While the physiological basis of cassava drought tolerance has been characterized, evaluation of the molecular responses to drought stress remains largely unexplored. This study provides an initial characterization of the molecular response of cassava to drought stress resembling field conditions. The candidate drought tolerance genes in cassava identified in this study can be used as expression-based markers of drought tolerance in cassava or be tested in the context of breeding and engineering drought tolerance in transgenics.
Keywords: Cassava, drought avoidance, drought tolerance, gene expression, osmotic adjustment, oxidative stress, real-time PCR
Abstract
Cassava is an important root crop to resource-poor farmers in marginal areas, where its production faces drought stress constraints. Given the difficulties associated with cassava breeding, a molecular understanding of drought tolerance in cassava will help in the identification of markers for use in marker-assisted selection and genes for transgenic improvement of drought tolerance. This study was carried out to identify candidate drought-tolerance genes and expression-based markers of drought stress in cassava. One drought-tolerant (improved variety) and one drought-susceptible (farmer-preferred) cassava landrace were grown in the glasshouse under well-watered and water-stressed conditions. Their morphological, physiological and molecular responses to drought were characterized. Morphological and physiological measurements indicate that the tolerance of the improved variety is based on drought avoidance, through reduction of water loss via partial stomatal closure. Ten genes that have previously been biologically validated as conferring or being associated with drought tolerance in other plant species were confirmed as being drought responsive in cassava. Four genes (MeALDH, MeZFP, MeMSD and MeRD28) were identified as candidate cassava drought-tolerance genes, as they were exclusively up-regulated in the drought-tolerant genotype to comparable levels known to confer drought tolerance in other species. Based on these genes, we hypothesize that the basis of the tolerance at the cellular level is probably through mitigation of the oxidative burst and osmotic adjustment. This study provides an initial characterization of the molecular response of cassava to drought stress resembling field conditions. The drought-responsive genes can now be used as expression-based markers of drought stress tolerance in cassava, and the candidate tolerance genes tested in the context of breeding (as possible quantitative trait loci) and engineering drought tolerance in transgenics.
Introduction
Improvement and expanded adoption of crops suited to growth with limited water resources on marginal lands is critical to ensuring food security, given the limited arable land and population growth, further compounded by the effects of climate change. In sub-Saharan Africa (SSA) and throughout much of the tropics and sub-tropics, the development and use of crop varieties with high water-use efficiency is particularly important for marginal areas with poor soils, unreliable rainfall and where irrigation is unavailable or unaffordable for resource-poor farmers. In this respect, cassava deserves particular attention because of its status and further potential as both a food security and a cash crop for most households living in marginal areas of the tropics and sub-tropics. In the tropics, cassava ranks third as a source of calories, and is typically grown by resource-poor smallholder farmers on marginal lands. A naturally drought-tolerant (DT) crop, it provides a critical staple to many of these populations vulnerable to food insecurity.
It has been estimated that moisture or drought stress is the most adverse crop environmental stress, accounting for over 70 % of potential agriculture yield losses worldwide (Boyer 1982). Drought is brought about by a shortage of rain or by a large variation in the amount of rainfall, and is the major abiotic stress limiting crop productivity worldwide (Saini and Westgate 2000). In Africa, the cassava growth cycle is typically interrupted by 3–6 months of drought, influencing various plant physiological processes resulting in depressed growth, development and economic yield (Pardales et al. 2001; Bakayoko et al. 2009). In general, cassava can withstand significant periods of drought stress. However, there is a range of drought-tolerance levels in available germplasm, and its growth and productivity in marginal areas are constrained by severe drought stress, especially during the earlier stages of growth (Pardales et al. 2001; Okogbenin et al. 2003; Bergantin et al. 2004; Perez et al. 2011). Development of cassava varieties with farmer-preferred traits and increased drought tolerance will allow its expanded cultivation and elevated yields in marginal areas.
Given the inherent challenges with cassava breeding, an understanding of the molecular basis of cassava drought responses and tolerance can help greatly in the development of appropriate varieties (Valliyodan and Nguyen 2006; El-Sharkawy 2007). Conventional breeding has been hindered by cassava's high heterozygosity, genotype by environment (G × E) interaction, long life cycle (Hahn et al. 1989; Fregene et al. 2001; Ceballos et al. 2004) and limited seed production, while molecular breeding is hindered by limited information on genomic regions and genes associated with drought tolerance in cassava. Efforts to improve cassava's water-use efficiency through conventional breeding have been limited in many parts of the world, including much of SSA. Breeding programmes in Latin America have successfully identified germplasm with increased levels of drought tolerance, with 2–3 times the yield of typical cassava genotypes in semi-arid conditions (El-Sharkawy 2007). A range of cassava drought-tolerance levels has also been characterized in West Africa (Okogbenin et al. 2003). Efforts in are now under way in eastern Africa to begin breeding for DT cassava.
Molecular breeding has already formed the basis of significant progress for other cassava traits. For instance, molecular markers tightly associated with the cassava mosaic disease (CMD) resistance gene CMD2 have been used in marker assistance breeding for CMD resistance (Akano et al. 2002). Breeding for tolerance to cassava postharvest physiological deterioration has also been reported (Morante et al. 2010). However, little progress has been made with respect to the development of DT cassava varieties. An understanding of the molecular response and basis of drought tolerance in cassava would significantly accelerate the production of DT varieties with farmer-preferred traits, through molecular breeding or genetic transformation, both of which have been successful in the development of DT plants. Utsumi et al. (2012) identified cassava genes responsive to drought treatment that consisted of wilting in vitro plantlets on a plastic plate for 1h under light. Further studies are required using drought stress methods more closely resembling drought stress conditions in the field in order to more confidently identify candidates appropriate for use in efforts to improve cassava drought tolerance.
Plant tolerance to drought stress is a complex trait with several interacting layers of molecular and physiological responses. Drought stress responses and tolerance genes have been well characterized in a number of plant species (Farooq et al. 2009; Gong et al. 2010), lending insight into the general pathways involved and potential tolerance mechanisms and genes in other species. Plant resistance to drought stress can be achieved through escape (e.g. early flowering time in drier environments), avoidance (e.g. transpiration control by stomata and development of extensive root systems), phenotypic flexibility, water conservation in tissues, antioxidant defences, plant growth regulation by hormones and osmotic adjustment (Farooq et al. 2009). Drought stress induces accumulation of metabolites and drought-related proteins (Ramachandra-Reddy et al. 2004). At the molecular level, the response to drought stress is a multi-genic trait. Through high-throughput microarray and real-time polymerase chain reaction (PCR) studies, a number of genes that respond to drought stress at the transcriptional level have been reported (Seki et al. 2002; Shinozaki and Yamaguchi-Shinozaki 2007; Talame et al. 2007; Guo et al. 2009). Some of these genes have been validated biologically and have been found to protect plants from desiccation through stress perception, signal transduction, transcriptional regulatory networks in cellular responses or tolerance to dehydration (Wang et al. 2005; Umezawa et al. 2006). Drought-stress-induced regulatory and functional genes have been used to increase drought tolerance through genetic engineering; for example, Vigna aconitifolia pyrroline-5-carboxylate synthetase (P5CS) has been used to engineer DT rice, and manganese superoxide dismutase (MnSOD) for DT alfalfa (McKersie et al. 1996; Zhu et al. 1998).
Cassava drought stress has been characterized physiologically and morphologically (reviewed by El-Sharkawy 2004; Setter and Fregene 2007; Okogbenin et al. 2011), and at the molecular level under conditions requiring further investigation to ensure their relevance to the context of field drought stress (Utsumi et al. 2012). Ecophysiologically, mechanisms of drought tolerance in cassava have been identified such as avoidance, through partial stomatal closure to reduce transpiration, development of extensive root systems and proportionally strategic reductions in leaf canopy (El-Sharkawy 2004, 2007); however, in some studies greater leaf retention has been correlated with drought tolerance (Lenis et al. 2006), so the relationship between leaf retention and drought tolerance depends on the genotype and probably on environmental factors (e.g. severity of drought). While a limited number of molecular studies have sequenced normalized expressed sequence tag libraries from cassava under drought stress (Lokko et al. 2007), no molecular studies have been conducted that quantify gene expression in single or contrasting cassava genotypes under conditions resembling those in the field, which would enable the identification of both drought-responsive and candidate drought-tolerance genes most relevant to cassava drought improvement efforts.
This study confirmed the DT and drought-susceptible (DS) status of improved and farmer-preferred cassava varieties, respectively, which are now part of the germplasm being integrated into the breeding programme at the National Crops Resources Research Institute (NACRRI) in Uganda to develop DT cassava with other farmer-preferred traits. The morphological and physiological responses of the two genotypes to drought stress were assessed. The relative expression levels of genes previously demonstrated to be functionally involved in, or associated with, drought stress responses in other species were also analysed. This study provides a general characterization of drought responses in cassava, yielding expression-based markers and candidate drought-tolerance genes for ongoing cassava improvement efforts. A molecular understanding of the drought responses of this DT species can also provide insights for increasing the drought tolerance of more drought-sensitive species.
