SUMMARY
Metabolic homeostasis is achieved by complex molecular and cellular networks that differ significantly among individuals and are difficult to model with genetically engineered lines of mice optimized to study single gene function. Here, we systematically acquired metabolic phenotypes by using the EUMODIC EMPReSS protocols across a large panel of isogenic but diverse strains of mice (BXD type) to study the genetic control of metabolism. We generated and analyzed 140 classical phenotypes and deposited these in an open-access web service for systems genetics (www.genenetwork.org). Heritability, influence of sex, and genetic modifiers of traits were examined singly and jointly by using quantitative-trait locus (QTL) and expression QTL-mapping methods. Traits and networks were linked to loci encompassing both known variants and novel candidate genes, including alkaline phosphatase (ALPL), here linked to hypophosphatasia. The assembled and curated phenotypes provide key resources and exemplars that can be used to dissect complex metabolic traits and disorders.
INTRODUCTION
Energy homeostasis is the result of a tight balance between energy intake and expenditure. Metabolic disorders, such as obesity and type 2 diabetes, often result when this equilibrium is disturbed by complex interactions between genetic and environmental factors (Auwerx, 2006). Predisposition to complex diseases such as the metabolic syndrome is inherited in a non-Mendelian fashion, emphasizing genetic heterogeneity and complex gene-by-environment interactions (GXE) in pathogenesis. Genetically engineered mouse models are not ideal for dissecting polygenic networks or GXE interactions precisely because they have been optimized to study actions of single genes on single genetic backgrounds (Auwerx et al., 2004). In contrast, studies in humans have identified risk factors for developing metabolic diseases with both environmental (e.g., lack of exercise) and genetic causes (e.g., mutations in the FTO locus [Dina et al., 2007]), but these studies typically fall short of defining GXE due to an inability to control environmental influences, cohort and admixture effects, difficulty in obtaining certain types of physiological and molecular data, and the inability to sample many individuals with identical genomes under different conditions. Effective population-based experimental methods to dissect intricate GXEs are needed to model complex genetically admixed human populations.
Over the past decades, study designs have been optimized to analyze genetic factors in large populations of naturally divergent strains, chiefly in Mus musculus, Saccharomyces cerevisiae (Brem et al., 2002; Ehrenreich et al., 2010), Drosophila (King et al., 2012), and more recently, Caenorhabditis elegans (Andersen et al., 2012). Murine genetic reference populations (GRPs) are among the best-established mammalian models with which to study GXE. These GRPs are typically sets of inbred strains that have been assembled to incorporate carefully titered levels of genetic complexity that model aspects of human populations. The recombinant inbred (RI) strain families are a type of GRP that enable tight experimental control, where each genotype is represented by an entire isogenic line, thereby enabling extensive replication studies (Singer et al., 2004; Williams et al., 2001). The BXD family, currently the largest and best characterized mouse GRP, is composed of ~160 lines that descend from crosses between C57BL/6J and DBA/2J, hereafter referred to as B and D, respectively (Peirce et al., 2004). GRPs such as the BXDs have been bred for quantitative trait loci (QTL) analyses, a suite of statistical genetic techniques that define regions of the genome (intervals or loci) and their modulating effects on phenotype. Another major advantage of GRPs is that high-density genotype data are publicly available. These genotypes can be combined with full-sequence data of the parental strains to simplify QTL mapping and identify causal sequence variants (Mozhui et al., 2008; Wang et al., 2010). Furthermore, owing to the relatively fixed genotypes of GRPs, massive databases of phenotypes and expression data can be assembled and shared across time, allowing for rapid multiscalar analyses. Over the last two decades, the BXD family has been exploited mainly to study the genetics of immune function and infectious disease (Bystrykh et al., 2005; Miyairi et al., 2007) and in behavioral and neuropharmacological research (Chesler et al., 2005; Gaglani et al., 2009; Laughlin et al., 2011; Philip et al., 2010). However, few metabolic phenotypes have been previously generated.
In the present metabolic survey, we systematically generated quantitative data for 140 standardized phenotypes, including glucose response, body weight change, physical activity, and oxygen consumption across a large subset of the BXD family by using adult males and females. All data are publicly available as a resource to the scientific community and complement massive expression data for key cells, tissues, and organs that we and others have deposited in the GeneNetwork database (www.genenetwork.org; see BXD phenotypes found under search term “LISP1”). These essential baseline clinical phenotypes are widely variable, often highly heritable, and in many cases can be linked to genetic loci encompassing known and novel candidate genes. Heritability and sex effects were highly variable among traits. Variation in one key trait, alkaline phosphatase, was linked to hypophosphatasia in lines containing the C57BL/6 allelic variant. Downstream of this hypophosphatasia, differences in bone structure were noted, as well as a significant increase in pyridoxal-phosphate (PLP) levels and a corresponding decrease in pyrixodal levels. This study validates the use of GRPs as a powerful resource to study metabolic homeostasis and identifies new candidate genes for disease.
RESULTS
Optimization Study Design for Genetics of Metabolism
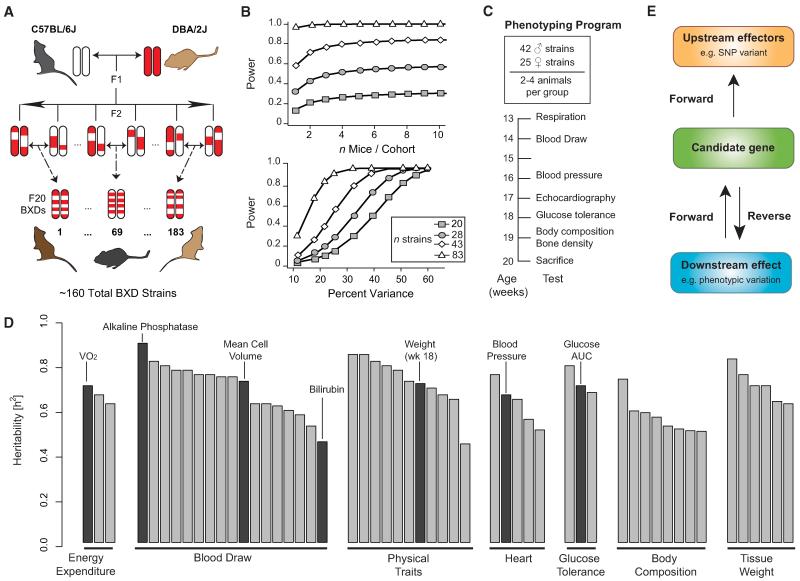
Mendelian traits such as coat color can be accurately measured with a sample size of one per genotype. In contrast, complex metabolic traits with lower heritability benefit greatly from larger sample sizes per genotype. One of the advantages of GRPs over noninbred yet genetically diverse strain populations (such as an F2 intercross) is that the heritability of a trait can be accurately measured and effectively boosted by repeatedly resampling cohorts of isogenic individuals of the same genotype and sex (Figure 1A). The observed variability within isogenic strains is thus primarily environmental, whereas differences among strains are largely genetic. A first step in experimental designs using GRPs is to estimate heritability and the sample size required to accurately measure a phenotype in a specific environment. Such calculations provide both a justifiable number of replicates per genotype and the total numbers of genotypes needed to reliably discriminate gene loci by using QTL-mapping methods. When traits have high heritability, increasing the number of genotypes (i.e., strains) greatly improves the power and precision with which QTLs can be mapped, whereas increasing the number of cases per genotype has almost no impact (Figure 1B, top). In the case of a Mendelian trait such as coat color, mutations in the brown (Tyrp1) and dilute (Myo5a) genes explain nearly all variation in pigmentation in the BXD family and map with peak LOD scores above 25 at these genes (Geisert et al., 2009). In contrast, complex diseases, including virtually all metabolic disorders, are polygenic, more dependent on environmental influences, and typically have a nearly normal distribution across genotypes. Power analysis shows that a study using 20 BXD strains with four animals per genotype will often detect loci that explain 50% or more of the genetic variance of a moderately heritable trait, whereas a study using ~40 strains will typically detect loci responsible for ~30% of the variance (Figure 1B, bottom).
Figure 1. Study Design for Identifying Genetic Loci that Control Metabolic Traits.
(A) The BXD lines were created by crossing B and D parents. The resulting heterozygous F1 mice were again crossed to generate genetically diverse but nonreproducible F2 animals. These F2 progeny were iteratively inbred until generation F20+, at which point the genome was 99.5+% isogenic and the strains are considered fully inbred and together constitute a GRP. The ~160 BXD strains are numbered 1–183.
(B) Top: power depends mainly on strain number instead on number of mice/strain. Calculations are for a trait variance fixed at 33%. Bottom: the power to detect a given fraction of variance with a single QTL. Power was calculated with cohort size fixed at four. Both calculations were done for a trait with heritability of 0.67.
