Abstract
The mitochondrial presequence translocase interacts with presequence-containing precursors at the intermembrane space (IMS) side of the inner membrane to mediate their translocation into the matrix. Little is known as too how these matrix-targeting signals activate the translocase in order to initiate precursor transport. Therefore, we analysed how signal recognition by the presequence translocase initiates reorganization among Tim-proteins during import. Our analyses revealed that the presequence receptor Tim50 interacts with Tim21 in a signal-sensitive manner in a process that involves the IMS-domain of the Tim23 channel. The signal-driven release of Tim21 from Tim50 promotes recruitment of Pam17 and thus triggers formation of the motor-associated form of the TIM23 complex required for matrix transport.
Keywords: import, mitochondria, presequence, TIM23 complex
Introduction
Mitochondrial functions rely on the constant import of proteins, which are synthesized on cytosolic ribosomes. The majority of ∼1000 mitochondrial proteins are transported into mitochondria post-translationally. Targeting signals within these precursor proteins direct the newly synthesized proteins to mitochondria and to their final destination within the organelle. In recent years, five distinct import pathways into the various mitochondrial subcompartments have been established: the outer and inner membranes, the intermembrane space (IMS) and the matrix (for review see Dolezal et al, 2006; van der Laan et al, 2006a; Mokranjac and Neupert, 2007; Chacinska et al, 2009; Endo and Yamano, 2010). The ‘classical’ import pathway into mitochondria pertains to proteins with N-terminal targeting signals, so-called presequences. A recent proteomic analysis revealed that ∼70% of mitochondrial proteins utilize presequences as targeting signals (Vögtle et al, 2009). Presequences are characterized as amphipathic α helical segments at the precursor’s N-terminus that are rich in positively charged amino acids, which facilitate passage of the precursor protein to and across the outer and inner membranes of mitochondria (Hurt et al, 1984; Heijne, 1986; Vögtle et al, 2009). The TOM (Translocase of the Outer Mitochondrial membrane) and TIM23 (Translocase of the Inner Mitochondrial membrane) complexes mediate the transfer of precursors across both mitochondrial membranes in a concerted action. Usually, presequences direct the precursor protein towards the mitochondrial matrix. However, some presequence-containing precursors possess hydrophobic sorting signals that stall precursor translocation across the inner membrane within the TIM23 complex and initiate the lateral release of the polypeptide into the lipid phase of the inner membrane (Glick et al, 1992; van der Laan et al, 2007).
The presequence translocase consists of five integral membrane proteins: Tim17, Tim23, Mgr2, Tim50, and Tim21. The multispanning membrane proteins Tim23 and Tim17 form the core of the TIM23 complex (Dekker et al, 1997; Moro et al, 1999). In contrast to Tim17, Tim23 exposes a domain of ∼100 amino acids into the IMS (Tim23IMS), which recognizes presequences in a process that stimulates channel activity (Bauer et al, 1996; Truscott et al, 2001; de la Cruz et al, 2010; Schulz et al, 2011). Tim50 acts as a presequence receptor of the complex and binds to Tim23 in order to promote channel closure in the inactive translocase (Geissler et al, 2002; Yamamoto et al, 2002; Meinecke et al, 2006; Marom et al, 2011; Qian et al, 2011; Schulz et al, 2011). Tim21 interacts with the IMS domain of Tom22 to promote presequence release from TOM and thus transfer from the TOM to the TIM23 complex (Chacinska et al, 2005; Mokranjac et al, 2005). In human mitochondria, TIM21 mediates the transfer of newly imported subunits of respiratory chain complexes I and IV to assembly intermediates (Mick et al, 2012).
The presequence translocase mediates the lateral release of precursors into the inner membrane in a mechanism that is referred to as the sorting process. This sorting process requires a hydrophobic sorting signal in the precursor, which is located downstream of the presequence. In addition, the presequence translocase mediates the full translocation of precursor proteins into the matrix. For both precursor classes, the membrane potential (Δψ) provides the initial electrophoretic driving force for presequence translocation across the inner membrane. While the Δψ, in principle, suffices for the transport of sorted precursors (Gambill et al, 1993), full translocation into the matrix requires the activity of the presequence translocase associated motor complex (PAM). This complex, with its core subunit mitochondrial Hsp70, provides a directional driving force at the expense of ATP. Other subunits of the PAM complex function in the positioning of Hsp70 to the exit of the translocation pore (Tim44) or regulate the ATPase cycle of Hsp70. The latter function has been attributed to the ADP/ATP exchange factor Mge1 and the Pam16/Pam18 J-protein complex.
Presequence translocase association with the PAM complex is dynamic in nature. While a Tim21-containing form of the presequence translocase (TIM23SORT) remains motor free and is able to promote inner membrane sorting, PAM association to the translocase requires the release of Tim21 and concomitant generation of TIM23MOTOR (Chacinska et al, 2005, 2010; van der Laan et al, 2007). It is currently unknown how this reorganization of the translocase is regulated during precursor import. However, Pam17, a protein that associates with the TIM23MOTOR complex, has been suggested to play a role in the subsequent translocase association of the Pam16/Pam18 complex (van der Laan et al, 2005; Popov-Čeleketić et al, 2008).
In order to understand how presequences provide directionality to protein transport and activate the translocase during the transport process, it is essential to obtain a comprehensive understanding of protein interactions and their dynamics. Here, we identified an interaction between the IMS-domains of Tim21 (Tim21IMS) and Tim50 (Tim50IMS). Our analyses revealed that the IMS-domains of Tim50 and Tim21 interact in vitro with an affinity 10-fold greater than that of Tim50 to presequences. In mitochondria, presequence docking at the cis-face of the presequence translocase confers internal rearrangements within the TIM23 complex leading to the release of Tim21 from Tim50. Subsequently, motor association is promoted via the recruitment of Pam17 to the translocase. Taken together, the dissection of the early steps in presequence transport reveals the long elusive function of targeting signal recognition, enabling translocase reorganization prior to membrane translocation.
Results
Tim50 interacts with Tim23 and Tim21 of the presequence translocase
Tim50 is the central receptor of the presequence translocase and interacts with the channel-forming subunit, Tim23 (Geissler et al, 2002; Yamamoto et al, 2002; Meinecke et al, 2006; Gevorkyan-Airapetov et al, 2009; Schulz et al, 2011). Structural analyses of a portion of Tim50’s IMS-domain (aa 164–361) revealed a binding site for Tim23IMS in an exposed β-hairpin (Qian et al, 2011). While the presence of a presequence-binding site in a negatively charged groove of the structure was suggested, photocrosslinking analysis revealed a presequence receptor site (aa 395–476) outside of this groove and the crystallized fragment (aa 164–361) (Figure 1A) (Schulz et al, 2011). Therefore, we aimed to identify a Tim50-ligand that would bind the orphan binding groove. Taking advantage of a cysteine residue in the binding groove (Figure 1B), we incubated isolated mitochondria in the presence of the oxidizing reagent CuSO4 to crosslink binding partners through disulphide-bond formation. Compared to the untreated control, crosslinked samples displayed several Tim50-adducts in Western blot analyses (Figure 1C). As a specificity control for the detected adduct bands, we utilized mitochondria containing an HA-tagged version of Tim50 (Tim50HA) for crosslinking and used antisera against Tim50 or the HA-tag for detection. The majority of adducts detected in wild-type mitochondria were shifted in size due to the presence of the tag in Tim50HA-mitochondria (Figure 1C, white circle, black circle and white diamond).
Figure 1.
Identification of Tim50 crosslinking partners. (A) Schematic representation of Tim50 and Tim50 fragments used in this study. Presequence (PS), transmembrane domain (TM), NIF-domain, presequence-binding domain (PBD), and the cysteine position (C268) are indicated. (B) Structure of Tim50C (Protein Data Bank entry 3QLE) with the Tim23-binding site, large negatively charged groove and the cysteine (C268, in green) positions indicated. Positively charged residues are coloured in blue, negative are red. (C) Wild-type and Tim50HA-mitochondria were subjected to CuSO4 crosslinking and analysed by western blotting using either anti-Tim50 or anti-HA antibodies. Selected shifted crosslinking products are marked with circles or a diamond (unknown Tim50 crosslinking partner). (D) Crosslinking was performed as in (C) using Tim21ProtA-, Tim23ProtA- and Tim23ProtA-mitochondria. Samples were analysed by western blotting using anti-Tim50 antiserum. (E) CuSO4 crosslinking was performed in mitochondria from Tim21ProtA and tim21Δ strains, transformed with either an empty pFL39 (control) or a Tim21-expressing plasmid (TIM21). Samples were analysed by western blotting using an antiserum against Tim50.
To identify the interaction partners of Tim50 in the crosslink-adducts, we tested for components of the presequence translocase. Since Tim23 and Tim21 expose soluble domains into the IMS, we performed Cu2+-induced crosslinking in wild-type mitochondria and mitochondria containing tagged versions of Tim21 or Tim23.
The observed 100 kDa Tim50-adduct was specifically shifted in Tim23ProtA- and Tim23HA-mitochondria (Figure 1D, white circle). Therefore, we assigned these crosslinks to Tim23–Tim50-adducts previously identified in other studies (Alder et al, 2008; Tamura et al, 2009).
