingual mesenchyme, epithelium and developing taste papillae and taste buds. Dev Biol. 2012;368:294–303. doi: 10.1016/j.ydbio.2012.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamauchi Y, Abe K, Mantani A, Hitoshi Y, Suzuki M, Osuzu F, et al. A novel transgenic technique that allows specific marking of the neural crest cell lineage in mice. Dev Biol. 1999;212:191–203. doi: 10.1006/dbio.1999.9323. [DOI] [PubMed] [Google Scholar]
- 50.Feltri ML, D’antonio M, Quattrini A, Numerato R, Arona M, Previtali S, et al. A novel P0 glycoprotein transgene activates expression of lacZ in myelin-forming Schwann cells. Eur J Neurosci. 1999;11:1577–1586. doi: 10.1046/j.1460-9568.1999.00568.x. [DOI] [PubMed] [Google Scholar]
- 51.Kawakami M, Umeda M, Nakagata N, Takeo T, Yamamura KI. Novel migrating mouse neural crest cell assay system utilizing P0-Cre/EGFP fluorescent time-lapse imaging. BMC Dev Biol. 2011;11:68. doi: 10.1186/1471-213X-11-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Couly G, Le Douarin NM. Head morphogenesis in embryonic avian chimeras: evidence for a segmental pattern in the ectoderm corresponding to the neuromeres. Development. 1990;108:543–558. doi: 10.1242/dev.108.4.543. [DOI] [PubMed] [Google Scholar]
- 53.Rothova M, Thompson H, Lickert H, Tucker AS. Lineage tracing of the endoderm during oral development. Dev Dyn. 2012;241:1183–1191. doi: 10.1002/dvdy.23804. [DOI] [PubMed] [Google Scholar]
- 54.Soukup V, Epperlein HH, Horácek I, Cerny R. Dual epithelial origin of vertebrate oral teeth. Nature. 2008;455:795–798. doi: 10.1038/nature07304. [DOI] [PubMed] [Google Scholar]
- 55.Soukup V, Horácek I, Cerny R. Development and evolution of the vertebrate primary mouth. J Anat. 2012 doi: 10.1111/j.1469-7580.2012.01540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mistretta CM, Liu HX. Development of fungiform papillae: Patterned lingual gustatory organs. Arch Histol Cytol. 2006;69:199–208. doi: 10.1679/aohc.69.199. [DOI] [PubMed] [Google Scholar]
- 57.Oakley B, Witt M. Building sensory receptors on the tongue. J Neurocytol. 2004;33:631–646. doi: 10.1007/s11068-005-3332-0. [DOI] [PubMed] [Google Scholar]
- 58.Hall JM, Hooper JE, Finger TE. Expression of sonic hedgehog, patched, and Gli1 in developing taste papillae of the mouse. J Comp Neurol. 1999;406:143–155. doi: 10.1002/(sici)1096-9861(19990405)406:2<143::aid-cne1>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 59.Athar M, Tang X, Lee JL, Kopelovich L, Kim AL. Hedgehog signalling in skin development and cancer. Exp Dermatol. 2006;15:667–677. doi: 10.1111/j.1600-0625.2006.00473.x. [DOI] [PubMed] [Google Scholar]
- 60.Harfe BD, Scherz PJ, Nissim S, Tian H, McMahon AP, Tabin CJ. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 2004;118:517–528. doi: 10.1016/j.cell.2004.07.024. [DOI] [PubMed] [Google Scholar]
- 61.Guth L. Taste buds on the cat’s circumvallate papilla after innervation by glossopharyngeal, vagus and hypoglossal nerves. Anat Rec. 1958;130:25–37. doi: 10.1002/ar.1091300104. [DOI] [PubMed] [Google Scholar]
- 62.May RM. The relation of nerves to degenerating and regenerating taste buds. J Exp Zool. 1925;42:371–410. [Google Scholar]
- 63.Barlow LA, Chien CB, Northcutt RG. Embryonic taste buds develop in the absence of innervation. Development. 1996;122:1103–1111. doi: 10.1242/dev.122.4.1103. [DOI] [PubMed] [Google Scholar]
- 64.Stone LS. Independence of taste organs with respect to their nerve fibers demonstrated in living salamanders. Proc Soc Exp Biol Med. 1932;30:1256–1257. [Google Scholar]
- 65.Fritzsch B, Sarai P, Barbacid M, Silos-Santiago I. Mice lacking the neurotrophin receptor trkB lose their specific afferent innervation but do develop taste buds. Intl J Dev Neurosci. 1997;15:563–576. doi: 10.1016/s0736-5748(96)00111-6. [DOI] [PubMed] [Google Scholar]
