Abstract
Wt1 regulates the epithelial-mesenchymal transition (EMT) in the epicardium and the reverse process (MET) in kidney mesenchyme. The mechanisms underlying these reciprocal functions are unknown. Here, we show in both embryos and cultured cells that Wt1 regulates Wnt4 expression dichotomously. In kidney cells, Wt1 recruits Cbp and p300 as coactivators; in epicardial cells it enlists Basp1 as a corepressor. Surprisingly, in both tissues, Wt1 loss reciprocally switches the chromatin architecture of the entire CTCF-bounded Wnt4 locus, but not the flanking regions; we term this mode of action “chromatin flip-flop”. CTCF and cohesin are dispensable for Wt1-mediated chromatin flip-flop but essential for maintaining the insulating boundaries. This work demonstrates that a developmental regulator coordinates chromatin boundaries with the transcriptional competence of the flanked region. These findings also have implications for hierarchical transcriptional regulation in development and disease.
Introduction
Some transcription factors (TFs) show dichotomous behaviour by activating target genes in one cellular context but repressing them in another (Roberts and Green, 1995), but the molecular mechanism behind this is mostly unclear. Wt1 is an archetypical dichotomous TF in development and disease. The gene was originally found to be inactivated in 15-20% of Wilms’ tumours, paediatric kidney tumours caused by a disturbance of normal renal development (Hohenstein and Hastie, 2006). The Wt1 gene encodes 36 different isoforms of proteins all carrying four C-terminal Zn-fingers (ZFs) which can function as transcriptional regulators or putatively in splicing regulation. Isoforms inserting three amino acids (KTS) between ZFs three and four have been proposed to mainly function in RNA metabolism, whereas isoforms lacking these residues are biased to function as TFs (Hohenstein and Hastie, 2006). During development Wt1 is expressed in a variety of tissues and cells, most of which are either going through an epithelial-to-mesenchymal transition (EMT) or the opposite MET (Moore et al., 1998). Wt1 is essential for embryogenesis, as Wt1 knockout mice die mid-gestation with, amongst other phenotypes, disturbed heart development and renal agenesis (Kreidberg et al., 1993). We have recently shown that Wt1 is essential for the epicardial EMT that generates vascular progenitors by controlling the expression of Snai1 and Cdh1 (Martinez-Estrada et al., 2010), but its role in the developing kidneys and Wilms’ tumourigenesis remains an enigma.
Renal development is characterized by the interaction of mesenchymal cells originating from the intermediate mesoderm with epithelial cells from the invading ureteric bud. The mesenchymal cells condense around the bud tips to form a pre-tubular aggregate and go through an MET to form nephrons, the filtering units in adult kidneys. For this MET the expression of Wnt4 is necessary (Stark et al., 1994) and sufficient (Kispert et al., 1998). WT1-mutant Wilms’ tumours, which are predominantly stromal and lack epithelialized structures, are believed to result from disturbance of this MET, a hypothesis supported by genome-wide expression (Li et al., 2002) as well as epigenetic analysis (Aiden et al., 2010). We previously demonstrated that Wt1 is essential for the nephron MET in organ culture, and showed that the knockdown of this gene phenocopies the loss of Wnt4 in the same ex vivo RNAi assay (Davies et al., 2004) . However, the mechanism by which Wt1 controls this essential step in nephrogenesis and Wilms’ tumour formation is unknown.
Here we show in vitro and in vivo that Wt1 directly activates the expression of Wnt4 in kidney mesenchyme but actively represses it in the epicardium. Elongating RNA polymerase II (RNAPII) occupies the Wnt4 transcriptional start site (TSS) in the kidney, while stalling RNAPII is detected in the epicardium. We demonstrate how Wt1-recruited cofactors are involved in either activation or repression in vitro and in vivo. Moreover, we identify a mechanism we designate ‘chromatin flip-flop’ by which Wt1 switches the chromatin state of the complete CCCTC-binding factor (CTCF)-delimited Wnt4 locus in a context-specific manner, thereby coupling the action of a dichotomous, DNA-binding TF to the CTCF-mediated-insulation. Finally, we show that CTCF and cohesin are required for insulation of the Wnt4 locus but are dispensable for regulating Wnt4 transcription. We propose that chromatin flip-flop is a common mechanism linking different modes of transcriptional regulation during development, steady-state cell maintenance and disease.
Results
Wt1 directly controls Wnt4 expression in the kidney mesenchyme in vitro and in vivo
We recently described a conditional Wt1 knockout mouse carrying a floxed exon 1, thereby mimicking the allele from the conventional knockout in a conditional manner (Martinez-Estrada et al., 2010). The expression of Nestin in kidney mesenchyme is controlled by Wt1 (Wagner et al., 2006), so to overcome the kidney agenesis phenotype from the conventional knockout we used a Nestin-Cre (Nes-Cre) allele (Tronche et al., 1999) to knockout Wt1 in the renal mesenchyme. In other Nes-Cre models, Cre toxicity has been demonstrated due to prolonged nuclear localization of the recombinase, either through use of a nuclear localization signal (NLS), in which case it was only observed in homozygous state, or through use of tamoxifen-induced nuclear translocation in the case of CreERT2 alleles (Forni et al., 2006). This however is not the case in our model. Our Nes-Cre allele is a different construct from the one used in the earlier studies, does not contain an NLS, is used in hemizygous state and is not tamoxifen-controled. We have not observed any of the abnormalities described by Forni et al, and the latter study did not report any kidney phenotype. Moreover, our Nes-Cre mice without any conditional allele have normal kidneys and a normal life span. The phenotype in our model can therefore be solely attributed to the loss of Wt1 in the kidney mesenchyme.
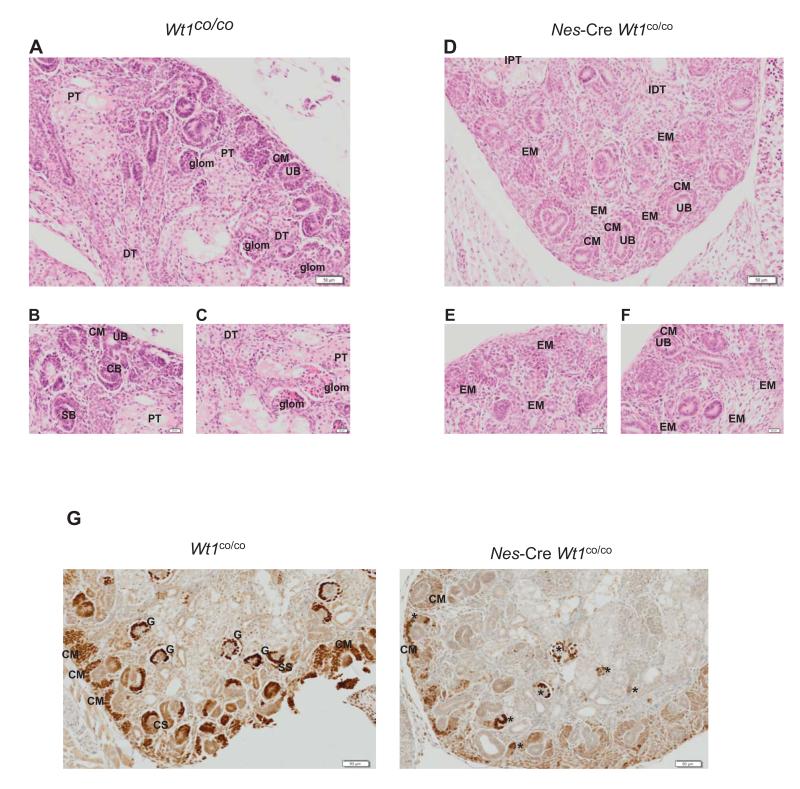
Analysis of kidneys at E18.5 showed a disturbance of nephron development (Figure 1A-F). Nes-Cre mediated loss of Wt1 results in a relative paucity of nephrogenesis at all cortical levels in the Nes-Cre mutants. In the early nephrogenic outer cortex there appear to be fewer ureteric buds, suggesting inhibited ureteric branching, and less condensation of metanephric mesenchyme into periureteric caps resulting in an overabundance of mesenchyme. In the intermediate cortex there is reduced aggregation, less epithelialisation of aggregates, and reduced formation of renal vesicles, comma-shaped bodies, and S-shaped bodies. In the inner cortex there is a marked reduction in tubulogenesis, glomerulogenesis, and tubular maturation (Figure 1A-F). The overall net effect is a relative abundance of mesenchyme over epithelial structures. All this is in accordance with an important role for Wt1 in the renal MET.
Figure 1.
Wt1 is essential for nephron formation at the MET stage.
A-F. H&E stained section of E18.5 control (A-C) and mutant (D-F) kidneys. Each section is taken from a different kidney.
G. Wt1 immunohistochemistry of E18.5 control (left panel) and mutant (right panel) kidneys. Abbreviations are: UB: ureteric bud; CM: condensed mesenchyme; EM expanded mesenchyme; CS: comma-shaped body; SS: S-shaped body; PT: proximal tubule; DT: distal tubule;G: glomerulus; IPT: immature proximal tubule; IDT: immature distal tubule.*: denotes epithelialised but abnormal structures that have partially lost Wt1.
We analysed Wt1 expression via immunohistochemistry in mutant and control kidneys to confirm the loss of the protein. We found in mutant kidneys that in most cases Wt1 is already lost from the cap mesenchyme. However, in few instances Wt1 was lost slightly later, which coincided with further developed, though abnormal, structures (Figure1G). This may account for the slight variation observed in the phenotype as described above.
