Abstract
Staphylococcus aureus is a Gram-positive bacterial pathogen that causes serious infections which have become increasingly difficult to treat due to antimicrobial resistance and natural virulence strategies. Bacterial sortase enzymes are important virulence factors and good targets for future antibiotic development. It has recently been shown that sortase enzymes are integral to bacterial survival of phagocytosis, an underappreciated, but vital, step in S. aureus pathogenesis. Of note, the reaction mechanism of sortases relies on a solvent-accessible cysteine for transpeptidation. Due to the common strategy of oxidative damage employed by professional phagocytes to kill pathogens, it is possible that this cysteine may be oxidized inside the phagosome, thereby inhibiting the enzyme. This study addresses this apparent paradox by assessing the ability of physiological reactive oxygen species, hydrogen peroxide and hypochlorite, to inhibit sortase A (SrtA) from S. aureus. Surprisingly, we found that SrtA is highly resistant to oxidative inhibition, both in vitro and in vivo. The mechanism of resistance to oxidative damage is likely mediated by maintaining a high reduction potential of the catalytic cysteine residue, Cys184. This is due to the unusual active site utilized by S. aureus SrtA, which employs a reverse protonation mechanism for transpeptidation, resulting in a high pKa as well as reduction potential for Cys184. The results of this study suggest that S. aureus SrtA is able to withstand the extreme conditions encountered in the phagosome and maintain function, contributing to survival of phagocytotic killing.
Keywords: Sortase, Staphylococcus aureus, phagocytosis, oxidation, virulence, reverse protonation, reactive oxygen species
Gram-positive bacteria are a major cause of infectious disease, posing a serious healthcare threat. Staphylococcus aureus is a major pathogen, responsible for pathologies such as blood stream infections, surgical site infections, prosthesis infections, skin infections, and pneumonia(1, 2). Antibiotic resistance among pathogenic bacteria like methicillin-resistant Staphylococcus aureus (MRSA) is surging, with few new antibacterials expected in the near future(3). As such, new antibiotic targets are needed to combat this growing epidemic. The strategy of targeting virulence factors produced by pathogens has gained interest as a way to reduce potential resistance to new antibiotics, as well as the possibility for use in combination therapies(4). The Gram-positive bacterial extracellular transpeptidase, sortase, is an attractive target for such an approach.
Sortase enzymes are localized to the outer face of the bacterial membrane. These transpeptidases recognize secreted proteins with a cell wall sorting sequence and covalently attach these proteins to cell wall peptidoglycan substrates. This includes virulence factors known as Microbial Surface Components Recognizing Adhesive Matrix Molecules (MSCRAMMs) and pilin proteins(5, 6). Sortase A (SrtA), the “housekeeping” sortase, recognizes an LPXTG sequence in its substrates where X is any amino acid. Using a reverse protonation mechanism, the peptide bond between the threonine and glycine is cleaved by nucleophilic attack from an active site cysteine thiolate(7). The resulting thioester intermediate is resolved by a deprotonated amine of the crossbridge peptide in branched lipid II, covalently attaching the protein substrate to the cell wall precursor(8). This product is then incorporated into the peptidoglycan by transpeptidases and transglycosylases such that the anchored protein is displayed to the extracellular environment.
The tertiary structures of sortases are remarkably similar despite their highly varied substrate specificity(9). There is a conserved core structure containing the substrate binding cleft and conserved active site residues cysteine, histidine, and arginine(10). It was observed in a recent crystal structure of the Streptococcus pyogenes SrtA that the catalytic Cys208 in the active site (Cys184 in S. aureus SrtA) was oxidized to a stable sulfenic acid(11). Based on the importance of this residue for catalysis, the enzyme's extracellular location, and the fact that pathogens encounter reactive oxygen species (ROS) during the host immune response to infection, we hypothesized that this modification might bear physiological relevance. Oxidation of some proteins is a normal process that regulates enzyme activity(12, 13). However, in the case of extracellular enzymes there are no redox systems to reduce oxidized residues, implying that oxidation of SrtA is a form of damage that will inhibit the enzyme to the detriment of S. aureus.
It is well established that sub-populations of S. aureus are able to survive phagocytosis, and building evidence suggests that this is important for staphylococcal pathogenesis(14-17). Intracellular survival within nonphagocytic cells as well as professional phagocytes has been demonstrated, and this provides a vehicle for S. aureus to remain cloaked from the immune system and antibiotics, as well as disseminate throughout the host(14, 17, 18). However, without a functioning sortase this ability is lost(14). To investigate whether oxidative modification of SrtA is important during the clearance of an infection by phagocytes, we assessed the ability of ROS to inhibit S. aureus SrtA in vitro and in vivo. We determined that SrtA is highly resistant to inhibition by oxidation from hydrogen peroxide and hypochlorite, prevalent ROS in the phagosome, beyond concentrations where staphylococcal growth becomes inhibited. The combination of the high reduction potential of cysteine in the thiol form and the reverse protonation mechanism provides SrtA with a mechanism of resistance to oxidation, allowing SrtA to be active in the phagosomal environment. Accordingly, S. aureus SrtA function in vivo is not observably affected by high concentrations of ROS. The data presented here demonstrates that S. aureus SrtA Cys184 is intrinsically resistant to oxidation, which contributes to the ability of S. aureus to survive phagocytotic killing.
Experimental Procedures (Materials and Methods)
Protein and Peptide Production and Purification
Recombinant S. aureus SrtAΔN24 was expressed and purified as previously described with minor alterations(19). An Abz-LPETGG-Dap(DNP)-NH2 peptide substrate was synthesized and purified as previously described(20), except synthesis was performed using a CEM Liberty microwave peptide synthesizer.