(C) All animals underwent the same phenotyping programs as specified in the result section. The age corresponds to the timing of the phenotype experiment. BXD60 was only phenotyped in females—all other female strains overlapped with male strains.
(D) Heritability of select traits from each phenotyping group. Traits discussed in-depth in this resource paper are indicated in black. Traits are grouped according to phenotyping test. “Body Composition” represents echocardiography phenotypes, bone size, bone density, and body weight.
(E) The analysis flow-chart. The cause-to-consequence effect reads from top to bottom. A SNP induces a change in the expression or function of a candidate gene, which has an impact on a given downstream effect or phenotype.
Related to Figure S1.
We phenotyped a set of 43 strains—24 strains with male and female animals, 18 strains only with males, and 1 strain only with females (Figure 1C). Phenotyping was initiated at 13 weeks of age, and all cases underwent seven testing categories (Figures 1C and 1D). Mapping power was estimated as a function of heritability and sample size. We defined heritability as the ratio of the within-strain variance to total sample variance (Figure 1D). Most parameters have moderate to high heritability (h2), averaging 0.67. The more heritable a trait, the higher the likelihood it will map to significant QTLs (Lynch and Walsh, 1998). Under the tightly controlled environmental conditions in our vivarium, both glucose tolerance and blood pressure had h2 ~0.7. Consistent with these moderately high values, each of these traits was associated with at least one significant locus. In the case of blood pressure, we have defined a strong candidate gene—UBP1—by using a human genome-wide association study (GWAS) as a cross validation cohort (Koutnikova et al., 2009).
The BXD GRP allows complementary forward and reverse genetic methods to analyze metabolic traits (Figure 1E). Conventional forward-mapping methods start with heritable differences in phenotype and define loci and sequence variants (Figures 1E and S1A available online). Reverse genetics starts from defined candidate genes and identifies downstream phenotypic effects (Figures 1E and S1B) (Thomas et al., 2009; Yamamoto et al., 2011). Because full-sequence data are available for both parental strains of the BXD family, reverse methods can now be applied genome-wide by using a standard mapping population. In this resource, we demonstrate both approaches to the data.
The BXD Family Has Wide Variation in Metabolic Traits
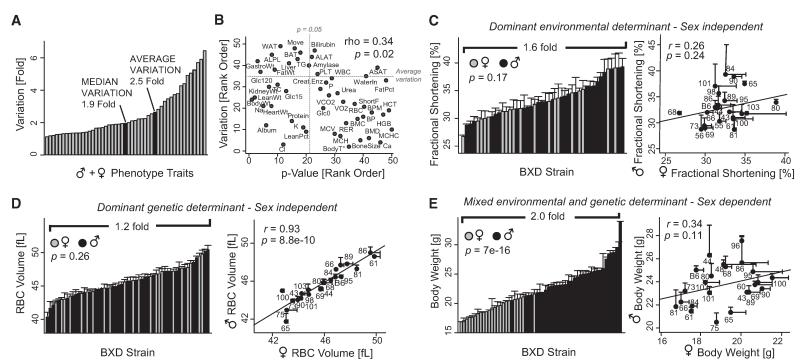
Metabolic traits vary to a remarkable degree among the BXD strains, with every trait showing significant variation (Figure 2A). The least variable trait, basal body temperature, varied significantly (from 35.8 ± 0.3 °C in BXD34 to 38.9 ± 0.2 °C in BXD95) despite an overall variation of less than 1.1-fold. The most variable trait, alkaline phosphatase (ALPL) protein activity level, varied 6.5-fold between BXD31 (44 ± 11 U/l) and BXD90 (287 ± 18 U/l). For the majority of traits, the distribution of phenotypes is continuous and approximately normal in both sexes.
Figure 2. Strain and Sex Influences Differ Widely between Parameters.
(A) Metabolic phenotype variation for all cohorts.
(B) Traits with higher variation were more likely to have significant differences between males and females; 41% of traits are significantly different (traits on the x axis lower than rank 22).
(C) Example of a sex-independent trait that is mostly determined by environment: the ventricular shortening fraction.
(D) Example of a sex-independent trait that is mostly determined by genetics: the mean cell volume of red blood cells.
(E) Example of a sex-influenced trait that is explained by a mix of genetic and environmental factors: body weight at 18 weeks. Bar graphs and X-Y plots are expressed as mean + SEM.
As expected, sex differences are a significant source of variance (Champy et al., 2004), with the more variable traits more likely to exhibit significant sexual dimorphism (Figure 2B). Traits can be divided into four categories in terms of the significance of variation and correlations between sexes. Nearly 60% of traits had no significant sex differences. These traits fall into two categories: those without tight male-female correlation (e.g., shortening fraction in Figure 2C) and those with it (e.g., red blood cell volume in Figure 2D). The first category, with no significant sex differences and without strong correlation between males and females, indicates phenotypes where variation cannot be reliably attributed to genetic factors (i.e., variation is due to environmental or technical influence) or that there is a strong sex-by-strain interaction effect. In the case of the shortening fraction, we suspect the former factor as the cause of the low heritability (h2 = 0.5) of this trait (Figure 2C). In contrast, the traits with strong correlations between sexes indicate that the genetic variability is not affected greatly by the environment, and thus the slope of the correlation is close to 1 (Figure 2D).
The remaining 40% of traits exhibit significant differences between males and females, and again with or without strong male-female correlation. The most striking example is body weight, with a dramatic difference between males and females, but without consistency across strains: e.g., BXD75 males are the leanest, whereas the BXD75 females are in the middle range of body weights (Figure 2E). Lastly, a few traits had both significant sex-associated differences and strong genetic correlation—one of them being discussed in Figure 4.
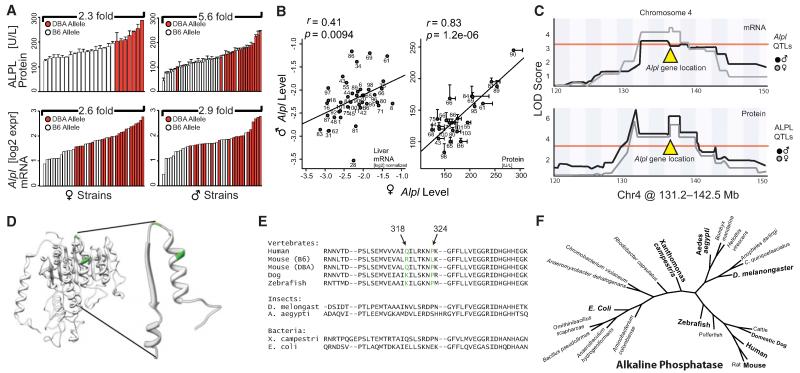
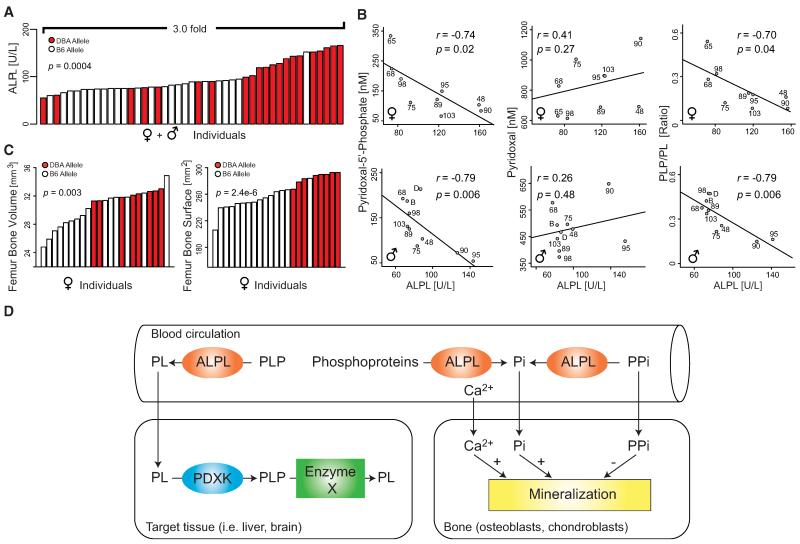
Figure 4. Reducing the ALPL QTL to Its Causative Mutation between DBA and C57 Alleles.
(A) Expression variation of serum ALPL activity and mRNA levels from liver transcriptome data. Strains with the D allele (red) have significantly more ALPL protein and Alpl mRNA than strains with the B allele (white) for all comparisons (p < 0.005). Males have significantly less ALPL protein than females, but there is no difference in mRNA levels.
(B) Correlation between male and female Alpl mRNA and ALPL protein levels demonstrate high trait heritability (Figure 1D).
