Abstract
The Enzyme-linked Immunosorbant Assay (ELISA) is a method commonly used to measure proteins in various biological matrices, due to its ease of performance and relatively low cost. In order for quantitative data to be generated, a reference standard curve must be prepared for each assay; however, due to investigator error or standard protein degradation, otherwise representative experimental sample data are rendered useless. Herein, we describe a protocol by which sample concentrations can be recovered from assays in which the standard curve fails. The ΔOD values of the experimental samples are used to generate a new standard curve, which is applied back to the original plate. For validation of this method, experimental sample concentrations obtained using acceptable standard curves were potted against those calculated using this new method. Using linear regression analysis, we show a near 1:1 correlation between sample concentrations, with r2 values between 0.98 – 0.99 and slopes between 0.97 – 1.10. This method demonstrates that assays resulting in unusable standard curves do not require re-assay of all samples. Instead, the experimental sample concentrations can be retrieved saving the investigator the time and resources required to rerun samples or repeat entire experiments.
1. Introduction
The Enzyme-linked Immunosorbant Assay (ELISA) is often the preferred method used in laboratories to measure protein concentrations in various biological matrices, such as serum, plasma, urine and cerebro-spinal fluid (1). One advantage of this technique is its high sensitivity, which allows accurate measurement of protein concentrations from as little as 10 µL of sample. This offers researchers the power to expand in vivo applications. For example, disease progression can be monitored over time by repeated sampling of small volumes of biological fluid such as blood from animal subjects (2, 3). Recovery of small volumes also highlights the preciousness of these samples, as investigators generally have one opportunity to measure target proteins. Unfortunately, investigator error, standard protein degradation and other intangible factors can result in assays in which a reliable standard curve is not generated. Normally, this would render otherwise accurate experimental sample readings useless, requiring the investigator to spend the time and resources to redo experiments in order to generate more sample.
We have developed a protocol by which experimental sample concentrations from assays that do not generate a usable standard curve can be recovered. This method involves selection of 8–10 of the original samples which are rerun after the assay has been re-optimized and can be run reliably. The experimental samples themselves are used to generate a new standard curve that is applied to the original assay plate. This method can be used when the assay defect is determined to be with the standard curve only. Failed standard curves will meet the following criteria: r2<0.97, greater than five samples have ΔOD higher than the highest concentration standard and a 4-parameter regression equation cannot be generated because the ΔOD does not decrease as the standard concentration decreases.
Investigators who are experienced in the ELISA method can generally determine the step in the assay at which the defect occurred. Technical problems involving the capture antibody usually result in faint color or no color development at all, while problems with the detection antibody usually result in uniform color development in all wells, including blanks. When the standard curve fails, experimental sample ΔOD values generally span a wide range and can be deemed usable. Technical mistakes that result in a failed standard curve may be the use of human recombinant standard when trying to measure mouse proteins, improper dilution of the standard, or use of degraded standard protein. Our results show a nearly 1:1 correlation between experimental sample concentrations calculated using a successful standard curve compared to the method described herein. Further, r2 values ranged from 0.98 – 0.99 and slopes of the regression lines fell between 0.97 – 1.10 for all cytokines measured.
2. Methods
2.1. Initial Assay
Experimental samples are run as described in the previously optimized standard ELISA protocol (4, 5), with a minor modification. Immediately prior to stopping the color reaction with 1.5M sulfuric acid, the OD590 is recorded.
The color reaction is stopped with sulfuric acid and the plate is read at OD590 and OD465. The ΔOD values are calculated and used to generate the standard curve and determine experimental sample concentrations. If a working standard curve results, experimental sample concentrations can be calculated as normal. However, if the standard curve is deemed to have failed, the subsequent protocol can be used to rescue the experimental sample data.
2.2. Troubleshoot standard curve
Fresh dilution buffers should be prepared and the standard curve rerun. The assay must be re-optimized so that successful standard curves can be reliably and reproducibly generated. This may require preparation of fresh stock solution of the standard protein.
2.3. New standard assay
To generate the new standard curve, 8–10 experimental samples are selected from the initial assay plate. The experimental samples with highest and lowest ΔOD are always chosen, along with experimental samples that are distributed evenly between the highest and lowest ΔOD.
These samples are then rerun according to the standard ELISA protocol.
After addition of the colorimetric reagent, the OD590 is monitored by repeated measurement by the microplate reader. The reaction is stopped when the OD590 values are equivalent to those measured in the initial assay. For ease, the plate can be left to develop in the microplate reader and re-read until the desired level of color development is obtained. Because the new standard is generated by running a new assay, the ability to directly compare the ΔOD values between plates must be preserved. The OD590 is thus monitored in each assay to prevent the extent of color development from affecting the corresponding concentrations.
