Abstract
In eukaryotes, permanent inhibition of the non-homologous end joining (NHEJ) repair pathway at telomeres ensures that chromosome ends do not fuse. In budding yeast, binding of Rap1 to telomere repeats establishes NHEJ inhibition. Here, we show that the Uls1 protein is required for the maintenance of NHEJ inhibition at telomeres. Uls1 protein is a non-essential Swi2/Snf2-related translocase and a Small Ubiquitin-related Modifier (SUMO)-Targeted Ubiquitin Ligase (STUbL) with unknown targets. Loss of Uls1 results in telomere–telomere fusions. Uls1 requirement is alleviated by the absence of poly-SUMO chains and by rap1 alleles lacking SUMOylation sites. Furthermore, Uls1 limits the accumulation of Rap1 poly-SUMO conjugates. We propose that one of Uls1 functions is to clear non-functional poly-SUMOylated Rap1 molecules from telomeres to ensure the continuous efficiency of NHEJ inhibition. Since Uls1 is the only known STUbL with a translocase activity, it can be the general molecular sweeper for the clearance of poly-SUMOylated proteins on DNA in eukaryotes.
Keywords: NHEJ, STUbL, SUMO, telomere
Introduction
Inhibition of the non-homologous end joining (NHEJ) repair pathway at telomeres ensures that chromosome ends do not fuse (Jain and Cooper, 2010). The necessity to be continuously efficient is a key feature of NHEJ inhibition at telomeres since a temporary lapse at two telomeres could result in a telomere fusion. Although this accident is sometimes reversible (Pobiega and Marcand, 2010), it poses a major challenge to genome stability. Protection against telomere fusions depends mostly on preventing fusions from occurring. Proteins bound to telomeric repeated sequences establish this strong cis-inhibition. Its strength and reliability are the result of in part the multiplicity of DNA-bound molecules at each telomere and in part the synergy between several pathways of inhibition. In fission yeast, telomere repeats are bound by multiple Taz1 proteins that recruit Rap1 proteins. Both factors are required for NHEJ inhibition (Miller et al, 2005; Fujita et al, 2012). In mammals, NHEJ inhibition is established by the binding of multiple TRF2 proteins to the double-stranded telomere repeats (van Steensel et al, 1998; Celli and de Lange, 2005; Bae and Baumann, 2007; Sfeir and de Lange, 2012). Mammalian RAP1, recruited to telomeres by TRF2, is not essential for NHEJ inhibition (Sfeir et al, 2010). Nevertheless, an artificial targeting of RAP1 to telomeres lacking TRF2 is sufficient to inhibit NHEJ, suggesting that TRF2 establishes several redundant pathways including one involving RAP1 (Sarthy et al, 2009). In the budding yeast Saccharomyces cerevisiae, Rap1 binds telomeric DNA directly and establishes three genetically separable pathways to inhibit NHEJ (Marcand et al, 2008). Rap1 also protects telomere ends from 5′ to 3′ degradation and checkpoint signalling, and limits telomere elongation by telomerase (Teixeira et al, 2004; Negrini et al, 2007; Bonetti et al, 2010; Vodenicharov et al, 2010; Anbalagan et al, 2011; McGee et al, 2011; Ribeyre and Shore, 2012). In addition to its role at telomeres, Rap1 binds a large number of gene promoters where it plays an essential role in transcription (Lickwar et al, 2012).