The 80-kDa Tim50-adduct was not affected in mitochondria with tagged versions of Tim23, but was selectively shifted in Tim21ProtA-mitochondria (Figure 1D, black circle). This result indicated that the disulphide-bonded adduct represented a thus far unidentified protein interaction of Tim50 with Tim21, both of which contain a single cysteine in the IMS. In a complementary approach to support the specificity of the crosslink-adduct, we used tim21Δ-mitochondria for crosslinking. As predicted from the previous experiment, the 80-kDa crosslink-adduct was selectively missing in mutant mitochondria and shifted in Tim21ProtA-mitochondria (Figure 1E). The expression of TIM21 from a plasmid reestablished the crosslink in both cases. These analyses demonstrated that within the mitochondrial presequence translocase the IMS-domain of Tim50 is in close contact with Tim23 and Tim21, and that these interactions can be stabilized by disulphide bridge formation (∼2 Å in distance).
The IMS-domains of Tim50 and Tim21 interact with high affinity
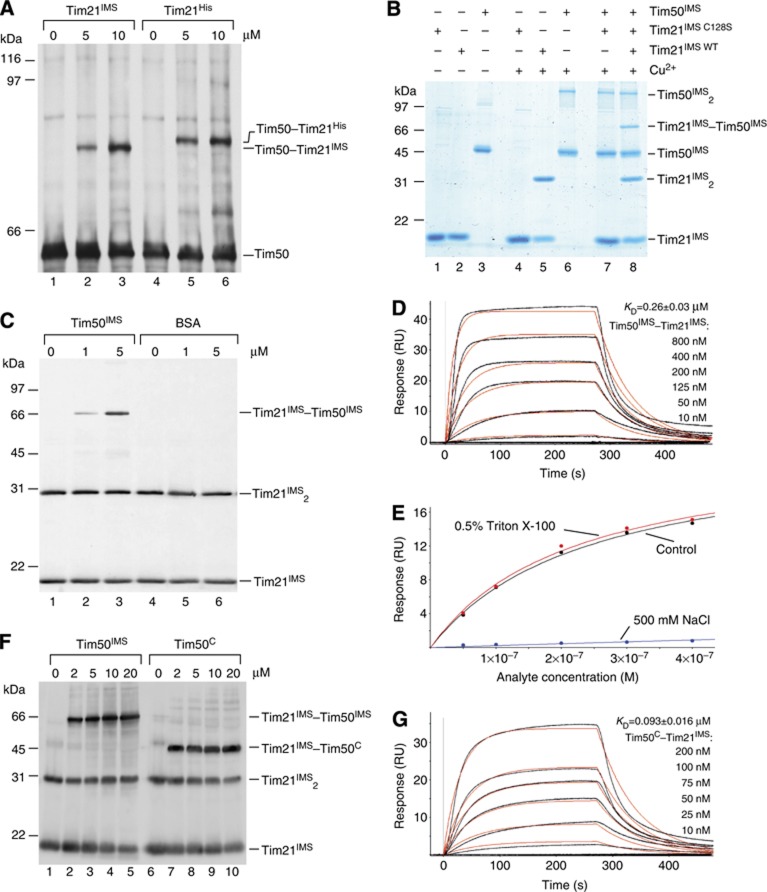
Tim50 and Tim21 are single spanning membrane proteins of the inner mitochondrial membrane and are both constituents of the presequence translocase. We asked if the positioning of the two proteins in the translocase was a prerequisite for their interaction or whether the soluble domains alone would suffice. To address this experimentally, we established an in organello assay. Mitochondria were converted to mitoplasts (disrupted outer membrane) by osmotic swelling to allow access to the IMS side of the inner membrane. Subsequently, mitoplasts were incubated with increasing amounts of purified IMS-domain of Tim21 (aa 103–225), both with and without a 6 × His tag (Tim21His/Tim21IMS), prior to Cu2+-induced crosslinking. Western blot analyses revealed that a Tim50–Tim21IMS-adduct was efficiently formed with increasing amounts of Tim21IMS (Figure 2A). Similarly, a Tim50–Tim21His-adduct was formed, which migrated slower in the gel than the Tim50–Tim21IMS-adduct due to the presence of the tag. Thus, we concluded that the IMS-domain of Tim21 was sufficient to interact with Tim50 in mitoplasts. Moreover, the experiment indicated that a pool of Tim50 existed in mitochondria, which was available to exogenously added Tim21IMS.
Figure 2.
Tim21 interacts directly with Tim50 in organello and in vitro. (A) Wild-type mitochondria were osmotically swollen and subjected to CuSO4 crosslinking in the presence of indicated concentrations of purified Tim21IMS or its His-tagged form Tim21His. Crosslinking-adducts were detected by immunoblotting against Tim50. (B) Purified recombinant proteins (25 μM Tim21IMS or Tim21C128S and 10 μM Tim50IMS) were crosslinked in the presence of 5 mM CuSO4. Crosslinking-adducts were visualized by colloidal Coomassie staining. (C) Indicated amounts of purified Tim50IMS or bovine serum albumin (BSA) were mixed with recombinant Tim21IMS (final concentration 1 μM) and crosslinked by adding of CuSO4, followed by immunoblotting and decoration with anti-Tim21 antibody. (D) Tim50IMS-His was immobilized on a chip and the SPR response was recorded after adding indicated concentrations of Tim21IMS. A typical sensogram is shown. Black lines, observed binding; red, calculated ka/kd fitting of kinetic data. KD is presented as mean±s.e.m. (n=3 for each measurement). (E) The SPR response was recorded as in (D) in three different buffers: control (50 mM HEPES, pH 7.4, 150 mM NaCl, and 50 μM EDTA); 0.5% Triton X-100 (50 mM HEPES, pH 7.4, 150 mM NaCl, 50 μM EDTA, and 0.5% Triton X-100); 500 mM NaCl (50 mM HEPES, pH 7.4, 500 mM NaCl, and 50 μM EDTA). Titration isotherms of maximal response versus analyte concentration are shown (data fitting is based on a model of a simple bimolecular interaction). (F) In all, 1 μM Tim21IMS was mixed with indicated amounts of Tim50IMS or Tim50C and crosslinked by CuSO4. Immunoblotting with anti-Tim21 antibody was used to detect crosslinking-adducts. (G) Interaction between Tim50C-His immobilized on a Ni2+-chelator chip and Tim21IMS was analysed by SPR, as in (D).
Based on the finding that the transmembrane domain of Tim21 was dispensable for its association with Tim50, we applied an in vitro interaction assay to address the association of Tim50IMS with Tim21IMS directly. Therefore, we purified Tim50IMS (aa 132–476), Tim21IMS, and Tim21IMS-C128S, in which the single cysteine 128 was exchanged with a serine (Figure 2B, lanes 1–3). Purified proteins were mixed in equimolar amounts and subjected to crosslinking prior to SDS–polyacrylamide gel electrophoresis (PAGE) analyses. Tim21IMS formed a dimer upon Cu2+-induced crosslinking, while Tim21IMS-C128S did not, due to the absence of the cysteine residue (Figure 2B, lanes 4 versus 5). Similarly, Tim50IMS formed dimers upon crosslinking (Figure 2B, lane 6), consistent with previous findings using homo-bifunctional crosslinkers (DSS) and two-hybrid analyses (Geissler et al, 2002; Gevorkyan-Airapetov et al, 2009). When we combined the Tim21IMS with Tim50IMS, homo-dimers as well as a Tim21IMS–Tim50IMS hetero-oligomer were efficiently formed (Figure 2B, lane 8). As expected, Tim21IMS-C128S did not form a crosslink-adduct with Tim50IMS. To ascertain that the observed disulphide crosslinks were specific, we performed crosslinking analyses of Tim21IMS, while titrating the amount of Tim50IMS or albumin (exemplifying a protein with multiple surface-exposed cysteine residues) as a control. While Tim21IMS dimers and Tim21IMS–Tim50IMS-adducts were efficiently formed, no crosslinks to BSA were detected nor did BSA affect Tim21IMS dimerization (Figure 2C). In summary, we found that the IMS-domains of Tim21 and Tim50 are able to dimerize and that we can stabilize these dimers for analyses through induction of disulphide bridge formation. Moreover, the IMS-domains of Tim50 and Tim21 are able to interact in the absence of the membrane span. Since both proteins contain a single cysteine, we conclude that both homo- and hetero-oligomerization interfaces localize to the same surface of the proteins.
So far, our analyses addressed the physical interaction between Tim21 and Tim50 solely with crosslinking experiments. However, this approach could not provide quantitative data on the affinity of the interaction. Therefore, we utilized surface plasmon resonance (SPR) in order to confirm the interactions and obtain quantitative data. His-tagged Tim50IMS was immobilized on a Ni2+ chelator sensorchip and Tim21IMS association was monitored over a wide range of concentrations. Figure 2D presents a typical sensogram. Mathematical fitting of the association and dissociation rates revealed fast association kinetics, with a ka rate of ∼65 per mM per second (Figure 2D; Table I).
