Abstract
BACKGROUND
Gastroesophageal reflux is a risk factor for esophageal adenocarcinoma and bile acid and its farnesoid X receptor (FXR) have been implicated in esophageal tumorigenesis. We investigated the role of FXR expression and activity in esophageal cancer initiation and growth.
METHODS
FXR expression in esophageal adenocarcinoma tissues was assessed by immunohistochemistry. Knockdown of FXR expression in esophageal cancer cells in vitro and in nude mice xenografts was suppressed by FXR shRNA and guggulsterone (a natural FXR inhibitor). Esophageal cancer cells were treated with bile acids to show their effects on growth-promoting genes.
RESULTS
FXR was expressed in 48 of 59 esophageal adenocarcinoma tissues (81.3%), and this overexpression was associated with higher tumor grade, greater tumor size, and lymph node metastasis, but was inversely associated with RAR-β2 expression. Knockdown of FXR expression suppressed tumor cell growth in vitro and in nude mouse xenografts. Guggulsterone reduced viability of esophageal cancer cells in a time- and dose-dependent manner, whereas this effect was diminished after knockdown of FXR expression. Guggulsterone induced apoptosis through activation of caspases-8, -9, and -3 in tumor cells. FXR mediated bile acid–induced alterations of gene expression, e.g., RAR-β2 and COX-2.
CONCLUSION
Inhibition of FXR by FXR shRNA or guggulsterone suppressed tumor cell viability and induced apoptosis in vitro and reduced tumor formation and growth in nude mouse xenografts. FXR mediated bile acid–induced alterations of cell growth-related genes in esophageal cancer cells.
Keywords: bile acids, farnesoid X-activated receptor, retinoic acid receptor, cyclooxygenase 2, guggulsterone, esophageal cancer
INTRODUCTION
Esophageal cancer is a lethal disease with increasing incidence and high mortality in the world.1,2 Frequent gastroesophageal reflux carrying bile, hydrochloric acid, and proteases insults esophageal epithelial cells, leading to formation of premalignant Barrett esophagus and esophageal adenocarcinoma.1–4 Bile acids are normally produced in liver cells by the cytochrome P450-mediated oxidation of cholesterol. The main function of bile acid is to facilitate the formation of micelles for the processing and absorption of dietary fat.5–7 As surfactants or detergents, bile acids are potentially toxic to the cells, so their concentrations in the small intestine are tightly regulated. However, patients with frequent gastroesophageal reflux will have damage caused by gastric acid and bile acid–containing juice in the distal esophagus, with the result that normal squamous cells around the gastroesophageal junction will change to a new cell phenotype, incomplete intestinal metaplasia, which is more resistant to injury by acid and bile; this is how Barrett esophagus develops.8 Bile and gastric acid reflux variably affect Barrett esophagus and may cause dysplasia or adenocarcinoma.3,4,8–10 Indeed, bile acids can activate the epidermal growth factor receptor pathway and induce expression of activating factor-1 (AP-1), cyclooxygenase-2 (COX-2), and other genes, such as matrix metalloproteinase 9 (MMP-9) and nuclear factor kappaB (NF-κB)10–15 that play antiapoptotic roles in cells and in turn increase cell proliferation and/or promote the invasiveness potential of Barrett metaplasia and neoplasia. Nevertheless, it remains to be determined how bile acids induce malignant phenotypes and altered gene expression in esophageal cells. Previous studies demonstrated that bile acid farnesoid X receptor (FXR), a nuclear hormone receptor, is overexpressed in Barrett esophagus tissues and could mediates carcinogenic effects of bile acids in esophageal tumorigeneiss.5,6,16 Furthermore, expression of nuclear retinoic acid receptor-β2 (RAR-β2) was reduced in different human cancers, including esophageal cancer. Induction of RAR-β2 expression in esophageal cancer cells suppressed tumor cell growth and colony formation and induced apoptosis.7,15 In contrast, esophageal, lung, and breast cancer cell lines that do not express RAR-β2 were resistant to retinoid treatment.5,17,18 Our previous studies also linked RAR-β2 expression with downregulation of COX-2 expression in vitro and ex vivo.15,19,20 However, both FXR and RAR-β2 need to bind to RXRs (retinoid X receptors) for their functioning13,14,21 and bile acid-reduced RAR-β2 may be mediated by FXR-RXR heterodimerization to compete with RXR-RAR-β2 binding (RXR-RAR-β2 heterodimer is needed to induce RAR-β2 expression, a positive feedback mechanism21). Thus, in this study, we hypothesized that carcinogenic effects of bile acid could be through FXR-mediated downregulation of RAR-β2 expression, whereas inhibition of FXR expression or activity will effectively control esophageal cancer.
MATERIALS AND METHODS
Tissue specimens and Immunohistochemistry
Our institutional review board approved our protocol for the use of patient samples in this study, and all patients agreed to participated this study. This study included paraffin block samples from 59 consecutive patients with esophageal adenocarcinoma who had undergone esophagectomy without preoperative chemotherapy or radiotherapy between the years 1986 and 1997 at The University of Texas MD Anderson Cancer Center.
The immunohistochemical analysis followed our previously described methodology.19, 22,23 The anti-FXR antibody was obtained from Santa Cruz Biotechnology (Santa Cruz, CA) and applied at 1:50 dilution. The sections were reviewed and scored under a microscope as positively or negatively stained (≥10% tumor cells with positive nuclear staining counted as positive staining).
Cell culture and drug treatment
Esophageal squamous cell cancer cell lines TE-3 and TE-12 and adenocarcinoma cell lines SKGT-4 and SKGT-5 were used in our previous studies20,24 and grown in Dulbecco modified essential medium (DMEM) with 10% fetal bovine serum at 37°C in a humidified atmosphere of 95% air and 5% CO2. For bile acid or guggulsterone treatment, the cells were plated for 24 h in regular medium and then replaced either with control medium (containing the same volume of dimethyl sulfoxide [DMSO]) or with medium containing a bile acid (chenodeoxycholic acid, deoxycholic acid, or lithocholic acid) or guggulsterone (all from Sigma Chemical Co., St. Louis, MO, and dissolved in DMSO before use) for qRT-PCR, Western blot, and the methylthiazolyl tetrazolium (MTT) assays. After the addition of bile acid, the pH of the growth medium was adjusted to 7.2–7.4.