Wt1 expression in the kidney mesenchyme precedes Wnt4 expression, and Wnt4 expression has previously been suggested to be downstream (direct or indirect) of Wt1. (Sim et al., 2002). We therefore took M15 mouse embryonic kidney mesenchymal cells expressing endogenous Wt1 (Larsson et al., 1995) and transiently silenced Wt1 using three independent RNAi constructs. All constructs led to almost complete loss of the Wt1 as well as Wnt4 transcripts (Figure 2A). We confirmed the physiological relevance of this by combining the Nes-Cre and Wt1co alleles with a Wt1GFP knock-in model (Hosen et al., 2007) and FACS-sorting GFP+ cells from control (Wt1co/GFP) and Wt1-deficient (Nes-Cre Wt1co/GFP) e12.5 kidney rudiments (Figure 2B). The expression of Wnt4 was restricted to the GFP+ cells (i.e. Wt1+ cells, Figure S1A), and was lost upon Wt1 knockout (Figure 2B and S1A). This loss in Wnt4 expression was not due to GFP+ cells apoptosis. GFP+ cell numbers and percentages were unchanged upon Wt1 knockout (Figure S1B) confirming the survival of kidney mesenchymal cells at e12.5 when Wt1 is deleted in the Nes-cre conditional background. Moreover, the expression levels of housekeeping genes (actin and Gapdh) as well as the pro-survival gene Bcl2 were unchanged upon Wt1 knockout in the GFP-sorted cells (Figure S1C), further supporting the survival of GFP+ cells upon Wt1 knockout. Additionally, using two methods, propodium and Annexin V staining we observe no difference in apoptosis between control and Nes-cre e12.5 GFP-sorted kidney mesenchyme cells (Fig. S1D-G). This is in sharp contrast to the apoptosis found in the intermediate mesoderm in the conventional Wt1 knockout (Kreidberg et al., 1993). Furthermore, staining with cleaved caspase 3 apoptosis marker confirmed the extremely low level of apoptosis in e18.5 mutant kidneys and showed that it is comparable to that observed in controls (data not shown). Moreover, the absence of apoptotic cells at e12.5 (Figure S1A-G) and Wt1 mosaicism observed at e18.5 (Figure 1G) argues against compensatory proliferation of the Wt1+ population to replace the recombined tissue (i.e. Cre+ and Wt1−).
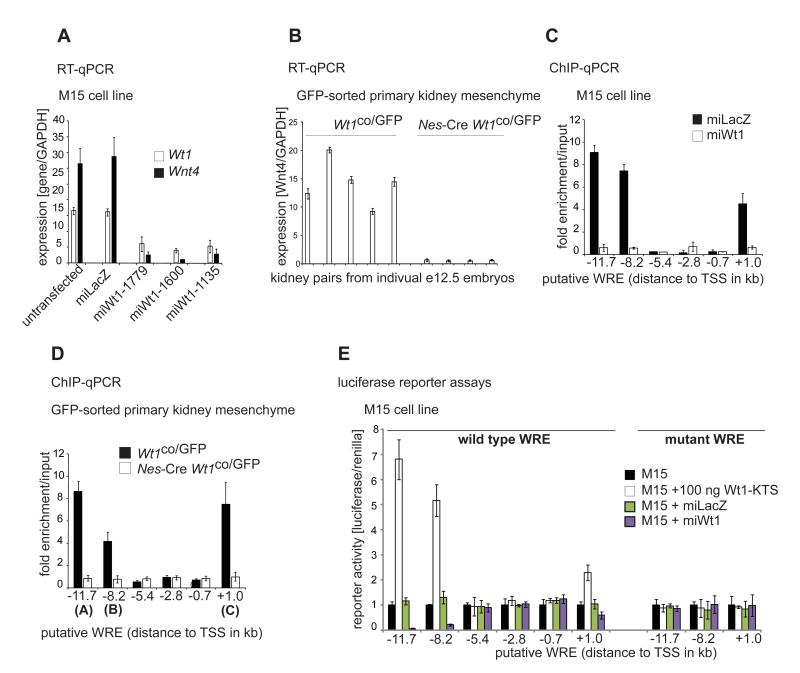
Figure 2.
Wnt4 is directly activated by Wt1 in kidney mesenchyme in vivo and in vitro.
A. Wt1 and Wnt4 mRNA in control and Wt1 knockdown M15 cells.
B. Wnt4 mRNA expression in FACS sorted control (Wt1co/GFP) and Wt1-deficient (Nes-Cre Wt1co/GFP) e12.5 kidney mesenchyme cells.
C. ChIP of putative WREs using Wt1 antibodies in control and Wt1 knockdown M15 cells.
D. ChIP of putative WREs using Wt1 antibodies in FACS sorted control (Wt1co/GFP) and Wt1-deficient (Nes-Cre Wt1co/GFP) e12.5 kidney mesenchyme cells.
E. Luciferase reporter assays of wild type and mutant putative WREs in M15 cells.
To further confirm the direct regulation of Wnt4 by Wt1, we looked for evolutionary conserved Wt1 response elements (WREs) (Bejerano et al., 2005) in the Wnt4 gene with 9-12 bp G/C-rich sequence harbouring the core motif T/GGGG/CCT/A , which was recently confirmed in ChIP-chip studies (Hartwig et al., 2010; Kim et al., 2009). We identified 6 putative WREs, five upstream of the TSS (−11.7 (called A), −8.2 (B), −5.4, −2.8, −0.7 kb) and one in intron 1 (+1.0 kb, C). Only A, B and C bound Wt1 using chromatin immunoprecipitation (ChIP) in the M15 cell line (Figure 2C) and in vivo GFP-sorted primary cells (Figure 2D). Only the three ChIP+ WREs but not the three ChIP− elements could confer transcriptional activity in in vitro reporter assays (Figure 2E). The ChIP+ WREs responded with a decrease in activity when co-transfected with Wt1 RNAi but an increase with a Wt1-KTS cDNA construct (Figure 2E), but not a Wt1+KTS construct (data not shown). Mutation of the WREs in the ChIP+ fragments resulted in complete loss of luciferase activity (Figure 2E). Together these data show that Wnt4 expression in kidney mesenchyme, and therefore the induction of nephron MET, is directly activated by Wt1.
We further analysed the absolute levels of Wnt4 in the M15 cell line and the GFP-sorted cells in order to compare the transcript expression in the in vivo context (primary GFP-sorted cells) and our in vitro model (M15). The expression of Wnt4 in absolute terms was 50% lower in vivo compared to M15 cells (Figure S2). However, upon the loss of Wt1 (miWt1 in M15 and conditional knockout in GFP-sorted cells), the loss in Wnt4 expression levels is comparable and statistically not different between M15 and GFP-sorted cells (Figure S2).
Wt1 directly represses Wnt4 expression in epicardial cells in vitro and in vivo
Direct Wt1 regulation of Wnt4 in nephron MET results in kidney mesenchymal cells terminally differentiating. This is in contrast to the situation in the epicardium where Wt1 controls an EMT and is responsible for the generation of cardiovascular progenitor cells (Martinez-Estrada et al., 2010). This provided us with a system to study bidirectional regulation by Wt1 in two physiologically relevant systems. In immortalized epicardial MEEC cells (Martinez-Estrada et al., 2010) using Wt1-specific RNAi, we observed an upregulation of Wnt4 transcript levels (Figure 3A). This was confirmed in vivo as the loss of Wt1 from FACS-sorted Wt1GFP-positive epicardial cells (Martinez-Estrada et al., 2010) resulted in a strong increase in Wnt4 expression (Figure 3B). ChIP confirmed binding of Wt1 to the same WREs (A, B and C) but not the other putative WREs (Figure 3C). Finally, the luciferase reporter constructs showed opposite behaviour in MEEC cells compared to M15; activity of the reporters was increased by Wt1 RNAi but decreased by co-transfection of Wt1-KTS, but not Wt1+KTS cDNAs, both effects being lost when the Wt1 binding sequences were mutated (Figure 3D).
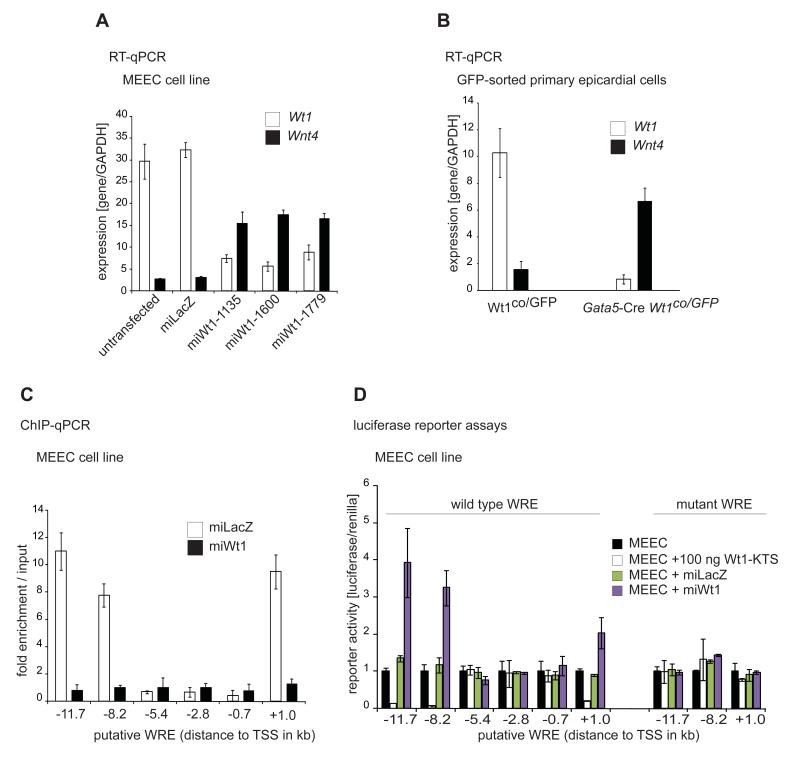
Figure 3.
