Abstract
Chronological aging of budding yeast cells results in a reduction in subsequent replicative life span through unknown mechanisms. Here we show that dietary restriction during chronological aging delays the reduction in subsequent replicative life span up to at least 23 days of chronological age. We further show that among the viable portion of the control population aged 26 days, individual cells with the lowest mitochondrial membrane potential have the longest subsequent replicative lifespan. These observations demonstrate that dietary restriction modulates a common molecular mechanism linking chronological and replicative aging in yeast and indicate a critical role for mitochondrial function in this process.
Keywords: chronological lifespan, replicative lifespan, caloric restriction, calorie restriction, dietary restriction, glucose, mitochondria
1. INTRODUCTION
The molecular and genetic mechanisms of aging have been studied extensively in the budding yeast Saccharomyces cerevisiae (Kaeberlein, 2010). Two distinct types of aging are commonly modeled in yeast: replicative and chronological (Longo et al., 2012). Replicative life span (RLS) is defined by the number of daughter cells produced by a mother cell prior to irreversible arrest (Mortimer and Johnston, 1959). Chronological life span (CLS) is defined as the length of time that a quiescent yeast cell can retain the capacity to re-enter mitotic growth upon appropriate nutritional cues (Fabrizio and Longo, 2003).
The determinants of chronological and replicative longevity in yeast appear to be both overlapping and distinct (Longo et al., 2012). Among those features that are shared between the two aging models is a robust lifespan extension in response to dietary restriction (DR), accomplished by reducing the glucose concentration of the culture medium from 2% to 0.5% or lower (Jiang et al., 2000; Kaeberlein et al., 2004; Lin et al., 2000; Murakami et al., 2008; Smith et al., 2007). Several interventions that decrease signaling through the nutrient-responsive target of rapamycin (TOR) pathway also extend both CLS and RLS (Laun et al., 2006), including deletion of genes encoding the mechanistic target of rapamycin (mTOR) homolog Tor1 or the ribosomal S6 kinase homolog Sch9, as well as treating yeast cells with the mTOR complex 1 inhibitor rapamycin (Fabrizio et al., 2004b; Fabrizio et al., 2001; Kaeberlein et al., 2005; Powers et al., 2006). Each of these interventions has also been shown to extend lifespan in nematodes, fruit flies, and mice, demonstrating that both yeast aging paradigms share conservation with aging in evolutionarily divergent multicellular organisms (Longo et al., 2012).
Although both types of yeast aging are strongly influenced by glucose availability and nutrient signaling, it remains unclear to what extent aging is caused by similar downstream molecular events in each system. Both chronological and replicative aging are associated with increased accumulation of mitochondrial damage and oxidatively damaged/aggregated proteins (Kaeberlein, 2010). Although these correlated molecular factors are very likely to play a causal role in determining both CLS and RLS, this has been difficult to convincingly establish. Instead, some causes of aging in each system appear to be private for each type of yeast aging: genomic instability within the rDNA limiting RLS, and cell death arising from acidification of the culture medium limiting CLS.
Genomic instability within the rDNA array during replicative aging can be observed through an age-associated accumulation of extrachromosomal rDNA circles within the mother cell (Sinclair and Guarente, 1997). DNA episomes induce a similar life shortening stress (Falcon and Aris, 2003). This instability can be suppressed by overexpression of the histone deacetylase Sir2 or deletion of the replication fork block protein Fob1, which both extend RLS (Defossez et al., 1999; Kaeberlein et al., 1999). Deletion of SIR2, on the other hand, increases rDNA instability and dramatically shortens RLS, but does not shorten CLS, and actually extends CLS under certain conditions (Fabrizio et al., 2005; Kaeberlein et al., 1999; Smith et al., 2007; Wu et al., 2011). These observations have led to the idea that rDNA instability is specific to yeast replicative aging and does not influence chronological aging (Steinkraus et al., 2008).
Acidification of the culture medium is a limiting factor for CLS when performed by quantifying survival of yeast cells aged in expired synthetic complete (SC) medium, conditions employed in the majority of published CLS studies (Longo et al., 2012). Buffering the culture medium to pH 6.0, addition of NaOH, or a shift to water after acid production during aging is sufficient to dramatically increase CLS (Burtner et al., 2009b; Burtner et al., 2011; Fabrizio et al., 2004a; Fabrizio et al., 2005; Murakami et al., 2011). Since RLS analysis is performed by isolation and microdissection of individual mother cells on the surface of an agar-based rich growth medium, medium acidification does not occur during this assay and buffering the medium does not extend RLS (our unpublished data).