(C) QTL map for liver Alpl mRNA and blood ALPL levels in males and females. All four QTLs (male and female traits are mapped separately) mapped to the same location and crossed the significance threshold of p < 0.05, indicated by the red horizontal line. The Alpl gene is located below the rectangular peak at right.
(D) 3D model of ALPL from Phyre2, based on the crystal structure of the placental form of ALP (ALPP). Enlarged on the right are the structure of the amino acids flanking and including the two missense mutations (noted in green).
(E) Forty-four amino acid excerpt of sequence comparison of orthologs of ALPL, the amino acid sequence of the region modeled in (D) is shown here. The full protein sequence is highly conserved in mammals (>90% homology) and moderately from mammals to bacteria (>30% homology).
(F) Phylogenetic tree of Alpl. The gene has orthologs in all known species, notably including invertebrates. Bar graphs and X-Y plots are expressed as mean+SEM. Related to Figure S2.
QTLs Link Phenotypes to Causal Loci
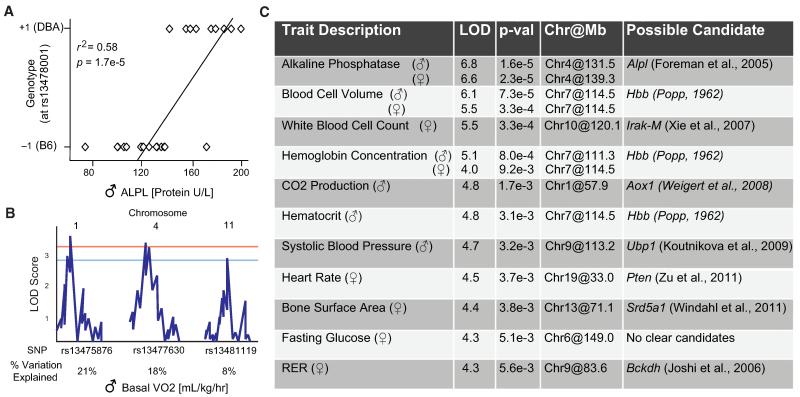
The data sets generated consist of both general blood parameters, such as hematocrit and iron levels, and physical phenotypes, such as heart rate and oxygen consumption. Suggestive or significant QTLs were detected for nearly 40% of all phenotypes (Table S1). Traits with the highest heritability and the most significant QTLs usually mapped to a single locus. These traits are exemplified by the tissue-nonspecific (liver/bone/kidney) ALPL (also commonly known as TNSALP) protein activity, a trait for which 58% of the variance is explained by the single peak QTL (Figure 3A). Many other traits, including most classical phenotypes, map to several suggestive or significant loci. In these cases, oligogenic or polygenic networks of unlinked loci influence trait variance. One good example is basal oxygen consumption (VO2), a trait for which three loci independently contribute to phenotypic variation—21% for the most significant locus on Chr1, 18% for the locus on Chr4, and 8% for the locus on Chr11 (Figure 3B).
Figure 3. Analysis of Peak Trait QTLs.
(A) The genetic variation attributable to a QTL is calculated by the correlation between the trait values (here, ALPL levels) and the genotype at the peak location. ALPL has only one QTL, which explains 58% of the genetic variation.
(B) The genetic variation attributable to each of the three QTLs mapped to oxygen consumption. Each QTL has a much smaller effect, but together account for a large amount of variation. Generalized linear modeling or ANOVA (used here) is necessary to calculate the variation independently attributable to each QTL for traits which map to multiple QTLs.
(C) The LOD score, corrected p value, and QTL peak location are given along with the trait for the top 10 distinct phenotypes. Of note, this is not all of the significant or suggestive peaks for each trait; only the most significant peak is listed. One positional candidate is given for traits when a literature analysis of every gene under each peak has yielded a linked result. Candidates not discussed in the text include the link between white blood cells and Irak-M (Xie et al., 2007), Pten and heart rate (Zu et al., 2011), and RER in females and Bckdh (Joshi et al., 2006). Systolic BP LOD score is for analysis as described in (Koutnikova et al., 2009). A list of all 54 traits with suggestive or significant peak QTLs is available in Table S1.
Genes located within QTLs were examined for known links to function by using PubMed and GeneRIF (Mitchell et al., 2003). All but one of the top ten QTLs contained at least one strong positional and functional candidate (Figure 3C). In some cases, the positional candidate is well established as the causative gene behind the QTL. This is best illustrated by hematocrit levels, which map to a locus containing the hemoglobin gene (Popp, 1962). In other cases, the candidates are linked more tentatively. For example, the QTL for bone surface area contains the steroid 5 alpha-reductase 1 (Srd5a1) gene, which has been linked to bone mass in a recent study on Srd5a1 knockout mice (Windahl et al., 2011).
Genetic Polymorphisms Underlying Variation in ALPL Levels Contribute to Changes in Bone and Vitamin B6 Homeostasis
Serum ALPL levels are strongly variable among BXD strains, with females having significantly higher expression (but less variation) than males (Figures 4A and S2A). Expression of Alpl mRNA in the liver is also highly variable but does not differ by sex (Figures 4A and S2B) (Gatti et al., 2007). Interestingly, both Alpl mRNA and ALPL protein activity are highly correlated by strain, regardless of the sex of the animal (Figure 4B), suggesting strong genetic modulation and a sex effect shared by all strains. Data for ALPL activity in males and females map to a common QTL: an 11 Mb region of Chr4 (Figure 4C) that has already been reported in an F2 cross between C57BL/6 and DBA/2 mice (Foreman et al., 2005). Alpl mRNA also map to the same region (Figure 4C). This QTL harbors ~100 genes, including the Alpl gene itself. We hypothesized that differences in ALPL activity are primarily and perhaps almost exclusively caused by one or more allelic differences in the cognate Alpl gene, i.e that this is a so-called cis-regulated expression QTL (cis eQTL). Rapid identification of all sequence variants in the region was performed by using a mouse SNP database (www.genenetwork.org). Of 11 exonic SNPs that differentiate B and D haplotypes of Alpl, nine cause synonymous mutations and two cause missense mutations in exon 9, resulting in amino acid changes of R318Q and L324P (Figure S2C). We modeled the impact of these mutations on ALPL protein structure by using Phyre2 (www.sbg.bio.ic.ac.uk/phyre2/) (Figure 4D). Although these mutations are not located in the active site of ALPL (around S110) (UniProt Consortium, 2012), they are predicted to change the β sheet/α-helix structure (R318Q and L324P are in green in Figure 4D, showing the described excerpt of ALPL ± 21 amino acids). Remarkably, mouse and human ALPL protein sequences share 93%/97% identity/homology. In particular, the region harboring the two SNPs is highly conserved in vertebrates—notably proline 324 is invariant in other vertebrates, except in C57BL/6 where it has mutated into leucine (Figure 4E, same excerpt as in 4D). Moreover, the high degree of conservation of the ALPL protein is also revealed by phylogenetic tree analysis—human ALPL has 77%/87% sequence identity/homology with zebrafish ALPL, 44%/61% with D. melanogaster, and 32%/45% with E. coli (Figure 4E). ALPL orthologs separate into three clusters: vertebrates, insects, and bacteria (Figure 4F). Given this high genetic conservation, it is likely ALPL has functions in vertebrates that are wholly unrelated to bone structure.
In humans, mutations in the ALPL gene can cause varying degrees of hypophosphatasia (Mornet, 2008), a disorder nearly always associated with the accumulation of phosphoethanolamine (PEA) in the urine, extracellular pyridoxal-5′-phosphate (PLP) in the blood, and extracellular inorganic pyrophosphate (PPi) in urine and blood. Downstream of these biomarkers are many pathways involved in bone remodeling (Whyte, 2000), amino acid metabolism, and neurotransmitter regulation (Lehninger et al., 2008). Therefore, we examined patterns of segregation of phenotypes among BXDs by testing several strains with either the B allele (C57BL/6, BXD48, BXD65, BXD68, BXD75, and BXD103) or the D allele of Alpl (DBA/2, BXD89, BXD90, and BXD95). We first reconfirmed that ALPL enzymatic activity was different among strains carrying the alleles (Figure 5A). As predicted, extracellular PLP levels were strongly negatively correlated with ALPL in both sexes, supporting the hypothesis that the BXDs are a good model for hypophosphatasia (Figure 5B). Furthermore, ALPL levels did not correlate with levels of calcium or phosphate (Pi) (Figure S3A), consistent with studies in humans differentiating hypophosphatasia from rickets and osteomalacia (Whyte, 2000). Femur bone structure evaluated via micro computed tomography (microCT) indicated decreased bone area and volume in females with the B allele of Alpl (Figure 5C), with no difference in overall animal weight between cohorts (p = 0.76). The same tendency was also noted for males (Figure S3B). Together these data suggest that the BXD strains are a useful model for hypophosphatasia.
