Abstract
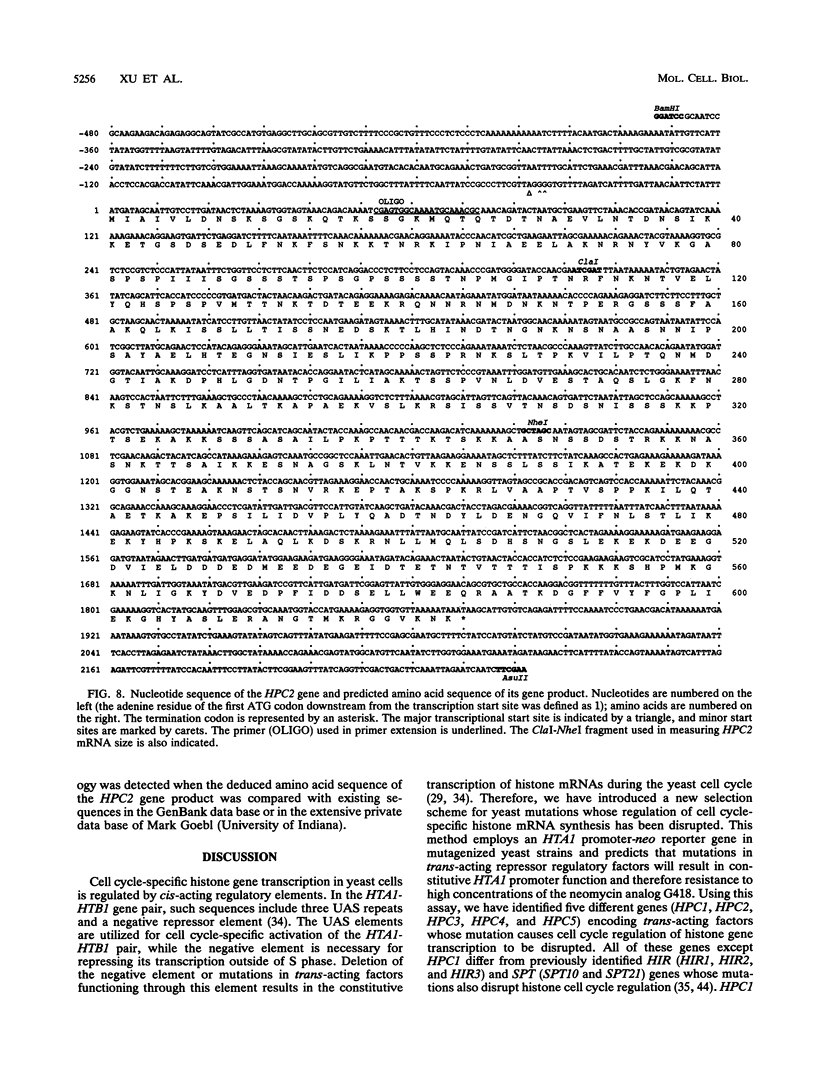
Histone mRNA synthesis is tightly regulated to S phase of the yeast Saccharomyces cerevisiae cell cycle as a result of transcriptional and posttranscriptional controls. Moreover, histone gene transcription decreases rapidly if DNA replication is inhibited by hydroxyurea or if cells are arrested in G1 by the mating pheromone alpha-factor. To identify the transcriptional controls responsible for cycle-specific histone mRNA synthesis, we have developed a selection for mutations which disrupt this process. Using this approach, we have isolated five mutants (hpc1, hpc2, hpc3, hpc4, and hpc5) in which cell cycle regulation of histone gene transcription is altered. All of these mutations are recessive and belong to separate complementation groups. Of these, only one (hpc1) falls in one of the three complementation groups identified previously by other means (M. A. Osley and D. Lycan, Mol. Cell. Biol. 7:4204-4210, 1987), indicating that at least seven different genes are involved in the cell cycle-specific regulation of histone gene transcription. hpc4 is unique in that derepression occurs only in the presence of hydroxyurea but not alpha-factor, suggesting that at least one of the regulatory factors is specific to histone gene transcription after DNA replication is blocked. One of the hpc mutations (hpc2) suppresses delta insertion mutations in the HIS4 and LYS2 loci. This effect allowed the cloning and sequence analysis of HPC2, which encodes a 67.5-kDa, highly charged basic protein.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alterman R. B., Ganguly S., Schulze D. H., Marzluff W. F., Schildkraut C. L., Skoultchi A. I. Cell cycle regulation of mouse H3 histone mRNA metabolism. Mol Cell Biol. 1984 Jan;4(1):123–132. doi: 10.1128/mcb.4.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews B. J., Herskowitz I. Regulation of cell cycle-dependent gene expression in yeast. J Biol Chem. 1990 Aug 25;265(24):14057–14060. [PubMed] [Google Scholar]
- Artishevsky A., Grafsky A., Lee A. S. Isolation of a mammalian sequence capable of conferring cell cycle regulation to a heterologous gene. Science. 1985 Nov 29;230(4729):1061–1063. doi: 10.1126/science.4059922. [DOI] [PubMed] [Google Scholar]
- Bilinski C. A., Miller J. J. Induction of normal ascosporogenesis in two-spored Saccharomyces cerevisiae by glucose, acetate, and zinc. J Bacteriol. 1980 Jul;143(1):343–348. doi: 10.1128/jb.143.1.343-348.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeden L. Cell cycle-regulated promoters in budding yeast. Trends Genet. 1988 Sep;4(9):249–253. doi: 10.1016/0168-9525(88)90031-5. [DOI] [PubMed] [Google Scholar]