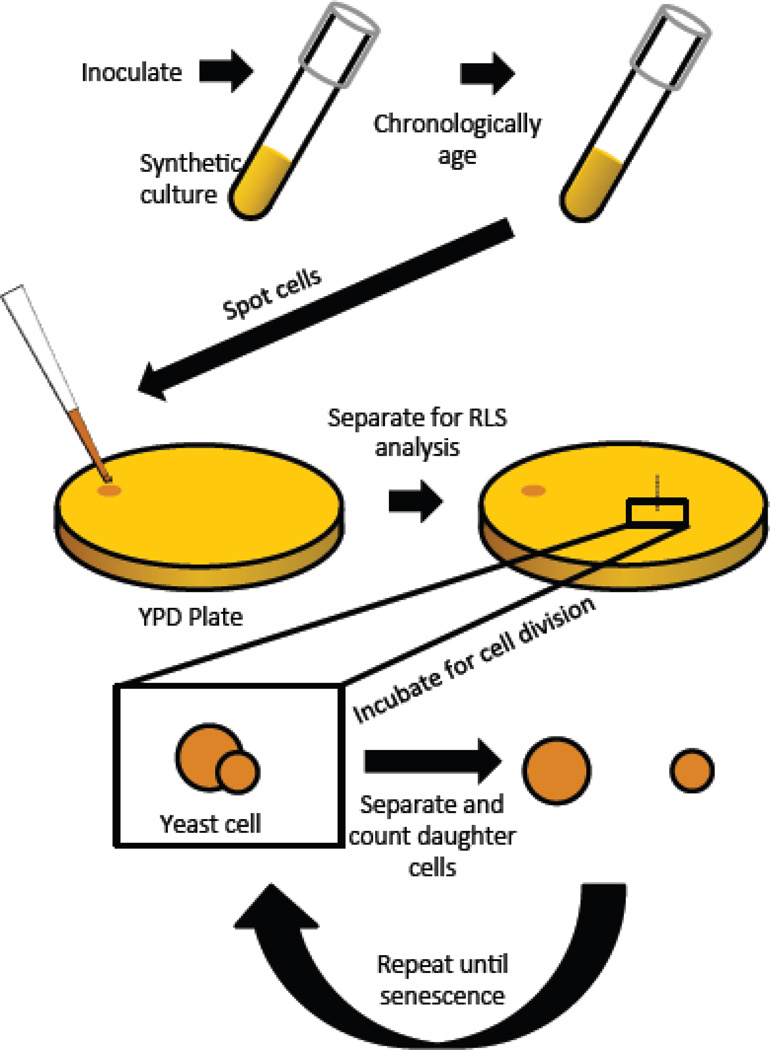
In order to better understand the shared molecular features of replicative and chronological aging in yeast, we have begun studying the effect of chronological aging on the subsequent RLS of mother cells. This is accomplished by aging yeast cells chronologically for various periods of time in liquid culture, removing an aliquot from the chronologically aging culture and plating the cells onto rich medium (2% yeast extract, 1% bacto peptone, YEP) agar plates with 2% glucose as the carbon source, arranging a sufficient number of cells for RLS analysis, and microdissecting daughter cells from mother cells to determine the RLS of those cells that are able to undergo at least one cell division (still chronologically alive). A schematic of this procedure is shown in Figure 1.
Figure 1. Diagram depicting the chronological lifespan to replicative lifespan assay.
Cells are aged chronologically in synthetic complete (SC) medium with either 2% glucose (control) or 0.05% glucose (DR). At different age points, an aliquot is removed from the chronologically aging culture and spotted onto rich medium (YPD) for replicative lifespan analysis. Individual cells are arrayed on the YPD plate and those replicative lifespan is determined.
Only a few studies have previously described this phenomenon of chronological aging reducing RLS. Ashrafi et al., (Ashrafi et al., 1999) reported that yeast cells aged chronologically in liquid YEP with 2% glucose as the carbon source show a reduction in RLS that is proportional to their chronological age. Importantly, this reduction in RLS did not involve rDNA instability as measured by accumulation of extrachromosomal rDNA circles. Piper et al. (Piper et al., 2006) showed a similar reduction in RLS when cells were aged chronologically in water at high temperature. More recently, we have confirmed these findings and have extended them to the more commonly used chronological aging protocol of culturing cells in liquid SC medium supplemented with 2% glucose (Murakami et al., 2012). In that study, we found that cells aged chronologically in SC 2% glucose medium show a more rapid reduction in RLS than cells aged chronologically in YEP 2% glucose medium. We also reported that buffering the SC 2% glucose culture medium to pH 6 dramatically attenuated the reduction in RLS associated with chronological age. This latter observation demonstrates that extracellular acidification of the culture medium drives intracellular changes that limit subsequent RLS.
Here we tested whether DR by reducing glucose from 2% to 0.05% could influence the effect of chronological age on subsequent RLS. Similar to the effect of buffering the medium, DR protected chronologically aging cells from a reduction in replicative lifespan. Among the cells aged in chronologically in 2% glucose, those that maintained the lowest mitochondrial membrane potential also retained the longest subsequent replicative lifespan. These observations suggest that metabolic state and mitochondrial function during the quiescent period determines subsequent replicative potential upon return to growth-promoting conditions.
2. MATERIALS AND METHODS
2.1 Yeast strains and media
All experiments were performed in the BY4743 strain obtained from Open Biosystems. Viability following chronological aging was obtained from the percentage of cells that were able to complete at least one mitotic division during the replicative aging assay. All chronological aging experiments were performed as previously described (Burtner et al., 2009a; Murakami and Kaeberlein, 2009; Murakami et al., 2008). Cultures were initiated by seeding a 5 ml liquid culture of YEPD with a single colony from a freshly streaked strain grown on YEPD agar at 30°C. A 1:100 dilution of the YEPD culture was made into SC medium, containing 2% glucose, unless otherwise noted. Basic medium is 1.7g/L Yeast Nitrogen Base (−AA/−AS) (BD Difco™,, Franklin Lakes, NJ, USA) and 5g/L (NH4)2SO4. Components of the SC medium used in this study have been described in detail (Murakami and Kaeberlein, 2009; Murakami et al., 2008). All strain auxotrophies for BY4743 (leucine, histidine, and uracil) were compensated with a fourfold excess of amino acids. Cultures were grown and aged in a roller drum enclosed in a water-jacketed incubator at 30°C. YEPD was 20g/L Bacto Peptone and 10g/L Yeast Extract (BD Difco™, Franklin Lakes, NJ, USA), supplemented with glucose at the indicated concentrations.
2.2 Replicative lifespan analysis
RLS was determined using a standard yeast tetrad dissection scope as previously described (Steffen et al., 2009), with the modification that virgin daughter cells were not specifically selected by allowing a cell division cycle to occur prior to initiating the experiment. Instead, 5 µL of the chronological aging culture was spotted onto 2% glucose YEPD plates, allowed to dry into the plates, and 60–1080 cells (depending on the predicted viability of the culture at that point) were randomly selected for RLS analysis. Selected cells were moved to the middle of the YEPD plates and the RLS of those cells was measured. Statistical significance for lifespan differences was determined using the Wilcoxon Rank-Sum test. Regression statistics were performed using Microsoft Excel Analysis Toolpak’s “Regression” function, and regression lines were compared using Microsoft Excel Analysis Toolpak’s “ANOVA: two factor without replication” function.