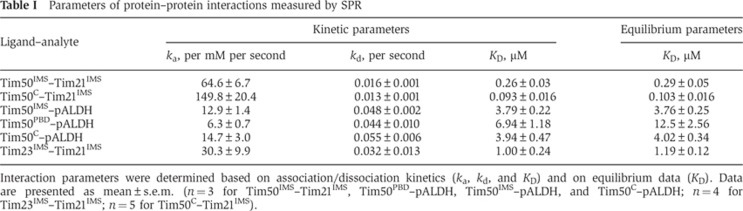
Table 1. Parameters of protein–protein interactions measured by SPR.
Interaction parameters were determined based on association/dissociation kinetics (ka, kd, and KD) and on equilibrium data (KD). Data are presented as mean±s.e.m. (n=3 for Tim50IMS–Tim21IMS, Tim50PBD–pALDH, Tim50IMS–pALDH, and Tim50C–pALDH; n=4 for Tim23IMS–Tim21IMS; n=5 for Tim50C–Tim21IMS).
To assess the nature of the molecular forces that stabilize the Tim21–Tim50 interaction, SPR measurements were carried out in the presence of salt or detergent (Figure 2E). Whereas addition of detergent had no effect on the SPR response, high-salt concentration dramatically reduced it, suggesting a predominantly ionic nature of interaction. In agreement with this observation, the area surrounding the cysteine in Tim21 is highly enriched in clusters of negatively charged amino acids (Albrecht et al, 2006).
Based on structural and functional analyses, two distinct domains have been attributed to the IMS portion of Tim50 (see above). Thus far, we had utilized the full-length IMS-domain for our analyses. Therefore, we addressed if the C-terminal presequence-binding domain was required for Tim21 association. We purified Tim21IMS and Tim50C (amino acids 164–361), corresponding to the crystallized portion of Tim50, and addressed protein interactions in vitro. Similar to Tim50IMS, Tim50C was able to dimerize and to hetero-oligomerize with Tim21IMS (Supplementary Figure S1A). Next, we directly compared the efficiency of oligomer formation between Tim21 and Tim50IMS or Tim50C to exclude that they displayed significantly altered affinities. Using identical amounts of Tim21IMS and increasing amounts of the two Tim50 forms, we performed in vitro crosslinking. Both Tim50IMS and Tim50C efficiently interacted with Tim21IMS; however, we reproducibly observed that the Tim21IMS–Tim50C-adduct formation reached saturation at lower concentrations than Tim21IMS–Tim50IMS (Figure 2F). To determine if this behaviour reflected different affinities of the two Tim50 constructs for Tim21, we analysed the Tim21IMS–Tim50C interaction by SPR. This method confirmed an approximately three times lower KD of the Tim21IMS–Tim50C interaction (∼100 nM), as compared with Tim21IMS–Tim50IMS (290 nM), largely due to its faster association kinetics (150 per mM per second for Tim21IMS-Tim50C versus 65 per mM per second for the Tim21IMS–Tim50IMS interaction) (Figure 2G; Table I). Hence, our data show that the presequence-binding domain of Tim50 is dispensable for the interaction of the IMS-domains of Tim50 and Tim21.
Presequence peptides interact with different regions of Tim50
The association kinetics and affinity of the Tim21–Tim50 interaction revealed by SPR were unexpectedly high. As a control, we compared Tim50’s affinity for Tim21 with its presequence interaction by SPR. Therefore, we again immobilized His-tagged Tim50IMS on a Ni2+ chelator sensorchip and recorded the SPR response after addition of rat ALDH presequence peptide (pALDH) or its inactive version pALDH-s (Schulz et al, 2011). Presequence peptides specifically bound to Tim50IMS with a ka value of ∼13 per mM per second and kd of 0.05 per second (Figure 3A; Table I), which reflected an approximately five-fold slower association and a three-fold faster dissociation as compared with the Tim21–Tim50 values. At the same time, the import-inactive peptide pALDH-s showed almost no binding (Figure 3B; Table I). In contrast to Tim50, Tim21IMS did not display significant specific interactions with pALDH (Supplementary Figure S1B). In conclusion, the IMS-domain of Tim50 binds presequences and additionally recognizes Tim21 with an order of magnitude higher affinity. Surprisingly, when we used SPR to compare Tim50IMS to the presequence-binding domain of Tim50 (Tim50PBD) (Schulz et al, 2011) with Tim50C as a control, we found that Tim50PBD as well as Tim50C specifically interacted with pALDH (Figure 3C–F). Previous structural analysis on Tim50C had speculated on a binding site in this region based on biophysical properties of presequences and Tim50 (Qian et al, 2011). However, this finding was surprising, since we had previously found that Tim50ΔPBD (aa 164–365) did not interact significantly with presequence peptides in crosslinking studies. To directly address these seemingly contradictory results, we performed in vitro photocrosslinking with two versions of ALDH peptides that carried the photoreactive amino-acid derivative para-benzoylphenylalanine (BPA) replacing either serine 16 (pS16B) or leucine 19 (pL19B) (Schulz et al, 2011). While Tim50IMS displayed efficient crosslinking to presequence peptides, as previously reported, both Tim50ΔPBD and Tim50C displayed very little photoadduct formation (Figure 3G). Similarly, while Tim50IMS could be efficiently crosslinked to Cox4 presequence peptides with the homo-bifunctional chemical crosslinker 1,5-Difluoro-2,4-dinitrobenzene (DFDNB), Tim50ΔPBD and Tim50C displayed no significant presequence crosslinks (Supplementary Figure S1C).
Figure 3.
Interaction of presequence peptides with Tim50. (A–F) Purified 6 × His-tagged Tim50IMS (A, B), Tim50PBD (C, D), or Tim50C (E, F) was immobilized on a Ni2+-chelator chip and the SPR response to the indicated concentrations of pALDH (A, C, E) or pALDH-s (B, D, F) was recorded. KD is presented as mean±s.e.m. (n=3 for each measurement). (G) Photocrosslinking was performed using purified 1 μM Tim50IMS, Tim50ΔPBD, Tim50C and biotinylated photopeptides pL19B and pS16B at indicated concentrations. After SDS–PAGE separation, crosslinking products were detected using streptavidin–horse radish peroxidase (SA-HRP) conjugate or by immunodecoration against Tim50. (H) HA-tagged full-length Tim50 (Tim50HA) and Tim50 fragments containing the first 365 or 361 amino acids (Tim501–365–HA, Tim501–361, and Tim501–361–HA) in pME2782 were used to replace the wild-type protein-encoding plasmid carrying URA3 in the gene deletion strain grown on 5-fluoroorotic acid-containing plates (SD-5FOA). Strains were grown on selective-Leu plates as a control.
Since the SPR data indicated that Tim50C interacts with presequences with similar affinities as Tim50IMS, we analysed if Tim50 (aa 1–361) was able to complement the lethal growth phenotype of a strain lacking Tim50. We expressed full-length Tim50, Tim50 (aa 1–361) and Tim50 (aa 1–365) in yeast cells in which a chromosomal deletion of TIM50 was complemented by a plasmid-encoded Tim50 expressed from a URA3-containing plasmid. Upon 5FOA treatment, expression of full-length Tim50 rescued the lethal phenotype, while both truncation constructs did not. Thus, the use of a complementation assay allowed us to collectively conclude that: (i) Tim50 possesses two binding domains for presequences, one in the C-terminal PBD and the second in the crystallized core and (ii) the presequence-binding site in the core of Tim50’s IMS-domain alone does not suffice for Tim50 function in mitochondria.
Presequences trigger Tim21–Tim50 dissociation
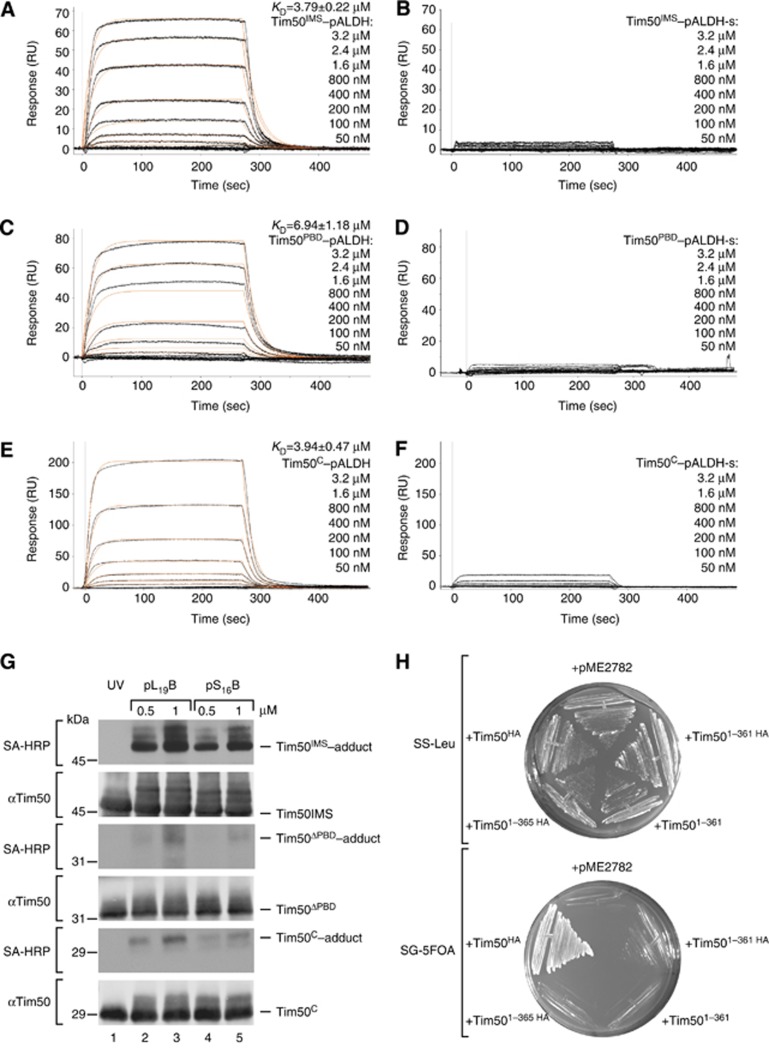
What is the role of the Tim21–Tim50 interaction during protein transport? To address this question, we incubated mitochondria in the presence of increasing amounts of a presequence peptide (pALDH) or its inactive variant (pALDH-s) and analysed Tim50 interactions under these conditions by Cu2+ crosslinking (Figure 4A and B). Interestingly, crosslinking between Tim21 and Tim50 was drastically reduced in the presence of presequence peptides in a concentration-dependent manner. In contrast, crosslinking between Tim50 and Tim23 was apparently promoted (Figure 4B).