Wnt4 is directly repressed by Wt1 in epicardium in vivo and in vitro.
A. mRNA of Wt1 and Wnt4 expression in control and Wt1 knockdown MEEC (epicardium) cells.
B. mRNA of Wt1 and Wnt4 expression in FACS sorted control (Wt1co/GFP) and Wt1-deficient (Gata5-Cre Wt1co/GFP) epicardium.
C. ChIP with Wt1 antibodies of putative WREs in control and Wt1 knockdown MEEC cells.
D. Luciferase reporter assays of wild type and mutant putative WREs in MEEC cells.
Wt1 controls Wnt4 expression via tissue-dependent recruitment of coactivators and corepressors
To understand how Wt1 bidirectionally controls Wnt4, we analysed whether specific cofactors could be involved in its dichotomous function. The CREB-binding protein (Cbp) coactivator has previously been shown to bind Wt1 (Wang et al., 2001). ChIP analysis revealed binding of Cbp and the associated p300 at the same three ChIP+ WREs in GFP-sorted cells (Figure 4A-B) and M15 (Figure 4C-D) in a Wt1-dependent manner. Loss of Cbp and/or p300 did not affect Wt1 binding to these WREs (Figure 4E). Thus, Wt1 is necessary to recruit Cbp and p300 to the Wnt4 locus. Knockdown of Cbp and/or p300 resulted in a loss of Wnt4 expression (Figure 4F), supporting their indispensable role in Wt1-mediated Wnt4 activation.
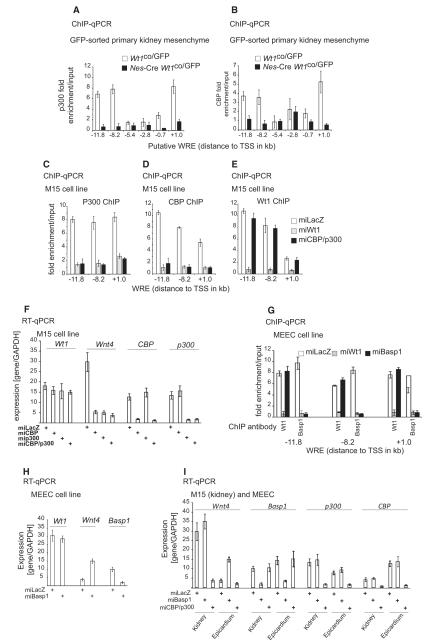
Figure 4.
In vitro and in vivo co-factors regulating Wnt4 expression in a tissue-specific manner downstream of Wt1.
A. ChIP using p300 antibodies in FACS sorted control (Wt1co/GFP) and Wt1-deficient (Nes-Cre Wt1co/GFP) e12.5 kidney mesenchyme cells.
B. ChIP using Cbp antibodies in FACS sorted control (Wt1co/GFP) and Wt1-deficient (Nes-Cre Wt1co/GFP) e12.5 kidney mesenchyme cells.
C-E. ChIP with p300 (C), Cbp (D) and Wt1 (E) antibodies in control (lacZ), Wt1 and a combined Cbp-p300 knockdown M15 cells.
F. mRNA of indicated genes in control, Cbp, p300 and Cbp/p300 knockdown M15 cells.
G. ChIP with Wt1 or Basp1 antibodies of WREs in control, Wt1 and Basp1 knockdown MEEC cells.
H. mRNA of indicated genes in control and Basp1 knockdown cells.
I. mRNA of indicated genes in control, Basp1 and Cbp/p300 knockdown M15 (kidney) and MEEC (epicardial) cells.
In epicardium, where Wnt4 is repressed by Wt1, we tested for the presence of potential corepressors. Brain abundant, membrane attached signal protein 1 (Basp1) has previously been identified as a Wt1 corepressor (Carpenter et al., 2004). In epicardial cells we found Basp1 bound to regions A, B and C in a Wt1-dependent fashion, but Wt1 binding was unaffected by Basp1 loss (Figure 4G). RNAi-mediated loss of Basp1 resulted in increased Wnt4 expression (Figure 4H). This supports an active role for Basp1 in Wt1-mediated repression of Wnt4 in epicardium.
We further tested the specificity of the cofactors by silencing Cbp, p300 and Basp1 in both kidney mesenchymal and epicardial cell lines (Figure 4I). Cbp/p300 knockdown only affected Wnt4 expression in kidney while Basp1 silencing only affected Wnt4 levels in epicardium (Figure 4I).
Wt1 bidirectionally controls the local Wnt4 chromatin in a tissue-specific manner
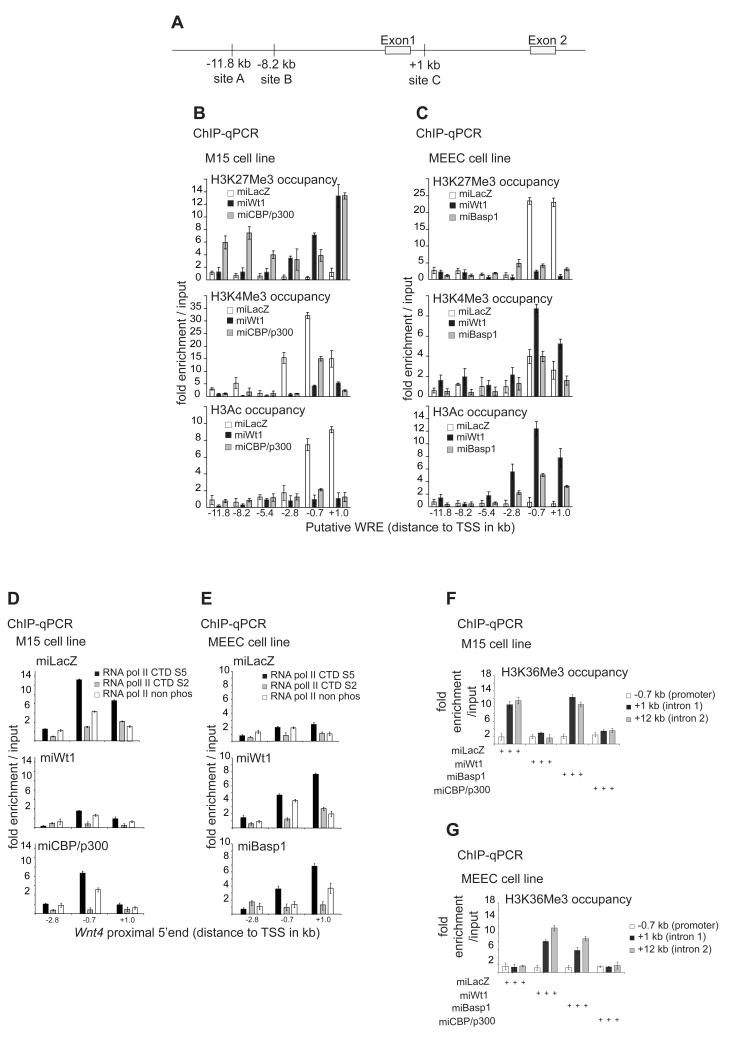
TFs regulate gene expression by partly modulating the chromatin structure either positively or negatively. However, this has not been studied previously on the same gene regulated by a dichotomous TF in two different tissues or in tissues undergoing opposite biological processes. To elucidate the molecular mechanism(s) by which Wt1 controls Wnt4 expression, we analysed the local chromatin state. The presence of H3K4me3 and H3K9/K14Ac (hereafter referred to as H3Ac) at 5′ ends of genes is indicative of gene activation whereas high polycomb-associated H3K27me3 and low H3Ac is a hallmark of repression (Kouzarides, 2007). Analysis of these marks near the Wnt4 TSS (Figure 5A) suggested an active chromatin conformation in kidney mesenchyme, which was switched to inactive upon Wt1 loss (Figure 5B). A similar outcome was found in vivo upon Wt1 knockout (Figure 6G). This switch of the Wnt4 local chromatin in kidney requires the Wt1-recruited coactivators Cbp and p300, as their knockdown led to high H3K27me3, a loss of H3Ac and a reduction in H3K4me3 (Figure 5B). Cbp/p300 silencing leads to an increase of H3K27me3 at the distant WREs A and B (Figure 5B). This might be due to Cbp’s roles in H3 acetylation that antagonises H3K27me3 deposition (Tie et al., 2009).
Figure 5.
Wt1 actively upregulates Wnt4 expression in kidney mesenchyme and actively downregulates its expression in epicardium via the local chromatin, RNAPII elongation and stalling.
A. A schematic representation of the 5′end of the Wnt4 gene.
B. ChIP with H3K27Me3 (top), H3K4Me3 (middle) and H3Ac (bottom) antibodies in control as well as Wt1 or Cbp/p300 knockdown M15 cells.
C. ChIP with H3K27Me3 (top), H3K4Me3 (middle) and H3Ac (bottom) antibodies in control as well as Wt1 or Cbp/p300 knockdown MEEC cells.
D. ChIP with RNApII CTDS5, RNAPII CTDS2 and CTDnon phos antibodies of control (top), Wt1 (middle) and Cbp/p300 (bottom) knockdown M15 cells.
E. ChIP with RNApII CTDS5, RNAPII CTDS2 and CTDnon phos antibodies of control (top), Wt1 (middle) and Basp1 (bottom) knockdown MEEC cells.