2.3 Data collection and analysis
RLS data for the cells aged chronologically under control conditions were presented in a prior report (Murakami et al., 2012). All of the data shown here for both control and DR cells were obtained from experiments performed at the same time, with all of the chronologically aged cultures for both control and DR experiments aged in medium prepared from the same batch and cultured in the same incubator. All of the RLS data presented in this study were obtained from cells aged on medium prepared from the same batch and dissected in parallel. All RLS experiments were performed blind, such that the identities of the strains being microdissected were unknown to the individuals performing the assay.
2.4 Flow cytometry
All flow cytometry experiments were performed in the University of Washington Nathan Shock Center of Excellence in the Basic Biology of Aging Imaging Core using a BD Biosciences Influx Cell Sorter. Cells were aged chronologically and then 5 × 107 cells were harvested for flow cytometric analysis. Cells were spun down (7k rpm 1 minute) and the supernatant removed. Cells were washed in sterile 50 mM Na citrate buffer, spun down again, and the supernatant discarded. Cells were resuspended in 50 mM Na citrate containing 5nM SYTOX Red (Invitrogen, S7020, Grand Island, NY, USA) and 17.5nM DiOC6. At a tenfold higher concentration, DiOC6 can stain the ER and plasma membrane, and this caveat must be kept in mind (Koning et al., 1993). Cells were kept on ice and immediately run through the flow cytometer. Mean readings for DiOC6 content were normalized to cell size by dividing by forward scatter measurements. 20,000 cells were used in each biological replicate measurement.
3. RESULTS
3.1 Dietary restriction protects against replicative life span shortening during chronological aging
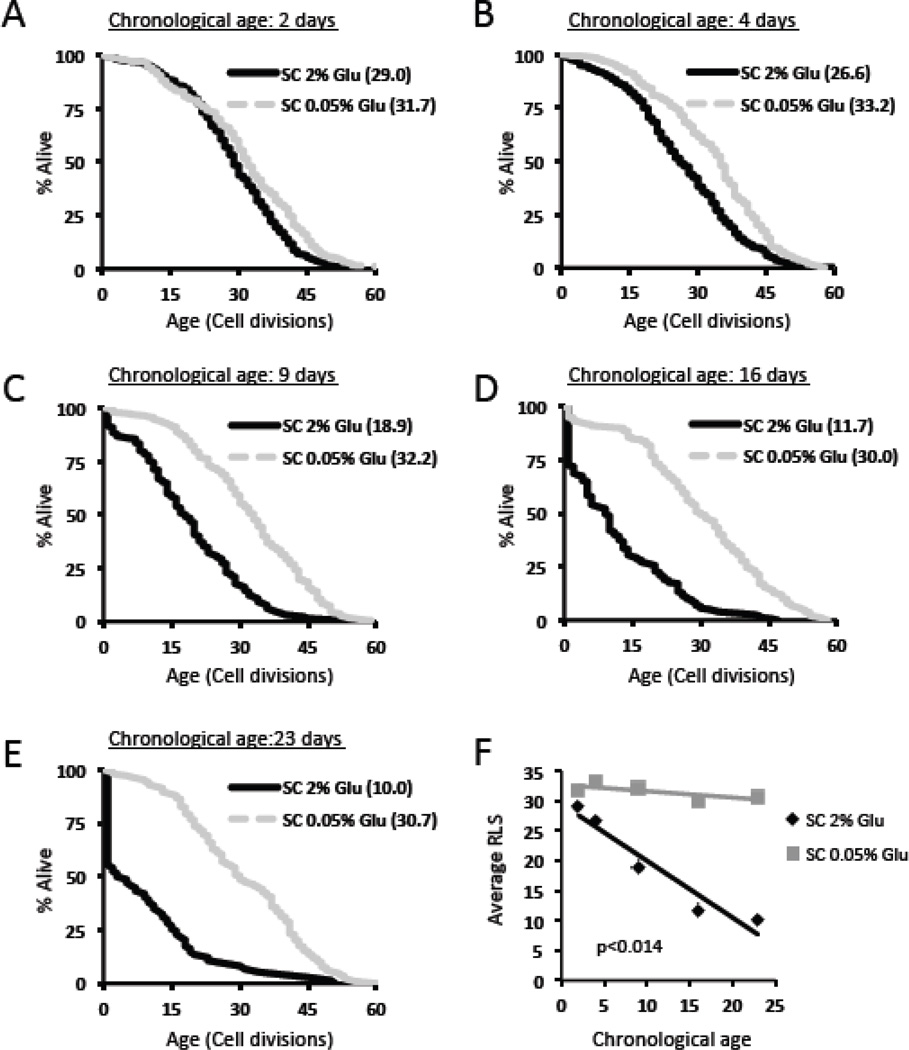
Cells were aged chronologically in SC 2% glucose medium (control) or SC 0.05% glucose medium (DR medium), and RLS analysis was performed on chronologically aged cells at day 2, 4, 9, 16, or 24 of the experiment (Figure 2a–e). Only those cells that were able to divide at least once were included in the RLS analysis. Cells aged chronologically under control conditions had a reduced subsequent RLS compared to cells aged chronologically under DR conditions at every age-point tested (Table 1). Over the entire course of the chronological aging experiment, cells aged under control conditions showed a significant inverse correlation between chronological age and subsequent RLS (Figure 2f, p<0.008). A similar correlation was not detected when cells were chronologically aged under DR conditions (p=0.2).
Figure 2. Dietary restriction slows the reduction in replicative lifespan following chronological aging.
Replicative lifespan curves of cells aged chronologically in SC 2% glucose (control) or SC 0.05% glucose (DR) for (a) 2 days, (b) 4 days, (c) 9 days, (d) 16 days, or (e) 23 days. Cells which did not divide are not included. Mean replicative lifespan is shown in parentheses. (f) Correlation plot of average replicative life span (RLS) with chronological age for cells aged chronologically in SC 2% glucose or SC 0.05% glucose.
Table 1.