In S. cerevisiae, the loss of the protein Uls1 results in a negative synthetic interaction with a rap1 hypomorphic allele, suggesting a link between Uls1 and Rap1 functions (Costanzo et al, 2010). Uls1 (also called Ris1, Dis1 or Tid4) is a non-essential Swi2/Snf2-related translocase exhibiting a DNA-dependent ATPase activity (Zhang and Buchman, 1997; Shah et al, 2010). It interacts genetically with several genes required for homologous recombination (Collins et al, 2007; Costanzo et al, 2010; Shah et al, 2010; Cal-Bakowska et al, 2011) and homologous recombination was proposed to be a target for Uls1 translocase (Chi et al, 2011). Uls1 is also a Small Ubiquitin-related MOdifier (SUMO)-Targeted Ubiquitin Ligase (STUbL) (Uzunova et al, 2007). SUMOylation is a regulatory reversible post-translational modification linking the SUMO carboxy-terminus and the ε-amino group of a lysine in the target protein. Poly-SUMO chains can be formed through the SUMOylation of SUMO monomers (Bylebyl et al, 2003; Ulrich, 2008; Sun and Hunter, 2012). In budding yeast, two STUbLs Uls1 and Slx5/Slx8 recognize and ubiquitinylate for proteosomal degradation proteins conjugated to poly-SUMO chains (Uzunova et al, 2007; Xie et al, 2007; Mullen and Brill, 2008; Nagai et al, 2008; Ulrich, 2008). Proteins targeted by Uls1 remain to be identified.
The genetic interaction between uls1 and rap1 encouraged us to address a potential function of Uls1 at telomeres. Several studies have linked SUMO and telomere functions (Askree et al, 2004; Zhao and Blobel, 2005; Potts and Yu, 2007; Xhemalce et al, 2007; Rog et al, 2009; Ferreira et al, 2011). For instance, SUMOylation of yeast telomere single-strand binding protein Cdc13 regulates its interaction with its partner Stn1 to limit telomere elongation by telomerase during S phase (Hang et al, 2011). Since cells lacking Uls1 display normal telomere length (Askree et al, 2004), we addressed a defect in NHEJ inhibition. Here, we show that Uls1 activities are required for the maintenance of NHEJ inhibition at telomeres through the clearance of non-functional poly-SUMOylated Rap1 molecules.
Results
Loss of Uls1 causes telomere fusions by NHEJ
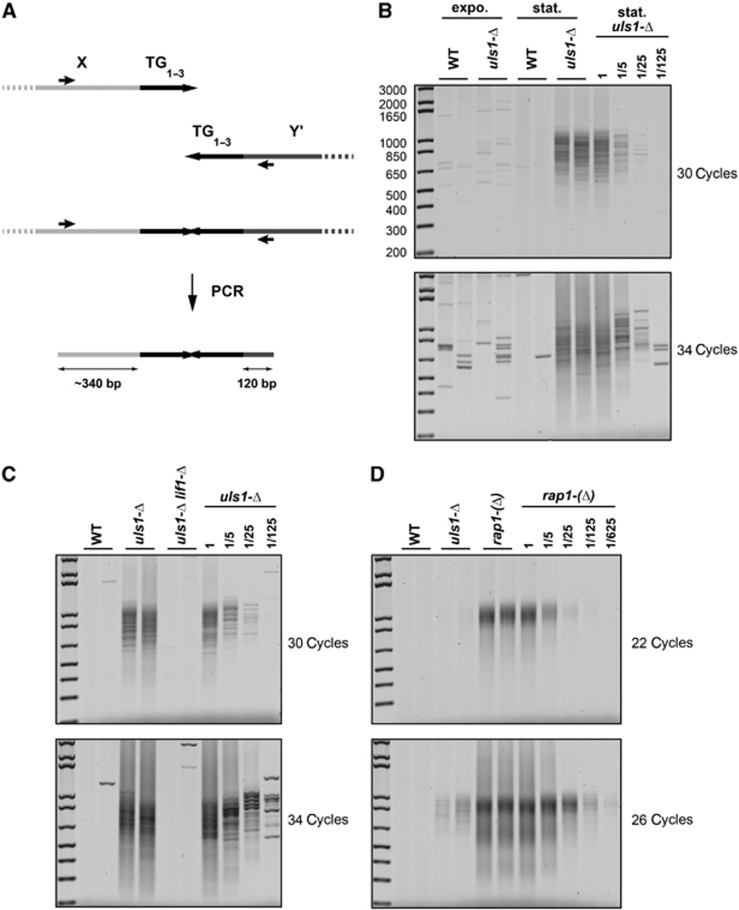
To address a defect in NHEJ inhibition at telomeres in cells lacking Uls1, we looked at the appearance of telomere fusions, which can to some extent be amplified by PCR despite their palindromic structures (Mieczkowski et al, 2003; Pardo and Marcand, 2005). All S. cerevisiae chromosome ends display a conserved X subtelomeric element. About half the chromosome ends contain one or several Y′ subtelomeric elements inserted between the X element and the telomere. We used two primers to amplify fusions between Y′ and X-only telomeres (Figure 1A). Cells were grown exponentially and then allowed to reach stationary phase. As shown in Figure 1B (top panel, 30 PCR cycles), telomere fusions are not detected in wild-type cells nor in uls1-Δ exponentially growing cells but are detected in stationary cells lacking Uls1. Telomere length being heterogeneous, amplified telomere fusions appear