F. ChIP with H3K36Me3 antibodies in control, Wt1, Basp1 and Cbp/p300 knockdown M15 cells.
G. ChIP with H3K36Me3 antibodies in control, Wt1, Basp1 and Cbp/p300 knockdown MEEC cells.
Figure 6.
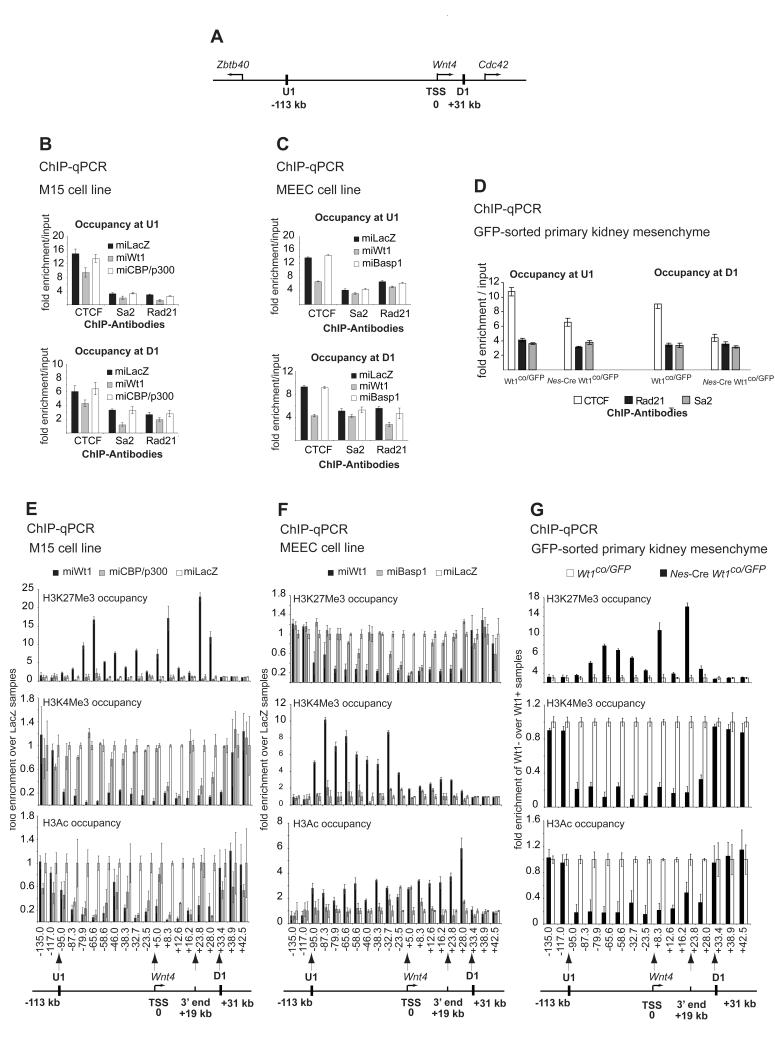
Wt1-mediated chromatin flip-flop.
A. A graphic illustration of the complete Wnt4 locus and neighbouring genes as demarcated by CTCF binding upstream (U1) and downstream (D1). Distances in kb from Wnt4 TSS are indicated.
B. ChIP with CTCF, Sa2 and Rad21 antibodies in control, Wt1 and Cbp/p300 knockdown M15 cells at U1 (left panel) and D1 (right panel) sites.
C. ChIP with CTCF, Sa2 and Rad21 antibodies in control, Wt1 and Cbp/p300 knockdown MEEC cells at U1 (left panel) and D1 (right panel) sites.
D. Occupancy of CTCF and cohesin subunits Rad21 and Sa2 at U1 and D1 in the primary GFP-sorted kidney mesenchymal cells.
E. ChIP with H3K27Me3 (top), H3K4Me3 (middle) and H3Ac (bottom) antibodies in control, Wt1 and Cbp/p300 knockdown M15 cells.
F. ChIP with H3K27Me3 (top), H3K4Me3 (middle) and H3Ac (bottom) antibodies in control, Wt1 and Basp1 knockdown MEEC cells.
G. ChIP with H3K27Me3 (top), H3K4Me3 (middle) and H3Ac (bottom) antibodies in FACS sorted control (Wt1co/GFP) and Wt1-deficient (Nes-Cre Wt1co/GFP) e12.5 kidney mesenchyme cells.
Conversely, in epicardial cells the 5′end of Wnt4 exhibits high levels of H3K27me3 and is devoid of any H3Ac (Figure 5C), indicative of a repressive chromatin structure. H3K4me3 is slightly enriched only around the TSS (Figure 5C). This confirms earlier studies that H3K4me3 occupies TSSs of both active and repressed genes, but is highly enriched at the former (Barski et al., 2007; Guenther et al., 2007). The loss of Wt1 switched the local chromatin from a repressive state to an active state (Figure 5C). Basp1 knockdown led to a switch of the repressed chromatin state (H3K27me3, Figure 5C) to an active confirmation, as supported by a marked increase in H3Ac but not corresponding changes in H3K4me3 (Figure 5C). Therefore, Basp1 is required for the control of Wt1-mediated Wnt4 repression via local chromatin modulation.
Wt1 controls the differential occupancy of RNAPII relative to the Wnt4 TSS
In the M15 kidney mesenchymal cell line, RNAPII occupies mostly the proximal upstream region (−0.7 kb) but also a region downstream of the TSS (+1 kb or C). Upon Wt1 loss the binding of RNAPII decreased but also switched mainly to C (Figure S3A). In the MEEC epicardial cells, the reverse was observed; RNAPII mostly occupies the downstream region, but binds upstream of the TSS upon Wt1 silencing (Figure S3B). The binding of RNAPII at both active and repressed genes is a known phenomenon and is linked to elongation and stalling, respectively (Buratowski, 2009). Our results indicated that higher RNAPII binding upstream of the TSS may reflect gene activation, while its increased binding downstream correlates with repression; and this switch between RNAPII upstream and downstream occupancy is dependent on Wt1 action and cofactor recruitment in a tissue-specific manner (Figure S3A-B).
Wt1 controls RNAPII elongation and stalling at the 5′end of Wnt4 in kidney versus epicardium
TFs mediate gene expression by either regulating the initiation phase of RNAPII-mediated transcription or, at a later step, RNAPII stalling. Each of these steps is associated with differential phosphorylation states of RNAPII and a change in H3K36me3, reflecting elongation or the lack of it (Buratowski, 2009).
The C terminal domain (CTD) of the RNAPII largest subunit, Rpb1, contains a conserved, tandemly repeated consensus sequence (YSPTSPS) Phosphorylation of serines at positions 2 (CTDS2) and 5 (CTDS5) is associated with RNAPII differential initiation, elongation, termination or stalling (reviewed in (Buratowski, 2009)). CTDS5 is associated with transcription initiation and is enriched at 5′ends of genes. CTDS2 enrichment, however, increases as CTDS5 levels start decreasing (0.6-1 kb downstream of yeast TSSs) coinciding with or following the exchanges of initiation for elongation factors. This transition from CTDS5 to CTDS2 is indicative of active RNAPII elongation (Buratowski, 2009). However, the existence of CTDS5 alone at the 5′end of genes, just after the TSS, is associated with RNAPII stalling and poised or repressed gene expression in ES cells (Stock et al., 2007) and yeast (Kim et al., 2010).
We examined RNAPII dynamics at the 5′end of the Wnt4 gene using isoform-specific antibodies that recognise CTDS5, CTDS2 and non-phosphorylated (CTDN-Ph) RNAPII isoforms which were validated previously (Stock et al., 2007). In kidney mesenchyme, the levels of CTDS5 are high 0.7kb upstream of the TSS but decrease at site C; this was accompanied by a significant increase in CTDS2 at C (Figure 5D). The transition from CTDS5 to CTDS2 supports RNAPII elongation. RNAPII elongation is associated with an increase in H3K36me3 in the body of active protein-coding genes (Barski et al., 2007). In kidney mesenchyme high levels of H3K36me3 occupy introns 1 and 2 of the Wnt4 gene in M15 and GFP-sorted cells (Figure 5F).
Upon Wt1 loss in the M15 cell line, CTDS5 levels are reduced by 80% at both −0.7 kb and site C , while CTDS2 is absent from the two regions (Figure 5D). This block in switch from CTDS5 to CTDS2 supports RNAPII stalling and gene repression (Buratowski, 2009). Consistently, the levels of H3K36me3 decreased 10 fold at intron 1 and below input enrichment at intron 2 (Figure 5F). A similar situation at the 5′end of Wnt4 occurs upon Cbp/p300 silencing, especially the lack of transition from CTDS5 to CTDS2 (Figure 5D) and the corresponding reductions in H3K36me3 intronic occupancy. (Figure 5F). However, the main difference is a two-fold decrease in CTDS5 and CTDS2 enrichment at a region upstream of the Wnt4 TSS (Figure 5E, lower panel as compared to Figure 5E, upper panel). Basp1 silencing in kidney mesenchyme had no effect on H3K36me3 levels compared to controls (Figure 5F). Combined, the data support the hypothesis that Wt1 and Cbp/p300 actively upregulate Wnt4 in kidney mesenchyme by promoting RNAPII elongation.