Summary of data replicative lifespan data for chronologically aged cells without censoring
| SC 2% glucose | |||||
| Chronological age (days) | 2 | 4 | 9 | 16 | 23 |
| # cells set up | 240 | 480 | 840 | 1200 | 1680 |
| # cells w/ 1+ divisions | 226 | 353 | 130 | 157 | 74 |
| 0 divisions (% of all cells) |
5.8 | 26.5 | 84.5 | 86.9 | 95.6 |
| 1 division (% of dividing cells) |
0.4 | 1.4 | 8.5 | 27.4 | 44.6 |
| 2+ divisions (% of dividing cells) |
99.6 | 98.6 | 91.5 | 72.6 | 55.4 |
| - | - | - | - | - | - |
| Mean RLS | 29.0 | 26.6 | 18.9 | 11.7 | 10.0 |
| SC 0.05% glucose | |||||
| Chronological age (days) | 2 | 4 | 9 | 16 | 23 |
| # cells set up | 240 | 240 | 240 | 240 | 240 |
| # cells w/ 1+ divisions | 224 | 237 | 222 | 226 | 175 |
| 0 divisions (% of all cells) |
6.7 | 1.3 | 7.5 | 5.8 | 27.1 |
| 1 division (% of dividing cells) |
0.9 | 0.0 | 0.0 | 4.9 | 0.6 |
| 2+ divisions (% of dividing cells) |
99.1 | 100 | 100 | 95.1 | 99.4 |
| - | - | - | - | - | - |
| Mean RLS | 31.7 | 33.2 | 32.2 | 30.0 | 30.7 |
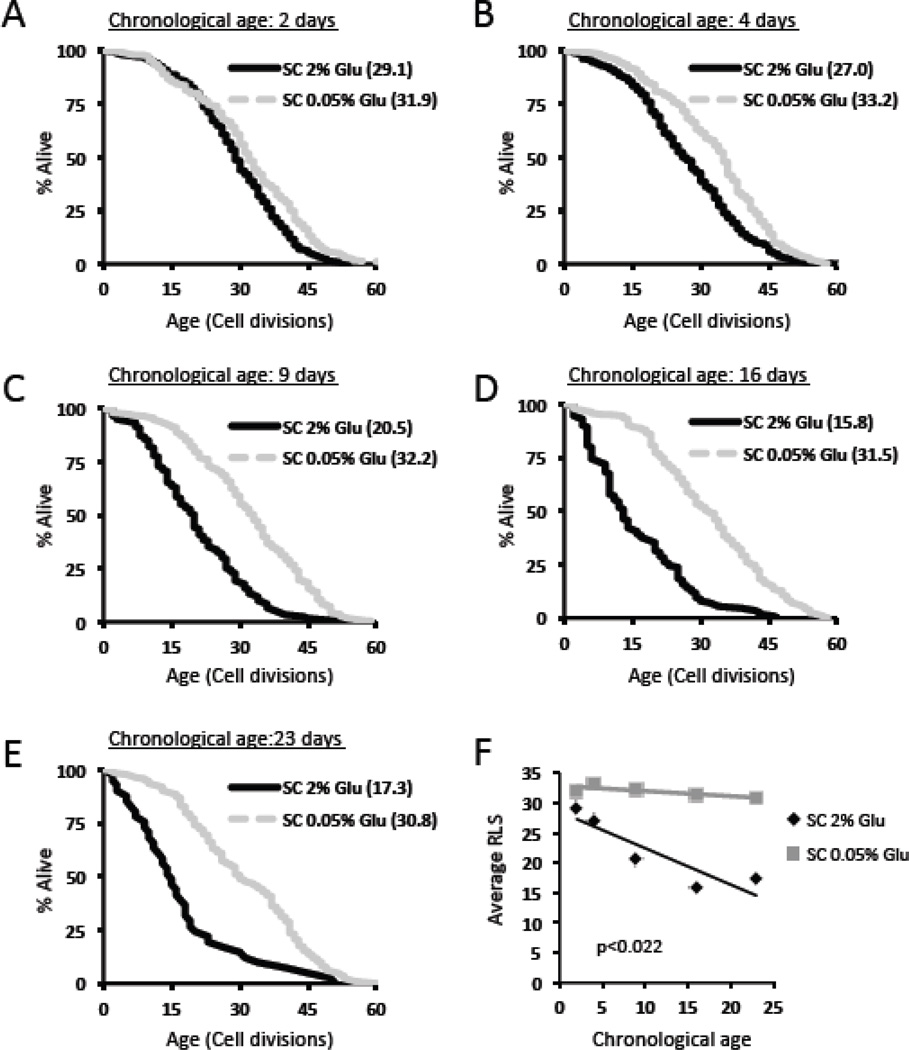
During the analysis, we noted an increase in the number of chronologically aged control cells that were only able to complete a single cell division during subsequent RLS analysis (Table 1). Even after censoring these cells from the RLS analysis, cells aged chronologically under control conditions were shorter-lived than cells aged chronologically under DR conditions at all age-points tested, and a similar significant inverse correlation between chronological age and subsequent RLS was detected for control (p<0.04) but not DR p=0.09) cells (Figure 3).
Figure 3. Censoring cells that only divide once does not significantly alter the effect of DR on replicative life span following chronological aging.
Replicative lifespan curves of cells aged chronologically in SC 2% glucose or SC 0.05% glucose (DR) for (a) 2 days, (b) 4 days, (c) 9 days, (d) 16 days, or (e) 23 days. Cells which did not divide are not included. Mean replicative lifespan is shown in parentheses. (f) Correlation plot of average replicative life span (RLS) with chronological age for cells aged chronologically in SC 2% glucose or SC 0.05% glucose.
As expected, we observed that more cells were able to resume mitotic growth during the RLS portion of the experiment when aged under DR conditions, relative to control conditions (Table 2). This was true regardless of whether chronologically viable cells are defined as being able to complete at least one (cells dividing only once would be counted as alive), or more than one, mitotic cycle. These data provide important confirmation for the positive effects of DR on CLS using a microscopy-based microdissection assay, rather than indirect colony forming unit or outgrowth kinetics based quantitation methods.
Table 2.