as a smeared signal. The lengths of the PCR products indicate that the mean length of telomeric repeats in the fusions is ∼500 bp, a length consistent with fusions occurring between telomeres close to the mean telomere length (∼300 bp). Dilution of the template DNA provides a semi-quantitative estimation of the method’s sensitivity (Figure 1B, second panel from top, 34 PCR cycles; rarer fusions are amplified as discrete bands). The relative lack of fusions in growing uls1-Δ cells could either be due to a continuous counter-selection of fused chromosomes or a reduced fusion frequency or both. In the following experiments, the presence of telomere fusions was tested in cells in stationary phase.
Figure 1.
Loss of Uls1 causes telomere fusions by NHEJ. (A) Schematic representation of the relative positions of the primers used for PCR amplification. The average wild-type telomere length is ∼300 bp. (B) Telomere fusions accumulate in stationary uls1-Δ cells. Two independent cultures of strains Lev346 (WT) and RL71 (uls1-Δ) were grown exponentially in rich medium (expo.) and allowed to reach stationary phase in 6 days (stat). Fusions between X and Y′ telomeres were amplified by PCR with 30 and 34 cycles. Genomic DNA from RL71 (uls1-Δ) cells in stationary phase was diluted serially to provide a semi-quantitative estimation of the method sensitivity. (C) Telomere fusions in uls1-Δ cells are NHEJ dependent. Strains 212-12a and 213-4a (WT), 213-7b and 213-9c (uls1-Δ) and 195-28a (uls1-Δ lif1-Δ) were grown to stationary phase. Telomere fusions were amplified with 30 and 34 cycles. Genomic DNA from 213-7b (uls1-Δ) cells was diluted serially. (D) Telomere fusions in uls1-Δ are less frequent than in rap1-(Δ) cells. Strains 210-3d and 211-1a (WT), 211-8d and 211-10b (uls1-Δ) and 209-1c and 209-2b (rap1-(Δ)) were grown to stationary phase. Telomere fusions were amplified with 22 and 26 cycles. Genomic DNA from 209-1c (rap1-(Δ)) cells was diluted serially to provide a semi-quantitative estimation of the relative difference of fusion frequencies.
To determine which repair pathway fuses telomeres, we combined the uls1-Δ mutant with an NHEJ defective lif1-Δ mutant. Lif1 loss suppresses the fusions caused by Uls1 loss (Figure 1C), indicating that fusions are produced by NHEJ. Thus, Uls1 is required for NHEJ inhibition at telomeres. To estimate how severely Uls1 loss challenges NHEJ inhibition, we compared the frequency of telomere fusions between uls1-Δ and rap1-(Δ) cells. The rap1-(Δ) mutant is a degron allele of RAP1. In rap1-(Δ) cells reaching stationary phase, Rap1 level drops and telomeres fuse at an approximate frequency of one fusion per cell (Pardo and Marcand, 2005; Pobiega and Marcand, 2010). As shown in Figure 1D, telomere fusions are detected in rap1-(Δ) cells with a lower number of PCR cycles than in uls1-Δ cells. A semi-quantitative estimation indicates that fusions are about a hundred times less frequent in uls1-Δ cells than in rap1-(Δ) cells in stationary phase, indicating that, relative to Rap1 loss, Uls1 loss causes a mild defect of NHEJ inhibition at telomeres.