Figure 5. Clinical Phenotyping of Mice with Extreme Alkaline Phosphatase Values.
(A) ALPL levels in all individuals in a further in-depth study in male and female mice for individuals from the 11 strains described in the text. The effect observed on serum ALPL levels shown in Figure 4A was highly reproducible in this independent study, even given a significant difference in the age of the animals in the two studies (19 weeks versus ~46 weeks).
(B) Extracellular PLP levels strongly negatively correlated with ALPL levels in males and females. No correlations were observed between ALPL and extracellular PL, but the ratio of extracellular PLP/PL was also strongly negatively correlated with ALPL.
(C) Bone volume and surface area were significantly decreased in animals with the B allele of Alpl, consistent with the hypothesis of hypophosphatasia in the strains with the B allele.
(D) Scheme summarizing the multiple roles of ALPL in metabolism. Before entering tissues, pyridoxal-5′-phosphate (PLP) in the plasma must be dephosphorylated by ALPL into pyridoxal (PL), which can traverse cell membranes. PL is converted back into PLP by the pyridoxine kinase (PDXK) in the target tissues, such as liver or brain. PLP is a cofactor in many enzymatic reactions. Circulating ALPL also converts pyrophosphate (PPi) into inorganic phosphate (Pi). Although PPi inhibits bone mineralization, Pi and calcium (Ca2+) stimulate it. Low levels of ALPL activity disrupt proper bone development and remodeling via this mechanism.
Related to Figure S3.
It is very plausible that the differential regulation of PLP (Figure 5B) by ALPL drives further phenotypic changes (Figure 5D). PLP acts as a cofactor in many important biochemical reactions: principally, it is a required prosthetic group for all aminotransferases, suggesting a critical role in amino acid metabolism (Lehninger et al., 2008), and it is also an essential coenzyme for biosynthesis of heme (Astner et al., 2005). Confirmation of this link between ALPL over PLP to amino acid and hematological parameters warrants future work.
Wide Variation in Glucose Response Identifies Genetic Factors of Diabetes
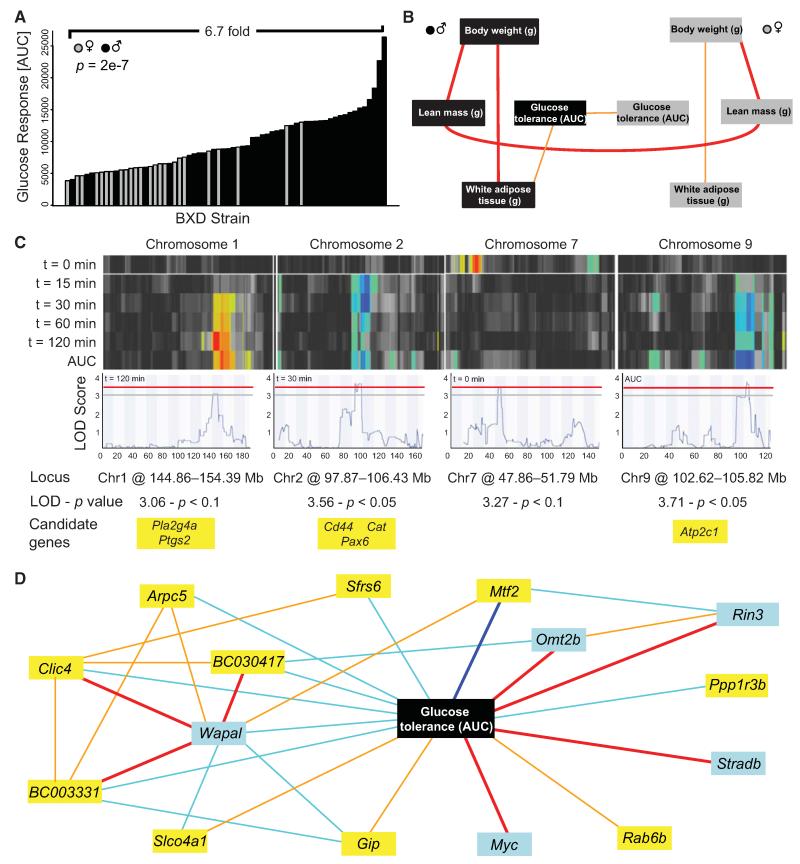
Analysis of complex metabolic phenotypes also yielded QTLs with promising candidates. One such phenotype is glucose response during intraperitoneal glucose tolerance test (IPGTT), which records glycaemia levels before and 15, 30, 60 and 120 min after glucose injection and the overall glucose excursion curve (AUC). The AUC is significantly higher in males (Figure 6A) but correlates between the sexes (Figure 6B and Table S2A). The AUC is also linked to physical parameters such as body weight and composition in males, but surprisingly, not in females (Figure 6B and Tables S2B and S2C), perhaps partly explained by the fact that fewer female strains were studied. These results show that variation in glucose response is significantly determined by genetics and partly sex independent. Consistent with these observations, mapping QTLs for glucose levels during IPGTT revealed both common and distinct loci in both sexes (Figure S4).
Figure 6. Analysis of Response to an Intraperitoneal Glucose Tolerance Test.
(A) Variation of the AUC of the glucose levels from 0 to 120 min.
(B) Network of glucose tolerance response determinants in males and females. Full details are given in Table S2.
(C) Multiple and single QTL heat map for glucose levels at each time point. Loci switch according to the feeding status of the mice. Fasting glucose (t = 0 min) maps appear only on Chr7. The early response to glucose injection (t = 15 min) maps on Chr2 only, whereas the rest of the time points also map on Chr1 and Chr9. Candidates under each QTL were selected according to existing literature showing their link with type 2 or type 1 diabetes. Further details are given in Figure S4.
(D) Network built around overall glucose response by using gene expression in the liver. Among the top 500 liver mRNA correlates with AUC, two were located on Chr1 QTL (Arpc5 and BC003331) and one on Chr9 QTL (Rab6b) (yellow boxes). One gene had a transQTL on the Chr1 QTL (Ppp1r3b), four had transQTLs on the Chr2 QTL (Sfrs6, Clic4, Slco4a1 and Mtf2), and one had a transQTL on the Chr9 QTL (Gip) (yellow boxes). Light blue boxes are attributed to the five genes that best correlated with AUC (Wapal, Rin3, Omt2b, Stradb and Myc). Bold dark blue lines represent −1 < r < −0.7, light blue lines −0.7 < r < −0.5, light orange lines 0.5 < r < 0.7 and bold red lines 0.7 < r < 1.
All genes under the four significant or suggestive QTLs for female glucose response on Chrs1, 2, 7 and 9 (Figure 6C) were investigated. The locus on Chr1 was associated with late glucose response, with glycaemia at 120 min giving the highest LOD score (Figure 6C). Within this locus, two genes are associated with type 2 diabetes susceptibility in humans: cytosolic phospholipase A2 group IV gene (Pla2g4a) and prostaglandinendoperoxide synthase 2 (Ptgs2, previously known as Cox2), both involved in prostaglandin metabolism (Konheim and Wolford, 2003; Wolford et al., 2003). A second QTL for early glucose response mapped on Chr2 (Figure 6C). Several genes known to be involved in insulin secretion or type 1 diabetes susceptibility are located under this peak, including paired box 6 (Pax6), Cd44, and catalase (Cat) (Chen et al., 2005; Jiang et al., 2011;Wen et al., 2009). When blood glucose is measured from mice under fasting conditions, only a single locus is mapped on Chr7 (Figure 6C). Although there are 91 genes under the locus, no gene was previously linked to fasting glucose levels in mice or in humans. Finally, the global AUC is strongly associated with the QTL located on Chr9 (Figure 6C). This region contains 31 genes, including one that has also been linked with insulin secretion: plasma membrane-related Ca2+-ATPase-1 (Atp2c1) (Mitchell et al., 2004).
Functional analysis of the three QTLs related to glucose response on Chr1, 2, and 9 uncovered six genes known to be involved in glucose regulation. However, these QTLs contain altogether 131 genes, many of which are not yet characterized. Therefore, we highlighted potential new candidates by finding the strongest mRNA correlates in liver (Figure 6D). From the list of the 500 best correlates, we selected candidates from three categories: (1) genes located under the QTLs on Chr1, 2, and 9; (2) genes that have trans-QTLs mapping to these loci; and (3) the five top gene correlates with glucose response. The most significant matches were integrated into the network (Figure 6D). Protein phosphatase 1 regulatory subunit 3B (Ppp1r3b), a protein that regulates hepatic glycogen synthesis (Kelsall et al., 2009), Myc, which plays a role in hepatic glucose uptake and utilization (Riu et al., 1996), STE20-related kinase adaptor beta (Stradb), and Rab6b, correlated exclusively with AUC (Figure 6D). In contrast, the rest of the candidates were entangled in an intricate network revolving around the AUC (Mtf2, Slco4a1, BC030417, Srfs6, Gip, Wapal, Clic4, Arpc5, Rin3, Omt2b, and BC003331) (Figure 6D). Among these genes, Sfrs6 and Gip have also been linked with glucose homeostasis (Fossey et al., 2001;Lyssenko et al., 2011).