Construction of FXR small hairpin RNA and gene transfection
FXR shRNA plasmids were purchased from OriGene Technologies (Rockville, MD). The plasmids were amplified and their sequences confirmed before use. The pRFP-CRS vector contains hairpin loop sequences and when used as a negative control is identical to the scrambled shRNA control used by some other companies. pRFP-CRS vector containing one of four different FXR shRNA constructs or control shRNA was transfected into SKGT-4 cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA), and the cells were treated with 0.25 μg/mL puromycin for 48 h. Sixteen hours before cells were harvested, 200 μM of chenodeoxycholic acid was added to the culture medium and subjected to qRT-PCR and Western blot.
RT-PCR and qRT-PCR
RNA from the cells was extracted and subjected to RT-PCR and qRT-PCR as described previously.20,23 GAPDH was used as a loading control. The primers for FXR expression were 5′-GGAAATGCAAAGAGATGGGA-3′ and 5′-AGACCCTTTCAGCAAAGCAA-3′, which generated a 416-bp band. The primers for COX-2 expression were 5′-CCTTCTGCCTGACACCTTTC-3′ and 5′-GGTCAATGGAAGCCTGTGAT-3′, which generated a 194-bp band. The primers for MMP-9 expression were 5′-GCACGACGTCTTCCAGTACC-3′ and 5′-GTTTGTATCCGGCAAACTGG-3′, which generated a 224-bp band. The primers for RAR-β2 expression were 5′-CAAACCGAATGGCAGCATCGG-3′ and 5′-GCGGAAAAAGCCCTTACATCCC-3′, which amplified a 195-bp band. GAPDH primers were 5′-CCCTTCATTGACCTCAACTACATGG-3′ and 5′-CATGGTGGTGAAGACGCCAG-3′, which generated 192-bp band.
Protein extraction and Western blot
Total cellular protein was isolated as described elsewhere.20, 24 Samples containing 50 μg of protein from control or treated cells were separated by 10–14% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and then transferred electrophoretically to a Hybond-C nitrocellulose membrane (GE Healthcare, Arlington Heights, IL) for Western blotting.20,24 The antibodies used were anti-RAR-β and anti-FXR (Santa Cruz Biotechnology), anti-COX-2 (BD Transduction Laboratories, Lexington, KY), anti–caspase-3, -8, and -9 and anti–activated caspase-3, -8, and -9 (Cell Signaling Technology, Danvers, MA), and anti-β-actin or GAPDH antibody (Sigma).
FXR shRNA transfection and immunocytochemical staining of Ki-67 protein
Esophageal cancer SKGT-4 and TE-12 cells were grown and transiently transfected with pCMS/EGFP (BD Clontech, San Diego, CA) plus either control shRNA (OriGene) or FXR shRNA (OriGene) using Lipofectamine 2000 and 36 h later, the cells were treated with 0.25 μg/mL puromycin for an additional 24 h. The cells were then fixed with 4% paraformaldehyde for 10 min and permeabilized in 0.5% Triton X-100 for 10 min at room temperature for immunostaining of proliferation marker Ki-67 as described elsewhere.19,22 Ki-67 antibody was purchased from Vector Laboratories (Burlingame, CA) and diluted to 1:50.
DNA fragmentation assay
The cells were treated with or without 25 μM guggulsterone for 3 days, and soluble DNA was extracted from both floating and attached cells and then subjected to DNA fragmentation assay as described previously.7
Nude mouse xenograft assay
The animal experiments were performed in accordance with an institution-approved Animal Care and Usage protocol. nu/nu nude mice (6 weeks of age) were each injected subcutaneously in the right flank through a 22-gauge needle with 2 × 106 stable control shRNA– or FXR shRNA–transfected SKGT-4 cells in a total volume of 200 μL per mouse. Other mice were given guggulsterone (50 mg/kg/day) by mouth for 2 days and then injected subcutaneously in the right flank through a 22-gauge needle with 3 × 106 SKGT-4 cells in a total volume of 200 μL per mouse and continued to receive guggulsterone (50 mg/kg) daily for an additional 20 days. Both groups of animals were monitored daily for tumor formation and growth. At the end of the experiment, the mice were killed and tumor xenografts were removed, fixed in 4% paraformaldehyde, weighed, and photographed.
Transient gene transfection and luciferase assay
The cells were seeded at a density of 1.5 × 105 per well in six-well plates and cultured overnight. The cells were then transfected with DNA (1 μg of RAR-β2-luciferase reporter plasmid, 1 μg of FXR shRNA, and 0.1 μg of pCH110) using 3 μL of Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol or with negative control vectors. The RAR-β2 promoter-luciferase reporter plasmid was provided by Dr. Reuben Lotan of our institution. pCH110, a β-galactosidase expression vector (GE Healthcare), was used as an internal control for assessing transfection efficiency. 36 h later, the cells were treated with or without 200 μM of chenodeoxycholic acid for an additional 24 h and harvested for analysis of β-galactosidase and luciferase activities. A Turner Designs luminometer (model TD-20/20, Promega) recorded the luciferase activity in relative light units that were normalized to β-galactosidase activity to correct for differences in transfection efficiency.20 All experiments were performed in triplicate and repeated at least twice to confirm the reproducibility of the results.
Statistical analysis
The effects of guggulsterone on tumor cell viability were summarized from three independent experiments. Results between groups were compared by the Student t-test. Error bars represent the standard deviation of the mean (± SD). Association of FXR with RAR-β2 expression in esophageal cancer tissues was analyzed by the McNemar test using SPSS 11.5 software (Chicago, IL, USA). A P-value less than 0.05 was considered statistically significant.