Uls1 translocase and ubiquitin ligase activities are involved in NHEJ inhibition at telomeres
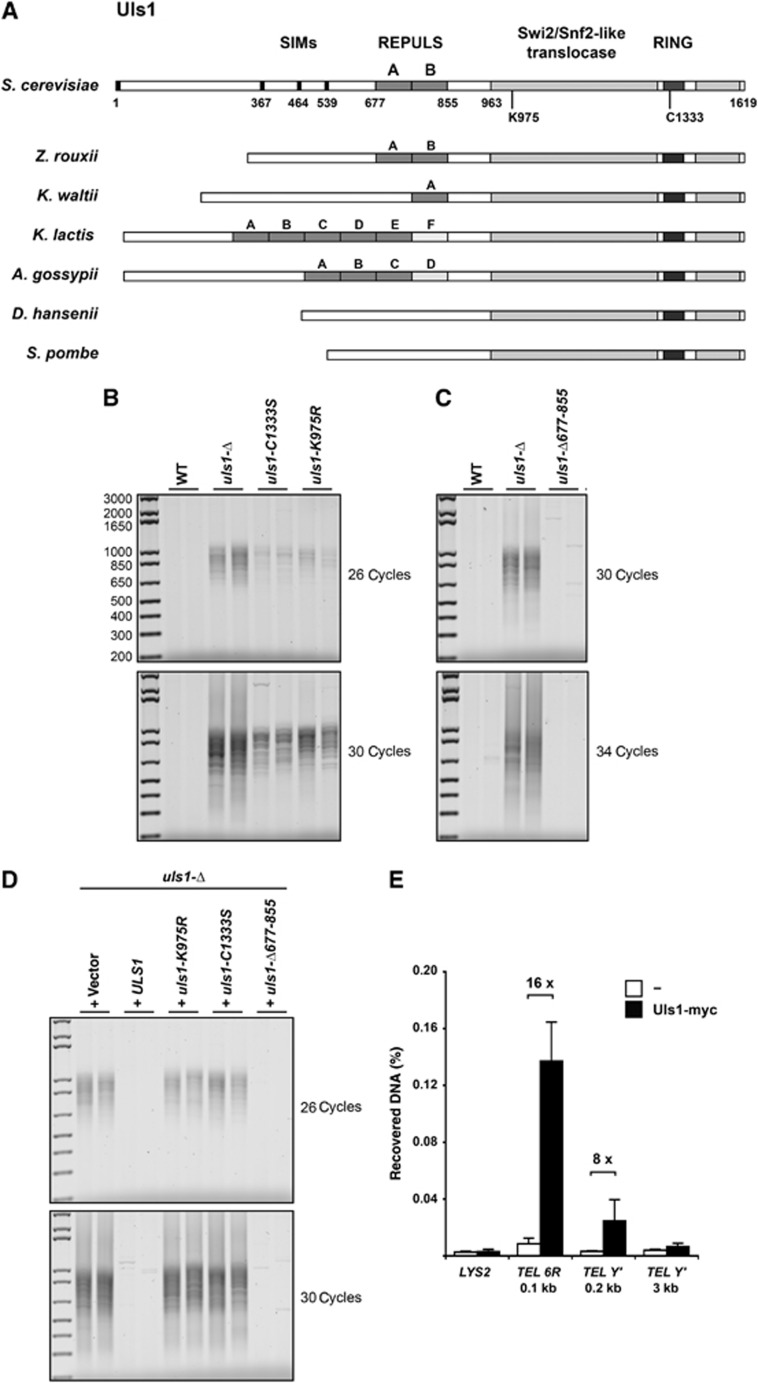
We next asked which activities of Uls1 are involved in NHEJ inhibition at telomeres (Figure 2A). The Uls1 protein possesses an ubiquitin E3 ligase RING domain and a Swi2/Snf2-like ATPase/translocase domain in its carboxy-terminal part (Zhang and Buchman, 1997; Uzunova et al, 2007; Shah et al, 2010). At least four SUMO-interacting motifs (SIM) are present in the amino-terminal region of the protein (Uzunova et al, 2007). An allele mutated within these SIMs still interacts with SUMO in a yeast two-hybrid assay (data not shown), suggesting that Uls1 may possess additional, less canonical SIMs that remain to be located (Sun and Hunter, 2012; Vogt and Hofmann, 2012). In the central region, we identified a new tandem domain that we propose to call REPULS for ‘Repeats in Uls1’. Variable number of this domain is present in Uls1 sequences from other hemisascomycetes including Kluyveromyces lactis and Ashbya gossypii but is missing in the putative Uls1 orthologues from the more distantly related species Debaromyces hansenii and Schizosaccharomyces pombe (Figure 2A; Supplementary data).
Figure 2.
Mutations in Uls1 translocase and ubiquitin ligase catalytic domains cause telomere fusions. (A) Schematic representation of S. cerevisiae Uls1 and Uls1 from other representative yeast species. REPULS domains are only present in Saccharomycetaceae, in tandem, isolated or in multiple copies, some of which being degenerated (light grey, see the multiple alignment in Supplementary Figure S1). REPULS domains are not