The 4-parameter standard curve is generated as normal and used to calculate the experimental sample concentrations. Any computer software capable of calculating a 4-parameter regression may be used. These experimental samples are then relabeled as standards, and their concentrations are entered into the computer program manually. These experimental samples are used to generate a new standard curve that can then be applied back to the initial assay plate and sample concentrations are interpolated.
3. Results and Discussion
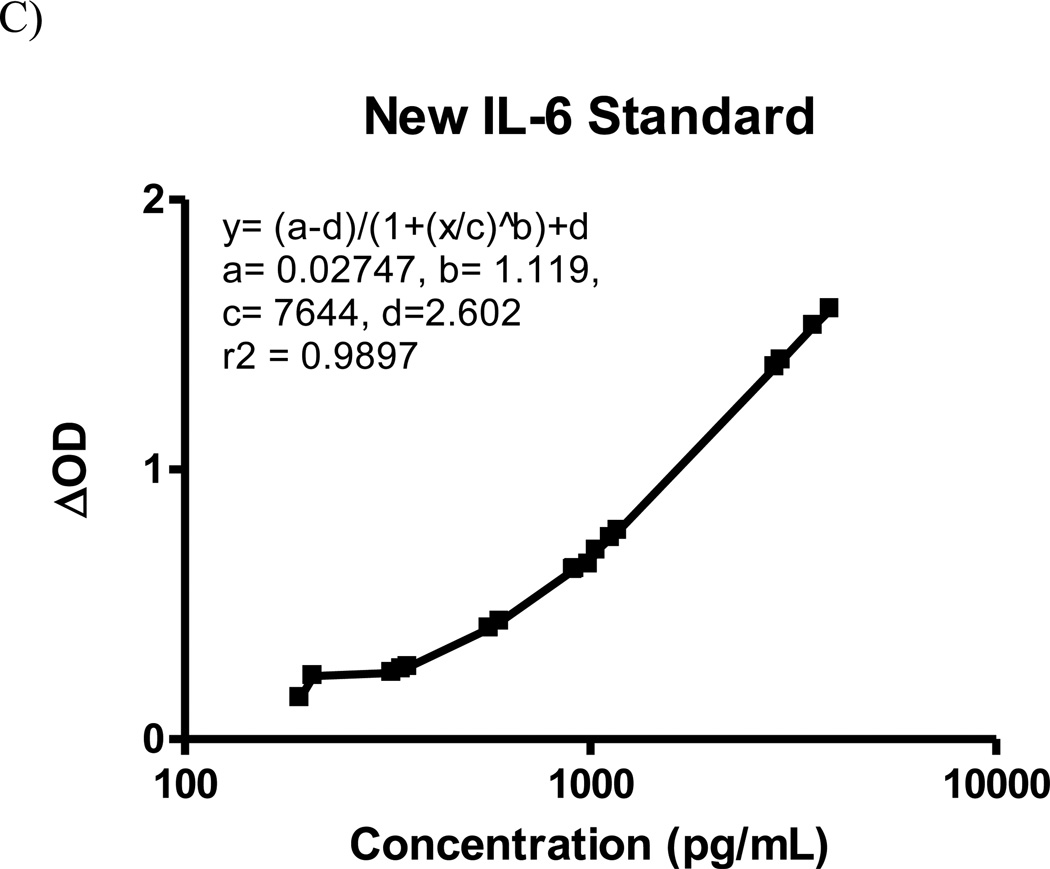
We use the OD590 as a surrogate for the ΔOD to monitor development of the colorimetric reaction. This step is key to ensuring that the new standard curve can be applied back to the initial assay plate. We verified that OD590 is an appropriate surrogate by plotting OD590 versus ΔOD. Linear regression analysis shows strong correlation between these values, with r2 = 0.98 (Figure 1). For clarity, we illustrate the relabeling of the experimental samples in the new standard assay as standards themselves in Figure 2A and B. Figure 2C shows a representative new standard curve and 4-parameter regression equation constructed from experimental samples derived from the initial assay. This new standard can then be applied back to the initial assay plate to accurately calculate the concentrations of those samples.
Figure 1.
Correlation of OD590 and ΔOD. Experimental samples were read before and after addition of acid to stop color development. OD 590nm was plotted against ΔOD to reveal a linear relationship1.
Figure 2.