RESULTS
Overexpression of FXR protein in esophageal adenocarcinoma tissues
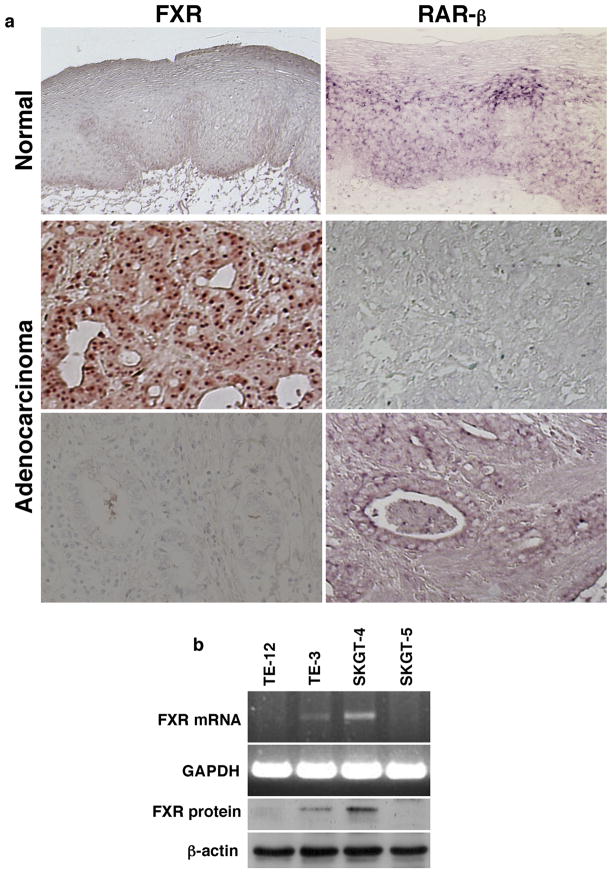
Expression of FXR protein was immunohistochemically stained in normal and cancerous esophageal tissue specimens and FXR was highly expressed in the nuclei of 48/59 esophageal adenocarcinoma tissues examined (81.3%). In contrast, none of 20 normal esophageal squamous epithelial specimens expressed FXR protein (0%; 0/20 cases; Figure 1a). FXR expression was associated with higher tumor grade, larger tumor size, and lymph node metastasis (Table 1). Expression of FXR protein was inversely associated with RAR-β2 expression (P = 0.0001; Table 2 and Figure 1a).
Figure 1.
Expression of FXR in esophageal cancer cells and tissue specimens. (a) Tissue samples from 59 esophageal adenocarcinoma patients were immunostained for FXR and hybridized in situ for RAR-β2 expression. The association of the expression of FXR with RAR-β2 was determined by the McNemar test. (b) Expression of FXR mRNA and protein in esophageal squamous cell carcinoma TE-3 and Te-12 cells and adenocarcinoma SKGT-4 and SKGT-5 cells using semi-quantitative RT-PCR and Western blot, respectively.
Table 1.
Association of FXR with clinicopathological data from esophageal adenocarcinoma patients (N = 59)
| N | FXR Association
|
P-value* | ||
|---|---|---|---|---|
| High | Low | |||
| Gender | ||||
| Male | 51 | 42 | 9 | |
| Female | 8 | 6 | 2 | 0.42 |
| Age (yrs) | ||||
| < 65 | 27 | 21 | 6 | |
| ≥ 65 | 32 | 27 | 5 | 0.25 |
| Tumor stage | ||||
| I–II | 17 | 11 | 6 | |
| III–IV | 42 | 37 | 5 | 0.018 |
| Lymph node metastasis | ||||
| + | 48 | 41 | 7 | |
| − | 11 | 7 | 4 | 0.047 |
| Distant metastasis | ||||
| + | 14 | 12 | 2 | |
| − | 45 | 36 | 9 | 0.31 |
| Tumor differentiation | ||||
| M to W | 22 | 17 | 5 | |
| P | 35 | 30 | 5 | 0.20 |
| N/A | 2 | 1 | 1 | |
| Tumor size | ||||
| ≤ 3 cm | 13 | 8 | 5 | |
| > 3 cm | 46 | 40 | 6 | 0.018 |
χ2 test.
M, medium; W, well; P, poorly; N/A, data not available
Table 2.
Association of FXR with RAR-β2 expression in esophageal adenocarcinoma tissue specimens (N = 59)
| FXR
|
P-value* | ||
|---|---|---|---|
| Positive | Negative | ||
| RAR-β2 | |||
| Positive | 20 | 3 | |
| Negative | 28 | 8 | 0.0001 |
McNammar test
FXR inhibition suppresses growth and proliferation of esophageal cancer cells
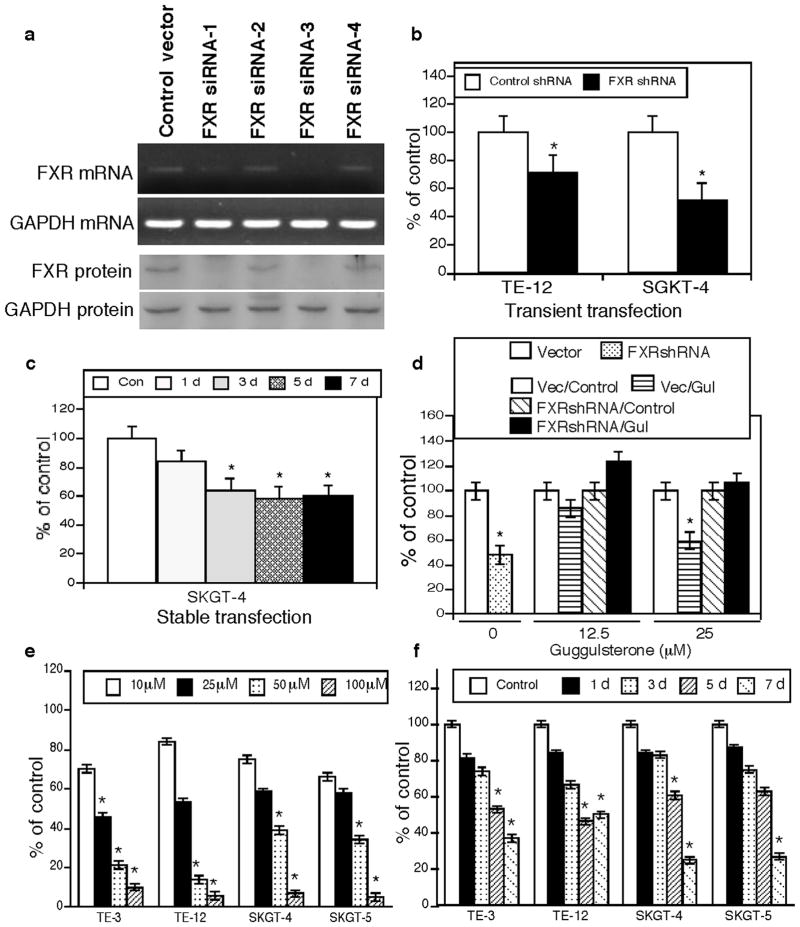
To knockdown FXR expression, we first assessed FXR expression in four different esophageal cancer cell lines and found that one of each esophageal squamous cell carcinoma and adenocarcinoma cell lines expressed FXR mRNA and protein (Figure 1b). After that we chose esophageal adenocarcinoma SKGT-4 cell line for transfection of four different FXR shRNA constructs and found that two of them reduced FXR expression (Figure 2a). We then transiently transfected a FXR shRNA vector into esophageal cancer cells and stained for Ki-67 expression and found that Ki-67 expression was significantly lower in cells transfected with the FXR shRNA constructs than in those transfected with control shRNA (Figure 2b). Moreover, stably control shRNA or FXR shRNA-transfected SKGT-4 cells showed that FXR knockdown significantly reduced tumor cell growth compared with the controls (Figure 2c).