Methods
Genotypes and treatments
Two cassava genotypes, DT MH96/0686 and DS Nyalanda, were used in this study. MH96/0686 is an improved cassava variety obtained from the cassava breeding programme at NACRRI, Uganda, while Nyalanda is a landrace obtained from farmers' fields in the Masindi district in western Uganda (1°40′28″ N, 31°42′54″ E). Previous field studies of 53 cassava genotypes in Uganda indicated that MH96/0686 was tolerant to drought and had a high harvest index, dry matter content, starch content, root yield and leaf retention under water stress compared with other genotypes, while Nyalanda was among the genotypes adversely affected by water stress and was significantly different from MH96/0686 in these phenotypes under drought stress (Turyagyenda et al. 2013). Based on the field data, these two genotypes were selected for detailed gene expression studies that were conducted in a glasshouse. The glasshouse conditions during the day were set at 25–30 °C (Alfredo and Setter 2004) with night-time temperatures typically ranging between 15 and 20 °C, and humidity typically at ∼50–80 %.
Cassava cuttings (30 cm in length) of these two genotypes were grown in 20-L plastic buckets in a randomized complete block design (RCBD) replicated three times. Before planting, each bucket was filled with 20 kg of sterilized soil (forest soil : river sand : ballast at 4 : 2 : 1 (v/v/v) respectively). One plant cutting was placed vertically in the soil in the middle of each bucket. To identify the effect of water stress on gene expression, plants were exposed to water stress by reducing soil moisture content (SMC). A similar number of plants per genotype remained watered at field capacity to act as a control. Three plants per genotype were included for each replication for each treatment. Before the application of treatments, all plants were watered with 1 L of water every 2 days until 60 days after planting (DAP). After 60 DAP, plants in the stress treatment were subjected to gradual drought stress conditions for a total of 10 days by withholding water to an SMC of ∼50 % for 5 days and then to ∼25 % for 5 days; gradual moisture stress was applied to mimic natural field drought conditions. Control (well-watered) plants were maintained at an SMC of ≥75 % by applying 1 L of water every 24 h. The SMC was monitored daily with a portable moisture meter (Delta Systems, UK).
Morphological and physiological drought stress measurements
During the water stress treatment period, physiological and morphological drought-stress-related traits were measured, including: leaf retention and plant height; and stomatal conductance, leaf relative water content (RWC) and leaf wilting. These measurements were collected from three plants per genotype from each replication for each treatment after 10 days of drought stress, just before leaf collection for gene expression analysis. Leaf wilting was scored on a scale of 1–3 modified from Bettina et al. (2007) (1 = no wilting; 2 = minimal wilting, where the plant showed leaf wilting only during hot hours and from which the leaves recovered; and 3 = severe wilting, where wilting leaves did not recover from wilting). For easy scoring, Bettina et al. (2007) scales 2 and 3 were combined to indicate minimal wilting, and 4 and 5 to indicate severe wilting. The stress treatment did not go as far as killing plants and therefore level 6 (death) (Bettina et al. 2007) was eliminated. The method of visual scoring of wilting is flexible as long as specific wilting categories are defined appropriately (Bettina et al. 2007). Stomatal conductance was measured on the third fully expanded leaf with an AP4 Porometer (Delta-T Devices, UK), while RWC was estimated on the fourth fully expanded leaf by following the procedure of Degenkolbe et al. (2009). Leaf retention was estimated as the percentage of the portion of stem height with leaves, and plant height was measured with a tape measure.
Leaf harvesting and RNA extraction
After 10 days of stress, the third fully expanded leaf was collected separately from three plants per genotype from each replication for each treatment. Leaf samples were collected between 12:00 and 12:30 p.m. To avoid taking material from the elongation zone at the base of the leaf blade or senescent tissue at the tip of the leaves, leaf samples were harvested from the middle section of the blades of fully expanded green leaves (Degenkolbe et al. 2009). The leaf samples were collected in Eppendorf tubes, immediately put in liquid nitrogen and stored at −80 °C until RNA extraction. Total RNA was extracted from the leaf samples (three biological replicates per genotype per treatment) using Concert Plant RNA Reagent (catalogue number 12322-012; Invitrogen) following the manufacturer's protocol. The concentration of RNA from each sample was determined by UV spectrophotometry at A260 using NanoDrop ND-1000 (NanoDrop Technologies, USA), while the integrity of total RNA was analysed by both Nanodrop (A260/A280) and 1.5 % 1× Tris-borate-EDTA (TBE) agarose gel electrophoresis (visualized with ethidium bromide; EtBr) after denaturation in 1×FDE (90 % v/v formamide, 1× TBE buffer, 0.5 % w/v bromophenol blue, 25 mM EDTA) at 65 °C for 5min and snap cooling on ice.
DNA contamination was removed using RNase-free DNaseI (Fermentas cat. no. EN0521) following the manufacturer's protocol. Two reverse transcription reactions (two technical replications) were prepared from each biological replicate. One microgram of total RNA was converted to cDNA by the GoScript reverse transcriptase system (Promega, USA) following the manufacturer's instructions. Briefly, 1 µg of total RNA was mixed with 10 pmol of random hexamer oligonucleotides to a final volume of 10 µL and the mixture incubated at 75 °C for 5 min. Then 1 µL of GoScript reverse transcriptase (Promega), 0.5 µL of RNasin RNase inhibitor (Promega), 4 µL of 5× PCR buffer, 1 µL of 10 mM dNTP and 1.2 µL (25 mM) of MgCl2 were added to the mixture on ice. Ribonuclease-free water was added to a final volume of 20 µL and the mixture incubated at an annealing temperature of 25 °C for 5 min. Extension was carried out at 42 °C for 60 min and the reaction was inactivated at 70 °C for 15 min. Two control reactions were included for each sample throughout this process: one without reverse transcriptase and one without RNA template.
Gene identification and PCR optimization
A literature survey was conducted to identify genes that have been functionally confirmed to confer drought tolerance in at least one plant species. The National Centre for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov/, accessed in June 2010) cDNA sequences of these genes were used as queries to identify cassava homologues through BLAST searches of the cassava genome database (http://www.phytozome.net, accessed in June 2010). This resulted in the identification of cassava gene homologues in cassava. When the query sequence was highly similar to several cassava genes (i.e. in a gene family), a multiple alignment of the highly similar cassava homologues was conducted with EBI clustalw2 tool (http://www.ebi.ac.uk/Tools/clustalw2/index.html, accessed in June 2010). Only homologues with high scores, percentage nucleotide identity to the gene of interest and high coverage of the query sequence were considered for clustering. Primers suitable for quantitative reverse transcription-PCR (qRT-PCR) (amplifying 200– to 350–bp products) and specific to the gene of interest (i.e. to a full-length coding sequence exhibiting the highest similarity to the query sequence; through maximum specificity at the 3′ end of the primer) were designed to amplify the identified cassava homologues.
The primers were optimized for target gene specificity with endpoint PCR using cassava cDNA. The endpoint PCR products were visualized by electrophoresis on a 2 % agarose 1× TBE gel, and visualized with EtBr. The PCR products for each primer pair were sequenced and compared with target genes (Table 2 ‘cassava homologues’ column) to confirm that the correct cassava genes were being amplified. All the designed primers (100 %) amplified cDNA to the expected product with a single band at an annealing temperature of 55 °C on an endpoint PCR system and thus this temperature was selected as the appropriate annealing temperature for qRT-PCR.
Table 2.
The primers designed for the 10 genes used in gene expression analysis. cDNA sequences of genes that confer drought tolerance in at least one plant species were used as queries to identify cassava homologues through BLAST searches of the cassava genome database. The cassava homologues were then used to manually design primers suitable for qRT-PCR (amplifying 200- to 350-bp products).
| Gene name | Accession | Mode of action | Cassava homologues (target genes)a | e-value (two species' genes) | Primer ID | Primer sequence (5′–3′) | Length | Expected size (bp) |
|---|---|---|---|---|---|---|---|---|
| Oryza sativa Japonica Group zinc finger protein ZFP252 | AY219847 | Osmotic adjustment through proline and sugars | cassava4.1_014662m.g (MeZFPa) | 0 | ZFP1F | CTC TAT TCT CAG CGC ACA TTC C | 22 | 245 |
| ZFP1R | AGC ATA ACG AGG CAG AGA GC | 20 | ||||||
| Arabidopsis thaliana amino acid transporter family II protein | NM_129684 | Likely role in osmotic adjustment | cassava4.1_007924m.g (MeATTF) | 2.7e-36 | ATTF1F | GTG GAA CTT TCT CCT CTC AGC A | 22 | 300 |
| ATTF1R | GCG TTA AAC TAC ATC CAT GGG C | 22 | ||||||
| Arabidopsis thaliana ALDH7B4 | NM_104287 | Antioxidant/ROS scavenging | cassava4.1_014540m.g (MeALDH) | 5.9e-43 | ALDH1F | GGA TGG AAT GCA TGC ATT GCA CTG | 24 | 263 |
| ALDH1R | CTG ATT CAC TGT TTG TTG CAC CAT C | 25 | ||||||
| Pisum sativum manganese superoxide dismutase | U30841 | Antioxidant/ROS scavenging/detoxication | cassava4.1_015272m.g (MeMSD) | 4.8e-40 | MSD1F | ATG AAT GCA GAA GGT GCT GCA | 21 | 269 |
| MSD1R | GAA GGG CAT TCT TTG GCA TAC | 21 | ||||||
| Arabidopsis thaliana GER3 (GERMIN 3) | NM_122070 | Regulation of plant growth | cassava4.1_016243m.g (MeGE3) | 2.4e-51 | GE31F | CGC TTG CAA GAA ACC TGC AG | 20 | 254 |
| GE31R | TGA ACC CAG CAC AGA TAG AC | 20 | ||||||
| Arabidopsis thaliana GBF3 (G-BOX BINDING FACTOR 3); | NM 180118 | Transcription factor and regulates alcohol dehydrogenase (Adh) via ABA | cassava4.1_008459m.g (MeGBF3) | 1.9e-18 | GBF32F | TGC ATC AAC TGT TGG GTG CG | 20 | 244 |
| GBF32R | ACC CAG AGC CAT GAG AAG GCT | 21 | ||||||
| Arabidopsis thaliana 14-3-3 protein GF14 lambda (GRF6) | AF145298 | Signalling factor/Delay leaf senescence (stay green trait) | cassava4.1_014556m.g (MeGF14) | 1.3e-104 | GF141F | AGC ACG CTT CTC TCT CTC TC | 20 | 261 |
| GF141R | AGG AAA CGA TCC TCC AAG CG | 20 | ||||||
| Arabidopsis thaliana RD28 | NM_129274 | Turgor responsive/transport of small molecules across membranes | cassava4.1_013192m.g (MeRD28) | 7.6e-64 | RD282F | TGC ACT GCT GGT ATC TCA GG | 20 | 237 |
| RD282R | GAT CTC AGC TCC CAA TCC AG | 20 | ||||||
| Arabidopsis thaliana MYC2 | NM_102998 | Transcription factor and regulates ABA-dependent RD22 and ADH1 | cassava4.1_002918m.g (MeMYC2) | 1.1e-40 | MYC21F | AGC GTC TCC AGA CCT TGA TC | 20 | 233 |
| MYC21R | AGT GGG ACC TGA GAT CAG C | 19 | ||||||
| Vigna aconitifolia pyrroline-5-carboxylate synthetase | M92276.1 | Osmotic adjustment | cassava4.1_002381m.g (MeP5CS) | 1.4e-78 | VAP1F | AGA CGT TAA GCG TAT CGT TG | 20 | 332 |
| VAP1R | CAA GAA GTT GAG CTG ATG TC | 20 |
aThe cassava homologues in parentheses were assigned gene names starting with ‘Me’ for Manihot esculenta.