Summary of data replicative lifespan data for chronologically aged cells with cells that only divided one time censored
| SC 2% glucose | |||||
| Chronological age (days) | 2 | 4 | 9 | 16 | 23 |
| # cells set up | 240 | 480 | 840 | 1200 | 1680 |
| # cells w/ 2+ divisions | 225 | 348 | 119 | 114 | 41 |
| Mean RLS | 29.1 | 27.0 | 20.5 | 15.8 | 17.3 |
| SC 0.05% glucose | |||||
| Chronological age (days) | 2 | 4 | 9 | 16 | 23 |
| # cells set up | 240 | 240 | 240 | 240 | 240 |
| # cells w/ 2+ divisions | 222 | 237 | 222 | 215 | 174 |
| Mean RLS | 31.9 | 33.2 | 32.2 | 31.5 | 30.8 |
3.2 Mitochondrial membrane potential during chronological aging determines subsequent RLS
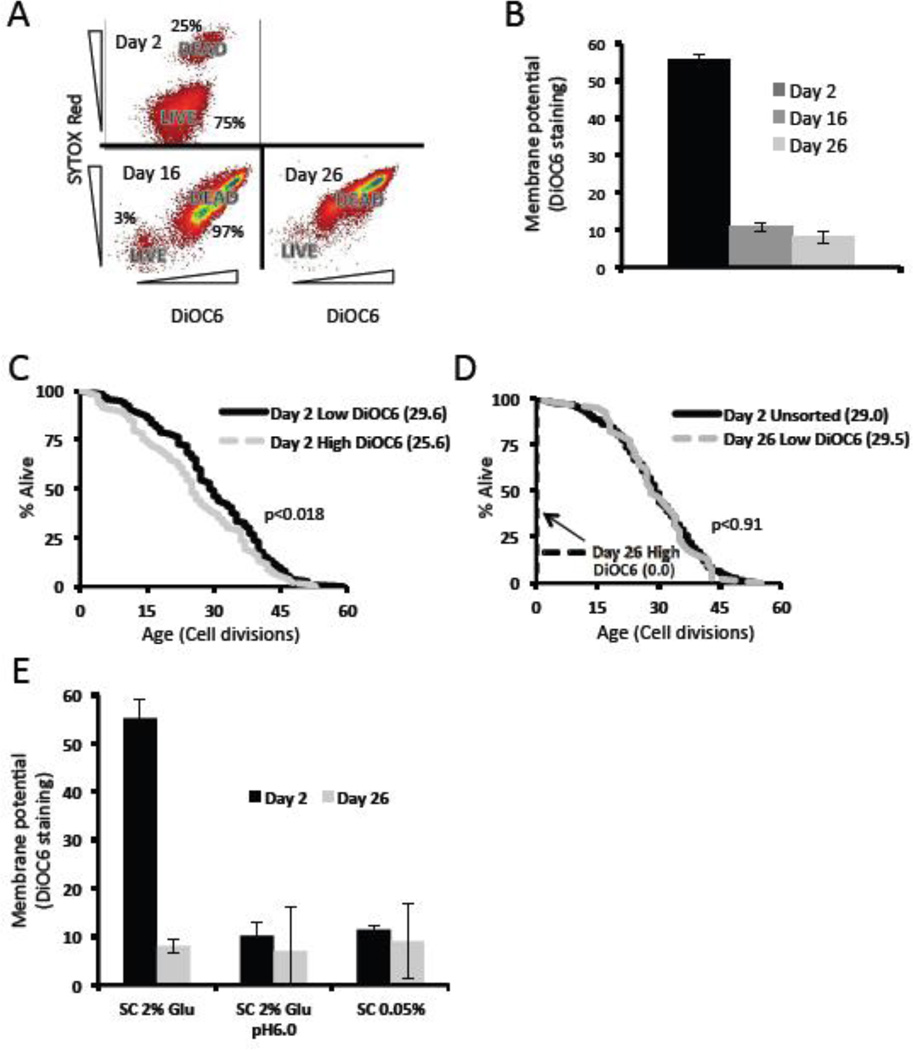
Mitochondrial dysfunction has been proposed to underlie both replicative and chronological aging, and degenerate mitochondria are asymmetrically inherited by the mother cell during cell division (Lai et al., 2002; Lam et al., 2011; McFaline-Figueroa et al., 2011). We therefore examined whether mitochondrial membrane potential during chronological aging could predict subsequent RLS. To address this question, we utilized the mitochondrial membrane potential dependent dye DiOC6 to measure membrane potential and the dye SYTOX Red to differentiate between live versus dead sub-populations during chronological aging. Cells were aged chronologically for 2, 16, or 26 days and sorted based on SYTOX Red staining. As expected, at day 2 nearly all cells were SYTOX Red negative (live sub-population); whereas by day 16 an increase in SYTOX Red positive cells was observed (Figure 4A). Membrane potential was reduced in the live cell sub-population by day 16 (Figure 4B).
Figure 4. Low mitochondrial membrane potential during chronological aging predicts longer replicative lifespan.
(a) Chronologically aged cells were co-stained for SYTOX Red (a vital dye) and DiOC6. Only cells with low SYTOX Red content were analyzed for DiOC6 levels (labeled LIVE). Relative percentages of live and dead gates are labeled for comparison. (b) Mean DiOC6 staining of live cells during chronological aging. (c,d) Replicative lifespans of cells co-stained for SYTOX Red and DiOC6. Days refer to the days spent chronologically in 2% glucose media, and “high” and “low” refer to the highest and lowest 5–10% DiOC6 stained cells. (e) Mean DiOC6 staining of live cells subjected to the indicated media conditions during chronological aging. The percent of the cell populations that were alive based on SYTOX Red at day 2 were: SC 2% glu = 75%, SC 2% glu pH 6 = 85%, SC 0.05% glu = 84%. The percent of the cell populations that were alive based on SYTOX Red at day 26 were: SC 2% glu = 2.8%, SC 2% glu pH 6 = 75%, SC 0.05% glu = 79%.