Figure 2.
Suppression of FXR expression or activity reduces growth and proliferation of esophageal cancer cells. (a) FXR expression in SKGT-4 cells was knocked down by one of four FXR shRNA constructs. (b) Ki-67 expression was detected by immunocytochemical staining in FXR shRNA-transfected esophageal cancer cell lines. *P <0.05 compared with control cells. (c) MTT assay. SKGT-4 cells stably transfected with FXR shRNA or negative control shRNA were subjected to cell viability MTT assay. *P <0.05 compared with control cells. (d) MTT assay. SKGT-4 cells stably transfected with control shRNA (Vector or Vec) or FXR shRNA were treated or not treated (Control) with 12.5 or 25 μM guggulsterone (Gul) for 5 days and subjected to MTT assay. The data showed that FXR knockdown antagonized the effects of FXR inhibitor guggulsterone on esophageal cancer cells. *P <0.05 compared with control cells. (e) MTT assay. TE-3, TE-12, SKGT-4, and SKGT-5 cells were grown and treated with different concentrations of guggulsterone for 5 days and then subjected to MTT assay. *P <0.05 compared with control cells. (f) MTT assay. TE-3, TE-12, SKGT-4, and SKGT-5 cells were grown and treated with 25 μM guggulsterone for up to 7 days and subjected to MTT assay. *P <0.05 compared with control cells.
In addition, we treated esophageal cancer cells with guggulsterone, a natural FXR inhibitor, for different durations at various concentrations and found that guggulsterone reduced tumor cell viability in a dose- and time-dependent manner (Figure 2e and f). However, this effect of guggulsterone was diminished after knockdown of FXR expression (Figure 2d), indicating that the effects of guggulsterone were mediated via FXR inhibition.
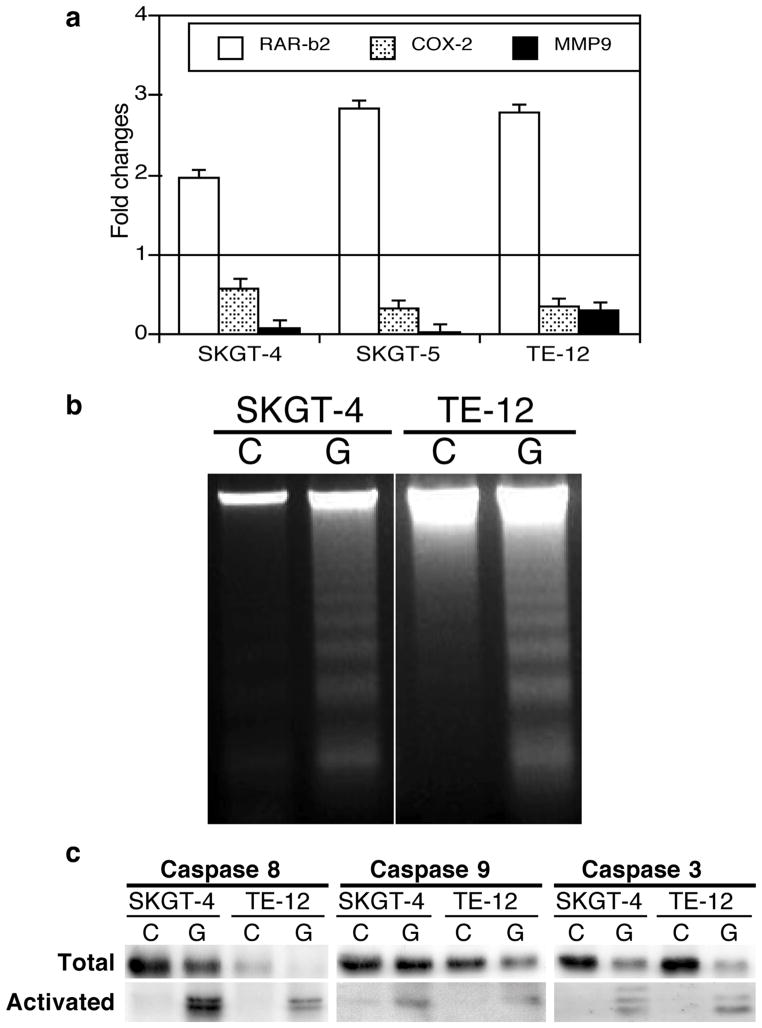
Guggulsterone treatment inhibited expression of the RAR-β2-led COX-2 and MMP-9 pathway genes (Figure 3a) and induced apoptosis and expression of apoptosis-related genes in esophageal cancer cells (Figure 3b and c).
Figure 3.