present in hypothetical proteins likely corresponding to the Uls1 orthologues in D. hansenii (UniProt Identifier Q6BMG3) and in S. pombe (two hypothetical orthologues: UniProt Identifier O60177 and O13762). (B) Telomere fusions occur in uls1-C1333S and uls1-K975R cells. Strains 206-2b and 205-9a (WT), 206-1d and 205-14c (uls1-Δ), 199-3a (uls1-C1333S) and 200-2c and 200-5d (uls1-K975R) were grown to stationary phase. Telomere fusions were amplified with 26 and 30 cycles. (C) Telomere fusions are undetectable in usl1-Δ677-855 cells. Strains 196-11c and 196-13b (WT), 196-5a and 196-6a (uls1-Δ) and Lev791 and Lev792 (uls1-Δ677-855) were grown to stationary phase. Telomere fusions were amplified with 30 and 34 cycles. (D) Complementation of uls1-Δ by ULS1 wild-type and mutant alleles. Strain 210-5b (uls1-Δ) was transformed with StuI-digested plasmids pRS406, pRS406-uls1-K975R, pRS406-uls1-C1333S and pRS406-uls1-Δ677-855 and with MfeI-digested plasmid pRS404-ULS1. Cultures of two independent transformants were grown to stationary phase in rich medium. Telomere fusions were amplified with 26 and 30 cycles. (E) Uls1 is present at telomeres. Uls1 was tagged with 13 myc epitopes in the C-terminal position (Uls1-myc). A wild-type strain with untagged Uls1 was used as a control (−). Distances of the primer pairs from the terminal TG1–3 repeats are indicated. LYS2 is an internal non-telomeric locus. ChIP assay was performed as described by Guglielmi et al (2007). Immunoprecipitated and input DNA were quantified by qPCR. ChIP signal was normalized to input DNA. Data shown are the mean and standard deviation of three independent experiments.