To determine whether elevated mitochondrial membrane potential might also have an effect on subsequent replicative lifespan, we sorted live (SYTOX Red negative) cells aged in SC 2% glucose by DiOC6 intensity and measured subsequent RLS. We predicted that among the cells aged in SC 2% glucose those with the highest membrane potential would have the longest replicative lifespan. Surprisingly, the chronologically aged cells with the highest mitochondrial membrane potential (top 5–10% of the live population) had a slightly reduced RLS even as early as day 2 (Figure 4C, p<0.02) and failed to divide even once by day 26 (Figure 4D). In contrast, cells with the lowest (bottom 5–10% of the population) mitochondrial membrane potential remarkably showed no decrease in RLS even after 26 days of chronological aging (Figure 4D, p=0.9). To address the possibility that the reduced DiOC6 intensity was caused by cells that had lost their ability to respire due to loss of mitochondrial DNA (rho0), we allowed these low staining cells to grow into colonies on YPD and replica plated these colonies to medium containing the non-fermentable carbon source glycerol as the sole carbon source. All of these colonies (20/20) were capable of growth under non-fermentable conditions from both day 2 and day 26 cultures, demonstrating that they retained functional mitochondria.
Since we have observed that buffering (Murakami et al., 2012) and DR (this publication) both attenuate the reduction in subsequent RLS which occurs during chronological aging, we wondered if these different culture conditions also influenced mitochondria membrane potential during chronological aging. Indeed, DiOC6 intensity was lower in both buffered and DR cultures at day 2 compared to SC 2% glucose cultures (Figure 4E). This supports the notion that DR and buffering may delay a reduction in subsequent RLS by maintaining a low mitochondria membrane potential throughout life.
4. DISCUSSION
It has been known for more than 10 years that chronological aging reduces RLS in budding yeast; yet, this fundamentally interesting observation has gone largely unstudied. Since DR is known to extend both CLS and RLS in yeast, we hypothesized that DR might also attenuate the reduction in RLS after chronological aging. Consistent with this hypothesis, we found that cells aged chronologically under DR conditions showed no significant decrease in RLS after up to 23 days of aging, while cells aged chronologically under control conditions showed a 65% reduction in mean RLS over this time course. Among the cells aged under control conditions, those that retained the lowest DiOC6 staining, a measure of mitochondrial membrane potential, retained a full RLS. Cells aged under DR conditions had low DiOC6 staining even as early as 2 days of chronological age. Taken together, these data demonstrate that DR attenuates the reduction in RLS caused by chronological aging in yeast. Further, among the cells aged chronologically in SC 2% glucose, reduced mitochondrial membrane potential predicts subsequent replicative longevity, suggesting that the molecular mechanisms linking chronological and replicative aging may center on preservation of mitochondrial function.
One mechanism by which DR might protect against reduced replicative capacity following chronological aging is by preventing acidification of the extracellular environment. We have shown that DR, accomplished either by reducing the glucose present in the medium or by utilizing alternative carbon sources, prevents medium acidification, likely due to the reduced metabolic production of acetic acid following alcoholic fermentation (Burtner et al., 2009b; Murakami et al., 2011). We have also recently reported that buffering the chronological culture medium to pH 6.0 attenuates the effect of chronological aging on RLS (Murakami et al., 2012), providing a potential link between these two sets of prior observations and this study. The mechanisms by which aging in an acidic environment can reduce subsequent RLS remain obscure, however.
It has been proposed that acidification during chronological aging accelerates damage to mitochondria and produces oxidative stress which also drives chronological aging in non-acidic environments (Longo et al., 2012). If correct, then these same forms of molecular damage may also drive the reduction in RLS following CLS. This makes intuitive sense, as both degenerate mitochondria and oxidatively damaged proteins are asymmetrically inherited by the mother cell during cell division (Aguilaniu et al., 2003; Erjavec et al., 2007; Lai et al., 2002; Lam et al., 2011; McFaline-Figueroa et al., 2011). Prior chronological aging under acidic conditions could therefore simply enhance the initial burden of such damage that each cell begins replicative life with, resulting in a reduced replicative capacity roughly proportional to the length of quiescence that cell had previously experienced.
DR also has effects on chronologically aging yeast cells that are at least partially independent of medium acidification and likely contribute to the maintenance of full RLS. DR can increase intracellular stores of trehalose and protect against oxidative and proteotoxic stress during chronological aging, responses which have been attributed to mitochondrial health (Ocampo et al., 2012). DR is known to inhibit mTOR activity, and reduced mTOR activity or DR are thought to increase CLS in part by an adaptive mitochondrial longevity signal involving increased superoxide and hydrogen peroxide levels (Mesquita et al., 2010; Pan et al., 2011). These effects are mediated by a metabolic shift toward mitochondrial respiration that is induced in cells subjected to DR (Lin et al., 2002). Consistent with this idea, overexpression of the Hap4-transcription factor is sufficient to induce a similar metabolic shift and to protect cells against the reduction in RLS associated with chronological aging in water at high temperature (Piper et al., 2006). In addition to promoting mitochondrial respiration, DR also induces autophagy via inhibition of mTOR (Stanfel et al., 2009). Genetic inhibition of autophagy shortens CLS and prevents CLS extension from rapamycin (Alvers et al., 2009a; Alvers et al., 2009b). It is plausible that enhanced autophagic degradation of damaged mitochondria, proteins, or other macromolecules in response to DR could also reduce the burden of such damage inherited by the mother cell upon resumption of vegetative growth.