Energy Expenditure Networks Link to Underlying mRNA Expression Differences
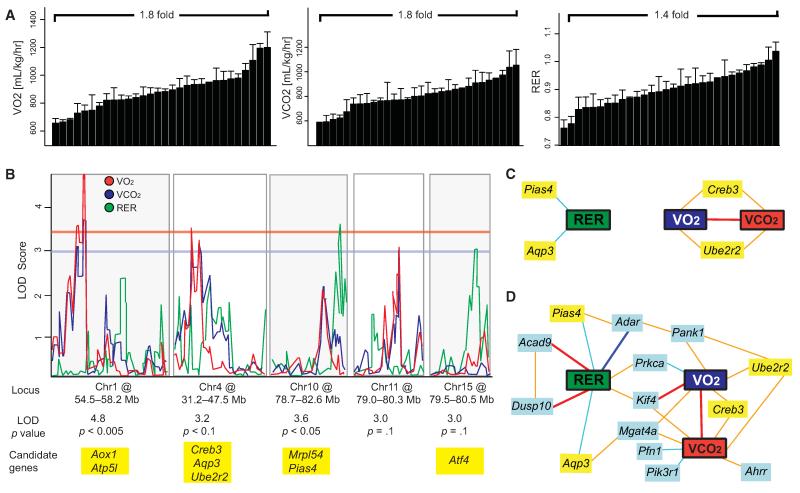
The volumes of oxygen consumed (VO2) and CO2 exhaled (VCO2) give an indirect measurement of mitochondrial function in vivo—another key complex metabolic phenotype (Auwerx, 2006; Tschöp et al., 2012). The respiratory exchange ratio (RER), defined as VCO2/VO2, is an indirect measurement of the primary energy substrate used by the organism, e.g., fats, proteins, or carbohydrates. Together, these parameters provide a window on global energy homeostasis. In contrast to ALPL and glucose response, the overall variation in these three parameters was quite modest (Figure 7A). Furthermore, these traits had only average heritability (Figure 1D). Males and females did not have significantly different energy expenditure, and the phenotypes were not significantly correlated by strain (Figures S5A and S5B). Despite these modest indicators of genetic influence, these traits mapped significantly and suggestively to five distinct QTLs (Figure 7B).
Figure 7. Regulatory Network Underlying Differences in Respiration.
(A) Variation among male strains of BXD mice in three parameters of respiration: VO2, VCO2, and RER. Variation across and correlation with female strains is given in Figure S3.
(B) QTL graphs of the three respiratory parameters, VO2 (blue), VCO2 (red), and RER (green). Significance is shown by the red horizontal line, suggestive by the blue line (LOD > 3.3 and 3.0, respectively). Candidate genes with established links to energy expenditure are listed below the locus (Aox1, Atp5l, Creb3, Ube2r2, Aqp3, Mrpl54, Pias4, Tab1, and Atf4).
(C) Network graph showing all positional candidates with mRNA expression that correlates significantly with the phenotypes in whole-eye tissue (Geisert et al., 2009).
(D) Expanded network graph using the same data set, including the top ten mRNA correlates of the phenotypes (light blue boxes). Interestingly, RER had the strongest top mRNA correlates (Acad9, Dusp10, and Adar), whereas Kif4 was the only significant correlate of all three parameters, and the only strong (|r| > 0.7) mRNA correlate to VO2 and none were observed for VCO2. Prkca and Mgat4a were shared significantly between two parameters. Despite the strong correlation between VO2 and VCO2, most top correlates were not shared. Bold dark blue lines represent −1 < r < −0.7, light blue lines −0.7 < r < −0.5, light orange lines 0.5 < r < 0.7 and bold red lines 0.7 < r < 1. Bar graphs are expressed as mean + SEM.
Related to Figure S5.
Similar to glucose response mapping, common QTLs were observed, with VO2 and VCO2 containing overlapping peaks on Chr1, 4, and 11. Somewhat surprisingly, RER maps to separate QTLs on Chr10 and Chr15 (Figure 7B). All 441 genes under the five peaks were analyzed for functional relationships to energy expenditure. Several genes closely related to mitochondrial function were identified under the QTLs (Figure 7B), for example ATP synthase H+ transporting mitochondrial F0 complex subunit G (Atp5l) and mitochondrial ribosomal protein 54 (Mrpl54) (Figure 7B) (Pagliarini et al., 2008). Several candidates related to fat metabolism were also identified, including aldehyde oxidase 1 (Aox1) and activating transcription factor 4 (Atf4, also known as Creb2) (Seo et al., 2009; Weigert et al., 2008).
All positional candidates were subsequently analyzed for covariation with VO2, VCO2, and RER in order to identify potential candidates that were not yet reported in literature. This was done by using an extensive BXD microarray database of whole-eye mRNA (Geisert et al., 2009), a tissue containing a variety of cell types, including neurons and both glycolytic and oxidative muscle. The network analysis of the 441 genes under the RER, VO2, and VCO2 QTLs narrowed the list to four genes tightly covarying with the phenotypes: aquaporin 3 (Aqp3), ubiquitin-conjugating enzyme E2R 2 (Ube2r2), cAMP responsive element binding protein 3 (Creb3), and protein inhibitor of activated STAT 4 (Pias4) (Figure 7C). Aqp3 and Pias4 are negative correlates of RER, whereas Ube2r2 and Creb3 are positive correlates of VO2 and VCO2 (Figure 7C). As of yet, none of these genes have been clearly linked to respiration or mitochondrial function. However, these candidates belong to gene families with clear ties, suggesting a functional, yet hitherto unknown relationship (further connections and references described in Figure S5C).
To expand the small network of positional candidates, the top 500 expression correlates with each trait were calculated and the top 10 correlates added into the network (Figure 7D). This network indicated that although VO2 and VCO2 are tightly correlated and share QTLs, several of the most significantly correlated genes, like profilin 1 (Pfn1), phosphatidylinositol 3-kinase, regulatory subunit 1 (Pik3r1), and aryl hydrocarbon receptor repressor (Ahrr), were linked to VCO2 exclusively (Figure 7D). RER also presented exclusive correlates, such as dual specificity phosphatase 10 (Dusp10) and acyl-CoA dehydrogenase family, member 9 (Acad9). Although on first thought it may be surprising that RER does not network closely with VO2 and VCO2, RER is a weight-independent measurement of metabolic flexibility. The common nodes that link VO2 or VCO2 with RER, such as protein kinase C alpha (Prkca), pantothenate kinase 1 (Pank1), and kinesin family member 4 (Kif4) have clear relevance to energy regulation (Leitges et al., 2002). Importantly, the only gene significantly linked to all energy expenditure phenotypes, Kif4, could affect metabolism due to its regulation of poly ADP-ribose polymerase 1 (Parp1) (Bai et al., 2011; Midorikawa et al., 2006). Parallel network analysis using the same analytical technique in liver showed many similar top candidates, notably including Pank1 and Kif4, which are also linked in the eye network to VO2 or VCO2 with RER (Figure S5D).
DISCUSSION
One of the main challenges in biomedical research is determining how genetic factors interact to influence human health-span and how to develop effective strategies for the diagnosis, prevention, and treatment of chronic multifactorial metabolic diseases. Studies in model organisms often focus on single genes, which allows for precise mechanistic dissection of individual pathways yet lacks the same level of real-world genetic complexity behind variation in metabolic traits. This can compromise the generality and the translational utility of findings. One of the first systems for studying complex systems was to use F2 intercrosses, which can be quickly generated. However, unless an F2 cross is large (n > 500) it will generally have modest mapping resolution (>20 Mb). Yet the more serious limitation is that F2 studies cannot be easily replicated or extended because each organism is genetically unique. To address this problem and to produce more robust models for systems genetics approaches, GRPs have been developed such as the BXD strains, currently the largest available mammalian GRP. The BXDs constitute a reproducible and high-resolution mapping panel with ~5 million segregating variants, similar to human populations (Altshuler et al., 2010). Perhaps the most important longterm advantage of RI families is that it is possible to accumulate huge electronic “health care” records for each of the genotypes under different conditions and across many scales of biological organization. Technical error, batch effects, laboratory environmental effects, and other issues can be detected, controlled, and even exploited simply by expanding the number of strains or animals used.