- Capasso O., Bleecker G. C., Heintz N. Sequences controlling histone H4 mRNA abundance. EMBO J. 1987 Jun;6(6):1825–1831. doi: 10.1002/j.1460-2075.1987.tb02437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capasso O., Heintz N. Regulated expression of mammalian histone H4 genes in vivo requires a trans-acting transcription factor. Proc Natl Acad Sci U S A. 1985 Sep;82(17):5622–5626. doi: 10.1073/pnas.82.17.5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Adams C. D., Norris D., Osley M. A., Fassler J. S., Winston F. Changes in histone gene dosage alter transcription in yeast. Genes Dev. 1988 Feb;2(2):150–159. doi: 10.1101/gad.2.2.150. [DOI] [PubMed] [Google Scholar]
- Clark-Adams C. D., Winston F. The SPT6 gene is essential for growth and is required for delta-mediated transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1987 Feb;7(2):679–686. doi: 10.1128/mcb.7.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey L., Hanly S. M., Roeder R. G., Heintz N. Distinct transcription factors bind specifically to two regions of the human histone H4 promoter. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7241–7245. doi: 10.1073/pnas.83.19.7241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey L., Roberts S. B., Heintz N. Purification of the human histone H4 gene-specific transcription factors H4TF-1 and H4TF-2. Genes Dev. 1988 Dec;2(12B):1700–1712. doi: 10.1101/gad.2.12b.1700. [DOI] [PubMed] [Google Scholar]
- Domdey H., Apostol B., Lin R. J., Newman A., Brody E., Abelson J. Lariat structures are in vivo intermediates in yeast pre-mRNA splicing. Cell. 1984 Dec;39(3 Pt 2):611–621. doi: 10.1016/0092-8674(84)90468-9. [DOI] [PubMed] [Google Scholar]
- Emr S. D., Vassarotti A., Garrett J., Geller B. L., Takeda M., Douglas M. G. The amino terminus of the yeast F1-ATPase beta-subunit precursor functions as a mitochondrial import signal. J Cell Biol. 1986 Feb;102(2):523–533. doi: 10.1083/jcb.102.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher C., Heintz N., Roeder R. G. Purification and characterization of OTF-1, a transcription factor regulating cell cycle expression of a human histone H2b gene. Cell. 1987 Dec 4;51(5):773–781. doi: 10.1016/0092-8674(87)90100-0. [DOI] [PubMed] [Google Scholar]
- Han M., Chang M., Kim U. J., Grunstein M. Histone H2B repression causes cell-cycle-specific arrest in yeast: effects on chromosomal segregation, replication, and transcription. Cell. 1987 Feb 27;48(4):589–597. doi: 10.1016/0092-8674(87)90237-6. [DOI] [PubMed] [Google Scholar]
- Hanly S. M., Bleecker G. C., Heintz N. Identification of promoter elements necessary for transcriptional regulation of a human histone H4 gene in vitro. Mol Cell Biol. 1985 Feb;5(2):380–389. doi: 10.1128/mcb.5.2.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz N. The regulation of histone gene expression during the cell cycle. Biochim Biophys Acta. 1991 Mar 26;1088(3):327–339. doi: 10.1016/0167-4781(91)90122-3. [DOI] [PubMed] [Google Scholar]
- Hereford L., Bromley S., Osley M. A. Periodic transcription of yeast histone genes. Cell. 1982 Aug;30(1):305–310. doi: 10.1016/0092-8674(82)90036-8. [DOI] [PubMed] [Google Scholar]
- Hereford L., Fahrner K., Woolford J., Jr, Rosbash M., Kaback D. B. Isolation of yeast histone genes H2A and H2B. Cell. 1979 Dec;18(4):1261–1271. doi: 10.1016/0092-8674(79)90237-x. [DOI] [PubMed] [Google Scholar]