Quantitative PCR
Quantitative RT-PCR was performed on a 7500HT standard Real-Time PCR system (ABI-PRISM®, USA) using SYBR Green JumpStart TaqReadyMix (Sigma, USA). The qRT-PCR was run on three biological replicates for each treatment for each genotype. Duplicate reactions were run for every biological replicate. The 20-µL reaction volume consisted of the following: 10 µL of 2× SYBR Green I ready mix, 0.02 µL of passive reference dye, 1 µL (10 pmoL) each of the forward (F) and reverse (R) gene-specific primers, 2 µL of template cDNA (50 ng) and 5.98 µL of distilled, deionized water (ddH2O). The PCR conditions were as follows: initial denaturation at 94 °C for 2 min; 40 cycles of denaturation at 94 °C for 15 s, annealing at 55 °C for 1 min and extension at 60 °C for 30 s. The dissociation curve analysis was carried out at the default setting of the 7500HT Real-Time PCR system to confirm the specificity of each reaction. A subset of the amplification products was also run on a 1× TBE agarose gel, stained with EtBr to ensure that each primer pair had one specific product.
The qRT-PCR reactions were normalized with the cassava actin gene (primers F: 5′-TGCAGACCGTATGAGCAAG-3′; R: 5′-CACCCTTGGAAATCCACATC-3′) as reference for all comparisons (Guo et al. 2009; Yang et al. 2011). The reference gene was expressed at similar levels in both well-watered and water-stressed treatments for both genotypes. The amplification efficiency of primers was determined by performing qRT-PCR on 1 : 2; 1 : 4, 1 : 8; 1 : 16; 1 : 32 and 1 : 64 dilutions of cDNA pooled from all experimental samples. All primer pairs amplified the genes with approximately the same efficiency as that of the reference gene actin and ranged from 1.96 to 2.01. The ΔΔCT method of relative gene quantification was used to conduct the various comparisons of relative gene expression from the qRT-PCR data, using the Relative Expression Software Tool (REST) (Pfaffl et al. 2002). A gene is significantly up-regulated or down-regulated when its expression in a treatment is higher or lower than that in a calibrator (standard/baseline), respectively, and when the t-test statistic is lower than 0.05 (at 95 % significant level). The expression in a calibrator is taken as unit (one), expression more than one is up-regulation and expression less than one is down-regulation. The t-statistic will show whether the up-regulation or down-regulation is significant or non-significant (NS).
Results
Morphological and physiological drought stress characteristics
At T0 (day 1/first day of drought treatment), the two genotypes were healthy and exhibited no readily observable symptoms of drought stress. After 10 days of reduced SMC, the two genotypes exhibited different levels of drought stress: the DS genotype Nyalanda showed severe wilting symptoms on the leaves, including mild chlorosis of upper leaves and senescence of many of the lower leaves, compared with more limited stress signs in the DT genotype MH96/0686 (Fig. 1). In order to assess the drought stress response in leaves that were at the onset of visible stress in the DS genotype at the 10-day time point, RWC and stomatal conductance were gathered from leaves just beginning to exhibit wilting symptoms. The mean SMC in stressed plants was significantly lower (28.80 ± 1.08) than that in well-watered plants (83.00 ± 2.45). Nyalanda lost 19 % of its leaves through shedding under stress while MH96/0686 retained almost all its leaves (over 99 %), (i.e. the ‘stay green’ trait) (Table 1 and Fig. 1). Nyalanda leaves were permanently wilted, compared with minimal leaf wilting of MH96/0686 genotype leaves that was limited to hot hours (∼1200–1500 h), after which the leaves recovered from wilting. In MH96/0686, stomatal conductance was more than two times lower than in Nyalanda under stress.
Fig. 1.
Effect of drought stress on improved MH96/0686 cassava genotype and landrace Nyalanda. Stress treatment was gradually given to the plants 60 days after planting. Moisture stress was gradually applied to mimic natural field drought conditions. Improved MH96/0686 and farmer preferred landrace Nyalanda were differentially affected by drought stress conditions. After 10 days of gradual application of drought stress, MH96/0686 was less affected by water stress than Nyalanda, which exhibited marked wilting and other drought stress symptoms.
Table 1.
Physiological and morphological responses of the two genotypes after 10 days of moisture stress. Physiological and morphological drought-stress-related traits, measured on three plants per replication for each treatment (stressed and control) after 10 days of water stress (just before leaf sample collection for qRT-PCR). All values shown are mean values at P ≤ 0.05. ns, not significant; **significant at P ≤ 0.05; SBG, significance between genotypes (columns); significance between treatments is shown in the rows.
| Cultivar | Conductance (mmol m−2 s−1) |
Leaf retention (%) |
Relative water content (%) |
Number of leaves |
Plant height (cm) ns |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Control | Stressed | Control | Stressed | Control | Stressed | Control | Stressed | Control | Stressed | |
| MH96/0686 | 350.0 ± 37.21 | 168.9 ± 200** | 76.88 ± 3.65 | 76.33 ± 2.33ns | 97.3 ± 1.12 | 95.4 ± 2.92ns | 26.78 ± 1.27 | 24.89 ± 1.50 ns | 52.22 ± 2.86 | 52.11 ± 2.64 ns |
| Nyalanda | 492.5 ± 43.0 | 355 ± 21.2** | 63.33 ± 4.22 | 51.25 ± 2.48** | 94.95 ± 1.37 | 81.3 ± 2.92ns | 21.50 ± 1.56 | 13.37 ± 1.59** | 48.67 ± 3.51 | 47.50 ± 2.80 ns |
| SBG | ** | ** | ** | ** | ns | ** | ** | ** | ns | ns |
Cassava drought stress gene PCR assay validation
Of the 10 genes previously confirmed to have a functional role in drought stress that were selected for this study (Table 2), seven were previously reported from studies in Arabidopsis thaliana, one in V. aconitifolia, one in Pisum sativum and one in Oryza sativa. All designed primers (Table 2) amplified cDNA with a single band at the expected size and of the expected sequence, based on the cassava database sequences from which the primers were designed (data not shown).
Gene expression analysis
Several comparative analyses were conducted to determine the dynamics of the drought response gene expression changes in the two genotypes. In all analyses, the drought response gene expression levels were first normalized to the control gene (actin). The first question addressed was whether the selected genes are in fact drought responsive in cassava. For this, the expression of each of the 10 genes was compared between well-watered controls and drought-stress-treated plants within each of the two genotypes (Table 3). The expression of each of the 10 genes responded to water stress in one or both genotypes: nine of the genes were up-regulated in one or both genotypes, and one gene (MeGE3) was down-regulated in both. In the susceptible genotype Nyalanda, six genes were differentially expressed across treatments, five of which were up-regulated (MeATTF, MeGBF3, MeGF14, MeP5CS and MeMYC2) and one of which was down-regulated (MeGE3); four genes (MeALDH, MeMSD, MeRD28 and MeZFP) were not differentially expressed in response to water stress. In the tolerant genotype MH96/0686, seven genes were differentially expressed, of which six (MeALDH, MeATTF, MeGBF3, MeMSD, MeRD28 and MeZFP) were significantly up-regulated and one (MeGE3) was down-regulated by drought stress; three genes (MeGF14, MeMYC2 and MeP5CS) were not differentially expressed. All 10 genes responded to drought, with differences in which genes were responsive to drought in the two genotypes (Fig. 2).
Table 3.