- 66.Mistretta CM, Liu HX, Gaffield W, MacCallum DK. Cyclopamine and jervine in embryonic rat tongue cultures demonstrate a role for Shh signaling in taste papilla development and patterning: fungiform papillae double in number and form in novel locations in dorsal lingual epithelium. Dev Biol. 2003;253:1–18. doi: 10.1016/s0012-1606(02)00014-3. [DOI] [PubMed] [Google Scholar]
- 67.Hall JM, Bell ML, Finger TE. Disruption of sonic hedgehog signaling alters growth and patterning of lingual taste papillae. Dev Biol. 2003;255:263–277. doi: 10.1016/s0012-1606(02)00048-9. [DOI] [PubMed] [Google Scholar]
- 68.Stone LS. The origin and development of taste organs in salamanders observed in the living condition. J Exp Zool. 1940;83:481–506. [Google Scholar]
- 69.Ito A, Nosrat C. Gustatory papillae and taste bud development and maintenance in the absence of TrkB ligands BDNF and NT-4. Cell Tissue Res. 2009;337:349–359. doi: 10.1007/s00441-009-0833-7. [DOI] [PubMed] [Google Scholar]
- 70.Ito A, Nosrat I, Nosrat C. Taste cell formation does not require gustatory and somatosensory innervation. Neurosci Lett. 2010;471:189–194. doi: 10.1016/j.neulet.2010.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nosrat C, MacCallum D, Mistretta C. Distinctive spatiotemporal expression patterns for neurotrophins develop in gustatory papillae and lingual tissues in embryonic tongue organ cultures. Cell Tissue Res. 2001;303:35–45. doi: 10.1007/s004410000271. [DOI] [PubMed] [Google Scholar]
- 72.Krimm RF, Barlow LA. The development of the taste system. In: Shepard G, Firestein S, Smith DV, editors. The senses: a comprehensive reference. Taste and olfaction. Oxford: Elsevier; 2007. pp. 157–181. [Google Scholar]
- 73.Hosley MA, Hughes SE, Morton LL, Oakley B. A sensitive period for the neural induction of taste buds. J Neurosci. 1987;7:2075–2080. doi: 10.1523/JNEUROSCI.07-07-02075.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hosley MA, Hughes SE, Oakley B. Neural induction of taste buds. J Comp Neurol. 1987;260:224–232. doi: 10.1002/cne.902600206. [DOI] [PubMed] [Google Scholar]
- 75.Pispa J, Thesleff I. Mechanisms of ectodermal organogenesis. Dev Biol. 2003;262:195–205. doi: 10.1016/s0012-1606(03)00325-7. [DOI] [PubMed] [Google Scholar]
- 76.Chuong C, Patel N, Lin J, Jung H, Widelitz R. Sonic hedgehog signaling pathway in vertebrate epithelial appendage morphogenesis: perspectives in development and evolution. Cell Mol Life Sci. 2000;57:1672–1681. doi: 10.1007/PL00000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Noden D. The embryonic origins of avian cephalic and cervical muscles and associated connective tissues. Am J Anat. 1983;168:257–276. doi: 10.1002/aja.1001680302. [DOI] [PubMed] [Google Scholar]
- 78.Mackenzie S, Walsh FS, Graham A. Migration of hypoglossal myoblast precursors. Dev Dyn. 1998;213:349–358. doi: 10.1002/(SICI)1097-0177(199812)213:4<349::AID-AJA1>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 79.Beites CL, Hollenbeck PLW, Kim J, Lovell-Badge R, Lander AD, Calof AL. Follistatin modulates a BMP autoregulatory loop to control the size and patterning of sensory domains in the developing tongue. Development. 2009;136:2187–2197. doi: 10.1242/dev.030544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Petersen CI, Jheon AH, Mostowfi P, Charles C, Ching S, Thirumangalathu S, et al. FGF signaling regulates the number of posterior taste papillae by controlling progenitor field size. PLoS Genet. 2011;7:e1002098. doi: 10.1371/journal.pgen.1002098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Barlow LA. Specification of pharyngeal endoderm is dependent on early signals from axial mesoderm. Development. 2001;128:4573–4583. doi: 10.1242/dev.128.22.4573. [DOI] [PubMed] [Google Scholar]
- 82.Barlow LA, Northcutt RG. Taste buds develop autonomously from endoderm without induction by cephalic neural crest or paraxial mesoderm. Development. 1997;124:949–957. doi: 10.1242/dev.124.5.949. [DOI] [PubMed] [Google Scholar]
- 83.Kim M, Kusakabe Y, Miura H, Shindo Y, Ninomiya Y, Hino A. Regional expression patterns of taste receptors and gustducin in the mouse tongue. Biochem Biophys Res Commun. 2003;312:500–506. doi: 10.1016/j.bbrc.2003.10.137. [DOI] [PubMed] [Google Scholar]