When using GRPs, statistical power and mapping precision are mainly a function of the number of genotypes that are included in a study. For the metabolic phenotypes measured here, nongenetic variation within a genotype is generally low compared to variation across genotypes, demonstrating high heritability in a standard vivarium environment. Essentially all measured traits have sufficient heritable variation to motivate detailed genetic and QTL analyses. Nearly half of the metabolic phenotypes differed significantly between sexes, thus metabolic traits do not generalize reliably from one sex to the other. However, rather than confining analysis to only one sex, balanced and interleaved sampling of both sexes may offer more benefit.
A total of 140 metabolic traits were analyzed, of which 39 had significant peak QTLs and an additional 15 had suggestive peak QTLs. The strongest QTLs are for expression phenotypes and Mendelian-like classical phenotypes. The most significant QTL, ALPL expression, was clearly linked to the Alpl gene and caused by two amino acid changes in evolutionary conserved residues—R318Q and L324P—which are predicted to change the 3D structure and activity of the ALPL protein. Presence of these mutations in carriers of the B allele closely mimics downstream phenotypes of the human disorder hypophosphatasia, primarily the strong increase in extracellular PLP levels, which along with ALPL levels provides the most precise biomarker of hypophosphatasia (Whyte, 2000). One prominent downstream effect of ALPL is its effect on promoting bone mineralization by hydrolyzing inorganic pyrophosphate (PPi), an inhibitor of hydroxyapatite formation, into inorganic phosphate. Consistent with this hypothesis, mice with the B allele had significantly reduced bone volume and surface area. These observation have important translational value, as Alpl coding variants are known to contribute to heritable differences in bone metabolism in humans (Nielson et al., 2011).
Most metabolic traits are, however, complex and map to multiple loci. For example, glucose response levels are associated with three loci and collectively contain six established positional candidates. Three of these candidates—Cd44, Cat, and Pax6—are located in the Chr2 locus and linked to insulin secretion or type I diabetes. In line with this linkage, a large F2 cross study of basal insulin levels also revealed a QTL on the same region (Mehrabian et al., 1998). Studies in humans have identified only a single insulin sensitivity gene (PPARγ [Deeb et al., 1998]), yet GWAS have identified several genes that control insulin secretion (McCarthy, 2010). In addition to those identified genes that are clear candidates for glucose response, it is possible, even likely, that other genes under these loci also influence glucose/insulin response.
Genetic mapping of the energy expenditure traits yields five distinct QTLs containing over 400 positional candidate genes, including established genes in oxidative metabolism in two general categories: those tied to general mitochondrial function (e.g., Mrpl54 and Atp5l) and those tied to the regulation of lipid metabolism (e.g., Aox1 and Atf4). Atp5l is a component of the complex V ATP synthesis machinery in oxidative phosphorylation, whereas Mrpl54 is part of the mitochondrial ribosome responsible for the translation of the 13 proteins encoded by the mitochondrial genome that are key components of the electron transport chain (Pagliarini et al., 2008). From the other angle, Aox1 is involved in lipid metabolism and produces reactive oxygen species as byproducts (Weigert et al., 2008) that are known to affect respiration (Balaban et al., 2005). Atf4 (aka Creb2) is an established regulator of lipid and energy homeostasis (Seo et al., 2009).
By combining QTL mapping with network building, it becomes practical to expand the phenotype and positional candidate networks to highlight intriguing candidates genes that are good targets for mechanistic studies of glycaemia and energy expenditure. For example, the serine/arginine-rich splicing factor 6 (Sfrs6) is among the top correlates of glucose response, and it is associated with type 2 diabetes susceptibility in humans (Fossey et al., 2001). Many other relevant genes appear as top correlates when networks are extended further. The glucose-dependent insulinotropic polypeptide (Gip) is only vicariously connected to the phenotypes, but it has a strong functional relation: it promotes pancreatic islet function and exerts pro-survival actions in humans (Lyssenko et al., 2011).
As with glucose response, network identification of the top mRNA correlates with the phenotype revealed a mélange of genes involved in different aspects of energy regulation. For example, the only significant correlate of all three parameters, Kif4, regulates PARP1, the major NAD+ consuming enzyme in the cell (Houtkooper et al., 2010). PARP1 critically controls oxidative metabolism due to its competition with SIRT1 for a common and limiting NAD+ pool in the cell (Bai et al., 2011). Acad9, linked with RER, is essential for complex I assembly via its impact on Ndufa1 and Ecsit (Haack et al., 2010; Nouws et al., 2010). However, other genes without known ties to lipid or glucose metabolism also appeared as top correlates, for example Ahrr and Pfn1.
In terms of building and testing models of complex processes such as metabolism, even traits with low heritability or without strong QTLs provide great value. For instance, although VCO2 is not linked to unique QTLs, its inclusion in network analysis not only strengthens the common interacting genes with VO2, but also uncovers unique genes, which may be novel to the phenotype. Likewise, the integration of gene expression from liver to the glucose tolerance network increases the power to identify genes that are specific to one organ function—in this case hepatic glucose metabolism. For both analyses, it is likely that more novel genes involved in glucose response and energy expenditure remain under the loci described. Such network construction underscores the usefulness of the BXD GRP as a powerful resource for hypothesis generation and validation in a field as complex as metabolism. Further complementary phenotypic measurements of the BXDs under challenged conditions may help refine these regulatory pathways.
Most analyses in this resource—including all of Figures 6 and 7—can be generated entirely with public access to www.genenetwork.org. The phenotypes from this study can be found under BXD phenotypes by searching for “LISP1” and either analyzed on-site or downloaded for separate analysis. A wide variety of public microarray data and phenotype data from other studies on the same BXD strains are also available for translational analyses on GeneNetwork. Together this makes the BXDs among the largest and best-categorized family of isogenic strains, and we expect resources like this to serve as the back-bone for murine studies on complex disease for the coming decade.
EXPERIMENTAL PROCEDURES
Animals
We acquired 25 BXD strains of females and 42 BXD strains of males from the University of Tennessee Health Science Center (Memphis, TN, USA). Strains and sexes were housed together with two to four animals per cage under 12 hr light, 12 hr dark cycle with ad libitum access to water and chow diet food (D04, SAFE, Augy, France) at all times. The studies shown in Figure 5 were carried out in a subset of these strains at a significantly older age (40–52 weeks) than the animals in the primary study protocol (8–20 weeks).
General Study Plan and Phenotyping tests
The general outline of the phenotyping study is described in the results section and summarized in Figure 1C. All tests were carried out according to rigorous standard operating procedures (SOP) established and validated within the EUMODIC EMPReSS program (Champy et al., 2004; Champy et al., 2008).
Bioinformatics and Statistical Analysis
R was used for the analysis of phenotypic data, and GeneNetwork (www.genenetwork.org) was used for all genetic analyses (Chesler et al., 2004). Student’s t test was used to calculate significance between males and females. Pearson’s r and p values were calculated to determine magnitude and significance of correlations except when otherwise indicated. Correlation networks and QTL calculations are Bonferroni-corrected for multiple testing. The y axes of bar graphs are often cropped to highlight strain differences.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Laurent Pouilly, Tania Sorg, Hongbo Zhang, and Ulrike Kettenberger for technical assistance with various aspects of mouse phenotyping. Discussions with Thomas Vogt (Merck Research Laboratories), Carmen Argmann and Sander Houten (University of Amsterdam), Vincent Mooser (CHUV), Stephan Morgenthaler (EPFL), Dominique Pioletti (EPFL), Bernard Thorens (UNIL), Paul Franken (UNIL), and Ioannis Xenarios (UNIL) and the team members of the Auwerx lab are acknowledged. R.H.H. is supported by a Rubicon fellowship of the NWO. J.A. is the Nestlé Chair in Energy Metabolism and the J.A./K.S. laboratory is supported by grants of the Ecole Polytechnique Fédérale de Lausanne, the EU Ideas program (ERC-2008-AdG-23118), the Velux Stiftung, the Swiss National Science Foundation (31003A-124713, 31003A-125487, and the Sinergia grant CSRII3-136201). R.W.W. and GeneNetwork are supported by NIH (P20-DA 21131 UO1AA13499 and U01AA14425) and the UT Center for Integrative and Translational Genomics.
Footnotes
Supplemental Information includes Extended Experimental Procedures, five figures, and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2012.08.012.