Inhibition Kinetics
SrtA activity was measured using a previously developed HPLC-based assay(21). Briefly, dose-response inhibition assays (IC50 determination) were performed as follows: 1 μM SrtA and various concentrations of oxidant were preincubated for 2 h in 150 mM NaCl, 5 mM CaCl2, 300 mM Tris pH 7.5 buffer (final concentrations) at 37 °C. Oxidant concentrations were determined spectrophotometrically. NaOCl has an extinction coefficient of 350 M-1 cm-1 at 290 nm, and H2O2 has an extinction coefficient of 43.6 M-1 cm-1 at 240 nm(22, 23). The reaction was initiated by addition of Abz-LPETGG-Dap(DNP)-NH2 (1 mM final concentration) and H-(Gly)5-OH (2 mM final concentration) to obtain a final reaction volume of 100 μL. In the case of hypochlorite, sodium bisulfite (equal concentration as NaOCl) was also added to scavenge the residual hypochlorite. The reaction was quenched after 10 min by addition of half volume of 1.2 M HCl. Reactions were run on an Agilent 1200 HPLC with a Vydac reversed-phase C18 column (4.6 × 50 mm, 3 μm). Cleavage product and substrate peaks were measured using UV absorption of the DNP group (λmax = 355 nm). Peaks were integrated with PeakFit v4.11 (Systat Software Inc). Reaction rates were calculated and fit in GraFit 6.0.1 (Erathicus Software Limited) to obtain an IC50 value using equation 1:
| (1) |
The time-dependent inhibition kinetics experiments to determine KI and kinact were performed as follows: SrtA and various concentrations of oxidant were preincubated at 37 °C for various amounts of time and the reaction was initiated as above by addition of Abz-LPETGG-Dap(DNP)-NH2 and H-(Gly)5-OH (and sodium bisulfite for hypochlorite assays). Reactions were quenched after 10 min with 1.2 M HCl and analyzed by HPLC by the above method. Reaction rates were calculated and fit in GraFit 6.0.1. Due to the non-specific and complex reactions that ROS undergo with proteins, exact kinetic mechanism determination is extremely difficult. Thus, a more general approach to curve-fitting was employed. The first equation utilized is a shifted inverse logistic function:
| (2) |
Where ν is the velocity of the reaction, ν0 is the velocity of the uninhibited reaction, kobs is the apparent first order rate constant for the interconversion of νuninhibited and νinhibited, t is preincubation time, and x is a time offset. This equation was used to account for the sigmoidal shape of time-dependent inhibition of SrtA seen with H2O2 and NaOCl. The origin of the lag phase seen in these experiments is likely multifaceted and due to some combination of low levels of oxidation of Cys184 prior to assay initiation, conversion of Cys184 between thiol and thiolate, conversion of NaOCl between hypochlorite and hypochlorous acid, and protein dynamics inherent in the SrtA active site. The 100 mM H2O2 sample was instead fit to a shifted exponential decay function because the inhibition at this concentration was too rapid to observe a lag phase and accurately fit to the above logistic function:
| (3) |
Where the variables are the same as above(24). The kobs values obtained for each oxidant concentration were then plotted vs. oxidant concentrations and fit in GraFit 6.0.1 to:
| 4 |
Where kinact is the rate of inactivation of the enzyme by the inhibitor, [I] is the concentration of inhibitor, and is the apparent concentration of the inhibitor required to reach half maximal rate of inactivation of the enzyme(24).
S. aureus Minimal Inhibitory Concentration Measurement
The minimal inhibitory concentrations (MIC) of H2O2 and NaOCl were determined for S. aureus strain Newman. Tryptic Soy Broth (TSB) cultures (5 mL) were inoculated with 50 μL of an overnight stationary-phase culture. The cultures were grown to mid-log phase shaking at 37 °C until OD600 = 0.7. The culture (10 μL) was diluted into 190 μL of TSB in a 96-well plate with final concentrations of oxidant varying from 0-10 mM. Cultures were grown at 37 °C in a humidified shaker and checked for growth at 24 and 48 h. The MIC was defined as the lowest concentration of ROS at which no growth was observed.
LC/MS Analysis of Oxidation
Recombinant SrtAΔN24 was treated with excess Cleland's Reductacryl™ Reagent (Calbiochem) at 37 °C for 45 min to ensure Cys184 was fully reduced. Reductacryl™ was then removed by centrifugation. Samples were adjusted to 150 mM NaCl, 5 mM CaCl2, 300 mM Tris pH 7.5 and 1 mM H2O2 or 1 mM NaOCl, and incubated at 37 °C for 2 h to oxidize Cys184. Catalase beads and equimolar NaHSO3 were added at 37 °C for 30 min to remove excess H2O2 and NaOCl, respectively. Free thiols were blocked with 20 mM iodoacetamide at ambient temperature in the dark for 1 h to prevent further reaction. Samples were buffer exchanged into 50 mM NH4CO3 pH 8 (AmBic) and concentrations were determined by Bradford assay (Biorad) using BSA as calibration standard. 10 μg from each sample treatment were removed and volumes were normalized with AmBic. Waters RapiGest® MS compatible surfactant was added to 0.1 % w/v final for solubilization. Promega trypsin (sequencing grade) was added at 50:1 protein:trypsin and proteins were allowed to proteolytically digest at 37 °C overnight. Trifluoroacetic acid (TFA) and acetonitrile (ACN) were added to each digest the following morning to yield 1 % TFA/2 % ACN final.
LC/MS Data Collection
Peptide digests obtained from each of the 3 treatments were analyzed using a nano-Acquity UPLC system coupled to a Synapt G2 HDMS mass spectrometer (Waters Corp, Milford, MA). Approximately 5 ng of peptide material in 1 μL was first trapped at 5 μL/min for 3 min in 99.9 % water with 0.1 % v/v formic acid on a 20 μm × 180 mm Symmetry C18 column. Separations were then performed on a 75 μm × 250 mm column with 1.7 μm C18 BEH particles (Waters) using a 60-min gradient of 5 to 40% acetonitrile with 0.1 % formic acid at a flow rate of 0.4 μL/min and 55 °C column temperature. We conducted one data-dependent analysis (DDA) per sample in sensitivity mode (∼15,000 Rs) using a 0.6 sec MS scan followed by MS/MS acquisition on the top 3 ions with charge greater than 1. MS/MS scans for each ion used an isolation window of 2.3 Da, a maximum of 3 seconds per precursor, and dynamic exclusion for 120 seconds within 1.2 Da.
LC-MS/MS Data Interpretation
Raw LC-MS/MS data were processed in Mascot distiller v2.3.2.0 (Matrix Science, Inc) and then submitted to the Mascot v2.2 search engine. Data were searched against the NCBInr database with eubacteria taxonomy with variable modifications set to carbamidomethyl (C); deamidation (NQ); single, double, and triple oxidation (C); pyroglutamic acid (N-term); and oxidation (M). Data were searched with 10 ppm precursor mass tolerance and 0.04 Da mass tolerance for product ions. The maximum number of missed cleavages was set at 2 and enzyme specificity was trypsin. Database search results and spectra have been uploaded as a Scaffold 3 file (Proteome Software, Inc) at the following link https://discovery.genome.duke.edu/express/resources/1794/1794_042011_final.sf3. Peak intensities were manually extracted from the raw data using MassLynx 4.1 (Waters) with a 20 ppm window around precursors of interest, and plots were generated within Microsoft Excel (see Table S1).
Reduction Potential Determination
Electrodes for reduction potential measurements were prepared as described previously(25). Briefly, a glassy carbon stick electrode (Bioanalytical Systems) was polished with 0.3 μm alumina, sonicated in water and isopropanol, and primed with a dilute solution of single-walled carbon nanotubes (NanoC) in water and dihexadecylphosphate surfactant (Sigma) that increases electrode surface area, resulting in signal enhancement. Diffusional cyclic voltammetry was used to measure the reduction potential of L-cysteine. A 5 mM solution of L-cysteine in 50 mM sodium phosphate (pH 3-11) was used to fill the three-electrode electrochemical cell, utilizing Ag/AgCl as reference electrode, Pt as auxiliary electrode, and carbon nanotube-modified glassy carbon as working electrode. Electrochemical analysis was performed using a PAR 273A potentiostat/galvastat (Princeton Applied Research, Oak Ridge, TN). Buffers were degassed prior to use. Measurements were done at 4 ± 2 °C at scan rates of 50 mV/s in the oxidative direction with a scan range of 0 V to +1.3 V to observe conversion of cysteine thiol to cysteine radical. Electrode potentials were recorded vs. the reference Ag/AgCl and reduction potentials were reported relative to the standard reference of normal hydrogen electrode, +0.197 V.