- 84.Tizzano M, Dvoryanchikov G, Barrows JK, Kim S, Chaudhari N, Finger TE. Expression of Galpha14 in sweet-transducing taste cells of the posterior tongue. BMC Neurosci. 2008;9:110. doi: 10.1186/1471-2202-9-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nguyen HM, Barlow LA. Differential expression of a BMP4 reporter allele in anterior fungiform versus posterior circumvallate taste buds of mice. BMC Neurosci. 2010;11:129. doi: 10.1186/1471-2202-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Barlow LA. Taste buds in ectoderm are induced by endoderm: implications for mechanisms governing taste bud development. In: Olsson L, Jacobson CO, editors. Regulatory processes in development: the legacy of Sven Hörstadius; Proceedings of the Wenner Gren International Symposium; London: Portland Press; 2000. pp. 185–190. [Google Scholar]
- 87.Kapsimali M, Kaushik AL, Gibon G, Dirian L, Ernest S, Rosa FM. Fgf signaling controls pharyngeal taste bud formation through miR-200 and Delta-Notch activity. Development. 2011;138:3473–3484. doi: 10.1242/dev.058669. [DOI] [PubMed] [Google Scholar]
- 88.Angers S, Moon RT. Proximal events in Wnt signal transduction. Nat Rev Mol Cell Biol. 2009;10:468–477. doi: 10.1038/nrm2717. [DOI] [PubMed] [Google Scholar]
- 89.DasGupta R, Fuchs E. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development. 1999;126:4557–4568. doi: 10.1242/dev.126.20.4557. [DOI] [PubMed] [Google Scholar]
- 90.Iwatsuki K, Liu HX, Grónder A, Singer MA, Lane TF, Grosschedl R, et al. Wnt signaling interacts with Shh to regulate taste papilla development. Proc Natl Acad Sci U S A. 2007;104:2253–2258. doi: 10.1073/pnas.0607399104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu F, Thirumangalathu S, Gallant NM, Yang SH, Stoick-Cooper CL, Reddy ST, et al. Wnt-beta-catenin signaling initiates taste papilla development. Nat Genet. 2007;39:106–112. doi: 10.1038/ng1932. [DOI] [PubMed] [Google Scholar]
- 92.Bitgood MJ, McMahon PA. Hedgehog and Bmp genes are coexpressed at many diverse sites of cell-cell interaction in the mouse embryo. Dev Biol. 1995;172:126–138. doi: 10.1006/dbio.1995.0010. [DOI] [PubMed] [Google Scholar]
- 93.Jung H, Oropeza V, Thesleff I. Shh, Bmp-2, Bmp-4 and Fgf-8 are associated with initiation and patterning of mouse tongue papillae. Mech Dev. 1999;81:179–182. doi: 10.1016/s0925-4773(98)00234-2. [DOI] [PubMed] [Google Scholar]
- 94.Yamamoto Y, Byerly MS, Jackman WR, Jeffery WR. Pleiotropic functions of embryonic sonic hedgehog expression link jaw and taste bud amplification with eye loss during cavefish evolution. Dev Biol. 2009;330:200–211. doi: 10.1016/j.ydbio.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yamamoto Y, Stock DW, Jeffery WR. Hedgehog signalling controls eye degeneration in blind cavefish. Nature. 2004;431:844–847. doi: 10.1038/nature02864. [DOI] [PubMed] [Google Scholar]
- 96.Zouvelou V, Luder HV, Mitsiadis TA, Graf D. Deletion of BMP7 affects the development of bones, teeth, and other ectodermal appendages of the orofacial complex. J Exp Zool B Mol Dev Evol. 2009;312B:361–374. doi: 10.1002/jez.b.21262. [DOI] [PubMed] [Google Scholar]
- 97.Zhou Y, Liu HX, Mistretta CM. Bone morphogenetic proteins and noggin: inhibiting and inducing fungiform taste papilla development. Dev Biol. 2006;297:198–213. doi: 10.1016/j.ydbio.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 98.Kiernan AE, Pelling AL, Leung KKH, Tang ASP, Bell DM, Tease C, et al. Sox2 is required for sensory organ development in the mammalian inner ear. Nature. 2005;434:1031–1035. doi: 10.1038/nature03487. [DOI] [PubMed] [Google Scholar]