Figure 4.
Presequence peptides affect interactions between Tim21, Tim23, and Tim50. (A) Mitochondria were pre-treated with indicated concentrations of pALDH or its inactive version pALDH-s (reaction volumes were matched by adding corresponding volumes of solvent), crosslinked by 1 mM CuSO4 for 30 min on ice and analysed by non-reducing SDS–PAGE and immunodecoration against Tim50. (B) Signals of Tim23–Tim50 (upper plot) and Tim21–Tim50 (lower plot) crosslinking-adducts (as in A) were quantified, normalized to buffer-treated controls (set as 100%), and presented as mean±s.e.m. (n=6). (C) Experiment (as in A) was performed in osmotically swollen mitochondria (mitoplasts, n=4), mitochondria with a dissipated membrane potential (–Δψ, n=4), tom22-2 (n=4) and Tim501–361–HA (n=3) mitochondria. The intensity of the Tim21–Tim50 crosslink-adducts after treatment with 50 μM pALDH was quantified and normalized to control (set to 100%). Data are presented as mean±s.e.m. (D) Purified Tim21IMS and Tim50IMS (1 μM each) were crosslinked in the presence of increasing amounts of pALDH or pALDH-s and analysed by immunoblotting against Tim21.
As Tim21 was found to interact with the IMS-domain of Tom22, in a process considered to assist in presequence transfer from the TOM to TIM23 complex (Chacinska et al, 2005; Mokranjac et al, 2005; Albrecht et al, 2006), we addressed if the presequence effect on the Tim21–Tim50 interaction occurred in mitoplasts and tom22-2 mutant mitochondria (lacking the Tom22 IMS-domain). In both cases, signal-mediated dissociation of Tim21 from Tim50 was observed (Figure 4C). Moreover, the effect was also present in mitochondria when Tim50 lacking the PBD (Tim501–361–HA) was expressed, replacing wild-type Tim50 that had been downregulated through the use of a GAL promoter (Figure 4C). In addition, we found that the presence of a membrane potential (Δψ) across the inner membrane was not a prerequisite for signal-induced protein dissociation, as dissipation of the Δψ through the addition of the ionophore valinomycin together with electron transport chain inhibitor antimycin and ATP synthase inhibitor olygomycin did not block dissociation (Figure 4C). This observation indicates that the presequence-mediated effect occurs on the cis side of the inner membrane, as an intact Δψ is required for channel activation and presequence transfer across the inner membrane (Bauer et al, 1996; Truscott et al, 2001; Schulz et al, 2011).
To directly test if the observed signal-triggered dissociation was mediated through signal recognition by Tim50, we analysed the Tim50IMS interaction with Tim21IMS in vitro in the presence or absence of presequence or control peptides. Interestingly, neither Tim21 dimerization nor Tim50–Tim21 interaction were affected by presequence peptides under these conditions (Figure 4D). This result indicated that presequences did not directly affect crosslinking of Tim50 to Tim21 in vitro, but rather that another translocase component plays a role in the presequence-triggered dissociation of Tim50 from Tim21 observed in organello.
The IMS-domain of Tim23 facilitates interactions between Tim21 IMS and Tim50
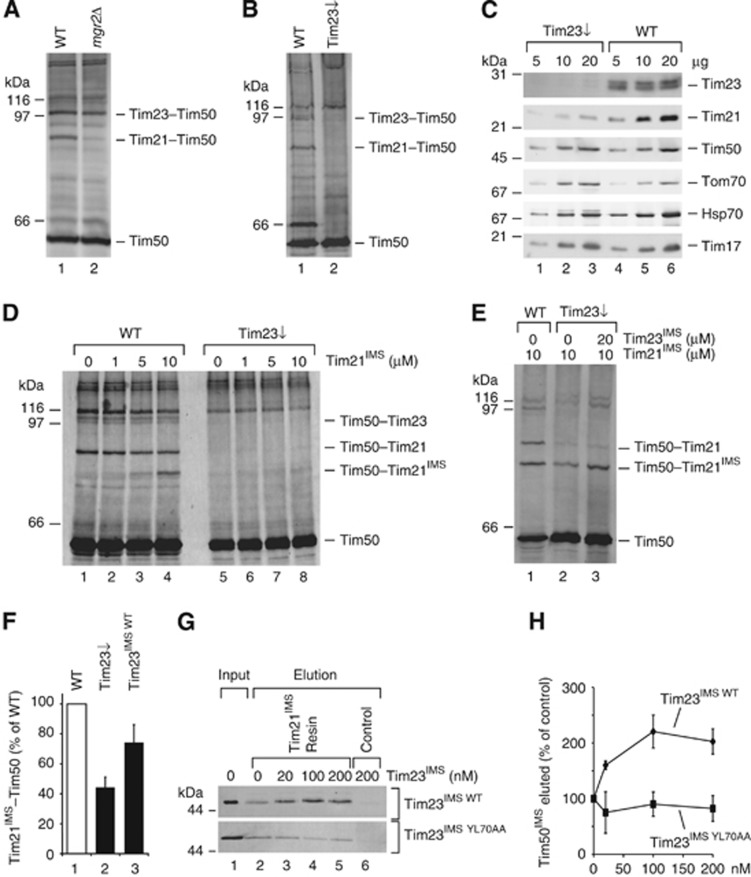
Do Tim21 and Tim50 interact under physiological conditions only within the TIM23 complex or can the interaction occur independently of their association with the presequence translocase? To address this question, Cu2+ crosslinking was performed in mgr2Δ mitochondria, in which Tim21 is not associated with the presequence translocase (Gebert et al, 2012). The Tim21–Tim50 crosslinking product was drastically reduced in mutant mitochondria, whereas the Tim23–Tim50 crosslinking product was not significantly affected (Figure 5A). Hence, the Tim21–Tim50 interaction in organello requires the presence of both components, Tim21 and Tim50, at the TIM23 complex, although in vitro the IMS-domains of both proteins can interact directly.
Figure 5.
The IMS-domain of Tim23 facilitates interaction between Tim21IMS and Tim50. (A, B) Cu2+ crosslinking was performed in wild-type and (A) mgr2Δ or (B) Tim23 shut down mitochondria (Tim23↓), followed by immunoblotting against Tim50. (C) Indicated amounts of mitochondria from wild-type and Tim23↓ strains were analysed by SDS–PAGE and immunoblotting. (D) Mitoplasts generated from wild-type or Tim23↓ mitochondria were subjected to crosslinking with Cu2+ after incubation with the indicated concentrations of Tim21IMS. (E) Mitochondria were treated like in (D), but Tim23IMS was added where indicated prior to crosslinking. (F) Tim50–Tim21IMS crosslink intensities from independent experiments (performed as in E) after treatment with 20 μM Tim23IMS (n=9) or buffer (n=11) were quantified. Data are presented as mean±s.e.m. (G) Purified Tim21IMS was immobilized on CNBr-activated Sepharose and incubated with 20 nM Tim50IMS in the presence of the indicated Tim23IMS WT or Tim23IMS YL70AA concentrations. Bound Tim50 was eluted with 0.1 M glycin, pH 2.5 and analysed by SDS–PAGE followed by immunoblotting against Tim50. (H) Quantification of Tim50IMS signal intensities, as in (G) (presented as mean±s.e.m., n≥3).