- Jimenez A., Davies J. Expression of a transposable antibiotic resistance element in Saccharomyces. Nature. 1980 Oct 30;287(5785):869–871. doi: 10.1038/287869a0. [DOI] [PubMed] [Google Scholar]
- Johnston L. H., White J. H., Johnson A. L., Lucchini G., Plevani P. The yeast DNA polymerase I transcript is regulated in both the mitotic cell cycle and in meiosis and is also induced after DNA damage. Nucleic Acids Res. 1987 Jul 10;15(13):5017–5030. doi: 10.1093/nar/15.13.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen R. A., Rothstein S. J., Reznikoff W. S. A restriction enzyme cleavage map of Tn5 and location of a region encoding neomycin resistance. Mol Gen Genet. 1979;177(1):65–72. doi: 10.1007/BF00267254. [DOI] [PubMed] [Google Scholar]
- Kim U. J., Han M., Kayne P., Grunstein M. Effects of histone H4 depletion on the cell cycle and transcription of Saccharomyces cerevisiae. EMBO J. 1988 Jul;7(7):2211–2219. doi: 10.1002/j.1460-2075.1988.tb03060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodkin A. L., Klar A. J., Stahl F. W. Double-strand breaks can initiate meiotic recombination in S. cerevisiae. Cell. 1986 Aug 29;46(5):733–740. doi: 10.1016/0092-8674(86)90349-1. [DOI] [PubMed] [Google Scholar]
- Kozak M. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol Cell Biol. 1987 Oct;7(10):3438–3445. doi: 10.1128/mcb.7.10.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeger P., Stewart C., Schaap T., van Wijnen A., Hirshman J., Helms S., Stein G., Stein J. Proximal and distal regulatory elements that influence in vivo expression of a cell cycle-dependent human H4 histone gene. Proc Natl Acad Sci U S A. 1987 Jun;84(12):3982–3986. doi: 10.1073/pnas.84.12.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lycan D. E., Osley M. A., Hereford L. M. Role of transcriptional and posttranscriptional regulation in expression of histone genes in Saccharomyces cerevisiae. Mol Cell Biol. 1987 Feb;7(2):614–621. doi: 10.1128/mcb.7.2.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Shore D. Transcriptional regulation in the yeast life cycle. Science. 1987 Sep 4;237(4819):1162–1170. doi: 10.1126/science.3306917. [DOI] [PubMed] [Google Scholar]
- Oechsner U., Magdolen V., Zoglowek C., Häcker U., Bandlow W. Yeast adenylate kinase is transcribed constitutively from a promoter in the short intergenic region to the histone H2A-1 gene. FEBS Lett. 1988 Dec 19;242(1):187–193. doi: 10.1016/0014-5793(88)81013-5. [DOI] [PubMed] [Google Scholar]
- Osley M. A., Gould J., Kim S., Kane M. Y., Hereford L. Identification of sequences in a yeast histone promoter involved in periodic transcription. Cell. 1986 May 23;45(4):537–544. doi: 10.1016/0092-8674(86)90285-0. [DOI] [PubMed] [Google Scholar]