Effect of water stress on mRNA levels, comparing stressed to control plants within a genotype. Quantitative RT-PCR was performed for each identified gene on three biological replicates for each treatment (stress and control) for each genotype (MH96/0686 and Nyalanda). Duplicate reactions were run for every biological replicate. The qRT-PCR reactions were normalized with the cassava actin gene as a reference for all comparisons. The ΔΔCT method of relative gene quantification was used to make the various comparisons of relative gene expression from the qRT-PCR data, using REST. For each genotype, the control plants were used as a calibrator. A gene is significantly up-regulated or down-regulated when its expression in a treatment is higher than or lower than that in a calibrator (standard/baseline), respectively, and when the t-test statistic is lower than 0.05 (at 95 % significance level). The expression in a calibrator is taken as unity (one), expression of more than one is up-regulation and expression less than one is down-regulation. The t-statistic will show whether the up-regulation or down-regulation is significant or non-significant (NS).
| Genotype | Gene | Expression | SE | 95 % CI | Probability | Result |
|---|---|---|---|---|---|---|
| MH96/0686 Stressed against well watered | MeALDH | 2.815 | 1.818–4.183 | 1.327–6.112 | 0.000 | Up-regulated |
| MeATTF | 3.245 | 1.444–9.582 | 1.069–11.530 | 0.000 | Up-regulated | |
| MeGBF3 | 3.241 | 2.221–5.467 | 1.688–9.982 | 0.000 | Up-regulated | |
| MeGE3 | 0.317 | 0.181–0.647 | 0.097–0.988 | 0.006 | Down-regulated | |
| MeGF14 | 1.303 | 0.963–1.768 | 0.844–2.344 | 0.095 | NS | |
| MeMYC2 | 1.350 | 0.718–2.196 | 0.608–3.336 | 0.204 | NS | |
| MeMSD | 3.148 | 2.316–4.431 | 1.897-6.394 | 0.001 | Up-regulated | |
| MeRD28 | 1.511 | 1.062–1.998 | 0.852–2.153 | 0.013 | Up-regulated | |
| MeP5CS | 1.425 | 0.686–3.784 | 0.384–4.745 | 0.301 | NS | |
| MeZFP | 4.043 | 2.869–5.828 | 2.164–8.014 | 0.000 | Up-regulated | |
| NYALANDA Stressed against well watered | MeALDH | 2.160 | 1.003–5.120 | 0.464–7.983 | 0.056 | NS |
| MeATTF | 2.671 | 1.902–3.812 | 1.393–4.954 | 0.001 | Up-regulated | |
| MeGBF3 | 1.875 | 1.161–3.028 | 0.733–3.979 | 0.018 | Up-regulated | |
| MeGE3 | 0.205 | 0.057–0.671 | 0.034–0.941 | 0.000 | Down-regulated | |
| MeGF14 | 2.285 | 1.094–6.336 | 0.826–18.602 | 0.031 | Up-regulated | |
| MeMYC2 | 2.201 | 1.391–3.371 | 0.976–4.183 | 0.006 | Up-regulated | |
| MeMSD | 1.506 | 0.983–2.255 | 0.815–4.129 | 0.063 | NS | |
| MeRD28 | 1.128 | 0.994–1.286 | 0.875–1.381 | 0.059 | NS | |
| MeP5CS | 1.662 | 1.125–2.414 | 0.977–3.496 | 0.007 | Up-regulated | |
| MeZFP | 1.578 | 0.806–3.368 | 0.525–5.198 | 0.159 | NS |
CI, confidence interval at 95 %; expression, fold change in the expression of a gene in water stress relative to control treatment (P = 0.05).
Fig. 2.
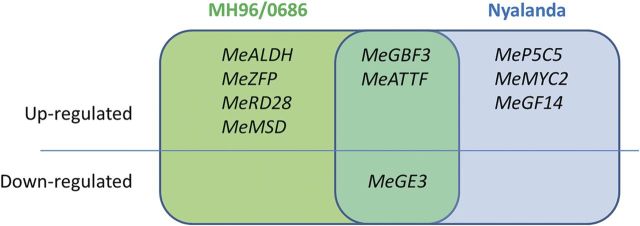
Gene expression changes induced by drought stress in the two genotypes. The relative changes in expression of each gene between the DT and DS genotypes are summarized according to the analysis of relative expression changes under drought stress compared with well-watered control conditions.
Given that the expression of the 10 previously identified plant drought-tolerance genes is responsive to drought in cassava, the next question was whether their expression can provide insight into the basis of the relative drought tolerance and susceptibility of the two genotypes; genes expressed at higher levels in MH96/0686 compared with Nyalanda are candidates for conferring drought tolerance. For this, two comparisons between genotypes were made: relative expression levels under non-stressed conditions (basal expression levels) and relative expression levels under drought stress.
To determine the relative expression levels under non-stressed conditions, baseline expression levels were compared under well-watered conditions for the two genotypes (Table 4). Even in the absence of drought stress, three genes exhibit different expression levels in the two genotypes. MeGF14 and MeMYC2 are expressed at approximately twice the level in MH96/0686 (DT) compared with Nyalanda (DS). Alternatively, MeMSD is expressed at approximately half the level in DT compared with DS.
Table 4.
Comparison of the baseline gene expressions of DT MH96/0686 and DS Nyalanda. Quantitative RT-PCR was performed for each identified gene on three biological replicates for well-watered plants (control) of each genotype (MH96/0686 and Nyalanda). Duplicate reactions were run for every biological replicate. The qRT-PCR reactions were normalized with the cassava actin gene as a reference for all comparisons. The ΔΔCT method of relative gene quantification was used to make the various comparisons of relative gene expression from the qRT-PCR data, using REST. During data collection, Nyalanda (DS landrace) was used as a calibrator. The DT genotype has several drought-tolerance genes whose levels are different from those in DS (under well-watered conditions). A gene is significantly up-regulated or down-regulated when its expression in a treatment is higher than or lower than that in a calibrator (standard/baseline), respectively, and when the t-test statistic is lower than 0.05 (at 95 % significant level). The expression in a calibrator is taken as unity (one), expression of more than one is up-regulation and expression less than one is down-regulation. The t-statistic will show whether the up-regulation or down-regulation is significant or non-significant (NS).
| Gene | Expression (fold difference in baseline expression levels, DT vs. DS) | SE | 95 % CI | Probability (P = 0.05) | Relative significant difference (DT vs. DS) |
|---|---|---|---|---|---|
| MeALDH | 1.379 | 0.598–2.832 | 0.370–4.163 | 0.319 | NS |
| MeATTF | 1.679 | 0.531–4.431 | 0.384–6.376 | 0.211 | NS |
| MeGBF3 | 1.094 | 0.570–1.789 | 0.272–2.591 | 0.746 | NS |
| MeGE3 | 1.959 | 0.501–6.700 | 0.270–10.729 | 0.221 | NS |
| MeGF14 | 2.332 | 1.202–5.717 | 0.824–17.293 | 0.013 | Up-regulation |
| MeMYC2 | 1.924 | 1.118–3.617 | 0.741–4.594 | 0.019 | Up-regulation |
| MeMSD | 0.584 | 0.429–0.837 | 0.366–1.282 | 0.014 | Down-regulation |
| MeRD28 | 1.120 | 0.990–1.297 | 0.867–1.409 | 0.075 | NS |
| MeP5CS | 1.934 | 0.746–3.786 | 0.657–6.197 | 0.054 | NS |
| MeZFP | 0.858 | 0.504–1.471 | 0.317–2.258 | 0.539 | NS |
The relative gene expression levels under drought stress were next compared between the tolerant and susceptible genotypes (Table 5). Under drought stress, 7 of the 10 genes were expressed at significantly higher levels in the tolerant genotype compared with the susceptible genotype. This included four that had been identified as exclusively up-regulated by drought stress in tolerant MH96/0686 (MeALDH, MeZFP, MeRD28 and MeMSD), two that were up-regulated in both genotypes (MeGBF3, MeATTF), and one that was exclusively up-regulated in susceptible Nyalanda (MeP5C5; however, not up-regulated enough to surpass expression in DT under drought stress). This also included the one gene down-regulated by drought stress in both genotypes (MeGE3).
Table 5.
Comparison of gene expression levels of DT and DS cassava genotypes after 10 days of stress. Quantitative RT-PCR was performed for each identified gene on three biological replicates for each genotype (MH96/0686 and Nyalanda) after 10 days of stress. Duplicate reactions were run for every biological replicate. The ΔΔCT method of relative gene quantification was used to make the various comparisons of relative gene expression from the qRT-PCR data, using REST. The DS is used as a calibrator, i.e. the relative expression level of each gene is shown as the relative times higher expression in DT (MH96/0686) than in DS (Nyalanda) under drought stress. The ‘Result’ column indicates whether each gene is significantly expressed at higher levels under drought in MH96/0686 versus Nyalanda. A gene is significantly up-regulated or down-regulated when its expression in a treatment is higher than or lower than in a calibrator (standard/baseline), respectively, and when the t-test statistic is lower than 0.05 (at 95 % significant level). The expression in a calibrator is taken as unity (one), expression of more than one is up-regulation and expression less than one is down-regulation. The t-statistic will show whether the up-regulation or down-regulation is significant or non-significant (NS).
| Gene | Fold higher in MH96/0686 (compared with Nyalanda) | SE | 95 % CI | Probability (P = 0.05) | Result |
|---|---|---|---|---|---|
| MeALDH | 1.797 | 1.038–3.258 | 0.6444.879 | 0.040 | Up-regulation |
| MeATTF | 2.040 | 1.585–2.669 | 1.377–3.090 | 0.000 | Up-regulation |
| MeGBF3 | 1.890 | 1.379–2.579 | 1.099–3.128 | 0.000 | Up-regulation |
| MeGE3 | 3.033 | 1.762–5.233 | 1.122–8.913 | 0.000 | Up-regulation |
| MeGF14 | 1.330 | 0.842–1.921 | 0.669–2.521 | 0.123 | NS |
| MeMYC2 | 1.180 | 0.767–1.775 | 0.634–2.532 | 0.350 | NS |
| MeMSD | 1.221 | 0.800–1.664 | 0.589–3.109 | 0.326 | NS |
| MeRD28 | 1.501 | 1.065–1.966 | 0.866–2.133 | 0.017 | Up-regulation |
| MeP5CS | 1.659 | 0.992–2.709 | 0.712–3.197 | 0.040 | Up-regulation |
| MeZFP | 2.197 | 1.115–3.590 | 0.815–3.900 | 0.017 | Up-regulation |
Discussion
Cassava is an essential crop for increasing food security in SSA and other food-insecure regions. Ranked highest in importance among tropical root crops, its roots are a remarkable food source for 500 million people globally. Subsistence farmers in eastern and central Africa rely heavily on it to survive periods of drought, general crop failure and food scarcity. Although cassava is a relatively DT crop, increased drought tolerance is nevertheless an important trait to consider for improving cassava for resource-poor farmers.