The observation that low mitochondrial membrane potential during chronological aging predicts longer RLS suggests that mitochondrial damage is an important link between CLS and RLS. One possible explanation for this observation could be that damage accumulated during chronological aging induces an increase in mitochondrial membrane potential, and by sorting for cells with lower potential we are also sorting for cells with the least amount of damage. This explanation seems unlikely, however, as there is a significant decrease in mitochondrial membrane potential between day 2 and day 26 of the chronological aging experiment. In fact, we initially expected that cells with high potential would live longer, since we predicted that damage to mitochondria would be reflected by reduced membrane potential. Recently, however, it was reported that DR cells exhibit a lower respiratory rate in stationary phase (Ocampo et al., 2012), supporting our observation that they also have reduced membrane potential at day 2 of the chronological aging experiment. A DR mimetic mutant, tor1Δ, has also been shown to maintain a low mitochondria potential during the diauxic shift and into stationary phase (Pan et al., 2011). Based on these data, we speculate that a low respiratory state during chronological aging, which is reflected by reduced membrane potential, leads to extension of CLS and preservation of RLS. This also suggests that the maintenance of replicative capacity following aging in SC 2% glucose is determined largely by the metabolic state adopted by each individual cell upon entry into stationary phase, with cells having a lower metabolic activity (and thus lower membrane potential) preserving replicative capacity the best. The events accounting for the cell-to-cell variability in stationary phase membrane potential will be of interest to determine in future studies and may account for a portion of the stochasticity associated with chronological aging. It will also be important to better understand the molecular details for how mitochondria are changing during chronological aging under different conditions and how these changes modulate mitotic capacity upon return to vegetative growth.
In this study and a prior one, we have shown that two interventions capable of robustly extending CLS, buffering and DR, also attenuate the reduction in RLS associated with chronological aging such that there is no significant trend toward reduced RLS through the first 24 days of chronological age. In both cases, preservation of RLS was associated with a reduced mitochondrial membrane potential early in the chronological aging experiment. Whether all interventions that extend CLS will have a similar effect on subsequent RLS and/or mitochondrial membrane potential remains to be determined. These findings provide a path toward understanding the fundamental link between mitotic and post-mitotic cellular aging in yeast and suggest a central role for mitochondrial metabolism and health in this process.
Highlights.
-
-
Dietary restriction delays the reduction in replicative lifespan associated with chronological age
-
-
Mitochondrial membrane potential during chronological aging predicts subsequent replicative lifespan
-
-
Among the metabolically viable cells in a chronologically aged population, those with the highest mitochondrial membrane potential are unable to undergo even a single replicative cell division, while those with the lowest potential maintain a full replicative lifespan
ACKNOWLDEGEMENTS
This work was supported by NIH Grant R01AG039390 to MK. Additional support for flow cytometry was provided by the University of Washington Nathan Shock Center of Excellence in the Basic Biology of Aging (NIH P30AG013280). JRD and GLS were supported by NIH Training Grant T32AG000057. JS and BMW were supported by NIH Training Grant T32ES007032.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
J.D., C.M., A.C., and M.K. jointly conceived this study. All except B.O. and M.K. performed experiments. B.O. performed computational analysis of survival data. J.D. and M.K. wrote the manuscript.
REFERENCES
- Aguilaniu H, Gustafsson L, Rigoulet M, Nystrom T. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science. 2003;299:1751–1753. doi: 10.1126/science.1080418. [DOI] [PubMed] [Google Scholar]
- Alvers AL, Fishwick LK, Wood MS, Hu D, Chung HS, Dunn WA, Jr, Aris JP. Autophagy and amino acid homeostasis are required for chronological longevity in Saccharomyces cerevisiae. Aging Cell. 2009a;8:353–369. doi: 10.1111/j.1474-9726.2009.00469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvers AL, Wood MS, Hu D, Kaywell AC, Dunn WA, Jr, Aris JP. Autophagy is required for extension of yeast chronological life span by rapamycin. Autophagy. 2009b;5:847–849. doi: 10.4161/auto.8824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi K, Sinclair D, Gordon JI, Guarente L. Passage through stationary phase advances replicative aging in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:9100–9105. doi: 10.1073/pnas.96.16.9100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtner CR, Murakami CJ, Kaeberlein M. A genomic approach to yeast chronological aging. Methods in molecular biology. 2009a;548:101–114. doi: 10.1007/978-1-59745-540-4_6. [DOI] [PubMed] [Google Scholar]
- Burtner CR, Murakami CJ, Kennedy BK, Kaeberlein M. A molecular mechanism of chronological aging in yeast. Cell Cycle. 2009b;8:1256–1270. doi: 10.4161/cc.8.8.8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtner CR, Murakami CJ, Olsen B, Kennedy BK, Kaeberlein M. A genomic analysis of chronological longevity factors in budding yeast. Cell Cycle. 2011;10:1385–1396. doi: 10.4161/cc.10.9.15464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defossez PA, Prusty R, Kaeberlein M, Lin SJ, Ferrigno P, Silver PA, Keil RL, Guarente L. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Molecular cell. 1999;3:447–455. doi: 10.1016/s1097-2765(00)80472-4. [DOI] [PubMed] [Google Scholar]
- Erjavec N, Larsson L, Grantham J, Nystrom T. Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes & development. 2007;21:2410–2421. doi: 10.1101/gad.439307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Battistella L, Vardavas R, Gattazzo C, Liou LL, Diaspro A, Dossen JW, Gralla EB, Longo VD. Superoxide is a mediator of an altruistic aging program in Saccharomyces cerevisiae. The Journal of cell biology. 2004a;166:1055–1067. doi: 10.1083/jcb.200404002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Gattazzo C, Battistella L, Wei M, Cheng C, McGrew K, Longo VD. Sir2 blocks extreme life-span extension. Cell. 2005;123:655–667. doi: 10.1016/j.cell.2005.08.042. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Longo VD. The chronological life span of Saccharomyces cerevisiae. Aging Cell. 2003;2:73–81. doi: 10.1046/j.1474-9728.2003.00033.x. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Pletcher SD, Minois N, Vaupel JW, Longo VD. Chronological aging-independent replicative life span regulation by Msn2/Msn4 and Sod2 in Saccharomyces cerevisiae. FEBS letters. 2004b;557:136–142. doi: 10.1016/s0014-5793(03)01462-5. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Pozza F, Pletcher SD, Gendron CM, Longo VD. Regulation of longevity and stress resistance by Sch9 in yeast. Science. 2001;292:288–290. doi: 10.1126/science.1059497. [DOI] [PubMed] [Google Scholar]