For SrtA reduction potential determination, recombinant SrtAΔN24 was mixed with CNTs and applied to electrodes prepared as above. SrtAΔN24 has only a single cysteine, the conserved active site nucleophile, Cys184. After drying at -20 °C, electrodes were sealed with 5 % Nafion in MeOH, which does not disrupt native protein structure(26). After drying again at -20 °C, measurements were performed in a three-electrode electrochemical cell as above.
S. aureus Protein A Anchoring
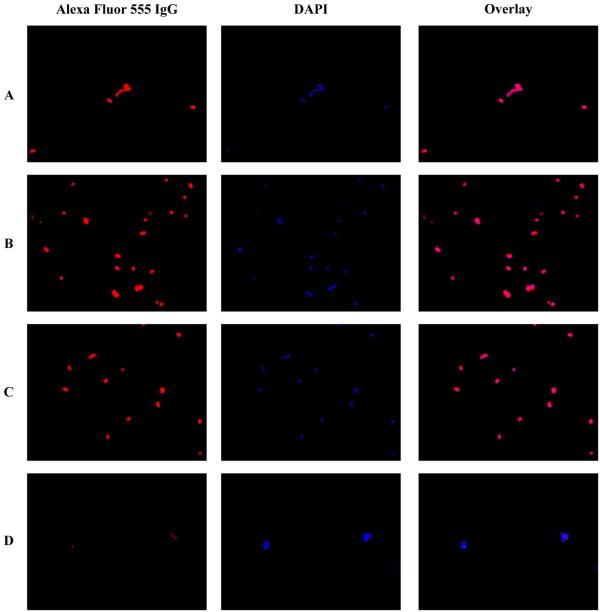
S. aureus strain Newman or ΔSrtA was grown in TSB media overnight. Cultures were diluted 1:100 in TSB that was adjusted to 1 mM H2O2, 1 mM NaOCl, pH 5, pH 7, or pH 9. Cultures were allowed to grow to mid-log phase, OD600 = 0.6, before harvesting. The concentration of H2O2 was confirmed via an Amplex Red hydrogen peroxide peroxidase assay and was not found to be significantly affected by the TSB media. Cells were pelleted, washed with PBS, and resuspended in PBS. Dilutions were plated on lysine-coated glass slides, spread, flamed, and allowed to dry. Slides were rinsed with PBS, blocked with 2 % BSA in PBS, and rinsed again with PBS. Finally, samples were incubated with 1:1000 dilution of Alexa Fluor 555 goat anti-mouse IgG (Invitrogen) in 2 % BSA for 1 h in the dark, rinsed with PBS, and allowed to dry. Slides were mounted with Dapi-Fluoromount-G (SouthernBiotech) and examined with a Zeiss Axio Imager widefield fluorescence microscope.
Results
Reactive Oxygen Species Inhibition
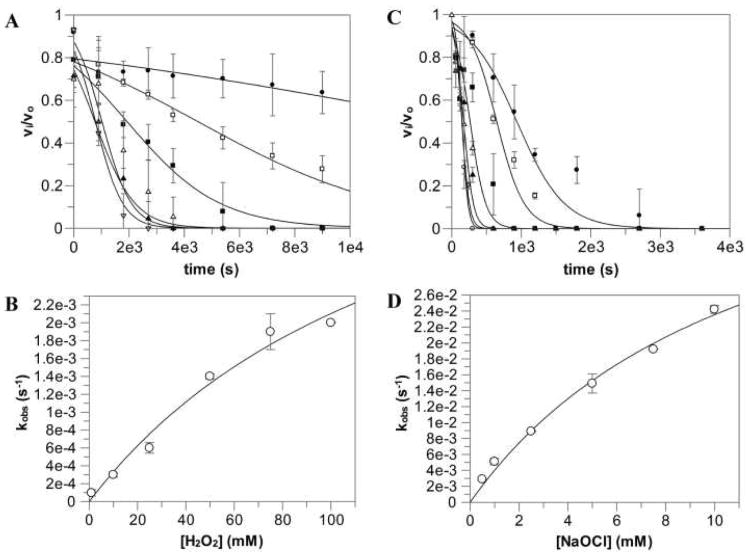
To assess the effects of the phagocyte oxidative burst on SrtA activity, purified S. aureus SrtAΔN24 was examined for sensitivity to reactive oxygen species (Figure 1). The kinetic inhibition values obtained can be found in Table 1. Somewhat surprisingly, SrtA is resistant to inhibition by ROS with a KI in the millimolar range for hydrogen peroxide and sodium hypochlorite, both found to be slow time-dependent inhibitors with kinact values of 5.2×10-3 and 5.1×10-2 s-1, respectively. Additionally, the KI value obtained for NaOCl is in the same range as its MIC of 7.5 mM for S. aureus, implying that S. aureus growth would be halted before detrimental inhibition of SrtA occurred (Table 1). Conversion of NaOCl and O2 into NaO3Cl could potentially contribute to inhibition seen by NaOCl. However, it was found that up to 5 mM NaO3Cl had no effect on SrtA activity even after 30 min (data not shown), indicating that inhibition was most likely entirely due to NaOCl. The KI for H2O2 is orders of magnitude higher than the MIC of 2.5 mM, as well as the micromolar concentrations that can be found in the phagosome(27). Likewise, the IC50 values imply that H2O2 is not a relevant inhibitor, while NaOCl is able to inhibit SrtA at physiological concentrations, but only after a prolonged incubation period (Figure S1, Table 1).
Figure 1.
Time-dependent inhibition kinetics of S. aureus SrtA. νi/ν0 vs time graphs were fit to obtain kobs values and kobs vs [ROS] graphs were fit to obtain K1 and kinact. H2O2 concentrations were 1 mM, 10 mM, 25 mM, 50 mM, 75 mM, and 100 mM. NaOCl concentrations were 500 μM, 1 mM, 2.5 mM, 5 mM, 7.5 mM, and 10 mM. (A) H2O2 kinetic data, (B) H2O2 kobs fit, (C) NaOCl kinetic data, (D) NaOCl kobs fit.
Table 1.