96-well plate layouts for generation of new standard curve. A) Plate layout with standard curve used to determine chosen experimental sample concentrations. Standard concentrations are entered manually based on 1:3 dilutions of top standard. Sample concentrations are determined using standard curve. B) Re-labeling of sample wells as standards by manually inputting average concentrations of experimental samples determined in the new standard assay. C) Example of the new standard curve constructed for mouse IL-61.
This method was validated by spiking 40 aliquots of normal mouse plasma with varying concentrations of mouse recombinant TNFα, IL-4 and IL-6. Sample concentrations of these cytokines were measured in assays that generated reliable standard curves, and the OD590 was recorded. The experimental samples with maximum and minimum ΔOD were chosen along with 6–8 other samples whose ΔOD values fell evenly within this range. These samples were then run in a new assay and the OD590 was monitored. The color reaction was stopped when the OD590 values were equivalent to those measured in the initial assay. The concentrations of these experimental samples were calculated, and the samples were relabeled as standards. The new standard curve was applied back to the initial assay plate to calculate concentrations of all experimental samples. Figure 3A shows a near 1:1 correlation between concentrations obtained in the initial assay, versus the new standard assay. An r2 ≥ 0.98 and slopes ranging from 0.92 and 1.1 were obtained for all cytokines used for validation, confirming that this is a highly effective method by which experimental sample concentrations can be recovered from ELISA assays with failed standard curves.
Figure 3.
Validation of the accuracy of the new standard curve. A) Sample concentrations determined in the initial sample plate were plotted against those determined using the newly generated standard. The graphs show a nearly 1:1 correlation for all cytokines used, indicating the accuracy of this method. B) r2 values and slopes indicate the new standard correctly calculates the concentrations1.
This recovery method is most valuable in instances where small sample volumes are collected. It is often the case for in vivo experiments that sample volumes are only sufficient to run 1–2 ELISAs. Therefore, having a method of data rescue in the event of technical mistakes or user error is of great value. This method can also be coupled with a previously reported sequential ELISA protocol (6), that further optimizes the generation of large amounts of data from small volumes of biological samples. Here we show that a failed standard curve does not render the entire ELISA assay useless. Assays in which sample concentrations span a wide range are especially suited to this application, as a wide range of concentrations will increase the dynamic range of the new standard curve.
This method assumes that only a portion of the total available experimental sample will be run in the initial assay. It is not possible to rerun the plated experimental samples from the initial run, as the protein of interest may have been depleted during the assay. To ensure the utility of this method, we recommend recording the OD590 of all ELISA assays with >20 samples. Because the investigator does not know whether the standard curve has failed until the end of the assay, recording the OD590 prophylactically gives the researcher the opportunity to use this method in the event of standard curve failure. This method is vulnerable to the relative stability of the proteins being assayed. Therefore, the method may be less effective when assaying more labile proteins. This protocol is targeted towards labs that are more experienced with the ELISA assay, which will be more familiar with proteins which are stable and suitable for the present assay, versus labile proteins which are not suitable. This method requires that only a small number of experimental samples (8–10) be rerun in order to rescue large amounts of data, which can save the investigator significant time and resources they would otherwise spend repeating and rerunning experiments.
Acknowledgements
This work was supported by NIH grants ES13528 and GM 67189.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reprinted from Journal of Immunological Methods, Vol. 336, Natarajan S and Remick D. The ELISA Standard Save: Calculation of sample concentrations in assays with a failed standard curve, pp. 242–245, 2008, with permission from Elsevier.
Contributor Information
Sudha Natarajan, Email: natarajs@bu.edu.
Daniel G. Remick, Email: remickd@bu.edu.
References
- 1.Salonen EM, Vaheri A. J Immunol Methods. 1981;41:95–103. doi: 10.1016/0022-1759(81)90277-5. [DOI] [PubMed] [Google Scholar]
- 2.O'Connor KA, Holguin A, Hansen MK, Maier SF, Watkins LR. Brain Behav Immun. 2004;18:274–80. doi: 10.1016/j.bbi.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 3.Osuchowski MF, Connett J, Welch K, Granger J, Remick DG. Crit Care Med. 2009;37:1567–73. doi: 10.1097/CCM.0b013e31819df06b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeForge LE, Remick DG. Immunol Invest. 1991;20:89–97. doi: 10.3109/08820139109054928. [DOI] [PubMed] [Google Scholar]
- 5.Nemzek JA, Siddiqui J, Remick DG. J Immunol Methods. 2001;255:149–57. doi: 10.1016/s0022-1759(01)00419-7. [DOI] [PubMed] [Google Scholar]
- 6.Osuchowski MF, Remick DG. Methods. 2006;38:304–11. doi: 10.1016/j.ymeth.2005.11.009. [DOI] [PubMed] [Google Scholar]