- Falcon AA, Aris JP. Plasmid accumulation reduces life span in Saccharomyces cerevisiae. J Biol Chem. 2003;278:41607–41617. doi: 10.1074/jbc.M307025200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang JC, Jaruga E, Repnevskaya MV, Jazwinski SM. An intervention resembling caloric restriction prolongs life span and retards aging in yeast. The FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2000;14:2135–2137. doi: 10.1096/fj.00-0242fje. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M. Lessons on longevity from budding yeast. Nature. 2010;464:513–519. doi: 10.1038/nature08981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Sir2-independent life span extension by calorie restriction in yeast. PLoS biology. 2004;2:E296. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes & development. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW, 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- Koning AJ, Lum PY, Williams JM, Wright R. DiOC6 staining reveals organelle structure and dynamics in living yeast cells. Cell Motil Cytoskeleton. 1993;25:111–128. doi: 10.1002/cm.970250202. [DOI] [PubMed] [Google Scholar]
- Lai CY, Jaruga E, Borghouts C, Jazwinski SM. A mutation in the ATP2 gene abrogates the age asymmetry between mother and daughter cells of the yeast Saccharomyces cerevisiae. Genetics. 2002;162:73–87. doi: 10.1093/genetics/162.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam YT, Aung-Htut MT, Lim YL, Yang H, Dawes IW. Changes in reactive oxygen species begin early during replicative aging of Saccharomyces cerevisiae cells. Free Radic Biol Med. 2011;50:963–970. doi: 10.1016/j.freeradbiomed.2011.01.013. [DOI] [PubMed] [Google Scholar]
- Laun P, Rinnerthaler M, Bogengruber E, Heeren G, Breitenbach M. Yeast as a model for chronological and reproductive aging - a comparison. Exp Gerontol. 2006;41:1208–1212. doi: 10.1016/j.exger.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez P, Culotta VC, Fink G, Guarente L. Calorie restriction extends Saccharomyces cerevisiae life span by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- Longo VD, Shadel GS, Kaeberlein M, Kennedy B. Replicative and Chronological Aging in Saccharomyces cerevisiae. Cell Metab. 2012;16:18–31. doi: 10.1016/j.cmet.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFaline-Figueroa JR, Vevea J, Swayne TC, Zhou C, Liu C, Leung G, Boldogh IR, Pon LA. Mitochondrial quality control during inheritance is associated with lifespan and mother-daughter age asymmetry in budding yeast. Aging cell. 2011;10:885–895. doi: 10.1111/j.1474-9726.2011.00731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesquita A, Weinberger M, Silva A, Sampaio-Marques B, Almeida B, Leao C, Costa V, Rodrigues F, Burhans WC, Ludovico P. Caloric restriction or catalase inactivation extends yeast chronological lifespan by inducing H2O2 and superoxide dismutase activity. Proc Natl Acad Sci U S A. 2010;107:15123–15128. doi: 10.1073/pnas.1004432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer RK, Johnston JR. Life span of individual yeast cells. Nature. 1959;183:1751–1752. doi: 10.1038/1831751a0. [DOI] [PubMed] [Google Scholar]
- Murakami C, Delaney JR, Chou A, Carr D, Schleit J, Sutphin GL, An EH, Castanza AS, Fletcher M, Goswami S, et al. pH neutralization protects against reduction in replicative lifespan following chronological aging in yeast. Cell Cycle. 2012;11 doi: 10.4161/cc.21465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami C, Kaeberlein M. Quantifying yeast chronological life span by outgrowth of aged cells. J Vis Exp. 2009 doi: 10.3791/1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami CJ, Burtner CR, Kennedy BK, Kaeberlein M. A method for high-throughput quantitative analysis of yeast chronological life span. The journals of gerontology Series A, Biological sciences and medical sciences. 2008;63:113–121. doi: 10.1093/gerona/63.2.113. [DOI] [PubMed] [Google Scholar]
- Murakami CJ, Wall V, Basisty N, Kaeberlein M. Composition and acidification of the culture medium influences chronological aging similarly in vineyard and laboratory yeast. PLoS One. 2011;6:e24530. doi: 10.1371/journal.pone.0024530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocampo A, Liu J, Schroeder EA, Shadel GS, Barrientos A. Mitochondrial respiratory thresholds regulate yeast chronological life span and its extension by caloric restriction. Cell Metab. 2012;16:55–67. doi: 10.1016/j.cmet.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Schroeder EA, Ocampo A, Barrientos A, Shadel GS. Regulation of yeast chronological life span by TORC1 via adaptive mitochondrial ROS signaling. Cell Metab. 2011;13:668–678. doi: 10.1016/j.cmet.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper PW, Harris NL, MacLean M. Preadaptation to efficient respiratory maintenance is essential both for maximal longevity and the retention of replicative potential in chronologically ageing yeast. Mech Ageing Dev. 2006;127:733–740. doi: 10.1016/j.mad.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Powers RW, 3rd, Kaeberlein M, Caldwell SD, Kennedy BK, Fields S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes & development. 2006;20:174–184. doi: 10.1101/gad.1381406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- Smith DL, Jr, McClure JM, Matecic M, Smith JS. Calorie restriction extends the chronological lifespan of Saccharomyces cerevisiae independently of the Sirtuins. Aging Cell. 2007;6:649–662. doi: 10.1111/j.1474-9726.2007.00326.x. [DOI] [PubMed] [Google Scholar]
- Stanfel MN, Shamieh LS, Kaeberlein M, Kennedy BK. The TOR pathway comes of age. Biochimica et biophysica acta. 2009;1790:1067–1074. doi: 10.1016/j.bbagen.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen KK, Kennedy BK, Kaeberlein M. Measuring replicative life span in the budding yeast. J Vis Exp. 2009 doi: 10.3791/1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinkraus KA, Kaeberlein M, Kennedy BK. Replicative aging in yeast: the means to the end. Annual review of cell and developmental biology. 2008;24:29–54. doi: 10.1146/annurev.cellbio.23.090506.123509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Song L, Liu SQ, Huang D. A high throughput screening assay for determination of chronological lifespan of yeast. Exp Gerontol. 2011;46:915–922. doi: 10.1016/j.exger.2011.08.002. [DOI] [PubMed] [Google Scholar]