To address the contribution of Uls1 ubiquitin E3 ligase activity, the second cysteine of the RING domain at position 1333 was mutated to a serine (Uzunova et al, 2007). To knocked down Uls1 translocase activity, the catalytic lysine at position 975 was mutated to an arginine. In cells carrying either mutation at the endogenous uls1 locus, telomere fusions are detected. Although an effect of these point mutations on protein folding cannot be ruled out, this result suggests that both enzymatic activities of Uls1 are required for efficient NHEJ inhibition at telomeres (Figure 2B). The slightly lower frequencies of fusions observed in both mutants relative to the frequency observed in uls1-Δ cells indicate that each knock-down activity may not be systematically employed by Uls1 or that the point mutants conserve some enzymatic activity. To investigate REPULS domains involvement in NHEJ inhibition at telomeres, an uls1-Δ677-855 allele lacking the tandem domains was created and integrated at the endogenous uls1 locus. This mutant displays normal growth and telomere length (data not shown) and telomere fusions remain undetectable (Figure 2C). The simplest interpretation of this result is that the REPULS domains are not involved or are facultative in Uls1 function at telomeres. A more complicated possibility we cannot rule out is that the truncated uls1-Δ677-855 protein displays an aberrant activity that suppresses the occurrence of telomere fusions indirectly.
To further address the phenotype of these uls1 alleles, we tested their ability to complement an uls1 deletion. The wild-type and mutant uls1 genes were inserted at an ectopic position in an uls1-Δ strain. The presence of a wild-type or an uls1-Δ677-855 sequence abolishes the telomere fusions caused by the uls1 deletion (Figure 2D). By contrast, the presence of an uls1-K975R or uls1-C1333S sequence has no obvious effect on the frequency of telomere fusions (a two-fold difference or less would be below the assay sensitivity). A synergy between these two point mutants and a mild expression defect caused by the ectopic position might explain the stronger phenotype observed in this situation compared to the situation where the same mutants are integrated at the endogenous ULS1 locus. Together, these results support the hypothesis that Uls1 function at telomeres involves its translocase and ubiquitin ligase activities and that the REPULS domains are facultative for this function.
Uls1 requirement for NHEJ inhibition suggests that the protein might be present at telomeres. We tested this possibility by cross-linked Chromatin Immuno-Precipitation (ChIP) directed against an epitope-tagged variant of Uls1. As shown in Figure 2E, sequences immediately adjacent to the X telomere TEL 6R and to the Y′ telomeres are specifically pulled down with Uls1. Sequences at an internal locus (LYS2) and 3 kb away from telomere within the Y′ subtelomeric elements are not significantly enriched. This shows that Uls1 is present at the tips of the chromosomes and suggests that Uls1 activities may target other proteins at telomeres.
The absence of poly-SUMOylation bypasses Uls1 requirement
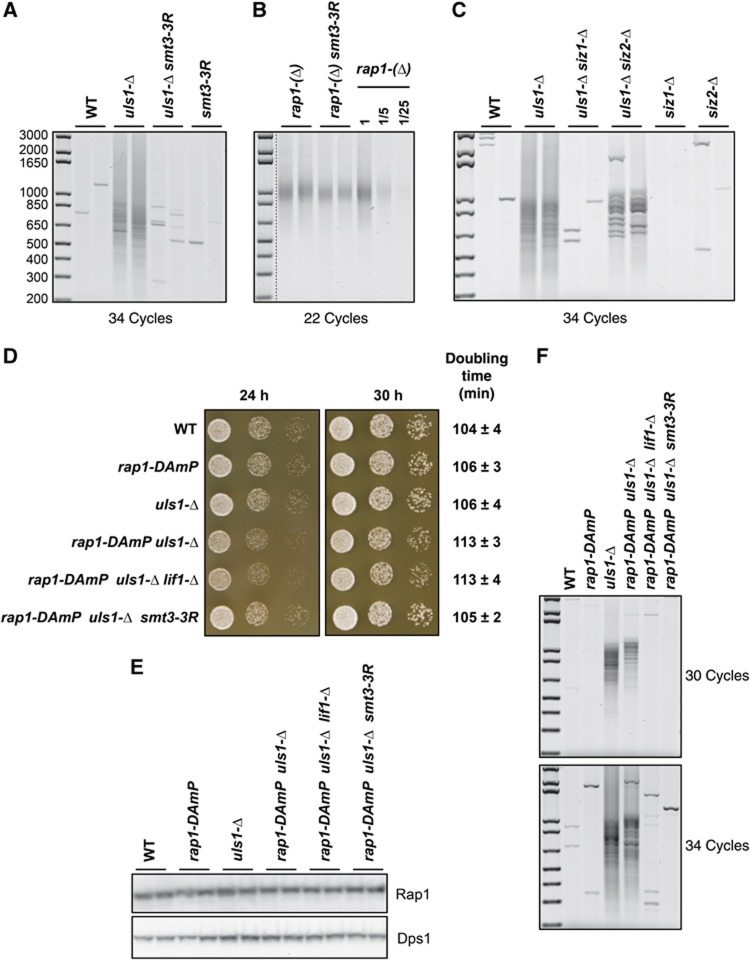