Inhibition Kinetics for S. aureus SrtA and Minimal Inhibitory Concentrations for S. aureus of H2O2 and NaOCl.
| KI (mM) | kinact (s-1) | kinact/KI (M-1s-1) | IC50 (mM) | MIC (mM) | |
|---|---|---|---|---|---|
| H2O2 | 145.0 ± 74.2 | (5.2 ± 1.7) × 10-3 | (3.5 ± 2.3) × 10-8 | 4.3 ± 0.3 | 2.5 |
| NaOCl | 11.8 ± 3.1 | (5.1 ± 0.8) × 10-2 | (4.3 ± 2.7) × 10-6 | (3.6 ± 0.3) × 10-2 | 7.5 |
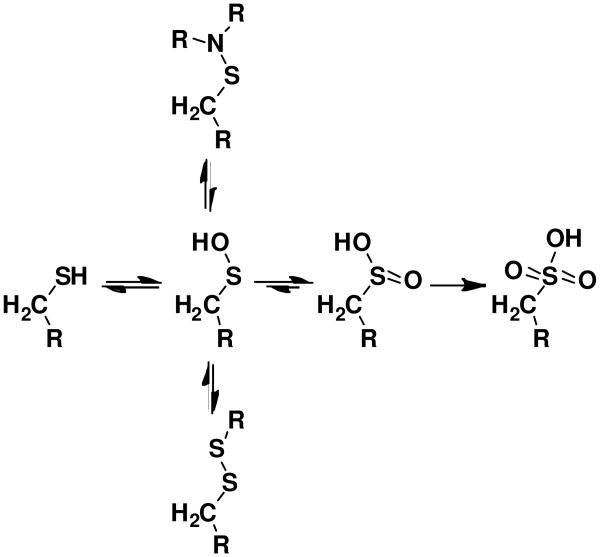
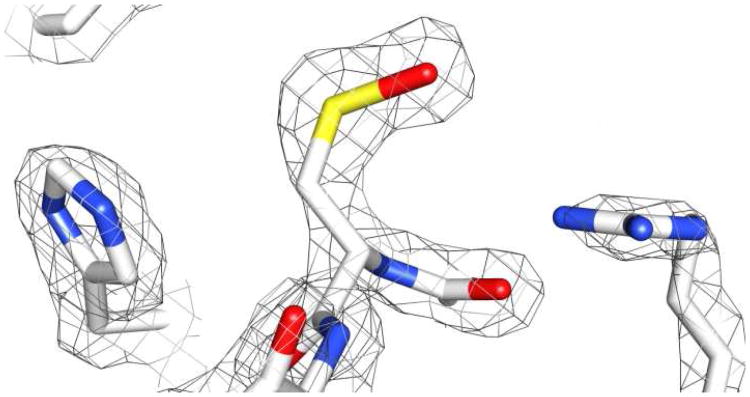
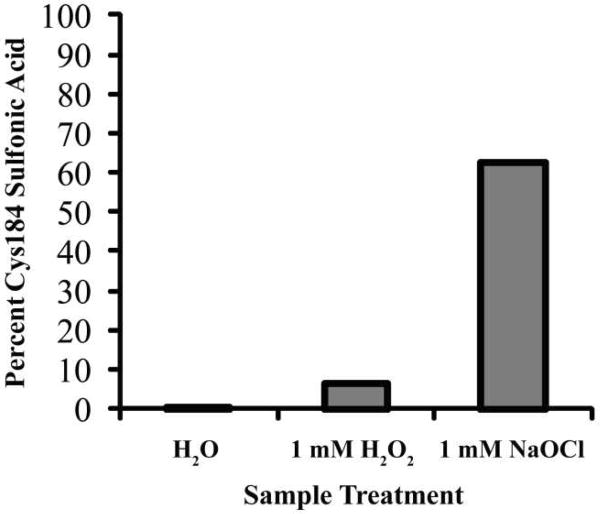
Cysteines can be oxidized to a number of states, including sulfenic, sulfinic, and sulfonic acid forms (Scheme 1). A recent crystal structure of Streptococcus pyogenes SrtA displayed a stable sulfenic acid at Cys208 (Cys184 in S. aureus SrtA) (Figure 2). To support our hypothesis that the inhibition of S. aureus SrtA is due to direct oxidation of Cys184, we incubated recombinant SrtA with 1 mM H2O2 or 1 mM NaOCl for 2 h and analyzed the tryptic digest by LC/MS. Sulfonic acid formation was observed at Cys184, implying oxidation at the active site cysteine can occur and is likely the cause of inhibition (Figure 3). NaOCl converted a higher fraction of Cys184 to sulfonic acid than H2O2, while a control sample showed no oxidation under these conditions. This supports a low rate of oxidation of Cys184 and a model of SrtA inhibition by Cys184 oxidation in the presence of NaOCl, with H2O2 being a largely ineffective physiological inhibitor. No sulfenic or sulfinic acid formation was observed under these conditions. While a stable sulfenic acid modification was discovered at S. pyogenes SrtA Cys208(11), this was likely an artifact of crystallization, and the fully oxidized cysteine observed at S. aureus SrtA Cys184 in this study is expected for unprotected cysteine oxidation. Of note, we did not observe any disulfide formation, discounting the possibility of intermolecular Cys184-Cys184 dimerization artifacts during treatment or analysis.
Scheme 1. Oxidation States of a Cysteine Residue.
Double arrows indicate physiogically reversible steps. Sulfinic acid formation has only been found to be reversible in a few specific cases when catalyzed by the enzyme sulfiredoxin(49). Disulfide formation is employed by enzymes like AhpC and sulfenyl amide formation by enzymes like PTP1B to protect the enzymes from irreversible oxidation. 82×77mm (600 × 600 DPI)
Figure 2.
Active site residues (His142, Cys208, and Arg216) of Streptococcus pyogenes SrtA crystal structure with Cys208 oxidized to a sulfenic acid (PDB ID 3FN6). 82 × 43mm (450 × 450 DPI)
Figure 3.
LC-MS/MS analysis of S. aureus SrtAΔN24 after oxidation by H2O2 and NaOCl. The percent of Cys184 oxidized to the sulfonic acid form was calculated based on the sum of all peak intensities containing free Cys 184 or triply-oxidized Cys184 for each of three treatment conditions (water, 1 mM H2O2, or 1 mM NaOCl for 2 h). Water showed no oxidation, H2O2 only 6 %, and NaOCl 63 % oxidation.
S. aureus Protein A Anchoring
In order to examine the effect on ROS on SrtA activity in vivo, we investigated the ability of H2O2 and NaOCl to inhibit SrtA activity in S. aureus strain Newman. An Alexa Fluor 555 conjugated IgG was used to detect the presence or absence of protein A on the surface of S. aureus. NaOCl and H2O2 had no observable effect on protein A anchoring, supporting our hypothesis that SrtA resistance in vitro may be paralleled by resistance to oxidation in vivo during infection (Figure 4).
Figure 4.
Analysis of Protein A anchoring in S. aureus strain Newman grown in TSB media at pH 7 under different conditions. Rows: (A) No ROS, (B) 1 mM H2O2, (C) 1 mM NaOCl, (D) S. aureus ΔSrtA.