In the MEEC epicardial cell line there is a lack of CTDS5 to CTDS2 transition (Figure 5E). Upon Wt1 knockdown, there is a marked increase in CTDS5 (2 and 3 fold increase at −0.7 and +1 kb, respectively; Figure 5E). Moreover, there is 3 a fold increase in CTDS2 occupancy downstream of the TSS (Figure 5E), supporting a CTDS5 to CTDS2 transition and RNAPII elongation. A less pronounced effect on RNAPII dynamics was seen upon Basp1 knockdown (Figure 5E). However, there is an increase in CTDS5 binding both upstream and downstream of the TSS (Figure 5E). These dynamics of RNAPII are unique for Basp1 loss in epicardial cells, and are found neither in the kidney mesenchyme nor epicardium, under any other condition. However, an increase in CTDS5 loading on both sides of the TSS in Basp1-silenced cells is likely linked to an exchange of initiation for elongation factors, as was recently shown in yeast, (Kim et al., 2010). H3K36me3 enrichment in regions downstream of the TSS (introns 1 and 2, Figure 5G) upon the loss of Wt1 and Basp1 further supports active elongation. Therefore, the loss of Wt1 or Basp1 in epicardial cells results in RNAPII elongation. To sum up, the data support an active role for Wt1 and Basp1 in repressing Wnt4 gene expression by promoting RNAPII intronic stalling.
Wt1 maintains CTCF and cohesin loading at the boundary elements in kidney mesenchyme and epicardium
So far we have described a Wt1-mediated local chromatin switching mechanism for a ~13kb region at the 5′end of Wnt4. However, we noticed that Wt1 knockdown led to a complete switch while the cofactor silencing did not. One possibility is that the cofactors only partially mediate the Wt1-mediated local chromatin switching. However, an intriguing alternative would be that unlike cofactors Wt1 loss affects not only the local chromatin but the extended Wnt4 locus architecture beyond the 13 kb. Recently, intergenic CTCF binding was shown to demarcate active and repressive regions and CTCF was hypothesised to act as an insulating barrier blocking chromatin extension to the flanking regions, but neither gain nor loss of function experiments were performed (Cuddapah et al., 2009; Negre et al., 2010). A recent study investigating the deletion of Ctcf in the mouse limb has reported massive apoptosis but only a weak effect on global gene expression. The authors concluded that these findings cast doubt on the insulator function of CTCF in mediating apoptosis, however they did not analyse the effect of Ctcf loss on chromatin demarcation (Soshnikova et al., 2010).We reasoned that this demarcation would be a good starting point to assess how far along the Wnt4 genomic locus the chromatin is switched by Wt1. We therefore delineated Wnt4’s closest intergenic CTCF binding sites (CBSs). Using ChIP-seq data from human (Barski et al., 2007) and murine cells (Huang et al., 2008), we identified U1 (located 113 kb upstream of Wnt4 TSS) and D1 (31 kb downstream) (Figure 6A). We confirmed that CTCF occupies U1 and D1 in both kidney and epicardial cells (Figure 6B-C). CTCF binding at the intergenic U1 and D1 (but not at a conserved Wnt4 intragenic CBS, data not shown) was significantly reduced in kidney and epicardial cell lines after Wt1 silencing, but not when Cbp/p300 or Basp1 were silenced, respectively (Figure 6B-C). Similarly, loss of Wt1 in kidney mesenchyme in vivo resulted in a marked reduction of CTCF binding at U1 and D1 (Figure 6D). This indicates a functional interaction between Wt1 and CTCF loading at U1 and D1.
Recently, it was shown that the multi-protein complex cohesin occupies the CBSs, and may directly mediate gene regulation via 3D modulation of chromosome domains and territories (Ohlsson et al., 2010). We therefore tested whether cohesin is also bound at U1 and D1, and whether its loading is affected by Wt1. Two cohesin subunits were shown to co-occupy CBSs, stromal antigen 2 (Sa2) and DNA repair exonuclease rad21 (Rad21). Both subunits bound U1 and D1 in kidney (Figure 6B) and epicardium (Figure 6C). The loss of Wt1, but not the associated cofactors, in both cell lines resulted in a significant decrease in Rad21 and Sa2 loading (Figure 6B-C). Again, loss of Wt1 in kidney mesenchyme in vivo resulted in lower binding of Rad21 and Sa2 at both sites (Figure 6D)., suggesting either a functional interaction between Wt1 and cohesin loading or of the requirement for CTCF in recruiting these cohesin subunits. Nevertheless, Wt1 loss reduces the loading of both CTCF and cohesin at U1 and D1.
Wt1-mediated chromatin flip-flop
To delineate the chromatin architecture within and beyond the U1-D1 domain, we designed primer sets that span a 177 kb around the Wnt4 gene (see the bottom of Figure 6E-G for a list of primer positions relative to the Wnt4 TSS and Supplementary Methods for the position of primers in the mouse genome). We performed ChIP-qPCR and present the data as follows: the fold of enrichment of a particular chromatin mark over input in any condition is always divided by the fold of enrichment of the same mark over input in control cells (controls are miLacZ in M15 and MEEC cell lines and Wt1co/GFP in GFP-sorted primary cells). In control M15 cells the complete U1-D1 locus is active (Figure 6E). Upon Wt1 knockdown, but not Cbp/p300, the complete U1-D1 domain was switched from active to repressive chromatin (Figure 6E). This is confirmed in vivo, as Wt1 knockout kidney mesenchyme switched only the U1-D1 domain, but not the flanking regions (Figure 6G). These data strongly support a domain-wide epigenetic regulation of Wnt4 by Wt1 in kidney through flipping the chromatin state of the U1-D1 domain and point to an insulation function for CTCF/cohesin.
The loss of Cbp/p300 in kidney mesenchyme had little effect on both the H3K27me3 and H3K4me3 marks within or outside the U1 and D1 (Figure 6E, top panel), but only a local effect around the 5′end of Wnt4. However, Cbp/p300 knockdown reduced histone acetylation within and outside the U1-D1 domain (Figure 6E, lower graph) as both Cbp and p300 modulate global histone acetylation (Kouzarides, 2007). Cells deficient in both Cbp and p300 did not show a reduction in H3K4me3 levels beyond the locale of the Wnt4 TSS (Figure 6E) and H3K27me3 levels were unchanged compared to control cells (Figure 6E). This confirms the specificity of Wt1 in regulating a chromatin switching mechanism that flips the complete CTCF-delimited Wnt4 locus in kidney in both GFP-sorted cells and M15 cell lines.
In the MEEC cell line, we see the reverse dynamics where Wt1 loss switched (or flopped) the chromatin from a repressive to an active state, but again the U1 and D1 barriers remain effective (Figure 6F). Basp1 loss in epicardial cells caused a change only to the local chromatin near the Wnt4 TSS (Figure 6F). This supports a local role for Basp1, but a U1-D1 domain-wide function for Wt1. Therefore, Wt1 regulates the reciprocal switching of the CTCF-demarcated Wnt4 chromatin locus and is required for its maintenance. We term this domain-wide reciprocal switch ‘chromatin flip-flop’ (Figure 8).
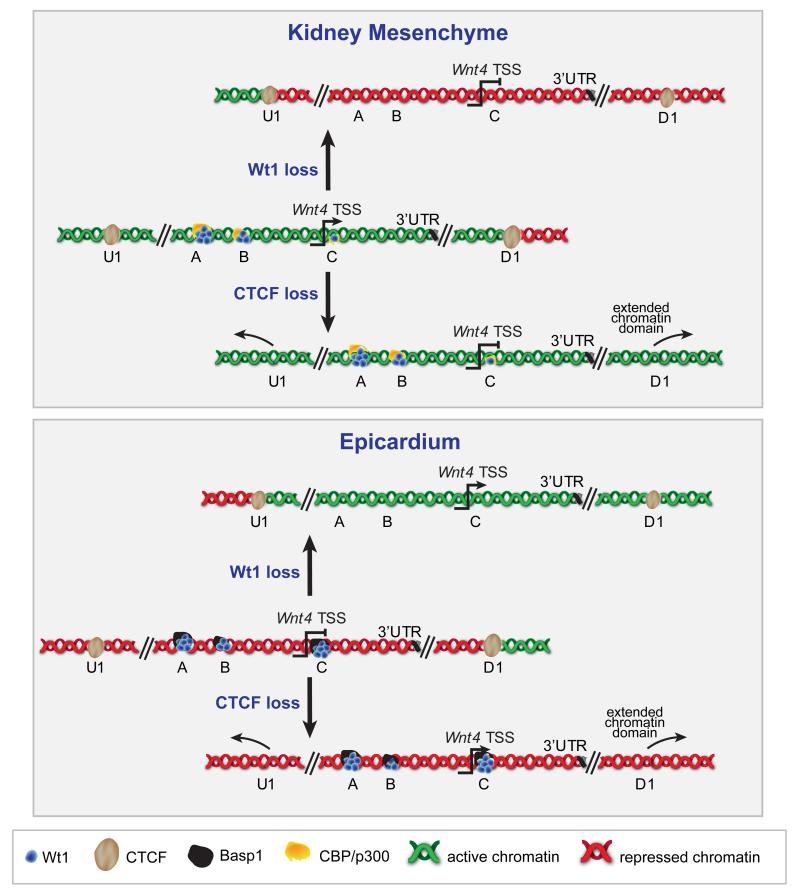
Figure 8.
Wt1-controled chromatin flip-flop in kidney mesenchyme and epicardium.
Upper panel: in kidney mesenchyme the Wnt4 locus extending to the CTCF sites U1 and D1 is active (green chromatin). Upon Wt1 loss, but not Cbp-p300 loss, the CTCF-delimited Wnt4 locus is switched to repressive chromatin (red). The flanking regions are unaffected. Upon Ctcf loss the active chromatin signature spreads to neighbouring loci from the Wnt4 locus.