In addition to Tim50, the IMS-domain of Tim23 recognizes presequences (Truscott et al, 2001; de la Cruz et al, 2010); therefore, we speculated that Tim23 could play an important role in the signal-stimulated dissolution of the Tim21–Tim50 interaction. Hence, we analysed the Tim21–Tim50 interaction in mitochondria with reduced levels of Tim23. These experiments were performed in wild-type mitochondria and mitochondria from a strain in which TIM23 was placed under the regulatable Gal promotor and reduced to <5% of the wild-type protein level (Schulz et al, 2011). While the Tim50–Tim23 and Tim50–Tim21 crosslink-adducts were readily detectable in wild-type mitochondria, both were significantly reduced when Tim23 levels were lowered (Figure 5B). As expected, a lack of Tim23 prevents crosslinking-adduct formation between Tim50 and Tim23. When we compared the steady-state levels of selected mitochondrial proteins in Tim23 downregulated mitochondria, we realized that Tim21, which is targeted to the inner membrane by a presequence and is a TIM23 substrate itself, was significantly reduced in these mitochondria (Figure 5C). In contrast, levels of Tim50, that is relatively stable in mitochondria (Geissler et al, 2002), were not affected. This finding explained the loss of Tim21–Tim50 crosslink-adducts in mutant mitochondria. At the same time, this loss of Tim21 allowed us to establish an in organello complementation assay to analyse the interaction between Tim21 and Tim50 as well as its dependency on Tim23IMS. Therefore, we generated mitoplasts from Tim23 downregulated mitochondria and supplemented purified Tim21IMS prior to Cu2+-induced crosslinking. While we observed efficient adduct formation between Tim21 and Tim50 as well as among Tim21IMS and Tim50 in wild-type mitoplasts, strongly reduced crosslinking was observed in the absence of Tim23 (Figure 5D). Thus, despite the addition of excess Tim21IMS, the interaction between Tim50 and Tim21IMS could not be efficiently reconstituted in the absence of Tim23 (Figure 5D, lanes 6–8). Based on this finding, we speculated that the interaction of Tim21 with Tim50 was dependent upon the presence of Tim23. We utilized our test system to directly assess this hypothesis and added Tim21IMS to Tim23 downregulated mitoplasts, where one sample received purified Tim23IMS and was omitted in the second (Figure 5E and F). Accordingly, addition of Tim23IMS stimulated the interaction between Tim50 and Tim21IMS. This finding indicated that the interaction between Tim50 and Tim21 required the receptor domain of Tim23 in mitoplasts, even though both proteins are capable of complex formation in vitro. Moreover, the membrane portion of the Tim23 channel was shown to be dispensable in promoting Tim21–Tim50 binding.
We then asked if Tim23IMS could mediate Tim50–Tim21 complex formation, as seen in in organello experiments, through its direct interaction with both Tim50 and Tim21. The interaction between the IMS-domains of Tim50 and Tim23 have previously been reported to exhibit an KD of ∼60 μM (Gevorkyan-Airapetov et al, 2009). Moreover, the IMS-domain of Tim23 was shown to be in close proximity to Tim21 in mitochondria (Tamura et al, 2009), but no quantitative data on the affinity of this interaction is available. Therefore, we analysed the binding affinity of Tim21IMS to immobilized Tim23IMS in an in vitro system via SPR (Supplementary Figure S2A). Our results confirmed their direct interaction with an KD of ∼1 μM (Table I), suggesting that Tim23 could exert its coordinating effect on Tim21–Tim50 complex formation via its direct interaction with both proteins.
To further investigate the molecular mechanisms responsible for this effect, we purified a Tim23IMS YL70AA mutant protein, which was previously shown to be deficient in its interaction with Tim50 (Gevorkyan-Airapetov et al, 2009). Indeed, a crosslinking-adduct was formed between Tim50IMS and Tim23IMS, but not with Tim23IMS YL70AA in an in vitro chemical crosslinking experiment (Supplementary Figure S2B). At the same time, SPR analysis showed that the mutant’s interaction with Tim21 was not affected (Supplementary Figure S2C). Therefore, we used Tim23IMS YL70AA in the above described in organello crosslinking assay. In contrast to Tim23IMS, Tim23IMS YL70AA did not significantly increase the amount of the Tim21IMS–Tim50 crosslink in Tim23 downregulated mitoplasts (Supplementary Figure S2D).
To address the effect of Tim23IMS on Tim21–Tim50 by an alternative approach, we performed pull-down experiments. Therefore, purified Tim50IMS was added to Tim21IMS immobilized on Sepharose beads in the presence of increasing concentrations of Tim23IMS. Addition of wild-type Tim23IMS significantly increased the binding of Tim50IMS to the Tim21 beads, whereas Tim23IMS YL70AA did not promote this binding (Figure 5G and H). Together, these data show that the IMS-domain of Tim23 positively affects the interaction between the IMS-domains of Tim21 and Tim50, and that Tim23 interaction with Tim50 is required for this effect.
Presequence recognition affects translocase organization in mitochondria
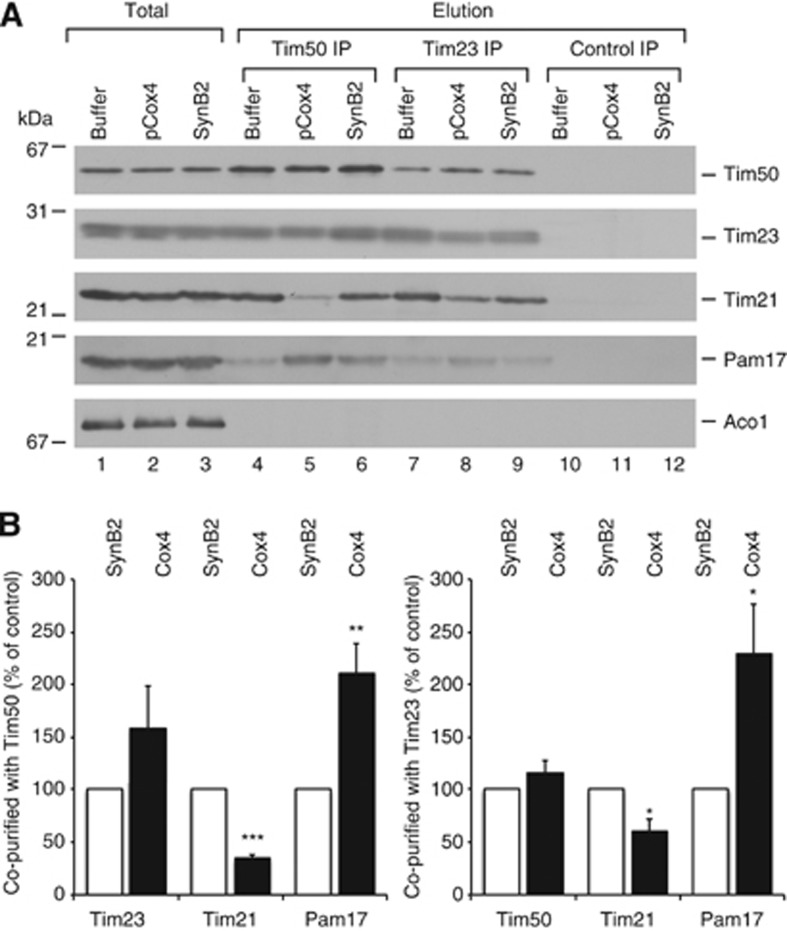
Our analyses revealed that an intricate connection between the IMS-domains of Tim21, Tim50, and Tim23 exists in mitochondria and that the interplay of these proteins maintains Tim21 in close proximity to the presequence receptor Tim50. The observed dissociation of Tim21 from Tim50 in mitochondria in response to targeting signals suggested that signal recognition could trigger thus far undefined downstream events at the translocase. To address this, we incubated mitochondria with presequence peptides (pCox4), an inactive variant thereof (SynB2) and a buffer control, and subsequently performed immunoprecipitation (IP) analyses using antisera against Tim50 or the Tim23 channel. This experimental set up enabled the comparison between primed (pCox4 incubated) and non-primed (SynB2 and buffer incubated) TIM23. Tim50 antiserum efficiently immunoprecipitated Tim50 together with Tim23 from mitochondrial extracts irrespective of the presence of presequence peptides (as shown by the lack of statistical significance of the large sample set, n=7). However, the co-precipitation of Tim21 was drastically affected by presequence peptide addition, while the inactive variant had no effect. Interestingly, the motor constituent Pam17 displayed the reciprocal phenotype, where Pam17 was enriched in the IP in the presence of presequence peptides, but not when the inactive variant was used (Figure 6A and B). As the Pam17 interaction site at the translocase is unknown, we aimed to ensure that the observed effect reflected alterations at the presequence translocase rather than on Tim50 alone. Thus, we performed IPs of the Tim23 channel under the identical conditions, and correspondingly found dissociation of Tim21 from the translocase in the presence of targeting signals, while at the same time Pam17 recruitment (Figure 6A and B). In conclusion, our analyses revealed a signal-mediated rearrangement of the presequence translocase in which a Tim21–Tim50 interaction is relieved leading to the dissociation of Tim21 from the translocase accompanied by Pam17 recruitment. Moreover, in organello crosslinking revealed that the presequence-triggered dissociation of the Tim21–Tim50 crosslink does not require the PBD-domain of Tim50 (Figure 4C). An IP experiment performed in mitochondria, in which wild-type Tim50 was downregulated and replaced by full-length (Tim50HA) or truncated Tim50 (Tim501–361–HA) additionally confirmed this observation (Supplementary Figure S2E).
Figure 6.
Presequences affect interactions at the TIM23 complex. (A) Co-IP was performed with Tim50 or Tim23 antisera from digitonin-solubilized mitochondria after treatment with pCox4, SynB2, or corresponding buffer. Control IP, unrelated antiserum was used as a specificity control. Eluted proteins were analysed by SDS–PAGE and immunoblotting. Total, 7%; Elution, 100%. (B) The results of seven independent IP experiments after pCox4 and SynB2 treatment utilizing Tim50 (left panel) or Tim23 (right panel) antisera (as in A) were quantified and normalized to the amount of precipitated Tim50 (for the Tim50 IP) or Tim23 (for the Tim23 IP); the SynB2 peptide control was set to 100%. Data are presented as mean±s.e.m. (n=7). The statistical significance of the changes was evaluated using a two-sided t-test. *P<0.05; **P<0.01; ***P<0.0001.