- 99.Wegner M, Stolt CC. From stem cells to neurons and glia: a Soxist’s view of neural development. Trends Neurosci. 2005;28:583–588. doi: 10.1016/j.tins.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 100.Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Okubo T, Pevny LH, Hogan BLM. Sox2 is required for development of taste bud sensory cells. Gen Dev. 2006;20:2654–2659. doi: 10.1101/gad.1457106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Böttcher RT, Niehrs C. Fibroblast growth factor signaling during early vertebrate development. Endocr Rev. 2005;26:63–77. doi: 10.1210/er.2003-0040. [DOI] [PubMed] [Google Scholar]
- 103.Crump JG, Maves L, Lawson ND, Weinstein BM, Kimmel CB. An essential role for Fgfs in endodermal pouch formation influences later craniofacial skeletal patterning. Development. 2004;131:5703–5716. doi: 10.1242/dev.01444. [DOI] [PubMed] [Google Scholar]
- 104.David NB, Saint-Etienne L, Tsang M, Schilling TE, Rosa FM. Requirement for endoderm and FGF3 in ventral head skeleton formation. Development. 2002;129:4457–4468. doi: 10.1242/dev.129.19.4457. [DOI] [PubMed] [Google Scholar]
- 105.Walshe J, Mason I. Fgf signalling is required for formation of cartilage in the head. Dev Biol. 2003;264:522–536. doi: 10.1016/j.ydbio.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 106.Guruharsha KG, Kankel MW, Artavanis-Tsakonas S. The Notch signalling system: recent insights into the complexity of a conserved pathway. Nat Rev Genet. 2012;13:654–666. doi: 10.1038/nrg3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yoon K, Gaiano N. Notch signaling in the mammalian central nervous system: insights from mouse mutants. Nat Neurosci. 2005;8:709–715. doi: 10.1038/nn1475. [DOI] [PubMed] [Google Scholar]
- 108.Seta Y, Seta C, Barlow LA. Notch-associated gene expression in embryonic and adult taste papillae and taste buds suggests a role in taste cell lineage decisions. J Comp Neurol. 2003;464:49–61. doi: 10.1002/cne.10787. [DOI] [PubMed] [Google Scholar]
- 109.Ota MS, Kaneko Y, Kondo K, Ogishima S, Tanaka H, Eto K, et al. Combined in silico and in vivo analyses reveal role of Hes1 in taste cell differentiation. PLoS Genet. 2009;5:e1000443. doi: 10.1371/journal.pgen.1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Seta Y, Oda M, Kataoka S, Toyono T, Toyoshima K. Mash1 is required for the differentiation of AADC-positive type III cells in mouse taste buds. Dev Dyn. 2011;240:775–784. doi: 10.1002/dvdy.22576. [DOI] [PubMed] [Google Scholar]
- 111.Miura H, Kusakabe Y, Harada S. Cell lineage and differentiation in taste buds. Arch Histol Cytol. 2006;69:209–225. doi: 10.1679/aohc.69.209. [DOI] [PubMed] [Google Scholar]
- 112.Brooker R, Hozumi K, Lewis J. Notch ligands with contrasting functions: Jagged1 and Delta1 in the mouse inner ear. Development. 2006;133:1277–1286. doi: 10.1242/dev.02284. [DOI] [PubMed] [Google Scholar]
- 113.Matsumoto I, Ohmoto M, Narukawa M, Yoshihara Y, Abe K. Skn-1a (Pou2f3) specifies taste receptor cell lineage. Nat Neurosci. 2011;14:685–687. doi: 10.1038/nn.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 115.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 116.Pauli A, Rinn JL, Schier AF. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet. 2011;12:136–149. doi: 10.1038/nrg2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kosik KS. The neuronal microRNA system. Nat Rev Neurosci. 2006;7:911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- 118.Peter ME. Let-7 and miR-200 microRNAs: guardians against pluripotency and cancer progression. Cell Cycle. 2009;8:843–852. doi: 10.4161/cc.8.6.7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 120.Kapsimali M, Kloosterman WP, de Bruijn E, Rosa F, Plasterk RHA, Wilson SW. MicroRNAs show a wide diversity of expression profiles in the developing and mature central nervous system. Genome Biol. 2007;8:173. doi: 10.1186/gb-2007-8-8-r173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 122.Lawson KA, Dunn NR, Roelen BA, Zeinstra LM, Davis AM, Wright CV, Korving JP, Hogan BL. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 1999;13:424–436. doi: 10.1101/gad.13.4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]