Lower panel: in epicardium the Wnt4 locus extending to the CTCF sites U1 and D1 is repressed (red chromatin). Upon Wt1 loss, but not Basp1 loss, the CTCF-delimited Wnt4 locus is switched to active chromatin (green). The flanking regions are unaffected. Upon Ctcf loss the repressive chromatin signature spreads to neighbouring loci from the Wnt4 locus.
CTCF and cohesin regulate the Wnt4 locus insulation but are dispensable for Wnt4 expression and Wt1-mediated chromatin flip-flop
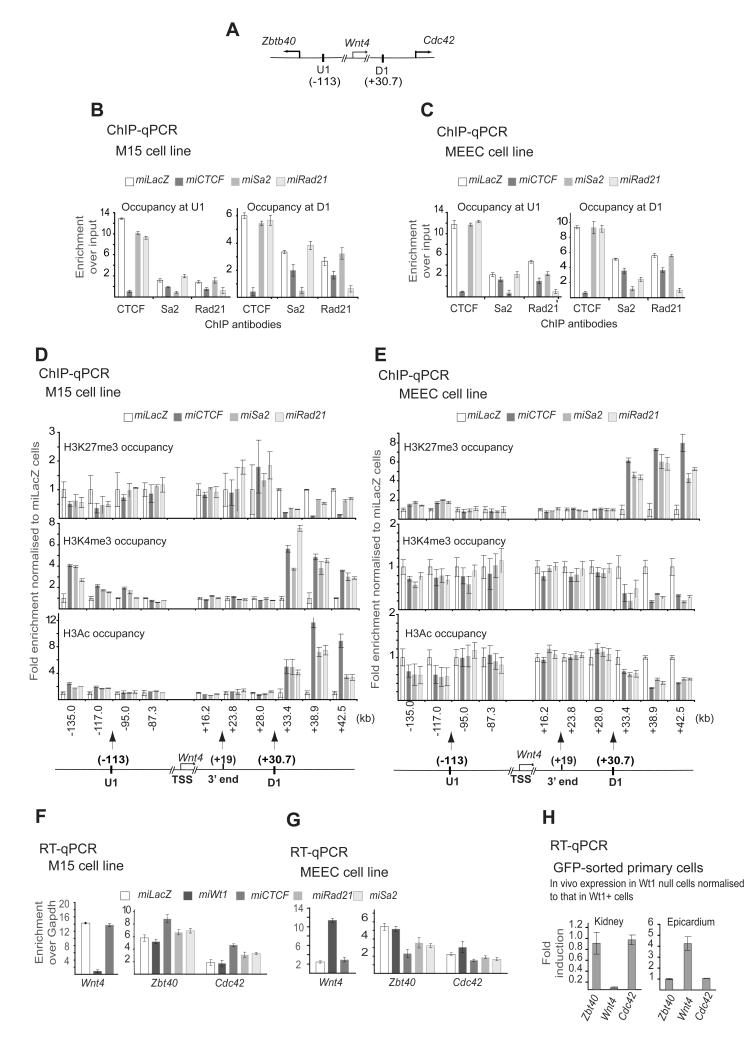
Our data so far suggest that CTCF, cohesin or both may act as barriers insulating the Wnt4 chromatin domain. Therefore, the loss of CTCF and/or cohesin may be sufficient to induce a chromatin flip-flop, while they are important for locking the U1-D1 domain (Figure 7A) in an active or repressed state in a tissue-specific manner. The evidence for CTCF function as an insulator is limited to special cases in specialised cells; its role in insulation of demarcated active and repressive domains remain circumstantial (Ohlsson et al., 2010) and cohesin’s role in this demarcation and insulation remains to be demonstrated. To address their role in insulation and chromatin flip-flop, we silenced CTCF, Rad21 and Sa2 in both kidney mesenchymal and epicardial cell lines. Use of CTCF-specific RNAi constructs resulted in an almost complete loss of CTCF protein at U1 and D1 in both tissues (Figure 7B-C), as well as a significant decrease in Rad21 and Sa2 loading at the two sites (Figure 7B-C). This suggests that Rad21 and Sa2 loading at U1 and D1 is partially mediated by CTCF, and therefore the reduction in their loading upon Wt1 loss may be indirect. Knockdown of Sa2 and Rad21 led to a complete loss of the appropriate protein from the U1 and D1 sites in both cell lines (Figure 7B-C) but did not reduce the levels of loaded CTCF at U1 and D1 (Figure 7B-C), indicating that CTCF loading does not require these cohesin subunits.
Figure 7.
CTCF and cohesin are essential for insulation but are dispensable for Wnt4 expression and Wt1-mediated chromatin flip-flop.
A. A graphic illustration of the complete Wnt4 locus and neighbouring genes as demarcated by U1 and D1.
B. ChIP with CTCF, Sa2 and Rad21 antibodies of control, Ctcf, Sa2 and Rad21 M15 cells at U1 (left panel) and D1 (right panel) sites.
C. ChIP with CTCF, Sa2 and Rad21 antibodies of control, Ctcf, Sa2 and Rad21 MEEC cells at U1 (left panel) and D1 (right panel) sites.
D. ChIP using H3K27Me3 (top), H3K4Me3 (middle) and H3Ac (bottom) antibodies in control, Ctcf, Sa2 and Rad21 knockdown M15 cells.
E. ChIP using H3K27Me3 (top), H3K4Me3 (middle) and H3Ac (bottom) antibodies in control, Ctcf, Sa2 and Rad21 knockdown MEEC cells.
F. mRNA of indicated genes in control, Wt1, Ctcf, Sa2 and Rad21 knockdown M15 cells.
G. mRNA of indicated genes in control, Wt1, Ctcf, Sa2 and Rad21 knockdown MEEC cells.
H. mRNA of indicated genes in Wt1-deficient kidney mesenchyme and epicardial cells in vivo normalized to control cells.
We next assessed whether the loss of each regulator affects the chromatin architecture within and outside the U1-D1 domain. The loss of CTCF, Sa2 and Rad21 did not affect the chromatin architecture between U1 and D1 in either kidney or epicardial cell lines (Figure 7D-E). This strongly suggests that CTCF and cohesin are dispensable for chromatin flip-flop. However, the flanking regions have acquired the Wnt4 chromatin in a tissue-specific manner upon CTCF, Rad21 or Sa2 silencing (Figure 7D-E). In other words, active chromatin now extended beyond U1 and D1 in kidney mesenchyme, while repressed chromatin extended beyond those CBSc in the epicardium, thereby spreading the U1-D1-delimited chromatin towards the neighbouring regions in both tissues. This is in full accordance with the hypothesized role for CTCF in insulating active and repressive domains and is direct evidence that CTCF controls insulation of a target chromatin domain.
The loss of Sa2 and Rad21 has a similar but less significant effect on the extension of the chromatin architecture outside U1-D1 domain (Figure 7D-E). This suggests a limited role for cohesin in insulation and chromatin boundary maintenance.
Loss of CTCF, Rad21 or Sa2, unlike Wt1 knockdown, had little effect on Wnt4 transcript levels in both cell lines (Figure 7F-G, left panels), supporting that fact these proteins are dispensable for the chromatin flip-flop (Figure 7D-E). However, the spread of the Wnt4 chromatin status beyond U1 and D1 upon CTCF and cohesin loss had functional implications for the neighbouring genes (Zbtb40 and Cdc42 at the 5′ and 3′ end of the Wnt4 locus, respectively). In kidney cells, Wnt4 and Zbt40 are active M15 and GFP-sorted cells, while Cdc42 is only expressed at low levels (Figure 7F-H). Upon Wt1 loss Wnt4 levels are down regulated, but the levels of Zbtb40 and Cdc42 remain unchanged (Figure 7F-H). However, upon the knockdown of CTCF, the transcript levels of Cdc42 and Zbtb40 are induced, but Wnt4 levels were unaffected (Figure 7F). Conversely, in epicardium, upon CTCF loss both Cdc42 and Zbtb40 are repressed further compared to controls or Wt1 knock-down cells, while Wnt4 is unaffected but induced in Wt1 silenced cells (Figure 7G). This confirms that the chromatin structure of the Wnt4 locus has spread beyond the U1 and D1 upon CTCF knockdown with corresponding changes in the expression of Cdc42 and Zbtb40.
Our analysis of the chromatin architecture assigned Rad21 and Sa2 a peripheral role in insulating the Wnt4 locus. We therefore tested their effect on Cdc42 and Zbtb40 expression. Silencing of either subunits led to perturbation of Cdc42 and Zbtb40 (Figure 7F-G). However, this was less pronounced than after CTCF knockdown. Together, the data suggest a partial role for Rad21 and Sa2 in Wnt4 domain insulation.
DISCUSSION
Here we illustrate how a tissue-specific transcriptional regulator (Wt1) controls the expression of a developmentally essential target gene (Wnt4) by modulation of the entire CTCF-defined locus. It is this domain-wide chromatin switch, which we term chromatin flip-flop, rather than differential binding of Wt1 to the Wnt4 regulatory sequences, that determines the outcome of Wt1 action in vitro and in vivo. This switch is reciprocal in a tissue-specific manner, underlying the bidirectional mode of gene regulation by Wt1. Wt1 is required for proper loading of CTCF and cohesin at the barrier sites. CTCF and cohesin binding at the barrier site is functional and responsible for insulating the chromatin milieu of the Wnt4 locus in an active state (kidney) or a repressive state (epicardium).