This study conducted morphological, physiological and molecular characterization of a DT improved variety (MH96/0686) and DS landrace (Nyalanda) from Uganda, both identified as such from previous observations in farmers' and research station fields. Characterization of cassava genotypes with disparate responses to drought allowed both characterization of the general cassava drought stress response and identification of the candidate genes that may be contributing to the increased drought tolerance in MH96/0686. These two genotypes represent ideal candidates for integration in the Ugandan cassava breeding programme; the farmer-preferred characteristics of Nyalanda and the drought tolerance and other improved characteristics of MH96/0686 can be combined to produce new varieties combining drought tolerance with high yield under non-drought conditions and other farmer-preferred traits. This study has established molecular tools that can be used to further characterize, understand and breed for drought tolerance in cassava through the inclusion of gene expression-based phenotyping using the drought stress expression-based markers for the identification of quantitative trait loci (QTLs), and finer molecular-level phenotyping of progeny to guide selection of the best genotypes during the breeding programme.
MH96/0686 resists drought by avoidance while Nyalanda is DS
Physiological and morphological analyses were conducted to assess the drought response of the two genotypes, and confirmed the relative tolerance of MH96/0686 and susceptibility of Nyalanda that had been observed in the field (Turyagyenda et al. 2013). Drought stress conditions were designed to more closely reflect those in the field, and those successfully used in other similar studies. Data were collected after 10 days of water stress, when the percentage SMC was close to field capacity (83.00 ± 2.45) in well-watered plants, while in stressed plants it was 28.80 ± 1.08, which is close to the 25 % SMC reported previously as a severe drought stress treatment for cassava (Aina et al. 2007). The third fully expanded leaf was beginning to exhibit visual drought stress symptoms in Nyalanda but not in the corresponding leaves in MH96/0686 (Fig. 1), making this leaf a good candidate for assessing the differential response to drought in the two genotypes at a relatively early stage of leaf response.
Measurements indicated that the tolerance of MH96/0686 was due to avoidance at the physiological level (Table 1). The lower stomatal conductance in MH96/0686 compared with Nyalanda is an indication of drought avoidance, reflecting its resistance to water loss through partial stomatal closure for increased water-use efficiency. This represents an effective adaptive response associated with drought tolerance in plants (Heschel and Riginos 2005). Rapid closing of stomata in response to reduced atmospheric humidity and soil moisture has been recognized as the principal mechanism of drought tolerance in cassava (El-Sharkawy 2004; Setter and Fregene 2007). Substantial variation in leaf conductance has been observed and this trait appears to be useful in pre-selecting DT germplasm (Iglesias et al. 1995). Lower stomatal conductance is an indication of reduced water loss through stomata for increased water-use efficiency. Wajid et al. (2011) also reported that cultivars susceptible to water stress have a higher stomatal conductance and transpiration rate than DT cultivars.
The RWC was significantly higher in MH96/0686 under drought stress (Table 1), a further reflection of drought avoidance achieved by the partial stomatal closure in MH96/0686. Cassava plants that can control leaf loss in response to drought are associated with increased drought tolerance (Lenis et al. 2006; El-Sharkawy 2007). As a result of its more limited capacity to cope with drought stress, Nyalanda lost significantly more leaves by shedding and senescence under drought stress (Table 1). Leaf wilting/folding and shedding, as exhibited by Nyalanda, has been described as a drought avoidance mechanism (Mitra 2001) for short-term drought, but this has serious consequences for photosynthesis, whole-plant physiology, productivity under prolonged water stress conditions. Retention of leaves or ‘stay green’ under water stress conditions, as exhibited by MH96/0686, has been correlated with drought tolerance and improved yields in cassava (Lenis et al. 2006) because this ‘stay green’ condition maintains photosynthesis. In tobacco, Rivero et al. (2007) were able to enhance drought tolerance by delaying drought-induced leaf senescence through transformation using the isopentenyl transferase gene. Similarly, the appearance and development of major damage symptoms such as wilting, dying of old leaves and necrosis of young leaves caused by the water stress conditions were delayed in the transgenic rice plants by HVA1 (group 3 LEA protein) (Xu et al. 1996).
Collectively, these data confirm field observations on relative levels of drought tolerance in the two genotypes, indicating that one of the mechanisms that MH96/0686 uses to cope with drought stress is avoidance. These observations support the suitability of these genotypes at this time point for further molecular characterization of drought stress responses in cassava.
Molecular characterization of cassava drought stress responses and identification of candidate tolerance genes
Other studies have also indicated that a common drought-tolerance mechanism at the physiological level is avoidance, as we have shown is the case for MH96/0686. Therefore, this genotype exhibits a common drought-tolerance mechanism for cassava. These other studies have highlighted the need for molecular analysis of the tolerance response, to help characterize the underlying molecular basis of the tolerance and to provide molecular tools to complement ongoing breeding efforts (El-Sharkawy 2007). Differences in gene expression can serve as expression-based markers for drought stress, and contribute to the drought tolerance of MH96/0686 and of cassava in general. We confirmed that 10 cassava genes are responsive to drought, and further identified the candidate genes underlying the tolerance, which can be used for cassava improvement. The molecular component of this investigation capitalized on the extensive molecular characterization and functional validation studies that have been conducted in other plant species; to optimize the chances of inclusion of functional drought response genes in cassava, a confirmed functional role in drought tolerance in other plant species served as the primary criterion for inclusion in this study.
Molecular characterizations were conducted on the same samples used for morphological and physiological characterizations, from the third fully expanded leaf with 10 days of gradually applied severe drought stress. Unlike recent gene expression studies in cassava by Utsumi et al. (2012) that applied a 1-h desiccation shock to identify genes differentially expressed in response to drought, the gradual drought stress used in this study more closely resembles field conditions in order to identify transcriptional changes crucial to adaptation under field conditions (Talame et al. 2007). Also, An et al. (2012), after subjecting cassava plants to cold stress at 7 °C for different periods, suggested that prolonged stress could trigger more stress-related gene expression. In addition, many molecular studies on drought responses have used a single genotype without comparing the expression of genes between contrasting genotypes (Sakurai et al. 2007; Guo et al. 2009; You-Zhi et al. 2010), making it difficult to separate drought-tolerance-related genes from drought-responsive genes. Genes differentially expressed in response to drought in a single genotype may not necessarily be responsible for enhancing drought tolerance (Guo et al. 2009). Contrasting gene expression changes induced by drought stress in tolerant versus susceptible genotypes allow better discrimination of those uniquely responsive in the tolerant genotype, representing candidate genes underlying the tolerance. On the flip side, gene expression changes that are significantly occurring uniquely in the susceptible genotype can serve as markers for drought stress and may contribute to its susceptibility.
When comparing disparately responding genotypes, genes up-regulated in both are less likely to play a significant functional role and are therefore termed ‘drought responsive’, whereas genes up- or down-regulated more significantly in a tolerant genotype represent candidate cassava ‘drought-tolerance’ genes (i.e. may underlie the tolerance of this genotype). Of course, these candidate drought-tolerance genes require further validation to confirm that they play a functional role in tolerance in cassava and specifically in the tolerant genotypes used in this study. Their identification is the first important step and provides insight into the molecular changes in response to drought in the tolerant genotype, and to cassava drought tolerance in general.
Cassava drought-responsive genes
First, it was assessed whether each of the genes selected from previous studies is responsive to drought in cassava (i.e. whether their expression levels change in response to drought stress). The expression of each of the 10 genes was significantly different when drought stress was applied to either one or both genotypes, compared with well-watered controls. This confirms that all 10 genes are also drought responsive in cassava. They can therefore be used as expression-based markers of drought stress and subjected to further study in the context of drought tolerance of MH96/0686 compared with susceptibility of Nyalanda. The genes commonly up- or down-regulated in both genotypes (MeGBF3, MeATTF and MeGE3; Table 3 and Fig. 2) may constitute part of the general response to drought stress that is commonly invoked within the range of tolerance/susceptibility represented by these two genotypes.