Discussion
The presequence translocase (TIM23 complex) is uniquely competent in both matrix translocation and lateral release of substrates into the inner mitochondrial membrane (for reviews see Mokranjac and Neupert, 2007; Chacinska et al, 2009; Endo and Yamano, 2010; Dudek et al, 2013). Previous work revealed functionally distinct forms of the presequence translocase (Chacinska et al, 2005, 2010; van der Laan et al, 2007; Popov-Čeleketić et al, 2008; Saddar et al, 2008); however, the mechanism by which targeting information is decoded, that is, the process in which TIM23 switches between its different functional states, has long been elusive. Recent works investigating TIM23’s presequence receptors have identified Tim50 as the primary inner membrane presequence receptor and have begun to breakdown its various functional domains, which have lead to the structural and functional characterization of both presequence and Tim23 binding (Marom et al, 2011; Qian et al, 2011; Schulz et al, 2011). These initial studies provide the basis of signal recognition at the TIM23 complex. Here, we built our analyses upon the available data on presequence receptors to understand how signal deciphering integrates into translocase dynamics (Figure 7). Thus, we set out to identify Tim50 interaction partners of the negatively charged binding groove (Qian et al, 2011). Our analyses identified a Tim23 coordinated Tim50–Tim21 interaction. The high affinity Tim50–Tim21 interaction is dynamic in nature as it responds to presequences. At the translocase, the presequence-mediated Tim50–Tim21 dissociation occurs in parallel with Pam17 recruitment. Previous analyses established that the Pam17-bound state of the translocase reflects the initial import motor recruitment stage: Pam17 binding is necessary for subsequent Pam16/Pam18 association (van der Laan et al, 2005; Popov-Čeleketić et al, 2008; Schiller, 2009). Hence, the observed antagonistic mode of TIM23 binding by Tim21 and Pam17 (van der Laan et al, 2006b; Popov-Čeleketić et al, 2008; Chacinska et al, 2010) can now be seen as signal triggered functional regulation, allowing for the initiation of matrix translocation. In this regard, our findings contribute quantitative mechanistic insight into the immediate early steps involved in TIM23 complex isoform switching and reveal that this switch represents a signal-mediated process.
Figure 7.
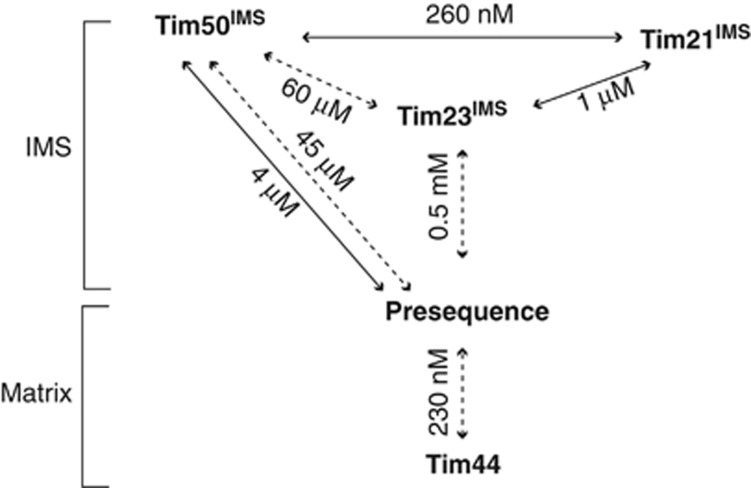
Affinities of protein–protein interactions within the presequence pathway. KD values of the interactions occurring in the mitochondrial IMS and on the matrix side of the inner mitochondrial membrane are indicated. Solid lines, affinities addressed in this study. Dashed lines, affinity data from Gevorkyan-Airapetov et al (2009); de la Cruz et al (2010); Marom et al (2011).
Integrating the results presented in this study with established work, we advocate the following TIM23 presequence priming model. Presequences are captured at the inner membrane by Tim50 (Marom et al, 2011; Schulz et al, 2011), leading to the release of Tim21 from Tim50. Tim50, with bound presequence, interacts with Tim23, an interaction seen here and previously in Alder et al (2008) to be dynamic. As Tim23’s presequence-binding site has been shown to overlap with its Tim50-binding site (Geissler et al, 2002; Yamamoto et al, 2002; Mokranjac and Sichting, 2009; Schulz et al, 2011), Tim50’s association with Tim23 causes presequence release from Tim50 and hand-over to the Tim23 channel in a membrane potential dependent manner (Bauer et al, 1996; Schulz et al, 2011). Tim50’s association with Tim23 results in Tim21 dissociation from the TIM23 complex and subsequent association of Pam17. This recruitment is a prerequisite for Pam16/18 complex association to regulate the ATPase cycle of mtHsp70 during matrix translocation of a precursor protein. As the initial stages of precursor transport addressed here are similar for all presequence-containing precursors irrespective of their target compartment, matrix or inner membrane, the changes in presequence translocase organization in response to presequence recognition should represent a general phenomenon. Investigation into additional signals encoded within the precursor will likely be required in order to understand further subunit exchange within the translocase during transport, which would then lead to lateral precursor release into the lipid phase or full matrix translocation.
Materials and methods
Yeast strains and preparation of mitochondria
Wild-type strains YPH499 and MB29 were used. Tim21ProtA, Tim23ProtA, Tim23HA, Tim50HA, Tim23↓, mgr2Δ, and tim21Δ strains have been published previously (Geissler et al, 2002; Chacinska et al, 2005; Schulz et al, 2011; Gebert et al, 2012). To restore Tim21 expression in the tim21Δ strain or to co-express it with Tim21ProtA, corresponding yeast strains were transformed with a pFL39-based vector expressing full-length Tim21 under the control of its native promoter (kindly provided by Dr B Guiard).
For expression of truncated and tagged versions of Tim50 from a plasmid, with simultaneous downregulation of wild-type Tim50 expression, the strain containing TIM50 under the control of the GAL1 promoter (Geissler et al, 2002) was transformed with plasmids encoding amino acids 1–361 of Tim50 (Tim501–361–HA) or full-length Tim50 (Tim50HA) with a single C-terminal HA-tag, under the control of the MET25 promotor (Mumberg et al, 1994). The resulting strains (yCS2 and yCS30) were precultured in selective lactate medium (0.67% YNB, 3% lactate, 0.003% lysine, 0.002% adenine, histidine, tryptophan, and uracil (pH 5.0)) with 1% galactose and 1% raffinose. Afterwards, the cells were grown in the same selective medium, however, containing 0.01% glucose, for 38 h at 30°C (Schulz et al, 2011).
For in vivo expression of different Tim50 constructs (Tim501–476–HA; Tim501–365–HA; Tim501–361–HA or Tim501–361), corresponding TIM50 fragments were cloned into pME2782 (Mumberg et al, 1994). These plasmids were used to replace the URA3 carrying plasmid encoding the wild-type protein in the gene deletion strain (Chacinska et al, 2005; Schulz et al, 2011).
All other yeast strains, if otherwise not stated, were grown at 30°C in YPG medium (1% yeast extract, 2% peptone, and 3% glycerol). Strains bearing the Tim21 expression plasmid were grown on synthetic medium containing 0.67% yeast nitrogen base and amino-acid drop-out mix without tryptophan (MP Biomedicals), supplemented with 3% glycerol. Mitochondria were isolated according to a conventional procedure (Meisinger et al, 2006) and stored at −80°C.
Protein expression and purification
Plasmids encoding N-terminally His-tagged Tim50IMS, Tim50PBD, Tim50ΔPBD (Schulz et al, 2011), Tim21IMS (Albrecht et al, 2006), and Tim23IMS (Truscott et al, 2001) were used. Tim21C128S was derived from a wild-type Tim21IMS-containing plasmid using the QuickChange site-directed mutagenesis kit (Stratagene) with forward primer 5′-ataagaagtttgttacagagcgacgatggcattacgg-3′ and reverse primer 5′-ccgtaatgccatcgtcgctctgtaacaaacttcttat-3′. The YL70AA substitution (Gevorkyan-Airapetov et al, 2009) was introduced into wild-type Tim23IMS by site-directed mutagenesis using primers 5′-gtctagacaagggtgtggaggcggcggatctggaagaagaacaac-3′ and 5′-gttgttcttcttccagatccgccgcctccacacccttgtctagac-3′ by QuickChange site-directed mutagenesis kit (Stratagene). To express Tim50C (Qian et al, 2011), a PCR-amplified fragment encoding amino acids 164–361 was cloned into pPROEX HTc (Invitrogen). All proteins were expressed in the Escherichia coli strain BL21 in LB medium at 37°C for 4 h after induction with 1 mM IPTG. Cells were opened using an EmulsiFlex-C3 homogenizer (Avestin) and proteins were purified from the soluble fraction by immobilized metal affinity chromatography on HisTrap HP columns (GE Healthcare). For crosslinking experiments, proteins were dialyzed against buffer containing 100 mM NaCl and 20 mM HEPES, pH 7.2 (for Tim21IMS, Tim21C128S, and Tim23IMS) or pH 8.0 (for Tim50IMS and Tim50C). For SPR measurements, all proteins were dialyzed against the SPR buffer (150 mM NaCl, 50 mM HEPES-NaOH, pH 7.4, and 50 μM EDTA). If necessary, the His tag was excised via TEV protease, which was removed, together with uncleaved protein, by Ni-NTA agarose beads (Qiagen). Purified proteins were snap-frozen in liquid nitrogen and stored at −80°C.