We propose chromatin flip-flop as a mechanism that might be generally used by transcriptional regulators in developmental processes with implications for hierarchical gene regulation. In this scenario, regulators like Wt1 control the accessibility of target loci to allow transcriptional control by other signals and pathways. In the kidney mesenchyme Wt1 would keep the Wnt4 locus open for activation by other signals, Wt1 loss would make the locus inaccessible for these signals. In the epicardium Wt1 could be keeping the Wnt4 locus closed, but here Wt1 loss could make it accessible for signals that are there to control loci other than Wnt4 in wild type epicardium. Our model accommodates the fact that other TFs may regulate Wnt4 through both Wt1-dependent and independent mechanisms in a context-dependent manner.
Wt1-mediated dichotomous Wnt4 gene regulation in space and time
Wt1 is not the only TF that regulates gene expression in a dichotomous fashion (Roberts and Green, 1995). For example, ERRα regulates osteopontin expression reciprocally in Hela versus ROS17/2.8 cells (Zirngibl et al., 2008). However, this mode of regulation was only shown in cell lines using in vitro assays and its role in vertebrate physiology and its implication for pathology remain unknown. Similarly, Wt1-mediated activation or repression of the same gene has previously only been described in vitro in ectopic over-expression systems (Hohenstein and Hastie, 2006). Here we show that in contrast to the direct activation of Wnt4 by Wt1 in the kidney mesenchyme, Wt1 loss in epicardial cells leads to ectopic activation of Wnt4 via the same WREs. Hence we conclude that Wt1 spatially regulates Wnt4 expression bidirectionally in kidney versus epicardium.
Additional data suggest that Wt1 regulates Wnt4 expression in a dichotomous manner even in different stages of nephron development (i.e. temporarily). Wt1 and Wnt4 expression briefly overlap during nephrogenesis; Wt1 is also expressed before and after the MET stage where Wnt4 expression is found, as we show here, under direct control of Wt1. We found that Basp1 occupies the Wnt4 A element (−11.7 kb) in embryonic kidneys isolated from E13.5 and E14.5 (data not shown). This would coincide with the stage Wnt4 expression is lost in the earliest (oldest) nephrons and suggests that Wt1 might switch to a Wnt4-repressing mode later during nephron development. However, as new nephrons in the mouse kidney keep on being induced even shortly after birth, at these later time points all possible stages of nephron development are found in the same kidney. Because of this we were unable to isolate material specifically from these later stages to characterise this further. This temporal regulation of gene expression would not be a unique feature of Wt1. Recently, it was shown that AML1 regulates PU.1 expression bidirectionally during haematopoietic system development so that AML1 represses PU.1 in embryonic development but activates it in the adult (Huang et al., 2008). Wt1 however could regulate reciprocal Wnt4 expression in vivo both spatially (kidney versus epicardium) and temporally (at different stages of kidney embryonic development).
Wt1-mediated chromatin flip-flop
In this study we propose chromatin flip-flop as reciprocal switching of chromatin structure between two insulating CTCF sites in a tissue specific manner and regulated by the same transcription factor (in this case Wt1). This mechanism has several unexpected features (Figure 8). First, Wt1 regulates this mechanism independent of differential DNA binding. Second, only Wt1, and not the associated cofactors that regulate Wnt4 expression and local chromatin, is necessary to switch the CTCF-demarcated Wnt4 locus in both tissues. Third, Wt1 interacts functionally with CTCF, and controls its loading and that of cohesin at the intergenic barrier sites. Fourth, CTCF and cohesin are functionally involved in maintaining the demarcated locus insulated. Fifth, CTCF and cohesin insulate the target locus directionally, so that upon their loss the neighbouring genes’ expression.
Our findings that the chromatin flip-flop is a switch from active to repressive chromatin (H3K27me3) or vice-versa point to a potential role for the polycomb-associated protein(s) that deposit or maintain the H3K27me3 mark. Recently, Xu et al have shown that Wt1 specifically binds and recruits Suz12 and EZH2 as well as DNA methylation regulator, DNMT1, to repress Pax2 expression (Xu et al., 2011).
In the past decade, our view of transcriptional regulation has changed from understanding mechanisms on a linear 2D scale to a role for chromosomal and nuclear organisation (or transcription in 3D). CTCF and cohesin have been implicated in regulating looping and localisation of chromosomal domains to different regions of the nucleus as well as domain insulation (Ohlsson et al., 2010). An interplay between nuclear organisation and gene expression mediated by CTCF and/or cohesin, is therefore a possibility (Ohlsson et al., 2010). However, our understanding of CTCF and cohesin function as insulators has not previously been validated and whether it is coupled to transcriptional regulation by TFs has not been elucidated. Here we showed a role for CTCF and cohesin in insulating of a locus to limit active and repressive chromatin spreading, and hence the locus accessibility with functional consequences for adjacent gene expression. A recent study investigating the effects of Ctcf loss in limb development (Soshnikova et al., 2010) supports our data. The authors note a slight change in genome-wide gene expression rarely beyond two fold increases or decreases. This would be consistent with our findings as the changes to Wnt4 and the neighbouring gene were below 2 fold either way. A close look at the list of genes they present in the supplementary tables S1A and S1B (Soshnikova et al., 2010) yield situations indicative of a chromatin insulation function for CTCF, where an expression of a gene is unchanged (e.g. Capn6 on chromosome (Chr) X and Uchl2 on Chr 1) but both of its neighbouring genes are either activated (Pak3 and Dcx on Chr X) or repressed (Troove2 and Rgs2 on Chr 1).
Chromatin flip-flop in physiology and pathology
Another implication of Wt1’s role in chromatin flip-flop is the potential activation of Wnt4 in epicardium upon Wt1 loss. Wnt4 is a signalling molecule that is essential for MET, and its ectopic activation in epicardium may affect the differentiation of epicardial progenitor cells. Our recent work has shown that Wt1 deletion in epicardium deregulates the EMT regulators Snai1 and Cdh1 leading to a block in cardiovascular differentiation (Martinez-Estrada et al., 2010), and the work described here suggests Wnt4 as another potential player in the epicardial phenotype in Wt1 knockout mice and human pathologies like Meacham syndrome (Suri et al., 2007).
Loss of Wt1 in mice results in disruption of multiple tissues and similarly inactivating mutations in humans leads to disease(s) associated with both kidney (Hohenstein and Hastie, 2006) and heart functions (Suri et al., 2007). At the same time, whereas loss of Wt1 helped identify its role as a tumour suppressor gene in Wilms’ tumours, other data suggests its ectopic activation in adult cancers could have oncogenic effects (Hohenstein and Hastie, 2006). The chromatin flip-flop mechanism could explain how loss or activation of Wt1 in different tissues might disrupt a delicate balance in the different bidirectional processes controlled by Wt1. This in turn results in the gene acting as a tumour suppressor gene in one context but oncogene in another.
EXPERIMENTAL PROCEDURES
Constructs
Wt1 expression constructs have been published previously (Martinez-Estrada et al., 2010). RNAi constructs for cell culture were made using the BLOCK-iT™ Pol II miR RNAi Expression Vector with EmGFP (Invitrogen) as per manufacturer’s instructions.
Cell culture
M15 Kidney mesenchymal cells, MEEC epicardial cells were described previously (Larsson et al., 1995; Martinez-Estrada et al., 2010). The CaCl2 transfection method was described previously (Essafi et al., 2005). For RNAi experiments GFP+ cells were isolated using a FACSaria II cell sorter (BD).
Mice
All animal experiments were approved by the University of Edinburgh ethical committee and all lines have been described previously (Martinez-Estrada et al., 2010; Tronche et al., 1999).
Reporter assays
ChIP+ fragments were cloned into a pGL4 plasmid and assayed and mutated as described previously (Essafi et al., 2005).
RNA isolation and analysis
RNA was isolated using Trizol (Invitrogen) following the manufacturers instructions. The cDNA synthesis was carried as described earlier (Martinez-Estrada et al., 2010) followed by SYBR green RTq-PCR analysis (Essafi et al., 2005).
ChIP
ChIP was carried as described before for cell lines (Essafi et al., 2005) and FACS-sorted cells (Goren et al., 2010), and quantified as described previously (Radoja et al., 2007).
Statistical error derivation
At least three biological replicates were used for each data point and were used for derivation of error bars as standard deviation from the mean for ChIP, reporter assays, RT-qPCR and FACS analysis experiments.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Elisabeth Freyer for FACS and Craig Nicol for illustrations. A.E. was funded by EuReGene, a Framework 6 program grant by the EU (05085) and is currently supported by a Beit Memorial fellowship for Medical Research. R.L.B. was supported by EuReGene and the Olson trust. S.F.B. was supported by Kidney Research UK. P.H. was supported by the Association for International Cancer Research and the MRC. J.A.D. was supported by the European Community’s 7th Framework program. HEALTH-F5-2008-223007 STAR-T-REK. All others were supported by the MRC.
Footnotes
SUPPLEMENTAL INFORMATION Supplemental Information includes Extended Experimental Procedures and Methods (including primers and oligos), 3 figures and is attached to this manuscript
COMPETING FINANCIAL INETERESTS The authors declare no competing financial interests.