Reduction Potential of Cysteine
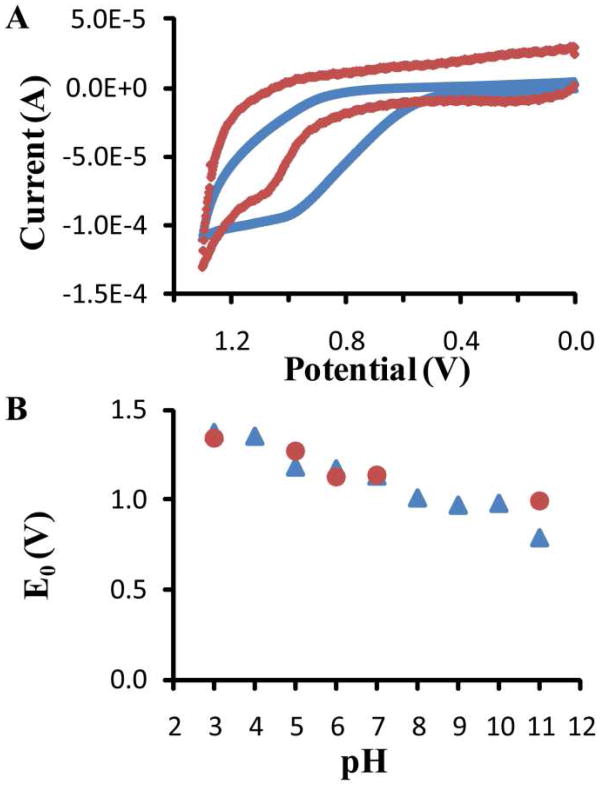
To understand why SrtA is resistant to ROS inhibition in vitro and in vivo even though Cys184 is a solvent-accessible cysteine, we measured the reduction potential. At pH 5, L-cysteine has a reduction potential of 1.18 V and SrtAΔN24 has a reduction potential of 1.27 V (Figure 5A). The reduction potential of SrtAΔN24 is likely in part due to Cys184, although deconvolution of the contribution to the reduction potential of different residues in SrtA is difficult. Indeed, the reduction potential of L-cysteine supports a reduction potential in this range for SrtAΔN24 Cys184. Hydrogen peroxide has a reduction potential of 1.77 V and hypochlorous acid has a reduction potential of 1.50 V at this pH(28). Since a compound with a higher reduction potential is able to oxidize a compound with a lower reduction potential, both NaOCl and H2O2 should be thermodynamically capable of oxidizing cysteine at pH 5. However, inhibition assays were performed at pH 7.5, which is the pKa of hypochlorite(29). Hypochlorite is a much less powerful oxidant than hypochlorous acid, with a reduction potential of 0.89 V(30). With that low of a reduction potential, hypochlorite would be thermodynamically unable to oxidize Cys184 of SrtA. However, inhibition assays at pH 6.5 and 8.5 showed no significant change in the IC50 of hypochlorite compared to at pH 7.5, implying that this does not fully account for the slow rate of inhibition (data not shown). Accordingly, hydrogen peroxide and hypochlorous acid are both two-electron oxidants, and for this class of oxidants, the oxidation rates are more indicative of physiological oxidation potential(28). While the kinact values for NaOCl and H2O2 inhibition are not large (Table 1), NaOCl has a kinact that is ten times larger than H2O2. This analysis predicts that NaOCl is a more rapid oxidizer of cysteine than H2O2, which is consistent with published data(31, 32).
Figure 5.
Cysteine reduction potential. (A) Representative cyclic voltammogram of L-cysteine (blue) and SrtAΔN24 (red) at pH 5 used to determine reduction potential. (B) Pourbaix diagram showing pH-dependence of the measured reduction potential of L-cysteine (blue ▲) and SrtAΔN24 (red ●) in 50 mM phosphate buffered solution at 4 °C over pH 3-11.
Part of the explanation of the resistance of SrtA to inhibition by oxidation may lie in the nature of cysteine. The pH dependence of the reduction potential observed in this study is expected as cysteine transitions from thiolate to thiol (Figure 5B). The rate of oxidation of a cysteine is known to depend on the protonation state of the thiol(33-35). Due to the increased reactivity of thiolates, they are more prone to oxidation than thiols. The measured reduction potentials at pH 7 were 1.13 V for L-cysteine and 1.14 for SrtAΔN24. Free L-cysteine has a pKa of 8.2, implying that a higher reduction potential for SrtA Cys184 is appropriate.
Discussion
Treatment of Staphylococcus aureus infections is complicated by issues of antimicrobial resistance and metastases in a significant portion of cases due to persistence. Persistence of S. aureus infections is linked to survival of intracellular subpopulations of bacteria, most notably within phagosomal compartments of phagocytes. Once intracellular, S. aureus is believed to undergo phenotypic switching to a small colony variant (SCV) persistent form(16, 17), which is associated with increased anchoring of fibronectin-binding proteins and clumping factors on the bacterial surface(36). These proteins are SrtA substrates, implying the requirement for an active sortase in order for a switching strategy to be effective.
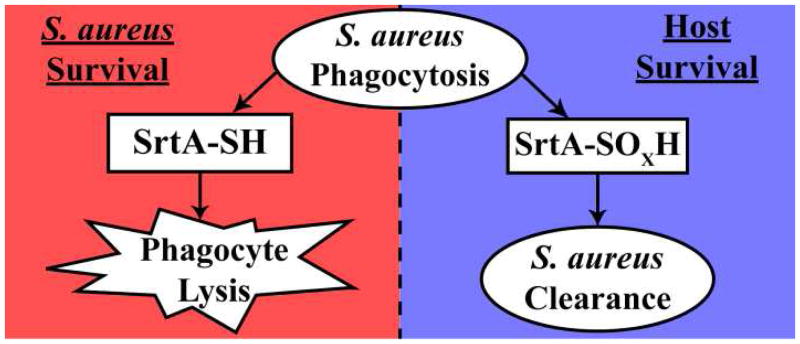
Accordingly, deletion of sortase enzymes results in drastically reduced virulence in multiple Gram-positive bacterial species(37). While S. aureus sortase activity is already known to be important at various stages of infection, only recently has its contribution to and the importance of phagocytosis survival become apparent(14, 38). In a recent study by Kubica et al., a SrtA knockout strain of S. aureus showed severely impaired ability to survive phagocytotic killing by macrophages(14). The anchoring of more than sixteen cell-surface substrates by SrtA in S. aureus appears to play a direct role in this phenomenon. Coupled with our observation of a stable Cys208 sulfenic acid in a recent crystal structure of the Streptococcus pyogenes SrtA enzyme (Figure 2)(11), we postulated that the interaction of reactive oxygen species and SrtA Cys184 within the phagosome might influence the intracellular survival of S. aureus (Figure 6).
Figure 6.
Model of SrtA oxidation during phagocytosis of S. aureus. Oxidation of SrtA (to sulfenic, sulfinic, or sulfonic acid) inside phagocytes results in inhibition of SrtA and death of S. aureus. To combat this, SrtA is resistant to oxidation, allowing S. aureus to survive within the phagocyte. 82×34mm (600 × 600 DPI)
In this study, we investigated the effect that the hostile environment of the phagosome has on S. aureus SrtA activity. LC/MS analysis of ROS-treated S. aureus SrtA supported our hypothesis that oxidation of Cys184 can be forced to occur, confirming sulfonic acid formation at Cys184 when treated with H2O2 or NaOCl (Figure 3). Surprisingly, SrtA is highly resistant to inhibition by the ROS H2O2 and NaOCl (Table 1), with a KI of 145.0 mM and 11.8 mM, respectively. The KI for these ROS are significantly higher than their MIC for S. aureus, rendering SrtA effectively resistant to oxidation by these species at meaningful concentrations (Table 1).