Candidate drought-tolerance genes
Genes underlying MH96/0686 tolerance, which is no doubt multi-genic, can be divided into two categories: those whose baseline expression levels are different in DT compared with DS (i.e. DT is primed to be less susceptible or responds more quickly to drought stress) and those whose expression levels under drought are significantly more changed/expressed at higher levels in DT compared with DS (i.e. they may play a role in the longer-term tolerance of MH96/0686 observed in the field). The more responsive genes (the second category) represent candidates for the adaptive response of MH96/0686 to drought and therefore important targets for further validation and for subsequent use in developing cassava varieties with enhanced drought tolerance.
In the first category of baseline tolerance genes, two genes were expressed at significantly higher levels in the DT compared with the DS genotype (MeGF14 and MeMYC2) and one at significantly lower levels (MeMSD). The second category, consisting of genes with higher expression levels under drought stress in DT compared with DS, included 7 of the 10 genes. The fact that so many of these genes, demonstrated to confer drought tolerance in other species, were up-regulated more significantly by drought in MH96/0686 demonstrates that overall the drought response in this genotype is more robust at a molecular level.
Genes exclusively up-regulated by drought stress in the DT MH96/0686
Four genes, MeALDH, MeZFP, MeRD28 and MeMSD, were exclusively up-regulated by water stress in the DT genotype. Under well-watered/control conditions, and using DS as a calibrator, MeALDH, MeZFP and MeRD28 were not differentially expressed between DT and DS (Table 4). Using the control treatment as a baseline, this means that the up-regulation of these genes in DT was due to water stress. The expression of MeMSD was lower in DT than in DS in the control treatment (well watered) but its expression in DT in response to stress was 2-fold higher than that in DS, indicating that water stress resulted in higher expression of this gene in the DT compared with the DS genotype. Therefore these four genes (MeALDH, MeZFP, MeRD28 and MeMSD) might be involved in drought adaptation, or the tolerance of MH96/0686 and potentially other tolerant cassava genotypes.
MeMSD encodes a manganese superoxide dismutase (MnSOD) enzyme that plays a role in oxidative stress tolerance in plants. Over-expression of superoxide dismutase (SOD) enzymes has been reported to increase oxidative stress tolerance in transgenics (Sen Gupta et al. 1993; Van Breusegem et al. 1999; Basu et al. 2001; Wang et al. 2005). For example, Sen Gupta et al. (1993) reported that a 3-fold increase in total pea Cu/ZnSOD activity resulted in a significant increase in resistance to methyl viologen-induced membrane damage in transgenic tobacco. Basu et al. (2001) showed that a 1.5- to 2.5-fold increase in total SOD activity in transgenic Brassica napus plants transformed with wheat MnSOD increased oxidative stress resistance compared with wild-type controls. Wang et al. (2005) reported that a 1.4-fold increase in total SOD activity in the MnSOD transgenic rice plants was enough to increase oxidative stress resistance and drought tolerance when the gene was fused with a chloroplast transit peptide sequence in order to target the MnSOD to the chloroplast. The 3.148-fold increase observed in DT genotype MH96/0686 therefore indicates a level that can plausibly confer increased oxidative stress and drought tolerance in cassava. Superoxide dismutase enzymes are involved in scavenging reactive oxygen species (ROS) that are produced in plants during water stress (Kendall and McKersie 1989; Price et al. 1989; Hernandez et al. 1995; McKersie et al. 1996; Fryer et al. 2002) and so we hypothesize that this gene confers drought tolerance through ROS scavenging in cassava.
The gene MeZFP that encodes a zinc finger protein (ZFP252) was also exclusively up-regulated in MH96/0686. This ZFP has been reported to confer drought tolerance in plants by maintaining cell membrane integrity during water stress. Xu et al. (2008) reported that the relative electrolyte leakage, an indicator of membrane injury (Morsy et al. 2005), was lower in O. sativa ZFP252-transformed rice plants than in untransformed and O. sativa ZFP252 knock-out plants under drought stress. This suggests that ZFP252 protects plants from stress by maintaining cell membrane integrity. The transformed rice plants also had higher free proline contents and soluble sugars than non-transgenic and ZFP252 knock-out transgenic rice plants (Xu et al. 2008). The findings of Xu et al. (2008) suggest that enhanced stress tolerance of ZFP252-transgenic plants might partly be through activation of proline synthesis and proline transport pathways by ZFP252 in rice under salt and drought stresses. Proline levels are known to contribute to drought tolerance through osmotic adjustment (Sanchez et al. 1998). In this study, MeZFP was over-expressed 4.043-fold under drought stress in DT (MH96/0686). It is therefore plausible that this gene is among those that enhance drought tolerance in cassava and specifically in MH96/0686 through increasing the free osmoprotectant proline and soluble sugars.
MeRD28 encodes the Responsive to Desiccation (RD28) gene. The expression of this gene was increased 1.511-fold by water stress, being exclusively upregulated in the DT genotype, suggesting that it plays a role in enhancement of drought tolerance in cassava. Daniels et al. (1994) reported that RD28 is a turgor-responsive, mercury-resistant plasma membrane aquaporin found in plasma membranes of all plant tissues except seeds. Earlier studies by Yamaguchi-Shinozaki et al. (1992) reported that RD28 enhances drought tolerance through an abscisic acid-independent pathway. It protects desiccated cells by transporting small molecules across cell membranes and it is believed here that it enhances the cells' desiccation tolerance in DT cassava through osmotic adjustment.
The fourth gene that was exclusively up-regulated in MH96/0686 is MeALDH, which encodes aldehyde dehydrogenase (ALDH7B4). It was up-regulated by 2.815-fold under drought stress and may therefore be involved in enhancement of drought tolerance in cassava and specifically MH96/0686. The findings of this study are in agreement with the studies by Kotchoni et al. (2006), who reported that transgenic A. thaliana plants with increased amounts of ALDH7B4 were more tolerant to dehydration and salt stress than wild-type plants. They further reported that over-expression of the ALDH7B4 gene in transgenic plants reduced the level of lipid peroxidation under drought and salt stress, suggesting that the gene confers tolerance towards both osmotic and oxidative stress in A. thaliana through ROS scavenging and reduction of lipid peroxidation. This gene had also been reported to be induced by pathogens (Zimmermann et al. 2004) and therefore might be a multi-stress-responsive gene. Over-expression by 2.815-fold and exclusive up-regulation of this gene in DT cassava are an indication that the gene may be involved in enhancement of drought tolerance in cassava, probably through ROS scavenging and reduced lipid peroxidation.
Genes up-regulated only in the DS genotype
Three genes (MeMYC2, MeP5CS and MeGF14) were exclusively up-regulated in the DS genotype and not in the DT genotype. These genes are therefore responding to the drought stress state in Nyalanda. However, they are not likely to be part of the genetic basis of drought tolerance in MH96/0686.
Genes up-regulated by water stress in both DT and DS genotypes
Two genes, MeATTF encoding an amino acid transporter family II protein and MeGBF3 encoding G-box binding factor 3 (GBF3), were up-regulated by stress in both tolerant and susceptible genotypes. They may therefore be among the general drought-responsive genes. GBF3 is one of the several G-box binding factors which are basic leucine zipper (bZIP) proteins (Schindler et al. 1992; Izawa et al. 1993). In arabidopsis, GBF3 is highly expressed in roots but not in leaves (Schindler et al. 1992) and is believed to be involved in the regulation of alcohol dehydrogenase (adh) through an ABA-dependent pathway. The adh gene is responsive to cold and dehydration. On the other hand, MeATTF has been reported to be up-regulated by water stress (Bray 2004; Guo et al. 2009), although its actual function in drought tolerance was not well demonstrated. Although these two genes were up-regulated in both tolerant and susceptible genotypes, their expression in the DT genotype was significantly higher than in the DS genotype (Table 5). Further studies using different genotypes and a large sample size may give insight into whether the two genes are associated with drought tolerance in cassava; however, this study establishes them as expression-based markers of drought stress in cassava that may be useful across a range of genotypes.
Genes down-regulated by water stress
MeGE3, encoding GERMIN 3 (GER3), was down-regulated by water stress treatment in both genotypes. This is in agreement with Bray (2004), who reported that GER3 (germin-like gene group 3) was repressed by drought stress in leaves of A. thaliana. Germin proteins were first identified in wheat as genes that were expressed during germination (Lane et al. 1991). The report by Bray (2004) suggested that a possible role for germin-like genes is alteration of cell wall properties that control growth. We speculate that GER3 down-regulation may contribute to, and is consistent with, the reduction in plant growth observed in both genotypes under drought stress (Table 1).
Conclusions and forward look
Further characterization of drought tolerance in cassava, especially at the molecular level, is necessary to improve understanding and enhancement of drought tolerance in this plant. The genetic and physiological basis of drought tolerance in cassava was investigated in two cassava genotypes with contrasting tolerance levels to drought stress. The improved, DT variety MH96/0686 exhibited physiological indicators of drought avoidance, through partial or complete closure of stomata to reduce loss of moisture through transpiration. At the molecular level, 10 cassava drought-responsive genes were identified, and further comparisons between the two genotypes helped in the identification of those that are most likely to play a role in drought tolerance in cassava. The genes exclusively up-regulated in DT MH96/0686 represent the most promising candidates for drought-tolerance genes of cassava. Based on these genes' known functions in other plant species, it is likely that MH96/0686 tolerance at the cellular level consists of a reduction of oxidative stress through ROS quenchers (MeMSD and MeALDH) and osmotic adjustment (MeZFP and MeRD28). Further research with more genotypes and at more time points, including analysis after re-watering, is warranted to determine whether these and other cassava genes represent drought tolerance QTLs for use in breeding, and for testing their efficacy in conferring drought tolerance on transgenic cassava.