REFERENCES
- Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, Peltonen L, et al. International HapMap 3 Consortium Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen EC, Gerke JP, Shapiro JA, Crissman JR, Ghosh R, Bloom JS, Félix MA, Kruglyak L. Chromosome-scale selective sweeps shape Caenorhabditis elegans genomic diversity. Nat. Genet. 2012;44:285–290. doi: 10.1038/ng.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astner I, Schulze JO, van den Heuvel J, Jahn D, Schubert WD, Heinz DW. Crystal structure of 5-aminolevulinate synthase, the first enzyme of heme biosynthesis, and its link to XLSA in humans. EMBO J. 2005;24:3166–3177. doi: 10.1038/sj.emboj.7600792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auwerx J. Improving metabolism by increasing energy expenditure. Nat. Med. 2006;12:44–45. doi: 10.1038/nm0106-44. [DOI] [PubMed] [Google Scholar]
- Auwerx J, Avner P, Baldock R, Ballabio A, Balling R, Barbacid M, Berns A, Bradley A, Brown S, Carmeliet P, et al. The European dimension for the mouse genome mutagenesis program. Nat. Genet. 2004;36:925–927. doi: 10.1038/ng0904-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai P, Cantó C, Oudart H, Brunyánszki A, Cen Y, Thomas C, Yamamoto H, Huber A, Kiss B, Houtkooper RH, et al. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab. 2011;13:461–468. doi: 10.1016/j.cmet.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- Bystrykh L, Weersing E, Dontje B, Sutton S, Pletcher MT, Wiltshire T, Su AI, Vellenga E, Wang J, Manly KF, et al. Uncovering regulatory pathways that affect hematopoietic stem cell function using ‘genetical genomics’. Nat. Genet. 2005;37:225–232. doi: 10.1038/ng1497. [DOI] [PubMed] [Google Scholar]
- Champy MF, Selloum M, Piard L, Zeitler V, Caradec C, Chambon P, Auwerx J. Mouse functional genomics requires standardization of mouse handling and housing conditions. Mamm. Genome. 2004;15:768–783. doi: 10.1007/s00335-004-2393-1. [DOI] [PubMed] [Google Scholar]
- Champy MF, Selloum M, Zeitler V, Caradec C, Jung B, Rousseau S, Pouilly L, Sorg T, Auwerx J. Genetic background determines metabolic phenotypes in the mouse. Mamm. Genome. 2008;19:318–331. doi: 10.1007/s00335-008-9107-z. [DOI] [PubMed] [Google Scholar]
- Chen H, Li X, Epstein PN. MnSOD and catalase transgenes demonstrate that protection of islets from oxidative stress does not alter cytokine toxicity. Diabetes. 2005;54:1437–1446. doi: 10.2337/diabetes.54.5.1437. [DOI] [PubMed] [Google Scholar]
- Chesler EJ, Lu L, Wang J, Williams RW, Manly KF. WebQTL: rapid exploratory analysis of gene expression and genetic networks for brain and behavior. Nat. Neurosci. 2004;7:485–486. doi: 10.1038/nn0504-485. [DOI] [PubMed] [Google Scholar]
- Chesler EJ, Lu L, Shou S, Qu Y, Gu J, Wang J, Hsu HC, Mountz JD, Baldwin NE, Langston MA, et al. Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat. Genet. 2005;37:233–242. doi: 10.1038/ng1518. [DOI] [PubMed] [Google Scholar]
- Deeb SS, Fajas L, Nemoto M, Pihlajamäki J, Mykkänen L, Kuusisto J, Laakso M, Fujimoto W, Auwerx J. A Pro12Ala substitution in PPARgamma2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nat. Genet. 1998;20:284–287. doi: 10.1038/3099. [DOI] [PubMed] [Google Scholar]
- Dina C, Meyre D, Gallina S, Durand E, Körner A, Jacobson P, Carlsson LM, Kiess W, Vatin V, Lecoeur C, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat. Genet. 2007;39:724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- Ehrenreich IM, Torabi N, Jia Y, Kent J, Martis S, Shapiro JA, Gresham D, Caudy AA, Kruglyak L. Dissection of genetically complex traits with extremely large pools of yeast segregants. Nature. 2010;464:1039–1042. doi: 10.1038/nature08923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman JE, Blizard DA, Gerhard G, Mack HA, Lang DH, Van Nimwegen KL, Vogler GP, Stout JT, Shihabi ZK, Griffith JW, et al. Serum alkaline phosphatase activity is regulated by a chromosomal region containing the alkaline phosphatase 2 gene (Akp2) in C57BL/6J and DBA/2J mice. Physiol. Genomics. 2005;23:295–303. doi: 10.1152/physiolgenomics.00062.2005. [DOI] [PubMed] [Google Scholar]
- Fossey SC, Mychaleckyj JC, Pendleton JK, Snyder JR, Bensen JT, Hirakawa S, Rich SS, Freedman BI, Bowden DW. A high-resolution 6.0-megabase transcript map of the type 2 diabetes susceptibility region on human chromosome 20. Genomics. 2001;76:45–57. doi: 10.1006/geno.2001.6584. [DOI] [PubMed] [Google Scholar]
- Gaglani SM, Lu L, Williams RW, Rosen GD. The genetic control of neocortex volume and covariation with neocortical gene expression in mice. BMC Neurosci. 2009;10:44. doi: 10.1186/1471-2202-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti D, Maki A, Chesler EJ, Kirova R, Kosyk O, Lu L, Manly KF, Williams RW, Perkins A, Langston MA, et al. Genome-level analysis of genetic regulation of liver gene expression networks. Hepatology. 2007;46:548–557. doi: 10.1002/hep.21682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisert EE, Lu L, Freeman-Anderson NE, Templeton JP, Nassr M, Wang X, Gu W, Jiao Y, Williams RW. Gene expression in the mouse eye: an online resource for genetics using 103 strains of mice. Mol. Vis. 2009;15:1730–1763. [PMC free article] [PubMed] [Google Scholar]
- Haack TB, Danhauser K, Haberberger B, Hoser J, Strecker V, Boehm D, Uziel G, Lamantea E, Invernizzi F, Poulton J, et al. Exome sequencing identifies ACAD9 mutations as a cause of complex I deficiency. Nat. Genet. 2010;42:1131–1134. doi: 10.1038/ng.706. [DOI] [PubMed] [Google Scholar]
- Houtkooper RH, Cantó C, Wanders RJ, Auwerx J. The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr. Rev. 2010;31:194–223. doi: 10.1210/er.2009-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Liang J, Noble PW. Hyaluronan as an immune regulator in human diseases. Physiol. Rev. 2011;91:221–264. doi: 10.1152/physrev.00052.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi MA, Jeoung NH, Obayashi M, Hattab EM, Brocken EG, Liechty EA, Kubek MJ, Vattem KM, Wek RC, Harris RA. Impaired growth and neurological abnormalities in branched-chain alpha-keto acid dehydrogenase kinase-deficient mice. Biochem. J. 2006;400:153–162. doi: 10.1042/BJ20060869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsall IR, Rosenzweig D, Cohen PT. Disruption of the allosteric phosphorylase a regulation of the hepatic glycogen-targeted protein phosphatase 1 improves glucose tolerance in vivo. Cell. Signal. 2009;21:1123–1134. doi: 10.1016/j.cellsig.2009.03.001. [DOI] [PubMed] [Google Scholar]
- King EG, Macdonald SJ, Long AD. Properties and power of the Drosophila synthetic population resource for the routine dissection of complex traits. Genetics. 2012;191:935–949. doi: 10.1534/genetics.112.138537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konheim YL, Wolford JK. Association of a promoter variant in the inducible cyclooxygenase-2 gene (PTGS2) with type 2 diabetes mellitus in Pima Indians. Hum. Genet. 2003;113:377–381. doi: 10.1007/s00439-003-1000-y. [DOI] [PubMed] [Google Scholar]
- Koutnikova H, Laakso M, Lu L, Combe R, Paananen J, Kuulasmaa T, Kuusisto J, Häring HU, Hansen T, Pedersen O, et al. Identification of the UBP1 locus as a critical blood pressure determinant using a combination of mouse and human genetics. PLoS Genet. 2009;5:e1000591. doi: 10.1371/journal.pgen.1000591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughlin RE, Grant TL, Williams RW, Jentsch JD. Genetic dissection of behavioral flexibility: reversal learning in mice. Biol. Psychiatry. 2011;69:1109–1116. doi: 10.1016/j.biopsych.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehninger AL, Nelson DL, Cox MM. Lehninger principles of biochemistry. Fifth Edition W.H. Freeman; New York: 2008. [Google Scholar]
- Leitges M, Plomann M, Standaert ML, Bandyopadhyay G, Sajan MP, Kanoh Y, Farese RV. Knockout of PKC alpha enhances insulin signaling through PI3K. Mol. Endocrinol. 2002;16:847–858. doi: 10.1210/mend.16.4.0809. [DOI] [PubMed] [Google Scholar]