In addition, inhibition by ROS is a slow reaction, with kinact of 5.2 × 10-3 s-1 and 5.1 × 10-2 s-1 for H2O2 and NaOCl, respectively. kinact/KI is often used as a measure of irreversible time-dependent inhibitor potency. By this measure, NaOCl is a 120-fold better inhibitor than H2O2, at 4.3 × 10-6 M-1s-1. For comparison, these values are 4-6 orders of magnitude less potent than phenyl vinyl sulfones, irreversible inactivators of SrtA(39), while the IC50 values are very similar (Figure S1, Table 1). The IC50 value of 36.3 μM for NaOCl indicates that given enough time, NaOCl is capable of inhibiting SrtA at a physiologically relevant concentration. More importantly, NaOCl is the predominant ROS in the phagosome, reaching levels capable of killing S. aureus(27). However, S. aureus treated with 1 mM NaOCl showed no observable defect in SrtA activity in vivo (Figure 4C), while S. aureus treated with 7.5 mM NaOCl were unable to grow (Table 1). It is not yet clear whether phagosomal NaOCl concentrations are maintained at or beyond the timeframe required by kinetic studies to oxidize SrtA at levels that affect its function in vivo. In addition, we observed that H2O2 also had no appreciable effect on SrtA activity in vivo (Figure 4B). These findings are supported by LC/MS analysis, demonstrating that NaOCl was able to partially convert Cys184 to the sulfonic acid form, whereas H2O2 was largely unable to do so (Figure 3). Thus, we conclude that NaOCl is a possible, if inefficient, inhibitor in the phagosome, while H2O2 most likely is ineffectual.
In addition to the resistance SrtA displays toward oxidation, S. aureus has many mechanisms to deal with ROS, including catalase, alkyl hydroperoxide reductase, and superoxide dismutases to detoxify hydrogen peroxide and superoxide anion (40, 41). S. aureus also produces a pigment molecule, staphyloxanthin, capable of scavenging ROS(42, 43). Additionally, the thick peptidoglycan layer, especially pronounced in vancomycin-resistant strains, and the multitude of proteins in the cell wall and membrane provide many targets more susceptible to oxidation than SrtA, in essence sacrificially titrating and diluting the killing potential of ROS by reducing their concentrations within the phagosome. Collectively, these data point to the maintenance of SrtA activity in host degradative environments.
Previous oxidation studies of enzymes with solvent exposed cysteines have focused on enzymes whose functions are regulated by ROS or involved in redox homeostasis. These proteins, such as PTP1B(13) and AhpC(12, 44), are highly susceptible to oxidation but have intrinsic protection mechanisms to avoid irreversible oxidation such as forming an intramolecular sulfenyl amide or disulfides (Scheme 1). Other cysteine hydrolases (e.g. caspases) are often potently inhibited by H2O2, sometimes with IC50 values in the low micromolar range(45). Furthermore, ROS exposure can inhibit cellular apoptosis through redox control of caspases via direct oxidation of the catalytic cysteine nucleophile(46).
In contrast, the catalytic residue Cys184 of SrtA appears to be unusually resistant to oxidation compared to related cysteine thiol-containing enzymes, even though solved structures show Cys184 to be readily accessible to solvent(10). We believe that this resistance is partially achieved by an unusually high reduction potential of Cys184 created by the unique protein environment. The measured reduction potential at pH 7 of free L-cysteine (1.13 V) and of SrtAΔN24 (1.14 V) are almost identical, implying that the reduction potential of Cys184 is not depressed, as is the case with active site cysteine residues in many other enzymes. While this reduction potential makes Cys184 more difficult to oxidize, it should be noted that the reduction potentials of 1.0-1.3 V of SrtA observed across the physiological pH range does not preclude oxidation by physiological oxidants, many of which have reduction potentials higher than 1.3 V. However, whereas Cys184 is thermodynamically capable of oxidation by H2O2 and NaOCl, it is clearly kinetically limited on meaningful physiological time scales for concentrations that do not drastically affect S. aureus viability.
It is interesting to note that cysteines that are easily oxidized tend to have an abnormally low pKa. The pKa of the catalytic cysteine of AhpC is 5.8(44), the pKa of PTP1B is 5.6(47), and caspases appear to have pKa around 6.5(48). This means that at physiological pH most catalytic cysteines are predominantly in the thiolate form, increasing their nucleophilicity and activity, but also making them more susceptible to oxidation(33, 34). This observation has two consequences for the current study. First, this is part of the explanation for the inverse relationship of pH and reduction potential for cysteine that predisposes cysteines to resist oxidation by ROS under normal conditions. Second, this supports our hypothesis that SrtA needs to be resistant to oxidation in order to survive in toxic host environments even though it has a solvent accessible catalytic cysteine. This is partially accomplished through the unusual reverse protonation mechanism employed by SrtA, where the catalytic cysteine (Cys184) has a pKa of 9.4 and the conserved histidine (His120) has a pKa of 6.2(7). Thus, Cys184 is predominantly in the thiol form and therefore has a high reduction potential (Figure 5). Incidentally, this also contributes to enzyme activity over a broad range of pH, another trait important for activity in multiple host environments including the phagosome. Indeed, S. aureus SrtA activity seems to be unaffected in vivo at pH 5 or 9 (Figure S2).
Our results provide for the first time a biochemical basis for SrtA contribution to S. aureus phagocytotic survival. By design, SrtA has acquired a combination of architectural and mechanistic elements that coordinately protect the enzyme from biological inhibition. Indeed, SrtA is intrinsically resistant to concentrations of ROS that approach or greatly exceed those that cause bacterial cell death in vitro, and S. aureus gains an evolutionary advantage by maintaining SrtA activity in the acidic environment of the phagosome in order to remodel the cell wall as part of an intracellular phenotypic switch. This remodeling is likely essential to the ability of S. aureus to persist intracellularly. It is unclear the scope to which phenotypic switching is beneficial, but nonetheless this survival behavior likely plays a role in avoidance of immune detection, protection against antibiotic exposure, and access to host cell nutrients. Certainly, many viruses such as herpes and HIV and bacteria such as Mycobacterium tuberculosis persist long term intracellularly within host immune defense cells, so it is not surprising that S. aureus may adopt a qualitatively similar persistence mechanism. With sortases like S. aureus SrtA playing such a critical role in Gram-positive bacterial virulence through cell wall protein anchoring of MSCRAMMs and virulence factors, resistance to phagocytotic killing, and putative phenotypic switching, this enzyme takes on additional importance as an anti-virulence antimicrobial target.