Sources of funding
This study was supported by funds from the Millennium Science Initiative through the National Council of Science and Technology, Uganda. Supplementary funds were obtained from the National Agricultural Research Organization (NARO), Uganda.
Contributions by the authors
L.T. conceived and conducted the research, collected the data, conducted experimental design, analysed the data and wrote the manuscript; E.B.K., M.F., Y.B. and D.S.O.O. conceived the research, analysed the data and reviewed the manuscript; M.A. conducted experimental design, analysed the data and reviewed the manuscript; and J.J.W.H. conducted experimental design, analysed the data and co-wrote the manuscript.
Conflict of interest statement
None declared.
Acknowledgements
We extend our sincere thanks to Bramwel Wanjala, Moses Ogugo, Moses Njahira, Martina Kyalo, Cyrus Too and members of the Segolip sequencing unit for their assistance during the implementation of laboratory and greenhouse work. We gratefully acknowledge the support and use of the BecA–ILRI Hub (http://hub.africabiosciences.org).
Literature cited
- Aina OO, Dixon AG, Akinrinde EA. Effect of soil moisture stress on growth and yield of cassava in Nigeria. Pakistan Journal of Biological Sciences. 2007;10:3085–3090. doi: 10.3923/pjbs.2007.3085.3090. [DOI] [PubMed] [Google Scholar]
- Akano AO, Dixon AGO, Mba C, Barrera E, Fregene M. Genetic mapping of a dominant gene conferring resistance to cassava mosaic disease. Theoretical and Applied Genetics. 2002;105:521–525. doi: 10.1007/s00122-002-0891-7. [DOI] [PubMed] [Google Scholar]
- Alfredo ACA, Setter TL. Response of cassava leaf area expansion to water deficit: cell proliferation, cell expansion and delayed development. Annals of Botany. 2004;94:605–613. doi: 10.1093/aob/mch179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An D, Yang J, Zhang P. Transcriptome profiling of low temperature-treated cassava apical shoots showed dynamic responses of tropical plant to cold stress. BMC Genomics. 2012;13:64. doi: 10.1186/1471-2164-13-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakayoko S, Tschannen A, Nindjin C, Dao D, Girardin O, Assa A. Impact of water stress on fresh tuber yield and dry matter content of cassava (Manihot esculenta Crantz) in Côte d'Ivoire. African Journal of Agricultural Research. 2009;4:021–027. [Google Scholar]
- Basu U, Good AG, Taylor GJ. Transgenic Brassica napus plants over expressing aluminum-induced mitochondrial manganese superoxide dismutase cDNA are resistant to aluminium. Plant, Cell and Environment. 2001;24:1269–1278. [Google Scholar]
- Bergantin VR, Yamauchi A, Pardales JR, Jr, Bolatete DM., Jr Screening cassava genotypes for resistance to water deficit during crop establishment. Philippine Journal of Crop Science. 2004;29:29–39. [Google Scholar]
- Bettina MJE, Melvin TT, Thomas AK. Visual assessment of wilting as a measure of leaf water potential and seedling drought survival. Journal of Tropical Ecology. 2007;23:497–500. [Google Scholar]
- Boyer JS. Plant productivity and environment. Science. 1982;218:443–448. doi: 10.1126/science.218.4571.443. [DOI] [PubMed] [Google Scholar]
- Bray EA. Genes commonly regulated by water-deficit stress in Arabidopsis thaliana. Journal of Experimental Botany. 2004;55:2331–2341. doi: 10.1093/jxb/erh270. [DOI] [PubMed] [Google Scholar]
- Ceballos H, Iglesias CA, Perez JC, Dixon AGO. Cassava breeding: opportunities and challenges. Plant Molecular Biology. 2004;56:503–516. doi: 10.1007/s11103-004-5010-5. [DOI] [PubMed] [Google Scholar]
- Daniels JM, Mirkov TE, Chrispeels MJ. The plasma membrane of Arabidopsis thaliana contains a mercury-insensitive aquaporin that is a homolog of the tonoplast water channel protein TIP. Plant Physiology. 1994;106:1325–1333. doi: 10.1104/pp.106.4.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenkolbe T, Thi DP, Zuther E, Repsilber D, Walther D, Hincha DK, Kohl KI. Expression profiling of rice cultivars differing in their tolerance to long-term drought stress. Plant Molecular Biology. 2009;69:133–153. doi: 10.1007/s11103-008-9412-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sharkawy MA. Cassava biology and physiology. Plant Molecular Biology. 2004;56:481–501. doi: 10.1007/s11103-005-2270-7. [DOI] [PubMed] [Google Scholar]
- El-Sharkawy MA. Physiological characteristics of cassava tolerance to prolonged drought in the tropics: implications for breeding cultivars adapted to seasonally dry and semiarid environments. Brazilian Journal of Plant Physiology. 2007;19:257–286. [Google Scholar]
- Farooq M, Wahid A, Kobayashi N, Fujita D, Basra SMA. Plant drought stress: effects, mechanisms and management. Agronomy for Sustainable Development. 2009;29:185–212. [Google Scholar]
- Fregene M, Okogbenin E, Mba C, Angel F, Suarez MC, Guiterez J, Chavarriaga P, Roca W, Bonierbale M, Tohme J. Genome mapping in cassava improvement: challenges, improvements and opportunities. Euphytica. 2001;120:159–165. [Google Scholar]
- Fryer MJ, Oxborough K, Mullineaux PM, Baker NR. Imaging of photo-oxidative stress responses in leaves. Journal of Experimental Botany. 2002;53:1249–1254. [PubMed] [Google Scholar]
- Gong P, Zhang J, Li H, Yang C, Zhang C, Zhang X, Khurram Z, Zhang Y, Wang T, Fei Z, Ye Z. Transcriptional profiles of drought-responsive genes in modulating transcriptional signal transduction, and biochemical pathways in tomato. Journal of Experimental Botany. 2010;61:3563–3575. doi: 10.1093/jxb/erq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P, Baum M, Grando S, Ceccarelli S, Bai G, Li R, Korff MV, Varshney RK, Graner A, Valkoun J. Differentially expressed genes between drought-tolerant and drought-sensitive barley genotypes in response to drought stress during the reproductive stage. Journal of Experimental Botany. 2009;60:3531–3544. doi: 10.1093/jxb/erp194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn SK, John C, Isoba G, Ikoun T. Resistance breeding in root and tuber crops at the International Institute for Tropical Agriculture (IITA), Ibadan Nigeria. Crop Protection. 1989;8:147–168. [Google Scholar]
- Hernandez JA, Olmos E, Corpas EF, Sevilla F, Del Rio LA. Salt-induced oxidative stress in chloroplasts of pea plants. Plant Science. 1995;105:151–167. [Google Scholar]
- Heschel MS, Riginos C. Mechanisms of selection for drought stress tolerance and avoidance in Impatiens capensis (Balsaminacae) American Journal of Botany. 2005;92:37–44. doi: 10.3732/ajb.92.1.37. [DOI] [PubMed] [Google Scholar]
- Iglesias C, Bonierbale M, El-Sharkawy M, Lozano C, Bellotti A, Wheatley C. Focusing basic research for cassava varietal improvement. In: Howeler RH, editor. Cassava breeding, agronomy research and technology transfer in Asia—Fourth Regional Workshop, Trivandrum, Kerala, India, 2–6 November 1993. Cali, Colombia: CIAT; 1995. pp. 40–60. [Google Scholar]
- Izawa T, Foster R, Chua NH. Plant bZlP protein DNA binding specificity. Journal of Molecular Biology. 1993;230:1131–1144. doi: 10.1006/jmbi.1993.1230. [DOI] [PubMed] [Google Scholar]
- Kendall EJ, McKersie BD. Free radical and freezing injury to cell membranes of winter wheat. Plant Physiology. 1989;76:86–94. [Google Scholar]
- Kotchoni OS, Kuhns C, Ditzer A, Kirch HH, Bartels D. Over-expression of different aldehyde dehydrogenase genes in Arabidopsis thaliana confers tolerance to abiotic stress and protects plants against lipid peroxidation and oxidative stress. Plant, Cell and Environment. 2006;2:1033–1048. doi: 10.1111/j.1365-3040.2005.01458.x. [DOI] [PubMed] [Google Scholar]
- Lane BG, Bernier F, Dratewka-Kos E, Shafai R, Kennedy TD, Pyne C, Munro JR, Vaughan T, Walters D, Altomare F. Homologies between members of the germin family in hexaploid wheat and similarities between these wheat germins and certain Physarum spherulins. Journal of Biological Chemistry. 1991;266:10461–10469. [PubMed] [Google Scholar]
- Lenis JI, Calle F, Jaramillo G, Perez JC, Ceballos H, Cock JH. Leaf retention and cassava productivity. Field Crops Research. 2006;95:126–134. [Google Scholar]
- Lokko Y, Anderson JV, Rudd S, Raji A, Horvath D, Mikel MA, Kim R, Liu L, Hernandez A, Dixon AGO, Ingelbrecht IL. Characterization of an 18,166 EST dataset for cassava (Manihot esculenta Crantz) enriched for drought-responsive genes. Plant Cell Reports. 2007;26:1605–1618. doi: 10.1007/s00299-007-0378-8. [DOI] [PubMed] [Google Scholar]
- McKersie BD, Bowley SR, Harjanto E, Leprince O. Water deficit tolerance and field performance of transgenic alfalfa overexpressing superoxide dismutase. Plant Physiology. 1996;111:1177–1181. doi: 10.1104/pp.111.4.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra J. Genetics and genetic improvement of drought resistance of crop plants. Current Science. 2001;80:758–763. [Google Scholar]