Co-immunoprecipitation
Co-IP experiments were performed according to Herrmann et al (2001). Rabbit Tim23 and Tim50 antisera or mouse monoclonal anti-HA IgG (12CA5) were coupled to Protein A Sepharose beads (GE Healthcare) by dimethyl pimelimidate crosslinking. Mitochondria were resuspended at a concentration of 1 mg protein/ml in solubilization buffer without digitonin (20 mM Tris–HCl, pH 7.4, 80 mM NaCl, 10% glycerol, 1 mM PMSF, and 0.5 mM EDTA), containing 50 μM peptide (pCox4 or SynB2) or an equal volume of 10 mM acetic acid, and incubated at 4°C for 20 min. Immediately following incubation, digitonin was added to a final concentration of 1%, and mitochondria were left to solubilize for 15 min at 4°C, followed by a clarifying spin at 20 000 g for 10 min. The clarified supernatant was incubated with the antibody-coupled beads for 1–3 h at 4°C. After washing the beads 10 times with 10 bed volumes of solubilization buffer, bound proteins were eluted (thrice with two bed volumes of 0.1 M glycine, pH 2.5, neutralized with Tris–HCl, pH 8, to a final concentration of 0.4 M for the Tim23 and Tim50 IP, or with four bed volumes of solubilization buffer containing HA peptide at a final concentration of 0.5 mg/ml for the HA IP), TCA precipitated and analysed by SDS–PAGE and immunoblotting.
In vitro pull-down
Tim21IMS-His was crosslinked to CNBr-activated Sepharose (GE Healthcare) in coupling buffer (0.1 M NaHCO3, pH 8.3, and 0.5 M NaCl) overnight at 4°C, and the resin was blocked with 0.1 M Tris–HCl pH 8.0 for 2 h at room temperature. The Tim21-Sepharose (bed volume 50 μl) was added to 20 nM Tim50IMS in the presence of different Tim23IMS concentrations in a binding buffer (20 mM HEPES, pH 7.4, 100 mM potassium acetate, 10 mM magnesium acetate, 10% glycerol, and 0.5% Triton X-100) at a final volume of 400 μl. After a 1-h incubation at 4°C and washing 10 times with 10 bed volumes of binding buffer, the bound Tim50 was eluted by 0.1 M glycin, pH 2.5. The eluate was TCA precipitated, analysed by SDS–PAGE and western blotting with anti-Tim50 immunodecoration.
In vitro crosslinking
Chemical crosslinking of recombinant proteins was performed in crosslinking buffer containing 20 mM HEPES-NaOH, pH 7.2, and 100 mM NaCl. Indicated amounts of purified protein or synthetic peptides were mixed, incubated for 10 min on ice, and a crosslinker (1 mM CuSO4 or 100 μM DFDNB) was added. After a 30-min incubation on ice, the reaction was quenched by the addition of 10 mM EDTA (for Cu2+ crosslinking) or 140 mM Tris, pH 7.4 (for DFDNB crosslinking) and SDS protein loading buffer. In all, 5% β-mercaptoethanol was added to non-crosslinked and DFDNB-crosslinked samples before loading the gel. Samples were analysed by SDS–PAGE and subsequent Coomassie staining or immunoblotting.
For in vitro photocrosslinking, photopeptides pL19B and pS16B were added to purified proteins, UV-irradiated for 30 min on ice and analysed by SDS–PAGE and western blotting, as previously described (Schulz et al, 2011).
In organello crosslinking
Isolated mitochondria or mitoplasts (obtained by osmotic swelling of mitochondria in 10 mM MOPS-KOH, pH 7.2, and 1 mM EDTA for 20 min on ice) were resuspended in isotonic SH buffer (containing 0.6 M sorbitol and 20 mM HEPES-KOH, pH 7.2) or hypotonic crosslinking buffer (20 mM HEPES-NaOH, pH 7.2, and 100 mM NaCl). Unless otherwise indicated, 100 μg of mitochondrial protein per sample was used. When required, the samples were treated with corresponding proteins, synthetic peptides or other substances for 10 min on ice. After that, crosslink formation was induced through the addition of 1 mM CuSO4. Samples were incubated for 30 min on ice, and subsequently quenched by adding 10 mM EDTA. Mitochondria were collected by centrifugation, resuspended in SDS protein loading buffer and analysed by conventional SDS–PAGE and immunoblotting.
Surface plasmon resonance
SPR measurements were done using a Reichert SPR Biosensor SR7000DC with a Ni2+ chelator sensorchip NiHC500m (XanTec bioanalytics), in running buffer containing 50 mM HEPES, pH 7.4, 150 mM NaCl, and 50 μM EDTA, at 20°C, at a flow rate of 40 μl/min. A 6 × His-tagged ligand (Tim23IMS, Tim50IMS, or Tim50C) was immobilized on the Ni2+-activated left (sample) channel of the chip at a concentration of 250 nM with a flow rate of 30 μl/min to response levels of 2500–3500 RU. The right channel served as a reference. Increasing concentrations of analyte (Tim21IMS, pCox4, SynB2, pALDH, or pALDH-s) in running buffer were injected for 270 s to both channels, and dissociation was followed for 12 min. The difference in response between sample and reference channels was recorded. Affinity and kinetic data analysis was performed using Scrubber 2.0 (BioLogic Software). The response was referenced to blank (buffer) injections and normalized using the molecular weight of the ligand (in kDa).
Miscellaneous
Presequence peptides pALDH (MLRAALSTARRGPRLSRLLSAA) and pCox4 (MLSLRQSIRFFKPATRTLSSSRYLL) and their inactive versions pALDH-s (MLRGKQPTKSLLPQRSPKLSAAA) and SynB2 (MLSRQQSQRQSRQQSQRQSRYLL) (Allison and Schatz, 1986; Abe et al, 2000; Schulz et al, 2011) were purchased from JPT Peptide Technologies. SDS–PAGE, immunoblotting and colloidal Coomassie staining were carried out according to the standard protocols. After immunoblotting, protein signals were detected by fluorescent dye-coupled secondary antibody (LI-COR) or HRP-coupled antibody and enhanced chemiluminescence (GE Healthcare). Fluorescent signals were detected using an FLA-9000 scanner (Fujifilm) and quantified by ImageQuant TL software (GE Healthcare). All quantitative data are presented as mean±standard error of the mean (s.e.m.). No image processing, with the exception of cropping and contrast adjustment using Adobe Photoshop CS3 and Adobe Illustrator CS3, was applied.
Supplementary Material
Acknowledgments
We are indebted to Drs Schwappach, Mick, Dudek, and Deckers for helpful discussion. We thank K Neifer and B Zapisek for expert technical support. We are grateful to Dr van der Laan for the mgr2Δ strain, Dr Wiedemann for constructs, and to Dr Geiss-Friedlander for antibodies and peptides. The study was supported by the Deutsche Forschungsgemeinschaft, SFB860, the Göttingen Graduate School for Neurosciences and Molecular Biosciences, the Max-Planck-Society (PR), and FOR 1086/2 (Grant SCHW 823/2-1) (MK, SPR Biosensor SR7000DC). OL, JM, and CS are doctoral students of the Ph.D. Programme ‘Molecular Biology’—International Max Planck Research School and the Göttingen Graduate School for Neurosciences and Molecular Biosciences (GGNB) (DFG Grant GSC 226/1) at the Georg August University Göttingen. CS is a recipient of a doctoral fellowship from the Boehringer Ingelheim Fonds.