The FXR inhibitor guggulsterone regulates gene expression and promotes apoptosis in esophageal cancer cells. (a) qRT-PCR. Esophageal cancer SGKT-4, SGKT-5, and TE-12 cell lines were grown and treated with 25 μM guggulsterone for 2 days for qRT-PCR. Bars below the horizontal 1-fold line indicate reduced expression; those above the line, increased expression induced by guggulsterone. (b) DNA fragmentation assay. SKGT-4 and TE-12 cells were grown and treated with 25 μM guggulsterone (G) or control (C) for 3 days and then subjected to the DNA fragmentation assay to measure apoptosis. (c) Western blot. Esophageal cancer SKGT-4 and TE-12 cells were grown and treated with 25 μM guggulsterone (G) or control (C) for 3 days, and total cellular protein was extracted and subjected to Western blotting.
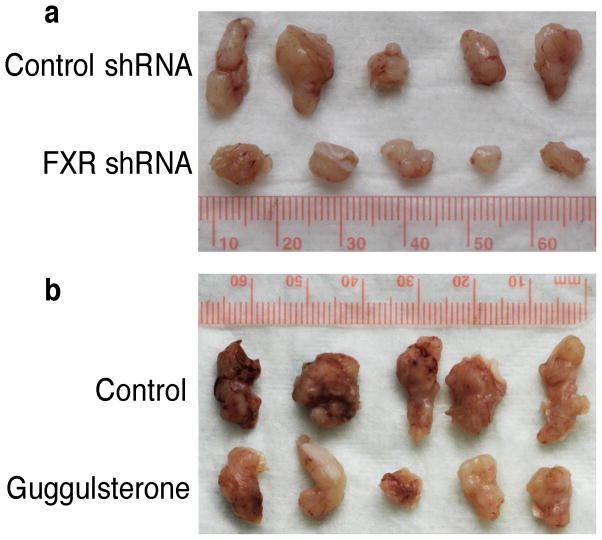
FXR knockdown inhibits growth of esophageal tumors in vivo
To confirm the effects of FXR inhibition in vivo, we first subcutaneously injected stable FXR shRNA– or control shRNA–transfected SKGT-4 cells into nude mice and monitored tumor formation and growth for 20 days. Both tumor size and weight were significantly reduced by FXR shRNA (Table 3 and Figure 4a). We also treated another group of nude mice with oral guggulsterone 2 days before subcutaneous injection of SKGT-4 cells and continuously treated for another 20 days after injection. Both FXR shRNA and guggulsterone suppressed tumor formation and growth in these mice (Table 3 and Figure 4b).
Table 3.
Results of the nude mouse SKGT-4 cells’ xenograft experiments
| Mouse body weight (g) | No. of mice | Tumor weight (g) | % of control | P-value* | |||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Before | After | Start | End | ||||
| Control | 20.4 ± 1.80 | 28.3 ± 1.98 | 5 | 5 | 0.47 ± 0.14 | ||
| Guggulsterone | 19.1 ± 1.74 | 28.7 ± 1.60 | 5 | 5 | 0.28 ± 0.09 | 59.6 | 0.10 |
| Control shRNA | 20.1 ± 1.15 | 27.3 ± 2.20 | 5 | 5 | 0.23 ± 0.09 | ||
| FXR shRNA | 19.9 ± 1.41 | 25.9 ± 1.46 | 5 | 5 | 0.10 ± 0.04 | 43.5 | 0.03 |
Student t-test compared with controls.
Figure 4.
FXR inhibition reduces growth of esophageal cancer cells in vivo. (a) SKGT-4 cells stably transfected with control shRNA or FXR shRNA were subcutaneously injected into nude mice. Tumor formation and growth were monitored daily. (b) Nude mice were treated with 50 mg/kg of guggulsterone orally for 2 days and then subcutaneously injected with SKGT-4 cells and continued to receive 50 mg/kg of guggulsterone daily for additional 20 days. Tumor formation and growth were monitored daily. At the end experiments, tumor xenografts were taken out, weighted, and photographed.
FXR mediates alteration of gene expression by bile acids
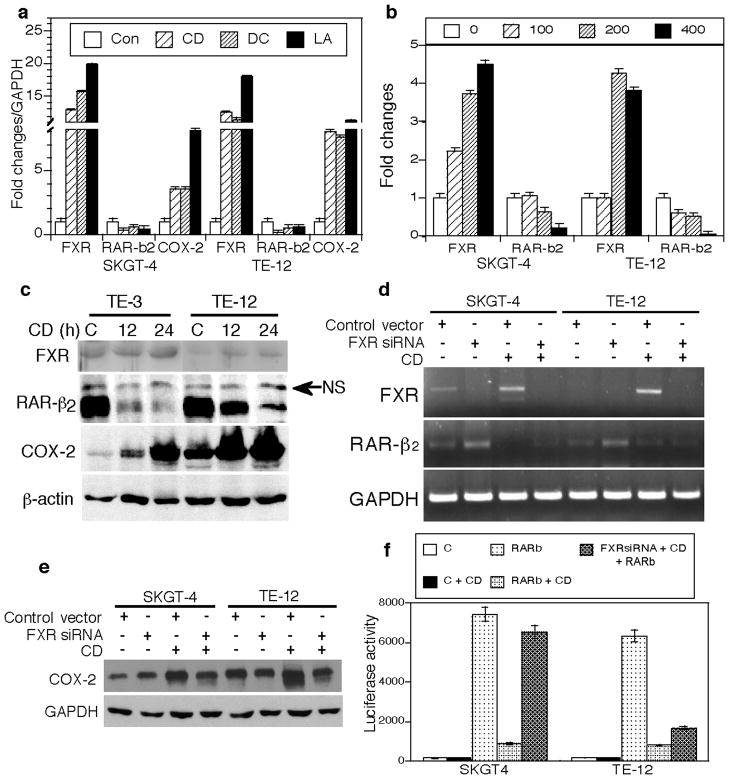
To better understand the role of FXR in the tumor-promoting effects of bile acids in esophageal cancer, we explored the effects of different bile acids on regulation of FXR, RAR-β2, and COX-2 expression in esophageal cancer cell lines. We found that the cells treated with a bile acid (chenodeoxycholic acid, deoxycholic acid, or lithocholic acid at a concentration of 200 μM) for 48 h had a decreased expression of RAR-β2 mRNA but induced expression of COX-2 and FXR mRNAs (Figure 5a). Chenodeoxycholic acid, a potent FXR ligand, inhibited RAR-β2 protein expression but induced FXR and COX-2 protein expression (Figure 5b and c).