References
- Aiden AP, Rivera MN, Rheinbay E, Ku M, Coffman EJ, Truong TT, Vargas SO, Lander ES, Haber DA, Bernstein BE. Wilms tumor chromatin profiles highlight stem cell properties and a renal developmental network. Cell Stem Cell. 2010;6:591–602. doi: 10.1016/j.stem.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bejerano G, Siepel AC, Kent WJ, Haussler D. Computational screening of conserved genomic DNA in search of functional noncoding elements. Nat Methods. 2005;2:535–545. doi: 10.1038/nmeth0705-535. [DOI] [PubMed] [Google Scholar]
- Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter B, Hill KJ, Charalambous M, Wagner KJ, Lahiri D, James DI, Andersen JS, Schumacher V, Royer-Pokora B, Mann M, et al. BASP1 is a transcriptional cosuppressor for the Wilms’ tumor suppressor protein WT1. Mol Cell Biol. 2004;24:537–549. doi: 10.1128/MCB.24.2.537-549.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JA, Ladomery M, Hohenstein P, Michael L, Shafe A, Spraggon L, Hastie N. Development of an siRNA-based method for repressing specific genes in renal organ culture and its use to show that the Wt1 tumour suppressor is required for nephron differentiation. Hum Mol Genet. 2004;13:235–246. doi: 10.1093/hmg/ddh015. [DOI] [PubMed] [Google Scholar]
- Essafi A, Fernandez de Mattos S, Hassen YA, Soeiro I, Mufti GJ, Thomas NS, Medema RH, Lam EW. Direct transcriptional regulation of Bim by FoxO3a mediates STI571-induced apoptosis in Bcr-Abl-expressing cells. Oncogene. 2005;24:2317–2329. doi: 10.1038/sj.onc.1208421. [DOI] [PubMed] [Google Scholar]
- Forni PE, Scuoppo C, Imayoshi I, Taulli R, Dastru W, Sala V, Betz UA, Muzzi P, Martinuzzi D, Vercelli AE, et al. High levels of Cre expression in neuronal progenitors cause defects in brain development leading to microencephaly and hydrocephaly. J Neurosci. 2006;26:9593–9602. doi: 10.1523/JNEUROSCI.2815-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goren A, Ozsolak F, Shoresh N, Ku M, Adli M, Hart C, Gymrek M, Zuk O, Regev A, Milos PM, et al. Chromatin profiling by directly sequencing small quantities of immunoprecipitated DNA. Nat Methods. 2010;7:47–49. doi: 10.1038/nmeth.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig S, Ho J, Pandey P, Macisaac K, Taglienti M, Xiang M, Alterovitz G, Ramoni M, Fraenkel E, Kreidberg JA. Genomic characterization of Wilms’ tumor suppressor 1 targets in nephron progenitor cells during kidney development. Development. 2010;137:1189–1203. doi: 10.1242/dev.045732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenstein P, Hastie ND. The many facets of the Wilms’ tumour gene, WT1. Hum Mol Genet. 2006;15(Spec No 2):R196–201. doi: 10.1093/hmg/ddl196. [DOI] [PubMed] [Google Scholar]
- Hosen N, Shirakata T, Nishida S, Yanagihara M, Tsuboi A, Kawakami M, Oji Y, Oka Y, Okabe M, Tan B, et al. The Wilms’ tumor gene WT1-GFP knock-in mouse reveals the dynamic regulation of WT1 expression in normal and leukemic hematopoiesis. Leukemia. 2007;21:1783–1791. doi: 10.1038/sj.leu.2404752. [DOI] [PubMed] [Google Scholar]
- Huang G, Zhang P, Hirai H, Elf S, Yan X, Chen Z, Koschmieder S, Okuno Y, Dayaram T, Growney JD, et al. PU.1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nat Genet. 2008;40:51–60. doi: 10.1038/ng.2007.7. [DOI] [PubMed] [Google Scholar]
- Kim H, Erickson B, Luo W, Seward D, Graber JH, Pollock DD, Megee PC, Bentley DL. Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat Struct Mol Biol. 2010;17:1279–1286. doi: 10.1038/nsmb.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MK, McGarry TJ, P OB, Flatow JM, Golden AA, Licht JD. An integrated genome screen identifies the Wnt signaling pathway as a major target of WT1. Proc Natl Acad Sci U S A. 2009;106:11154–11159. doi: 10.1073/pnas.0901591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kispert A, Vainio S, McMahon AP. Wnt-4 is a mesenchymal signal for epithelial transformation of metanephric mesenchyme in the developing kidney. Development. 1998;125:4225–4234. doi: 10.1242/dev.125.21.4225. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kreidberg JA, Sariola H, Loring JM, Maeda M, Pelletier J, Housman D, Jaenisch R. WT-1 is required for early kidney development. Cell. 1993;74:679–691. doi: 10.1016/0092-8674(93)90515-r. [DOI] [PubMed] [Google Scholar]
- Larsson SH, Charlieu JP, Miyagawa K, Engelkamp D, Rassoulzadegan M, Ross A, Cuzin F, van Heyningen V, Hastie ND. Subnuclear localization of WT1 in splicing or transcription factor domains is regulated by alternative splicing. Cell. 1995;81:391–401. doi: 10.1016/0092-8674(95)90392-5. [DOI] [PubMed] [Google Scholar]
- Li CM, Guo M, Borczuk A, Powell CA, Wei M, Thaker HM, Friedman R, Klein U, Tycko B. Gene expression in Wilms’ tumor mimics the earliest committed stage in the metanephric mesenchymal-epithelial transition. Am J Pathol. 2002;160:2181–2190. doi: 10.1016/S0002-9440(10)61166-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Estrada OM, Lettice LA, Essafi A, Guadix JA, Slight J, Velecela V, Hall E, Reichmann J, Devenney PS, Hohenstein P, et al. Wt1 is required for cardiovascular progenitor cell formation through transcriptional control of Snail and E-cadherin. Nat Genet. 2010;42:89–93. doi: 10.1038/ng.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AW, Schedl A, McInnes L, Doyle M, Hecksher-Sorensen J, Hastie ND. YAC transgenic analysis reveals Wilms’ tumour 1 gene activity in the proliferating coelomic epithelium, developing diaphragm and limb. Mech Dev. 1998;79:169–184. doi: 10.1016/s0925-4773(98)00188-9. [DOI] [PubMed] [Google Scholar]
- Negre N, Brown CD, Shah PK, Kheradpour P, Morrison CA, Henikoff JG, Feng X, Ahmad K, Russell S, White RA, et al. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 2010;6:e1000814. doi: 10.1371/journal.pgen.1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson R, Lobanenkov V, Klenova E. Does CTCF mediate between nuclear organization and gene expression? Bioessays. 2010;32:37–50. doi: 10.1002/bies.200900118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radoja N, Guerrini L, Lo Iacono N, Merlo GR, Costanzo A, Weinberg WC, La Mantia G, Calabro V, Morasso MI. Homeobox gene Dlx3 is regulated by p63 during ectoderm development: relevance in the pathogenesis of ectodermal dysplasias. Development. 2007;134:13–18. doi: 10.1242/dev.02703. [DOI] [PubMed] [Google Scholar]
- Roberts SG, Green MR. Transcription. Dichotomous regulators. Nature. 1995;375:105–106. doi: 10.1038/375105a0. [DOI] [PubMed] [Google Scholar]
- Sim EU, Smith A, Szilagi E, Rae F, Ioannou P, Lindsay MH, Little MH. Wnt-4 regulation by the Wilms’ tumour suppressor gene, WT1. Oncogene. 2002;21:2948–2960. doi: 10.1038/sj.onc.1205373. [DOI] [PubMed] [Google Scholar]
- Soshnikova N, Montavon T, Leleu M, Galjart N, Duboule D. Functional analysis of CTCF during mammalian limb development. Dev Cell. 2010;19:819–830. doi: 10.1016/j.devcel.2010.11.009. [DOI] [PubMed] [Google Scholar]
- Stark K, Vainio S, Vassileva G, McMahon AP. Epithelial transformation of metanephric mesenchyme in the developing kidney regulated by Wnt-4. Nature. 1994;372:679–683. doi: 10.1038/372679a0. [DOI] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- Suri M, Kelehan P, O’Neill D, Vadeyar S, Grant J, Ahmed SF, Tolmie J, McCann E, Lam W, Smith S, et al. WT1 mutations in Meacham syndrome suggest a coelomic mesothelial origin of the cardiac and diaphragmatic malformations. Am J Med Genet A. 2007;143A:2312–2320. doi: 10.1002/ajmg.a.31924. [DOI] [PubMed] [Google Scholar]
- Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, Diaz MO, Scacheri PC, Harte PJ. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development. 2009;136:3131–3141. doi: 10.1242/dev.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban PC, Bock R, Klein R, Schutz G. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- Wagner N, Wagner KD, Scholz H, Kirschner KM, Schedl A. Intermediate filament protein nestin is expressed in developing kidney and heart and might be regulated by the Wilms’ tumor suppressor Wt1. Am J Physiol Regul Integr Comp Physiol. 2006;291:R779–787. doi: 10.1152/ajpregu.00219.2006. [DOI] [PubMed] [Google Scholar]
- Wang W, Lee SB, Palmer R, Ellisen LW, Haber DA. A functional interaction with CBP contributes to transcriptional activation by the Wilms tumor suppressor WT1. J Biol Chem. 2001;276:16810–16816. doi: 10.1074/jbc.M009687200. [DOI] [PubMed] [Google Scholar]
- Xu B, Zeng DQ, Wu Y, Zheng R, Gu L, Lin X, Hua X, Jin GH. Tumor Suppressor Menin Represses Paired Box Gene 2 Expression via Wilms Tumor Suppressor Protein-Polycomb Group Complex. J Biol Chem. 2011;286:13937–13944. doi: 10.1074/jbc.M110.197830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirngibl RA, Chan JS, Aubin JE. Estrogen receptor-related receptor alpha (ERRalpha) regulates osteopontin expression through a non-canonical ERRalpha response element in a cell context-dependent manner. J Mol Endocrinol. 2008;40:61–73. doi: 10.1677/JME-07-0114. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.