- Morante N, Sanchez T, Ceballos H, Calle F, Perez JC, Egesi C, Cuambe CE, Escobar AF, Ortiz D, Chavez AL, Fregene M. Tolerance to post-harvest physiological deterioration in cassava roots. Crop Science. 2010;50:1333–1338. [Google Scholar]
- Morsy MR, Almutairi AM, Gibbons J, Yun SJ, de Los Reyes BG. The OsLti6 genes encoding low-molecular weight membrane proteins are differentially expressed in rice cultivars with contrasting sensitivity to low temperature. Gene. 2005;344:171–180. doi: 10.1016/j.gene.2004.09.033. [DOI] [PubMed] [Google Scholar]
- Okogbenin E, Ekanayake IJ, Porto MCM. Genotypic variability in adaptation responses of selected clones of cassava to drought stress in the Sudan savanna zone of Nigeria. Journal of Agronomy and Crop Science. 2003;189:376–389. [Google Scholar]
- Okogbenin E, Setter TL, Ferguson M, Mutegi R, Alves AC, Ceballos H, Fregene M. Phenotyping cassava for adaptation to drought. In: Monneveux P, Ribaut J-M, editors. Drought phenotyping in crops: from theory to practice. 2011. pp. 381–400. CGIAR Generation Challenge Programme, Texcoco, Mexico. [Google Scholar]
- Pardales JR, Jr, Yamauchi A, Belmonte DV, Jr, Esquibel CB. Dynamics of root development in root crops in relation to the prevailing moisture stress in the soil. Proceedings of the 6th symposium of the International Society of Root Research; Nagoya, Japan: 2001. pp. 72–73. November. [Google Scholar]
- Perez JC, Lenis JI, Calle F, Morante N, Sanchez T, Debouck D, Ceballos H. Genetic variability of root peel thickness and its influence in extractable starch from cassava (Manihot esculenta Crantz) roots. Plant Breeding. 2011 doi:10.1111/j.1439-0523.2011.01873.x. [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative Expression Software Tool (REST) for group-wise comparison and statistical analysis of relative expression results in Real-Time PCR. Nucleic Acids Research. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AH, Atherton N, Hendry GAF. Plants under drought stress generated activated oxygen. Free Radical Research Communications. 1989;8:61–66. doi: 10.3109/10715768909087973. [DOI] [PubMed] [Google Scholar]
- Ramachandra-Reddy A, Chaitanya KV, Vivekanandan M. Drought-induced responses of photosynthesis and antioxidant metabolism in higher plants. Journal of Plant Physiology. 2004;161:1189–1202. doi: 10.1016/j.jplph.2004.01.013. [DOI] [PubMed] [Google Scholar]
- Rivero MR, Kojima M, Gepstein A, Sakakibara H, Mittler R, Gepstein S, Blumwald E. Delayed leaf senescence induces extreme drought tolerance in a flowering plant. Proceedings of the National Academy of Sciences of the USA. 2007;104:19631–19636. doi: 10.1073/pnas.0709453104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini HS, Westgate ME. Reproductive development in grain crops during drought. Advances in Agronomy. 2000;68:59–96. [Google Scholar]
- Sakurai T, Plata G, Rodrıguez-Zapata F, Seki M, Salcedo A, Toyoda A, Ishiwata A, Tohme J, Sakaki Y, Shinozaki K, Ishitani M. Sequencing analysis of 20,000 full-length cDNA clones from cassava reveals lineage specific expansions in gene families related to stress response. BMC Plant Biology. 2007;7:66–83. doi: 10.1186/1471-2229-7-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez FJ, Manzanares M, De Andres EF, Tenorio JL, Ayerbe L. Turgor maintenance, osmotic adjustment and soluble sugar and proline accumulation in 49 pea cultivars in response to water stress. Field Crop Research. 1998;59:225–235. [Google Scholar]
- Schindler U, Menkens AE, Beckmann H, Ecker JR, Cashmore AR. Heterodimerization between light-regulated and ubiquitously expressed Arabidopsis thaliana GEF bZlP proteins. EMBO Journal. 1992;4:1261–1273. doi: 10.1002/j.1460-2075.1992.tb05170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, Satou M, Akiyama K, Taji T, Yamaguchi-Shinozaki K, Carninci P, Kawai J, Hayashizaki Y, Shinozaki K. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. The Plant Journal. 2002;31:279–292. doi: 10.1046/j.1365-313x.2002.01359.x. [DOI] [PubMed] [Google Scholar]
- Sen Gupta A, Heinen JL, Holaday AS, Burke JJ, Allen RD. Increased resistance to oxidative stress in transgenic plants that over-express chloroplastic Cu/Zn superoxide dismutase. Proceedings of the National Academy of Sciences of the USA. 1993;90:1629–1633. doi: 10.1073/pnas.90.4.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setter L, Fregene M. Recent advances in molecular breeding of cassava for improved drought stress tolerance. In: Jenks M, Hasegawa P, Jjain M, editors. Advances in molecular breeding towards drought and salt tolerant crops. Berlin, Germany: Springer; 2007. pp. 701–711. [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. Journal of Experimental Botany. 2007;58:221–227. doi: 10.1093/jxb/erl164. [DOI] [PubMed] [Google Scholar]
- Talame V, Ozturk NZ, Bohnert HJ, Tuberosa R. Barley transcript profiles under dehydration shock and drought stress treatments: a comparative analysis. Journal of Experimental Botany. 2007;58:229–240. doi: 10.1093/jxb/erl163. [DOI] [PubMed] [Google Scholar]
- Turyagyenda FL, Kizito EB, Baguma Y, Osiru D. Evaluation of Ugandan cassava germplasm for drought tolerance. International Journal of Agriculture and Crop Sciences. 2013 In press. [Google Scholar]
- Umezawa T, Fujita M, Fujita Y, Yamaguchi-Shinozaki K, Shinozaki K. Engineering drought tolerance in plants: discovering and tailoring genes to unlock the future. Current Opinion in Biotechnology. 2006;17:113–122. doi: 10.1016/j.copbio.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Utsumi Y, Tanaka M, Morosawa T, Kurotani A, Yoshida T, Mochida K, Matsui A, Umemura Y, Ishitani M, Shinozaki K, Sakurai T, Seki M. Transcriptome analysis using a high-density oligomicroarray under drought stress in various genotypes of cassava: an important tropical crop. DNA Research. 2012;19:335–451. doi: 10.1093/dnares/dss016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valliyodan B, Nguyen HT. Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Current Opinion in Biotechnology. 2006;9:189–195. doi: 10.1016/j.pbi.2006.01.019. [DOI] [PubMed] [Google Scholar]
- Van Breusegem F, Slooten L, Stassart J, Moens T, Botterman J, Van Motagu M, Inze D. Overproduction of Arabidopsis thaliana FeSOD confers oxidative stress tolerance to transgenic maize. Plant and Cell Physiology. 1999;40:515–523. doi: 10.1093/oxfordjournals.pcp.a029572. [DOI] [PubMed] [Google Scholar]
- Wajid AJ, Muhammad JB, Kumbhar MB, Khan NU, Kerio MI. Effect of water stress on physiological and yield parameters at anthesis stage in elite spring wheat cultivars. Sarhad Journal of Agriculture. 2011;27:59–65. [Google Scholar]
- Wang FZ, Wang QB, Kwon SY, Kwak SS, Su WA. Enhanced drought tolerance of transgenic rice plants expressing a pea manganese superoxide dismutase. Journal of Plant Physiology. 2005;162:465–472. doi: 10.1016/j.jplph.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Xu DQ, Huang J, Guo SQ, Yang X, Bao YM, Tang HJ, Zhang HS. Over-expression of a TFIIIA-type zinc finger protein gene ZFP252 enhances drought and salt tolerance in rice (Oryza sativa L.) FEBS Letters. 2008;582:1037–1043. doi: 10.1016/j.febslet.2008.02.052. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Koizumi M, Urao S, Shinozaki K. Molecular cloning and characterization of 9 cDNAs for genes that are responsive to desiccation in Arabidopsis thaliana: sequence analysis of one cDNA clone that encodes a putative transmembrane channel protein. Plant and Cell Physiology. 1992;33:217–224. [Google Scholar]
- Yang J, An D, Zhang P. Expression profiling of cassava storage roots reveals an active process of glycolysis/gluconeogenesis. Journal of Integrative Plant Biology. 2011;53:193–211. doi: 10.1111/j.1744-7909.2010.01018.x. [DOI] [PubMed] [Google Scholar]
- You-Zhi L, Ying-Hua P, Chang-Bin S, Hai-Tao D, Xing-Lu L, Zhi-Qiang W, Ji-Liang T, Baoshan C. An ordered EST catalogue and gene expression profiles of cassava (Manihot esculenta) at key growth stages. Plant Molecular Biology. 2010;74:573–590. doi: 10.1007/s11103-010-9698-0. [DOI] [PubMed] [Google Scholar]
- Zhu BC, Su J, Chan MC, Verma DPS, Fan YL, Wu R. Over-expression of a delta pyrroline-5-carboxylate synthetase gene and analysis of tolerance to water-stress and salt-stress in transgenic rice. Plant Science. 1998;139:41–48. [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann N, Henning L, Gruissem W. Genevestigator. Arabidopsis microarray database and analysis toolbox. Plant Physiology. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]