- Lynch M, Walsh B. Genetics and analysis of quantitative traits. Sinauer; Sunderland, Mass.: 1998. [Google Scholar]
- Lyssenko V, Eliasson L, Kotova O, Pilgaard K, Wierup N, Salehi A, Wendt A, Jonsson A, De Marinis YZ, Berglund LM, et al. Pleiotropic effects of GIP on islet function involve osteopontin. Diabetes. 2011;60:2424–2433. doi: 10.2337/db10-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MI. Genomics, type 2 diabetes, and obesity. N. Engl. J. Med. 2010;363:2339–2350. doi: 10.1056/NEJMra0906948. [DOI] [PubMed] [Google Scholar]
- Mehrabian M, Wen PZ, Fisler J, Davis RC, Lusis AJ. Genetic loci controlling body fat, lipoprotein metabolism, and insulin levels in a multifactorial mouse model. J. Clin. Invest. 1998;101:2485–2496. doi: 10.1172/JCI1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midorikawa R, Takei Y, Hirokawa N. KIF4 motor regulates activity-dependent neuronal survival by suppressing PARP-1 enzymatic activity. Cell. 2006;125:371–383. doi: 10.1016/j.cell.2006.02.039. [DOI] [PubMed] [Google Scholar]
- Mitchell JA, Aronson AR, Mork JG, Folk LC, Humphrey SM, Ward JM. Gene indexing: characterization and analysis of NLM’s GeneRIFs; AMIA Annu. Symp. Proc.; 2003; 2003. pp. 460–464. [PMC free article] [PubMed] [Google Scholar]
- Mitchell KJ, Tsuboi T, Rutter GA. Role for plasma membranerelated Ca2+-ATPase-1 (ATP2C1) in pancreatic beta-cell Ca2+ homeostasis revealed by RNA silencing. Diabetes. 2004;53:393–400. doi: 10.2337/diabetes.53.2.393. [DOI] [PubMed] [Google Scholar]
- Miyairi I, Tatireddigari VR, Mahdi OS, Rose LA, Belland RJ, Lu L, Williams RW, Byrne GI. The p47 GTPases Iigp2 and Irgb10 regulate innate immunity and inflammation to murine Chlamydia psittaci infection. J. Immunol. 2007;179:1814–1824. doi: 10.4049/jimmunol.179.3.1814. [DOI] [PubMed] [Google Scholar]
- Mornet E. Hypophosphatasia. Best Pract. Res. Clin. Rheumatol. 2008;22:113–127. doi: 10.1016/j.berh.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Mozhui K, Ciobanu DC, Schikorski T, Wang X, Lu L, Williams RW. Dissection of a QTL hotspot on mouse distal chromosome 1 that modulates neurobehavioral phenotypes and gene expression. PLoS Genet. 2008;4:e1000260. doi: 10.1371/journal.pgen.1000260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielson CM, Zmuda JM, Carlos AS, Wagoner WJ, Larson EA, Orwoll ES, Klein RF. Rare coding variants in ALPL are associated with low serum alkaline phosphatase and low bone mineral density. J. Bone Miner. Res. 2011;27:93–103. doi: 10.1002/jbmr.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouws J, Nijtmans L, Houten SM, van den Brand M, Huynen M, Venselaar H, Hoefs S, Gloerich J, Kronick J, Hutchin T, et al. Acyl-CoA dehydrogenase 9 is required for the biogenesis of oxidative phosphorylation complex I. Cell Metab. 2010;12:283–294. doi: 10.1016/j.cmet.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirce JL, Lu L, Gu J, Silver LM, Williams RW. A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genet. 2004;5:7. doi: 10.1186/1471-2156-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip VM, Duvvuru S, Gomero B, Ansah TA, Blaha CD, Cook MN, Hamre KM, Lariviere WR, Matthews DB, Mittleman G, et al. High-throughput behavioral phenotyping in the expanded panel of BXD recombinant inbred strains. Genes Brain Behav. 2010;9:129–159. doi: 10.1111/j.1601-183X.2009.00540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp RA. Studies on the mouse hemoglobin loci. II. Position of the hemoglobin locus with respect to albinism and shaker-1 loci. J. Hered. 1962;53:73–80. doi: 10.1093/oxfordjournals.jhered.a107127. [DOI] [PubMed] [Google Scholar]
- Riu E, Bosch F, Valera A. Prevention of diabetic alterations in transgenic mice overexpressing Myc in the liver. Proc. Natl. Acad. Sci. USA. 1996;93:2198–2202. doi: 10.1073/pnas.93.5.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J, Fortuno ES, 3rd, Suh JM, Stenesen D, Tang W, Parks EJ, Adams CM, Townes T, Graff JM. Atf4 regulates obesity, glucose homeostasis, and energy expenditure. Diabetes. 2009;58:2565–2573. doi: 10.2337/db09-0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer JB, Hill AE, Burrage LC, Olszens KR, Song J, Justice M, O’Brien WE, Conti DV, Witte JS, Lander ES, Nadeau JH. Genetic dissection of complex traits with chromosome substitution strains of mice. Science. 2004;304:445–448. doi: 10.1126/science.1093139. [DOI] [PubMed] [Google Scholar]
- Thomas C, Gioiello A, Noriega L, Strehle A, Oury J, Rizzo G, Macchiarulo A, Yamamoto H, Mataki C, Pruzanski M, et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009;10:167–177. doi: 10.1016/j.cmet.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschöp MH, Speakman JR, Arch JR, Auwerx J, Brüning JC, Chan L, Eckel RH, Farese RV, Jr., Galgani JE, Hambly C, et al. A guide to analysis of mouse energy metabolism. Nat. Methods. 2012;9:57–63. doi: 10.1038/nmeth.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2012;40:D71–D75. doi: 10.1093/nar/gkr981. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Agarwala R, Capra J, Chen Z, Church D, Ciobanu D, Li Z, Lu L, Mozhui K, Mulligan M, et al. High-throughput sequencing of the DBA/2J mouse genome. BMC Bioinformatics. 2010;11(Suppl 4):O7. [Google Scholar]
- Weigert J, Neumeier M, Bauer S, Mages W, Schnitzbauer AA, Obed A, Gröschl B, Hartmann A, Schäffler A, Aslanidis C, et al. Small-interference RNA-mediated knock-down of aldehyde oxidase 1 in 3T3-L1 cells impairs adipogenesis and adiponectin release. FEBS Lett. 2008;582:2965–2972. doi: 10.1016/j.febslet.2008.07.034. [DOI] [PubMed] [Google Scholar]
- Wen JH, Chen YY, Song SJ, Ding J, Gao Y, Hu QK, Feng RP, Liu YZ, Ren GC, Zhang CY, et al. Paired box 6 (PAX6) regulates glucose metabolism via proinsulin processing mediated by prohormone convertase 1/3 (PC1/3) Diabetologia. 2009;52:504–513. doi: 10.1007/s00125-008-1210-x. [DOI] [PubMed] [Google Scholar]
- Whyte M. Hypophosphatasia. In: Scriver, Beaudet, Valle, et al., editors. The Metabolic and Molecular Bases of Inherited Disease. Eighth Edition McGraw-Hill; New York: 2000. pp. 5313–5329. [Google Scholar]
- Williams RW, Gu J, Qi S, Lu L. The genetic structure of recombinant inbred mice: high-resolution consensus maps for complex trait analysis. Genome Biol. 2001;2:H0046. doi: 10.1186/gb-2001-2-11-research0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windahl SH, Andersson N, Börjesson AE, Swanson C, Svensson J, Movérare-Skrtic S, Sjögren K, Shao R, Lagerquist MK, Ohlsson C. Reduced bone mass and muscle strength in male 5a-reductase type 1 inactivated mice. PLoS ONE. 2011;6:e21402. doi: 10.1371/journal.pone.0021402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolford JK, Konheim YL, Colligan PB, Bogardus C. Association of a F479L variant in the cytosolic phospholipase A2 gene (PLA2G4A) with decreased glucose turnover and oxidation rates in Pima Indians. Mol. Genet. Metab. 2003;79:61–66. doi: 10.1016/s1096-7192(03)00051-9. [DOI] [PubMed] [Google Scholar]
- Xie Q, Gan L, Wang J, Wilson I, Li L. Loss of the innate immunity negative regulator IRAK-M leads to enhanced host immune defense against tumor growth. Mol. Immunol. 2007;44:3453–3461. doi: 10.1016/j.molimm.2007.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Williams EG, Mouchiroud L, Cantó C, Fan W, Downes M, Héligon C, Barish GD, Desvergne B, Evans RM, et al. NCoR1 is a conserved physiological modulator of muscle mass and oxidative function. Cell. 2011;147:827–839. doi: 10.1016/j.cell.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu L, Shen Z, Wesley J, Cai ZP. PTEN inhibitors cause a negative inotropic and chronotropic effect in mice. Eur. J. Pharmacol. 2011;650:298–302. doi: 10.1016/j.ejphar.2010.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.