Supplementary Material
Acknowledgments
We thank T. Prest for technical assistance with microscopy and E. J. Soderblom and McCafferty lab members for critical comments and advice during preparation of this manuscript.
Footnotes
This work was supported, in whole or in part, by National Institutes of Health Grant AI46611.
Supporting Information Paragraph: Figures S1 and S2 and Table S1. Figure S1 shows IC50 results for H2O2 and NaOCl for SrtA. Figure S2 shows Protein A anchoring by S. aureus strain Newman in pH 5 and pH 9. Table S1 contains the raw data from LC/MS analysis of oxidation of SrtA by H2O2 and NaOCl. This material is available free of charge online at http://pubs.acs.org.
References
- 1.Ippolito G, Leone S, Lauria FN, Nicastri E, Wenzel RP. Methicillin-resistant Staphylococcus aureus: the superbug. Int J Infect Dis. 2010;14(4):11. doi: 10.1016/j.ijid.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 3.Song JH. What's new on the antimicrobial horizon? Int J Antimicrob Agents. 2008;32(4):S207–213. doi: 10.1016/S0924-8579(09)70004-4. [DOI] [PubMed] [Google Scholar]
- 4.Clatworthy AE, Pierson E, Hung DT. Targeting virulence: a new paradigm for antimicrobial therapy. Nat Chem Biol. 2007;3:541–548. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- 5.Patti JM, Allen BL, McGavin MJ, Hook M. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu Rev Microbiol. 1994;48:585–617. doi: 10.1146/annurev.mi.48.100194.003101. [DOI] [PubMed] [Google Scholar]
- 6.Hendrickx AP, Budzik JM, Oh SY, Schneewind O. Architects at the bacterial surface - sortases and the assembly of pili with isopeptide bonds. Nat Rev Microbiol. 2011;9:166–176. doi: 10.1038/nrmicro2520. [DOI] [PubMed] [Google Scholar]
- 7.Frankel BA, Kruger RG, Robinson DE, Kelleher NL, McCafferty DG. Staphylococcus aureus sortase transpeptidase SrtA: insight into the kinetic mechanism and evidence for a reverse protonation catalytic mechanism. Biochemistry. 2005;44:11188–11200. doi: 10.1021/bi050141j. [DOI] [PubMed] [Google Scholar]
- 8.Schneewind O, Fowler A, Faull KF. Structure of the cell wall anchor of surface proteins in Staphylococcus aureus. Science. 1995;268:103–106. doi: 10.1126/science.7701329. [DOI] [PubMed] [Google Scholar]
- 9.Clancy KW, Melvin JA, McCafferty DG. Sortase transpeptidases: insights into mechanism, substrate specificity, and inhibition. Biopolymers. 2010;94:385–396. doi: 10.1002/bip.21472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ilangovan U, Ton-That H, Iwahara J, Schneewind O, Clubb RT. Structure of sortase, the transpeptidase that anchors proteins to the cell wall of Staphylococcus aureus. Proc Natl Acad Sci U S A. 2001;98:6056–6061. doi: 10.1073/pnas.101064198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Race PR, Bentley ML, Melvin JA, Crow A, Hughes RK, Smith WD, Sessions RB, Kehoe MA, McCafferty DG, Banfield MJ. Crystal structure of Streptococcus pyogenes sortase A: implications for sortase mechanism. J Biol Chem. 2009;284:6924–6933. doi: 10.1074/jbc.M805406200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parsonage D, Karplus PA, Poole LB. Substrate specificity and redox potential of AhpC, a bacterial peroxiredoxin. Proc Natl Acad Sci U S A. 2008;105:8209–8214. doi: 10.1073/pnas.0708308105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salmeen A, Andersen JN, Myers MP, Meng TC, Hinks JA, Tonks NK, Barford D. Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature. 2003;423:769–773. doi: 10.1038/nature01680. [DOI] [PubMed] [Google Scholar]
- 14.Kubica M, Guzik K, Koziel J, Zarebski M, Richter W, Gajkowska B, Golda A, Maciag-Gudowska A, Brix K, Shaw L, Foster T, Potempa J. A potential new pathway for Staphylococcus aureus dissemination: the silent survival of S. aureus phagocytosed by human monocyte-derived macrophages. PLoS One. 2008;3:e1409. doi: 10.1371/journal.pone.0001409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang JH, Zhou YJ, He P. Staphylococcus aureus induces apoptosis of human monocytic U937 cells via NF-kappaB signaling pathways. Microb Pathog. 2010;49:252–259. doi: 10.1016/j.micpath.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 16.Thwaites GE, Gant V. Are bloodstream leukocytes Trojan Horses for the metastasis of Staphylococcus aureus? Nat Rev Microbiol. 2011;9:215–222. doi: 10.1038/nrmicro2508. [DOI] [PubMed] [Google Scholar]
- 17.Tuchscherr L, Medina E, Hussain M, Volker W, Heitmann V, Niemann S, Holzinger D, Roth J, Proctor RA, Becker K, Peters G, Loffler B. Staphylococcus aureus phenotype switching: an effective bacterial strategy to escape host immune response and establish a chronic infection. EMBO Mol Med. 2011;3:129–141. doi: 10.1002/emmm.201000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graves SF, Kobayashi SD, DeLeo FR. Community-associated methicillin-resistant Staphylococcus aureus immune evasion and virulence. J Mol Med. 2010;88:109–114. doi: 10.1007/s00109-009-0573-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bentley ML, Lamb EC, McCafferty DG. Mutagenesis studies of substrate recognition and catalysis in the sortase A transpeptidase from Staphylococcus aureus. J Biol Chem. 2008;283:14762–14771. doi: 10.1074/jbc.M800974200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bentley ML, Gaweska H, Kielec JM, McCafferty DG. Engineering the substrate specificity of Staphylococcus aureus Sortase A. The beta6/beta7 loop from SrtB confers NPQTN recognition to SrtA. J Biol Chem. 2007;282:6571–6581. doi: 10.1074/jbc.M610519200. [DOI] [PubMed] [Google Scholar]
- 21.Kruger RG, Dostal P, McCafferty DG. Development of a high-performance liquid chromatography assay and revision of kinetic parameters for the Staphylococcus aureus sortase transpeptidase SrtA. Anal Biochem. 2004;326:42–48. doi: 10.1016/j.ab.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 22.van der Vliet A, Hu ML, O'Neill CA, Cross CE, Halliwell B. Interactions of human blood plasma with hydrogen peroxide and hypochlorous acid. J Lab Clin Med. 1994;124:701–707. [PubMed] [Google Scholar]