Author contributions: OL, JM, CS, MK, and DPH performed the experiments. PR led the project. OL, JM, and PR wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abe Y, Shodai T, Muto T, Mihara K, Torii H, Nishikawa S, Endo T, Kohda D (2000) Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell 100: 551–560 [DOI] [PubMed] [Google Scholar]
- Albrecht R, Rehling P, Chacinska A, Brix J, Cadamuro SA, Volkmer R, Guiard B, Pfanner N, Zeth K (2006) The Tim21 binding domain connects the preprotein translocases of both mitochondrial membranes. EMBO Rep 7: 1233–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alder NN, Sutherland J, Buhring A (2008) Quaternary structure of the mitochondrial TIM23 complex reveals dynamic association between Tim23p and other subunits. Mol Biol Cell 19: 159–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison DS, Schatz G (1986) Artificial mitochondrial presequences. Proc Natl Acad Sci USA 83: 9011–9015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer MF, Sirrenberg C, Neupert W, Brunner M (1996) Role of Tim23 as voltage sensor and presequence receptor in protein import into mitochondria. Cell 87: 33–41 [DOI] [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N (2009) Importing mitochondrial proteins: machineries and mechanisms. Cell 138: 628–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, Lind M, Frazier AE, Dudek J, Meisinger C, Geissler A, Sickmann A, Meyer HE, Truscott KN, Guiard B, Pfanner N, Rehling P (2005) Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell 120: 817–829 [DOI] [PubMed] [Google Scholar]
- Chacinska A, van der Laan M, Mehnert CS, Guiard B, Mick DU, Hutu DP, Truscott KN, Wiedemann N, Meisinger C, Pfanner N, Rehling P (2010) Distinct forms of mitochondrial TOM-TIM supercomplexes define signal-dependent states of preprotein sorting. Mol Cell Biol 30: 307–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz L, Bajaj R, Becker S, Zweckstetter M (2010) The intermembrane space domain of Tim23 is intrinsically disordered with a distinct binding region for presequences. Protein Sci 19: 2045–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker PJ, Martin F, Maarse AC, Bömer U, Müller H, Guiard B, Meijer M, Rassow J, Pfanner N (1997) The Tim core complex defines the number of mitochondrial translocation contact sites and can hold arrested preproteins in the absence of matrix Hsp70-Tim44. EMBO J 16: 5408–5419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezal P, Likic V, Tachezy J, Lithgow T (2006) Evolution of the molecular machines for protein import into mitochondria. Science 313: 314–318 [DOI] [PubMed] [Google Scholar]
- Dudek J, Rehling P, van der Laan M (2013) Mitochondrial protein import: Common principles and physiological networks. Biochim Biophys Acta 1833: 274–285 [DOI] [PubMed] [Google Scholar]
- Endo T, Yamano K (2010) Transport of proteins across or into the mitochondrial outer membrane. Biochim Biophys Acta 1803: 706–714 [DOI] [PubMed] [Google Scholar]
- Gambill BD, Voos W, Kang PJ, Miao B, Langer T, Craig EA, Pfanner N (1993) A dual role for mitochondrial heat shock protein 70 in membrane translocation of preproteins. J Cell Biol 123: 109–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebert M, Schrempp SG, Mehnert CS, Heißwolf AK, Oeljeklaus S, Ieva R, Bohnert M, von der Malsburg K, Wiese S, Kleinschroth T, Hunte C, Meyer HE, Haferkamp I, Guiard B, Warscheid B, Pfanner N, van der Laan M (2012) Mgr2 promotes coupling of the mitochondrial presequence translocase to partner complexes. J Cell Biol 197: 595–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler A, Chacinska A, Truscott KN, Wiedemann N, Brandner K, Sickmann A, Meyer HE, Meisinger C, Pfanner N, Rehling P (2002) The mitochondrial presequence translocase: an essential role of Tim50 in directing preproteins to the import channel. Cell 111: 507–518 [DOI] [PubMed] [Google Scholar]
- Gevorkyan-Airapetov L, Zohary K, Popov-Čeleketić D, Mapa K, Hell K, Neupert W, Azem A, Mokranjac D (2009) Interaction of Tim23 with Tim50 Is essential for protein translocation by the mitochondrial TIM23 complex. J Biol Chem 284: 4865–4872 [DOI] [PubMed] [Google Scholar]
- Glick B, Beasley E, Schatz G (1992) Protein sorting in mitochondria. Trends Biochem Sci 17: 453–459 [DOI] [PubMed] [Google Scholar]
- Heijne GV (1986) Mitochondrial targeting sequences may form amphiphilic helices. EMBO J 5: 1335–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann JM, Westermann B, Neupert W (2001) Analysis of protein-protein interactions in mitochondria by coimmunoprecipitation and chemical cross-linking. Methods Cell Biol 65: 217–230 [DOI] [PubMed] [Google Scholar]
- Hurt EC, Pesold-Hurt B, Schatz G (1984) The cleavable prepiece of an imported mitochondrial protein is sufficient to direct cytosolic dihydrofolate reductase into the mitochondrial matrix. FEBS Lett 178: 306–310 [DOI] [PubMed] [Google Scholar]
- Marom M, Dayan D, Demishtein-Zohary K, Mokranjac D, Neupert W, Azem A (2011) Direct interaction of mitochondrial targeting presequences with purified components of the TIM23 complex. J Biol Chem 286: 43809–43815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinecke M, Wagner R, Kovermann P, Guiard B, Mick DU, Hutu DP, Voos W, Truscott KN, Chacinska A, Pfanner N, Rehling P (2006) Tim50 maintains the permeability barrier of the mitochondrial inner membrane. Science 312: 1523–1526 [DOI] [PubMed] [Google Scholar]
- Meisinger C, Pfanner N, Truscott KN (2006) Isolation of yeast mitochondria. Methods Mol Biol 313: 33–39 [DOI] [PubMed] [Google Scholar]
- Mick DU, Dennerlein D, Wiese H, Reinhold R, Pacheu-Grau D, Lorenzi I, Sasarman F, Weraarpachai W, Shoubridge EA, Warscheid B, Rehling P (2012) MITRAC links mitochondrial protein translocation to respiratory-chain assembly and translational regulation. Cell 151: 1528–1541 [DOI] [PubMed] [Google Scholar]
- Mokranjac D, Neupert W (2007) Protein import into isolated mitochondria. Methods Mol Biol 372: 277–286 [DOI] [PubMed] [Google Scholar]
- Mokranjac D, Popov-Čeleketić D, Hell K, Neupert W (2005) Role of Tim21 in mitochondrial translocation contact sites. J Biol Chem 280: 23437–23440 [DOI] [PubMed] [Google Scholar]
- Mokranjac D, Sichting M (2009) Role of Tim50 in the Transfer of Precursor Proteins from the Outer to the Inner Membrane of Mitochondria. Mol Biol Cell 20: 1400–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro F, Sirrenberg C, Schneider HC, Neupert W, Brunner M (1999) The TIM17.23 preprotein translocase of mitochondria: composition and function in protein transport into the matrix. EMBO J 18: 3667–3675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Müller R, Funk M (1994) Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res 22: 5767–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov-Čeleketić D, Mapa K, Neupert W, Mokranjac D (2008) Active remodelling of the TIM23 complex during translocation of preproteins into mitochondria. EMBO J 27: 1469–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Gebert M, Höpker J, Yan M, Li J, Wiedemann N, van der Laan M, Pfanner N, Sha B (2011) Structural basis for the function of Tim50 in the mitochondrial presequence translocase. J Mol Biol 411: 513–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saddar S, Dienhart MK, Stuart RA (2008) The F1F0-ATP synthase complex influences the assembly state of the cytochrome bc1-cytochrome oxidase supercomplex and its association with the TIM23 machinery. J Biol Chem 283: 6677–6686 [DOI] [PubMed] [Google Scholar]
- Schiller D (2009) Pam17 and Tim44 act sequentially in protein import into the mitochondrial matrix. Int J Biochem Cell Biol 41: 2343–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz C, Lytovchenko O, Melin J, Chacinska A, Guiard B, Neumann P, Ficner R, Jahn O, Schmidt B, Rehling P (2011) Tim50’s presequence receptor domain is essential for signal driven transport across the TIM23 complex. J Cell Biol 195: 643–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y, Harada Y, Shiota T, Yamano K, Watanabe K, Yokota M, Yamamoto H, Sesaki H, Endo T (2009) Tim23-Tim50 pair coordinates functions of translocators and motor proteins in mitochondrial protein import. J Cell Biol 184: 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truscott KN, Kovermann P, Geissler A, Merlin A, Meijer M, Driessen AJM, Rassow J, Pfanner N, Wagner R (2001) A presequence- and voltage-sensitive channel of the mitochondrial preprotein translocase formed by Tim23. Nat Struct Biol 8: 1074–1082 [DOI] [PubMed] [Google Scholar]
- van der Laan M, Chacinska A, Lind M, Perschil I, Sickmann A, Meyer HE, Guiard B, Meisinger C, Pfanner N, Rehling P (2005) Pam17 is required for architecture and translocation activity of the mitochondrial protein import motor. Mol Cell Biol 25: 7449–7458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan M, Meinecke M, Dudek J, Hutu DP, Lind M, Perschil I, Guiard B, Wagner R, Pfanner N, Rehling P (2007) Motor-free mitochondrial presequence translocase drives membrane integration of preproteins. Nat Cell Biol 9: 1152–1159 [DOI] [PubMed] [Google Scholar]
- van der Laan M, Rissler M, Rehling P (2006a) Mitochondrial preprotein translocases as dynamic molecular machines. FEMS Yeast Res 6: 849–861 [DOI] [PubMed] [Google Scholar]
- van der Laan M, Wiedemann N, Mick DU, Guiard B, Rehling P, Pfanner N (2006b) A role for Tim21 in membrane-potential-dependent preprotein sorting in mitochondria. Curr Biol 16: 2271–2276 [DOI] [PubMed] [Google Scholar]
- Vögtle F-N, Wortelkamp S, Zahedi RP, Becker D, Leidhold C, Gevaert K, Kellermann J, Voos W, Sickmann A, Pfanner N, Meisinger C (2009) Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell 139: 428–439 [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Esaki M, Kanamori T, Tamura Y, Nishikawa SI, Endo T (2002) Tim50 is a subunit of the TIM23 complex that links protein translocation across the outer and inner mitochondrial membranes. Cell 111: 519–528 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.