Figure 5.
FXR mediates the effects of bile acids on regulation of gene expression in esophageal cancer cells. (a) qRT-PCR. SKGT-4 and TE-12 cells were grown and treated with 200 μM of chenodeoxycholic acid (CD), deoxycholic acid (DC), lithocholic acid (LA), or control (Con) for 48 h for qRT-PCR. (b) qRT-PCR. SKGT-4 and TE-12 cells were grown and treated with different doses of chenodeoxycholic acid (all, μM) for qRT-PCR. (c) Western blot. Cells treated with chenodeoxycholic acid or control (C) for 12 or 24 h for Western blotting. NS, nonspecific. (d) semiquantitative RT-PCR. SKGT-4 and TE-12 cells were grown and transiently transfected with FXR shRNA-3 and treated with 200 μM chenodeoxycholic acid for 24 h for semiquantitative RT-PCR. (e) Western blot. Cells treated as for (d) were subjected to Western blotting. (f) Luciferase assay. SKGT-4 and TE-12 cells were grown and transiently transfected with RAR-β2 gene promoter-driven luciferase reporter vector with or without FXR shRNA vector or empty vector. pCH110, a β-galactosidase expression vector, was used as an internal control for assessing transfection efficiency. Thirty-six hours after transfection, the cells were treated with chenodeoxycholic acid or left untreated for an additional 24 h. The cells were then harvested, and luciferase activities were measured. The data showed that luciferase activity was high after RAR-β2 promoter transfection, while CD treatment suppressed RAR-β2 luciferase activity. However, after co-transfection with FXR shRNA, RAR-β2 luciferase activity was significantly rescued in SKGT-4 cells compared to the CD treated cells but only faintly rescued in TE-12 cells (FXR expression in TE-12 cells is very low compared to SKGT-4 cells). C, negative control; CD, chenodeoxycholic acid.
Furthermore, the effects of chenodeoxycholic acid on RAR-β2 and COX-2 expression were significantly antagonized in cells transiently transfected with FXR shRNA (Figure 5d and e). Chenodeoxycholic acid acts at the transcriptional level through FXR to suppress RAR-β2 expression because knockdown of FXR expression abolished the suppressive effect of chenodeoxycholic acid on RAR-β2 promoter activity (Figure 5f). Together, these data indicate that FXR mediated the effects of bile acids in esophageal cancer cell lines (i.e., inhibition of RAR-β2 and induction of COX-2 expression).
DISCUSSION
FXR is activated by bile acids functioning as signaling molecules in the liver and intestines to regulates bile acid and lipid metabolism.5,6,16,25 FXR also plays roles in growth regulation, apoptosis, and cancer development.26–30 A number of recent publications have reported that FXR protects liver cells from apoptosis induced by serum deprivation in vitro and fasting in vivo.26 FXR was shown to protect against intestinal tumorigenesis.27 However, the expression and functions of FXR in organs other than the liver-intestine system potray a different picture. For example, an Oncomine database search identified more than 30 cDNA microarray studies showing that FXR expression was associated with breast and esophageal cancer development, higher tumor grade, metastasis, and patient survival (www.oncomine.com). FXR expression was significantly correlated with the proliferation marker Ki-67 and with positive nodal status in postmenopausal women with estrogen receptor–positive breast cancer, and in vitro experiments showed that bile acids stimulated the proliferation of estrogen receptor–positive cells in steroid-free medium.29 The FXR agonist chenodeoxycholic acid significantly increased endothelial cell motility and tube formation, and increased cell motility was associated with prominent increases in focal adhesion that were inhibited by FXR or MMP-9 siRNA.31 In esophageal cancer, FXR expression was higher in esophagitis, Barrett esophagus than in normal mucosa.30 In vitro treatment with guggulsterone was associated with a significant increase in apoptosis and caspase-3 activity in Barrett esophagus–derived cells.32 Bile acid–stimulated expression of FXR was shown to enhance the immune response in Barrett esophagus.32 Furthermore, FXR was overexpressed in pancreatic cancer and associated with lymph node metastasis in ex vivo and FXR expression promoted tumor cell migration and invasion.33 These data indicate that FXR plays a role in promoting the development or progression of different human cancers.
In this study, we demonstrated that guggulsterone induced expression of RAR-β2 but reduced expression of COX-2 and MMP-9, effects similar to those of FXR shRNA, in esophageal cancer cells. Guggulsterone, an ingredient in many nutritional supplements, acts in humans as a natural antagonist of FXR.34,35 It suppresses activation of the inflammatory transcription factor NF-κB, which is induced by various carcinogens and tumor promoters, including bile acids.34,35 Guggulsterone has been shown to inhibit the proliferation of a wide variety of tumor cells and to induce them to undergo apoptosis. It also has been shown to inhibit angiogenesis by blocking STAT3 and VEGF expression in colon cancer cells.36 Most importantly, guggulsterone is also a partial FXR agonist. Previous studies demonstrated that guggulsterone exerted antagonistic effects on FXR-induced recruitment of Src-1 and other genes, but it also induced expression of the bile salt export pump (BSEP) transporter, a known FXR-regulated gene.37,38 Furthermore, a number of studies showed that guggulsterone has a suppressive effect on xenograft formation and growth of different cancers in vivo.37,38 However, although the in vivo effect of guggulsterone was not statistically significant in the current study, future study will allow us to better define the experimental conditions, such as the dose and bioavailability of guggulsterone and the number of tumor cells injected.
In previous studies, gastric acid and bile acid were found to induce ERK activity, PPAR-γ expression, and cell proliferation in normal esophageal epithelial cells.12 Bile acid exposure causes phosphatidyl-inositol-3-kinase–mediated proliferation of Barrett adenocarcinoma cells,39 and deoxycholic acid at neutral pH activates NF-κB and induces interleukin-8 expression in esophageal cells in vitro.40 Our study demonstrated that inhibition of bile acid–induced COX-2 expression by retinoic acid was dependent on RAR-β2 expression.15 Together, the results from these studies suggest that bile acids have carcinogenic effects in gastrointestinal cancers, including esophageal adenocarcinoma.