- 23.Noble RW, Gibson QH. The reaction of ferrous horseradish peroxidase with hydrogen peroxide. J Biol Chem. 1970;245:2409–2413. [PubMed] [Google Scholar]
- 24.Copeland RA. Enzymes. 2nd. Wiley-VCH; New York: 2000. [Google Scholar]
- 25.Wang L, Yuan Z. Direct electrochemistry of xanthine oxidase at a gold electrode modified with single-wall carbon nanotubes. Anal Sci. 2004;20:635–638. doi: 10.2116/analsci.20.635. [DOI] [PubMed] [Google Scholar]
- 26.George S, Lee HK. Direct electrochemistry and electrocatalysis of hemoglobin in nafion/carbon nanochip film on glassy carbon electrode. J Phys Chem B. 2009;113:15445–15454. doi: 10.1021/jp905690a. [DOI] [PubMed] [Google Scholar]
- 27.Winterbourn CC, Hampton MB, Livesey JH, Kettle AJ. Modeling the reactions of superoxide and myeloperoxidase in the neutrophil phagosome: implications for microbial killing. J Biol Chem. 2006;281:39860–39869. doi: 10.1074/jbc.M605898200. [DOI] [PubMed] [Google Scholar]
- 28.Winterbourn CC. Reconciling the chemistry and biology of reactive oxygen species. Nat Chem Biol. 2008;4:278–286. doi: 10.1038/nchembio.85. [DOI] [PubMed] [Google Scholar]
- 29.Cherney DP, Duirk SE, Tarr JC, Collette TW. Monitoring the speciation of aqueous free chlorine from pH 1 to 12 with Raman spectroscopy to determine the identity of the potent low-pH oxidant. Appl Spectrosc. 2006;60:764–772. doi: 10.1366/000370206777887062. [DOI] [PubMed] [Google Scholar]
- 30.Cotton FA, Wilkinson G. Advanced Inorganic Chemistry. 5th. John Wiley and Sons; New York: 1988. [Google Scholar]
- 31.Winterbourn CC, Metodiewa D. Reactivity of biologically important thiol compounds with superoxide and hydrogen peroxide. Free Radic Biol Med. 1999;27:322–328. doi: 10.1016/s0891-5849(99)00051-9. [DOI] [PubMed] [Google Scholar]
- 32.Pattison DI, Davies MJ. Absolute rate constants for the reaction of hypochlorous acid with protein side chains and peptide bonds. Chem Res Toxicol. 2001;14:1453–1464. doi: 10.1021/tx0155451. [DOI] [PubMed] [Google Scholar]
- 33.Hung M, Stanbury DM. Catalytic and direct oxidation of cysteine by octacyanomolybdate(V) Inorg Chem. 2005;44:3541–3550. doi: 10.1021/ic050427c. [DOI] [PubMed] [Google Scholar]
- 34.Wang X, Stanbury DM. Direct oxidation of L-cysteine by [FeIII(bpy)2(CN)2]+ and [FeIII(bpy)(CN)4] Inorg Chem. 2008;47:1224–1236. doi: 10.1021/ic701891m. [DOI] [PubMed] [Google Scholar]
- 35.Ison A, Odeh IN, Margerum DW. Kinetics and mechanisms of chlorine dioxide and chlorite oxidations of cysteine and glutathione. Inorg Chem. 2006;45:8768–8775. doi: 10.1021/ic0609554. [DOI] [PubMed] [Google Scholar]
- 36.Vaudaux P, Francois P, Bisognano C, Kelley WL, Lew DP, Schrenzel J, Proctor RA, McNamara PJ, Peters G, Von Eiff C. Increased expression of clumping factor and fibronectin-binding proteins by hemB mutants of Staphylococcus aureus expressing small colony variant phenotypes. Infect Immun. 2002;70:5428–5437. doi: 10.1128/IAI.70.10.5428-5437.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paterson GK, Mitchell TJ. The biology of Gram-positive sortase enzymes. Trends Microbiol. 2004;12:89–95. doi: 10.1016/j.tim.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 38.Foster TJ. Colonization and infection of the human host by staphylococci: adhesion, survival and immune evasion. Vet Dermatol. 2009;20:456–470. doi: 10.1111/j.1365-3164.2009.00825.x. [DOI] [PubMed] [Google Scholar]
- 39.Frankel BA, Bentley M, Kruger RG, McCafferty DG. Vinyl sulfones: inhibitors of SrtA, a transpeptidase required for cell wall protein anchoring and virulence in Staphylococcus aureus. J Am Chem Soc. 2004;126:3404–3405. doi: 10.1021/ja0390294. [DOI] [PubMed] [Google Scholar]
- 40.Cosgrove K, Coutts G, Jonsson IM, Tarkowski A, Kokai-Kun JF, Mond JJ, Foster SJ. Catalase (KatA) and alkyl hydroperoxide reductase (AhpC) have compensatory roles in peroxide stress resistance and are required for survival, persistence, and nasal colonization in Staphylococcus aureus. J Bacteriol. 2007;189:1025–1035. doi: 10.1128/JB.01524-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karavolos MH, Horsburgh MJ, Ingham E, Foster SJ. Role and regulation of the superoxide dismutases of Staphylococcus aureus. Microbiology. 2003;149:2749–2758. doi: 10.1099/mic.0.26353-0. [DOI] [PubMed] [Google Scholar]
- 42.Liu GY, Essex A, Buchanan JT, Datta V, Hoffman HM, Bastian JF, Fierer J, Nizet V. Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J Exp Med. 2005;202:209–215. doi: 10.1084/jem.20050846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clauditz A, Resch A, Wieland KP, Peschel A, Gotz F. Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress. Infect Immun. 2006;74:4950–4953. doi: 10.1128/IAI.00204-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nelson KJ, Parsonage D, Hall A, Karplus PA, Poole LB. Cysteine pK(a) values for the bacterial peroxiredoxin AhpC. Biochemistry. 2008;47:12860–12868. doi: 10.1021/bi801718d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borutaite V, Brown GC. Caspases are reversibly inactivated by hydrogen peroxide. FEBS Lett. 2001;500:114–118. doi: 10.1016/s0014-5793(01)02593-5. [DOI] [PubMed] [Google Scholar]
- 46.Meissner F, Molawi K, Zychlinsky A. Superoxide dismutase 1 regulates caspase-1 and endotoxic shock. Nat Immunol. 2008;9:866–872. doi: 10.1038/ni.1633. [DOI] [PubMed] [Google Scholar]
- 47.Lohse DL, Denu JM, Santoro N, Dixon JE. Roles of aspartic acid-181 and serine-222 in intermediate formation and hydrolysis of the mammalian protein-tyrosine-phosphatase PTP1. Biochemistry. 1997;36:4568–4575. doi: 10.1021/bi963094r. [DOI] [PubMed] [Google Scholar]
- 48.Stennicke HR, Salvesen GS. Biochemical characteristics of caspases-3, -6, -7, and -8. J Biol Chem. 1997;272:25719–25723. doi: 10.1074/jbc.272.41.25719. [DOI] [PubMed] [Google Scholar]
- 49.Rhee SG, Jeong W, Chang TS, Woo HA. Sulfiredoxin, the cysteine sulfinic acid reductase specific to 2-Cys peroxiredoxin: its discovery, mechanism of action, and biological significance. Kidney Int Suppl. 2007:S3–8. doi: 10.1038/sj.ki.5002380. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.