In cells lacking Uls1, unidentified poly-SUMOylated proteins that would normally be degraded become detectable (Uzunova et al, 2007). We asked whether the role of Uls1 in NHEJ inhibition at telomeres is linked to its ability to downregulate poly-SUMOylated proteins. In S. cerevisiae, a single gene, SMT3, encodes SUMO. SUMO polymerization occurs on three amino-terminal lysines of Smt3 at positions K11, K15 and K19. In the smt3-3R allele, these SUMOylatable lysines of SUMO are mutated to arginine, preventing poly-SUMO chains but preserving the essential functions of SUMO (Bylebyl et al, 2003; Ulrich, 2008). Since the smt3-3R allele suppresses the accumulation of poly-SUMOylated proteins in cells lacking Uls1 (Uzunova et al, 2007), we looked at telomere fusions in cells carrying this allele, either alone or in combination with an uls1 deletion. As shown in Figure 3A, telomere fusions remain undetectable in smt3-3R cells. However, the smt3-3R allele abolishes the fusions caused by Uls1 loss. The smt3-3R allele could downregulate NHEJ globally. To address this possibility, we introduced the smt3-3R mutant in a rap1-(Δ) strain. As shown in Figure 3B, telomere fusions still occur in rap1-(Δ) smt3-3R double mutant cells. This indicates that NHEJ remains active in the smt3-3R context and that smt3-3R must suppress uls1 deletion by restoring NHEJ inhibition at telomeres. Hence in the absence of poly-SUMO chains Uls1 is no longer needed for NHEJ inhibition. This suggests that poly-SUMOylated proteins are in part responsible for the NHEJ inhibition defect caused by Uls1 loss.
Figure 3.
Suppression of uls1-Δ by smt3-3R. (A) Telomere fusions are undetectable in usl1-Δ smt3-3R cells. Two independent cultures of strains RL179 (WT), RL183 (uls1-Δ), RL185 (uls1-Δ smt3-3R) and RL181 (smt3-3R) were grown to stationary phase. Telomere fusions were amplified with 34 cycles. (B) Telomere fusions remain frequent in rap1-(Δ) smt3-3R cells. Strains 169-1c and 169-15c (rap1-(Δ)) and 169-6b and 169-11a (rap1-(Δ) smt3-3R) were grown to stationary phase. Telomere fusions were amplified with 22 cycles. Genomic DNA from 169-1c rap1-(Δ) cells was diluted serially to provide a semi-quantitative estimation of the method sensitivity. (C) Uls1 suppression by Siz loss. Two independent cultures of strains 168-3d (WT) and 168-3d (uls1-Δ) and strains RL266-1 and RL266-2 (uls1-Δ siz1-Δ), RL267-1 and RL267-2 (uls1-Δ siz2-Δ), RL268-1 and RL268-2 (siz1-Δ) and RL269-1 and RL269-2 (siz2-Δ) were grown to stationary phase. Telomere fusions were amplified with 34 cycles. (D) Slow growth of rap1-DAmP uls1-Δ cells is suppressed by smt3-3R. Freshly growing cells of strains 195-19d (WT), 195-3a (rap1-DAmP), 195-7b (uls1-Δ), 195-8a (rap1-DAmP uls1-Δ), 195-6b (rap1-DAmP uls1-Δ lif1-Δ) and 195-1d (rap1-DAmP uls1-Δ smt3-3R) were serially diluted by 10-fold in water and spotted on a rich medium plate. Pictures were taken after 24 and 30 h at 30°C. Doubling times are from exponential growth in liquid-rich medium at 30°C. (Data shown are the mean and standard deviation of five independent samples). (E) Cultures from the same strains were grown exponentially and total urea-extracted proteins were analysed by western blotting with polyclonal antibodies directed against Rap1 (upper panel) and against RNA synthetase Dps1 (an internal control, lower panel). (F) Cultures from the same strains were grown to stationary phase and telomere fusions were amplified with 30 and 34 cycles.