- Osley M. A., Lycan D. Trans-acting regulatory mutations that alter transcription of Saccharomyces cerevisiae histone genes. Mol Cell Biol. 1987 Dec;7(12):4204–4210. doi: 10.1128/mcb.7.12.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osley M. A. The regulation of histone synthesis in the cell cycle. Annu Rev Biochem. 1991;60:827–861. doi: 10.1146/annurev.bi.60.070191.004143. [DOI] [PubMed] [Google Scholar]
- Peterson T. A., Prakash L., Prakash S., Osley M. A., Reed S. I. Regulation of CDC9, the Saccharomyces cerevisiae gene that encodes DNA ligase. Mol Cell Biol. 1985 Jan;5(1):226–235. doi: 10.1128/mcb.5.1.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Novick P., Thomas J. H., Botstein D., Fink G. R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60(2-3):237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schümperli D. Multilevel regulation of replication-dependent histone genes. Trends Genet. 1988 Jul;4(7):187–191. doi: 10.1016/0168-9525(88)90074-1. [DOI] [PubMed] [Google Scholar]
- Seiler-Tuyns A., Paterson B. M. Cell cycle regulation of a mouse histone H4 gene requires the H4 promoter. Mol Cell Biol. 1987 Mar;7(3):1048–1054. doi: 10.1128/mcb.7.3.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood P. W., Osley M. A. Histone regulatory (hir) mutations suppress delta insertion alleles in Saccharomyces cerevisiae. Genetics. 1991 Aug;128(4):729–738. doi: 10.1093/genetics/128.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman S. J., Fink G. R. Effects of Ty insertions on HIS4 transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Jul;4(7):1246–1251. doi: 10.1128/mcb.4.7.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. M., Andrésson O. S. DNA sequences of yeast H3 and H4 histone genes from two non-allelic gene sets encode identical H3 and H4 proteins. J Mol Biol. 1983 Sep 25;169(3):663–690. doi: 10.1016/s0022-2836(83)80164-8. [DOI] [PubMed] [Google Scholar]
- Smith M. M., Murray K. Yeast H3 and H4 histone messenger RNAs are transcribed from two non-allelic gene sets. J Mol Biol. 1983 Sep 25;169(3):641–661. doi: 10.1016/s0022-2836(83)80163-6. [DOI] [PubMed] [Google Scholar]
- Wallis J. W., Rykowski M., Grunstein M. Yeast histone H2B containing large amino terminus deletions can function in vivo. Cell. 1983 Dec;35(3 Pt 2):711–719. doi: 10.1016/0092-8674(83)90104-6. [DOI] [PubMed] [Google Scholar]
- Winston F., Chaleff D. T., Valent B., Fink G. R. Mutations affecting Ty-mediated expression of the HIS4 gene of Saccharomyces cerevisiae. Genetics. 1984 Jun;107(2):179–197. doi: 10.1093/genetics/107.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H. X., Johnson L., Grunstein M. Coding and noncoding sequences at the 3' end of yeast histone H2B mRNA confer cell cycle regulation. Mol Cell Biol. 1990 Jun;10(6):2687–2694. doi: 10.1128/mcb.10.6.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijnen A. J., Wright K. L., Lian J. B., Stein J. L., Stein G. S. Human H4 histone gene transcription requires the proliferation-specific nuclear factor HiNF-D. Auxiliary roles for HiNF-C (Sp1-like) and HiNF-A (high mobility group-like). J Biol Chem. 1989 Sep 5;264(25):15034–15042. [PubMed] [Google Scholar]