Our current study demonstrated that FXR mediated the effects of bile acids on gene expressions in esophageal cancer cells and that inhibition of FXR not only suppressed tumor cell viability via induction of apoptosis in vitro but also reduced formation and growth of nude mouse xenograft tumors. These data, together with published data,10,28–31 indicate that targeting of FXR could be further evaluated as a novel strategy in prevention of esophageal adenocarcinoma.
Acknowledgments
Funding: This work was supported in part by a grant from the National Cancer Institute (R01 CA117895) and a grant from the Duncan Family Institute for Cancer Prevention and Risk Assessment at The University of Texas MD Anderson Cancer Center.
We thank the Department of Scientific Publications at MD Anderson Cancer Center for editing the manuscript.
Footnotes
The authors have declared no competing interests.
References
- 1.Blot W. Esophageal cancer trends and risk factors. Semin Oncol. 1994;21:403–410. [PubMed] [Google Scholar]
- 2.Spechler SJ. Barrett’s esophagus: a molecular perspective. Curr Gastroenterol Rep. 2005;7:177–181. doi: 10.1007/s11894-005-0031-z. [DOI] [PubMed] [Google Scholar]
- 3.Chen X, Yang CS. Esophageal adenocarcinoma: a review and perspectives on the mechanism of carcinogenesis and chemoprevention. Carcinogenesis. 2001;22:1119–1129. doi: 10.1093/carcin/22.8.1119. [DOI] [PubMed] [Google Scholar]
- 4.Barak N, Ehrenpreis ED, Harrison JR, Sitrin MD. Gastro-oesophageal reflux disease in obesity: pathophysiological and therapeutic considerations. Obes Rev. 2002;3:9–15. doi: 10.1046/j.1467-789x.2002.00049.x. [DOI] [PubMed] [Google Scholar]
- 5.Makishima M, Okamoto AY, Repa JJ, et al. Identification of a nuclear receptor for bile acids. Science. 1999;284:1362–1625. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- 6.Parks DJ, Blanchard SG, Bledsoe RK, et al. Bile acids: natural ligands for an orphan nuclear receptor. Science. 1999;284:1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- 7.Xu X-C, Liu X, Tahara E, Lippman SM, Lotan R. Expression and up-regulation of retinoic acid receptor-beta is associated with retinoid sensitivity and colony formation in esophageal cancer cell lines. Cancer Research. 1999;59:2477–2483. [PubMed] [Google Scholar]
- 8.Smith KJ, O’Brien SM, Smithers BM, et al. Interactions among smoking, obesity, and symptoms of acid reflux in Barrett’s esophagus. Cancer Epidemiol Biomarkers Prev. 2005;14:2481–2486. doi: 10.1158/1055-9965.EPI-05-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindblad M, Rodriguez LA, Lagergren J. Body mass, tobacco and alcohol and risk of esophageal, gastric cardia, and gastric non-cardia adenocarcinoma among men and women in a nested case-control study. Cancer Causes Control. 2005;16:285–294. doi: 10.1007/s10552-004-3485-7. [DOI] [PubMed] [Google Scholar]
- 10.Xu X-C. Risk factors and altered gene expression in esophageal cancer. In: Verma M, editor. Cancer Epidemiology. New York: Humana Press; 2009. pp. 335–360. [Google Scholar]
- 11.Jaiswal K, Lopez-Guzman C, Souza RF, Spechler SJ, Sarosi GA., Jr Bile salt exposure increases proliferation through p38 and ERK MAPK pathways in a non-neoplastic Barrett’s cell line. Am J Physiol Gastrointest Liver Physiol. 2006;290:G335–342. doi: 10.1152/ajpgi.00167.2005. [DOI] [PubMed] [Google Scholar]
- 12.Looby E, Abdel-Latif MM, Athie-Morales V, Duggan S, Long A, Kelleher D. Deoxycholate induces COX-2 expression via Erk1/2, p38-MAPK and AP-1-dependent mechanisms in esophageal cancer cells. BMC Cancer. 2009;9:190. doi: 10.1186/1471-2407-9-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernstein H, Bernstein C, Payne CM, Dvorak K. Bile acids as endogenous etiologic agents in gastrointestinal cancer. World J Gastroenterol. 2009;15:3329–3340. doi: 10.3748/wjg.15.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu J, Gong J, Geng J, Song Y. Deoxycholic acid induces the overexpression of intestinal mucin, MUC2, via NF-kB signaling pathway in human esophageal adenocarcinoma cells. BMC Cancer. 2008;8:333. doi: 10.1186/1471-2407-8-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li M, Song S, Lippman SM, et al. Induction of retinoic acid receptor-β suppresses cyclooxygenase-2 expression in esophageal cancer cells. Oncogene. 2002;21:411–418. doi: 10.1038/sj.onc.1205106. [DOI] [PubMed] [Google Scholar]
- 16.Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: opening the X-files. Science. 2001;294:1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- 17.Houle B, Leduc F, Bradley WE. Implication of RARB in epidermoid (squamous) lung cancer. Genes Chromosomes Cancer. 1991;3:358–366. doi: 10.1002/gcc.2870030506. [DOI] [PubMed] [Google Scholar]
- 18.Ren M, Pozzi S, Bistulfi G, et al. Impaired retinoic acid (RA) signal leads to RARbeta2 epigenetic silencing and RA resistance. Mol Cell Biol. 2005;25:10591–10603. doi: 10.1128/MCB.25.23.10591-10603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song S, Guan B, Men T, Hoque A, Lotan R, Xu X-C. Antitumor effect of retinoic acid receptor-β2 associated with suppression of cyclooxygenase-2. Cancer Prevention Res. 2009;2:274–280. doi: 10.1158/1940-6207.CAPR-08-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song S, Lippman SM, Zou Y, Ye X, Ajani JA, Xu X-C. Induction of cyclooxygenase-2 by benzo[a]pyrene diol epoxide through inhibition of retinoic acid receptor-β2 expression. Oncogene. 2005;24:8268–8276. doi: 10.1038/sj.onc.1208992. [DOI] [PubMed] [Google Scholar]
- 21.Chambon P. A decade of molecular biology of retinoic acid receptors. FASEB J. 1996;10:940–954. [PubMed] [Google Scholar]
- 22.Liang ZD, Lippman SM, Wu TT, et al. RRIG1 mediates effects of retinoic acid receptor-β2 on tumor cell growth and gene expression through binding to and inhibiting RhoA. Cancer Res. 2006;66:7111–7118. doi: 10.1158/0008-5472.CAN-06-0812. [DOI] [PubMed] [Google Scholar]
- 23.Huang J, Liang ZD, Wu TT, et al. Tumor-suppressive effect of retinoid receptor-induced gene-1 (RRIG1) in esophageal cancer. Cancer Res. 2007;67:1589–1593. doi: 10.1158/0008-5472.CAN-06-2472. [DOI] [PubMed] [Google Scholar]
- 24.Song S, Xu X-C. Effect of benzo[a]pyrene diol epoxide on expression of retinoic acid receptor-beta in immortalized esophageal epithelial cells and esophageal cancer cells. Biochem Biophys Res Comm. 2001;281:872–877. doi: 10.1006/bbrc.2001.4433. [DOI] [PubMed] [Google Scholar]
- 25.Lefebvre P, Cariou B, Lien F, Kuipers F, Staels B. Role of bile acids and bile acid receptors in metabolic regulation. Physiol Rev. 2009;89:147–191. doi: 10.1152/physrev.00010.2008. [DOI] [PubMed] [Google Scholar]
- 26.Wang YD, Yang F, Chen WD, et al. Farnesoid X receptor protects liver cells from apoptosis induced by serum deprivation in vitro and fasting in vivo. Mol Endocrinol. 2008;22:1622–1632. doi: 10.1210/me.2007-0527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Modica S, Murzilli S, Salvatore L, Schmidt DR, Moschetta A. Nuclear bile acid receptor FXR protects against intestinal tumorigenesis. Cancer Res. 2008;68:9589–9594. doi: 10.1158/0008-5472.CAN-08-1791. [DOI] [PubMed] [Google Scholar]
- 28.Swales KE, Korbonits M, Carpenter R, Walsh DT, Warner TD, Bishop-Bailey D. The farnesoid X receptor is expressed in breast cancer and regulates apoptosis and aromatase expression. Cancer Res. 2006;66:10120–10126. doi: 10.1158/0008-5472.CAN-06-2399. [DOI] [PubMed] [Google Scholar]
- 29.Journe F, Durbecq V, Chaboteaux C, et al. Association between farnesoid X receptor expression and cell proliferation in estrogen receptor-positive luminal-like breast cancer from postmenopausal patients. Breast Cancer Res Treat. 2009;115:523–535. doi: 10.1007/s10549-008-0094-2. [DOI] [PubMed] [Google Scholar]
- 30.De Gottardi A, Dumonceau JM, Bruttin F, et al. Expression of the bile acid receptor FXR in Barrett’s esophagus and enhancement of apoptosis by guggulsterone in vitro. Mol Cancer. 2006;5:48. doi: 10.1186/1476-4598-5-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Das A, Yaqoob U, Mehta D, Shah VH. FXR promotes endothelial cell motility through coordinated regulation of FAK and MMP-9. Arterioscler Thromb Vasc Biol. 2009;29:562–570. doi: 10.1161/ATVBAHA.108.182725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Capello A, Moons LM, Van de Winkel A, et al. Bile acid-stimulated expression of the farnesoid X receptor enhances the immune response in Barrett esophagus. Am J Gastroenterol. 2008;103:1510–1516. doi: 10.1111/j.1572-0241.2008.01908.x. [DOI] [PubMed] [Google Scholar]
- 33.Lee JY, Lee KT, Lee JK, et al. Farnesoid X receptor, overexpressed in pancreatic cancer with lymph node metastasis promotes cell migration and invasion. Br J Cancer. 2011;104:1027–1037. doi: 10.1038/bjc.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shishodia S, Sethi G, Ahn KS, Aggarwal BB. Guggulsterone inhibits tumor cell proliferation, induces S-phase arrest, and promotes apoptosis through activation of c-Jun N-terminal kinase, suppression of Akt pathway, and downregulation of antiapoptotic gene products. Biochem Pharmacol. 2007;74:118–130. doi: 10.1016/j.bcp.2007.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Urizar NL, Moore DD. GUGULIPID: a natural cholesterol lowering agent. Annu Rev Nutr. 2003;23:303–313. doi: 10.1146/annurev.nutr.23.011702.073102. [DOI] [PubMed] [Google Scholar]
- 36.Kim ES, Hon SY, Lee HK, et al. Guggulsterone inhibits angiogenesis by blocking STAT3 and VEGF expression in colon cancer cells. Oncol Report. 2008;20:1321–1327. [PubMed] [Google Scholar]
- 37.Urizar NL, Liverman AB, Dodds DT, et al. A natural product that lowers cholesterol as an antagonist ligand for FXR. Science. 2002;296:1703–1706. doi: 10.1126/science.1072891. [DOI] [PubMed] [Google Scholar]
- 38.Cui J, Huang L, Zhao A, Lew JL, Sahoo S, Meinke PT. Guggulsterone is a farnesoid X receptor antagonist in coactivator association assays but acts to enhance transcription of bile salt export pump. J Biol Chem. 2003;278:10214–10220. doi: 10.1074/jbc.M209323200. [DOI] [PubMed] [Google Scholar]
- 39.Jaiswal K, Tello V, Lopez-Guzman C, et al. Bile salt exposure causes phosphatidyl-inositol-3-kinase-mediated proliferation in a Barrett’s adenocarcinoma cell line. Surgery. 2004;136:160–168. doi: 10.1016/j.surg.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 40.Jenkins GJ, Harries K, Doak SH, et al. The bile acid deoxycholic acid (DCA) at neutral pH activates NF-kappaB and induces IL-8 expression in oesophageal cells in vitro. Carcinogenesis. 2004;25:317–323. doi: 10.1093/carcin/bgh032. [DOI] [PubMed] [Google Scholar]