SUMOylation requires an E1 activating enzyme, an E2 conjugating enzyme and one of several SUMO E3 ligases (Ulrich, 2008). We tested whether the SUMO E3 ligases Siz1 and Siz2 might be part of the activities counteracted by Uls1. As shown in Figure 3C, Siz1 loss suppresses the occurrence of telomere fusions in cells lacking Uls1. Siz2 loss causes a more limited suppression. Thus, a SUMOylation defect bypasses the need for Uls1 at telomeres, further suggesting that SUMOylated proteins perturb NHEJ inhibition in the absence of Uls1.
A synthetic negative interaction between uls1-Δ and a rap1 hypomorphic allele is suppressed by smt3-3R
A systematic study revealed previously a negative synthetic interaction between an uls1 deletion and a hypomorphic allele of rap1 created by the ‘Decreased Abundance by mRNA Perturbation’ (DAmP) method (Yan et al, 2008; Costanzo et al, 2010). In this rap1-DAmP allele, RAP1 coding sequence is separated from its native 3′ UTR. As shown in Figure 3D, rap1-DAmP and uls1-Δ single mutant cells grow as well as wild-type cells but double mutant rap1-DAmP uls1-Δ cells grow slightly slower, reproducing in our W303 genetic background the negative synthetic interaction initially observed in a BY4147 background. This growth defect is unaffected by an NHEJ-defective lif1-Δ mutant but is suppressed by the smt3-3R allele. Thus, the negative interaction between uls1 and rap1 requires poly-SUMOylated proteins. The rap1-DAmP allele does not significantly reduced relative Rap1 protein level (Figure 3E). Loss of the native 3′ UTR may alter translation quality. We checked the occurrence of telomere fusions in these cells once they reached stationary phase (Figure 3F). Telomere fusions remain undetectable in rap1-DAmP single mutant cells and the frequency of fusions caused by Uls1 loss is not increased by the rap1-DAmP allele. Their occurrence is suppressed by Lif1 loss and by the smt3-3R allele. The average telomere length remains unchanged in these mutant cells (data not shown).
The negative interaction between uls1-Δ and rap1-DAmP is not a consequence of telomere fusions since fusions only occur in a small fraction of cells in stationary phase. Furthermore, this negative interaction is not suppressed by a loss of NHEJ. Its cause might be an unidentified telomere defect or a perturbation of transcription from one or several of the many essential genes whose promoter is bound and regulated by Rap1 (Lickwar et al, 2012). In both scenarios and in agreement with the telomere fusions caused by Uls1 loss, Rap1 function might be perturbed in the absence of Uls1 either directly or indirectly through poly-SUMOylation. Rap1 is SUMOylated at a low level in normal growth condition (Hang et al, 2011). This encouraged us to address the hypothesis of a direct perturbation of Rap1 by SUMO.
Suppression of uls1-Δ by rap1 alleles
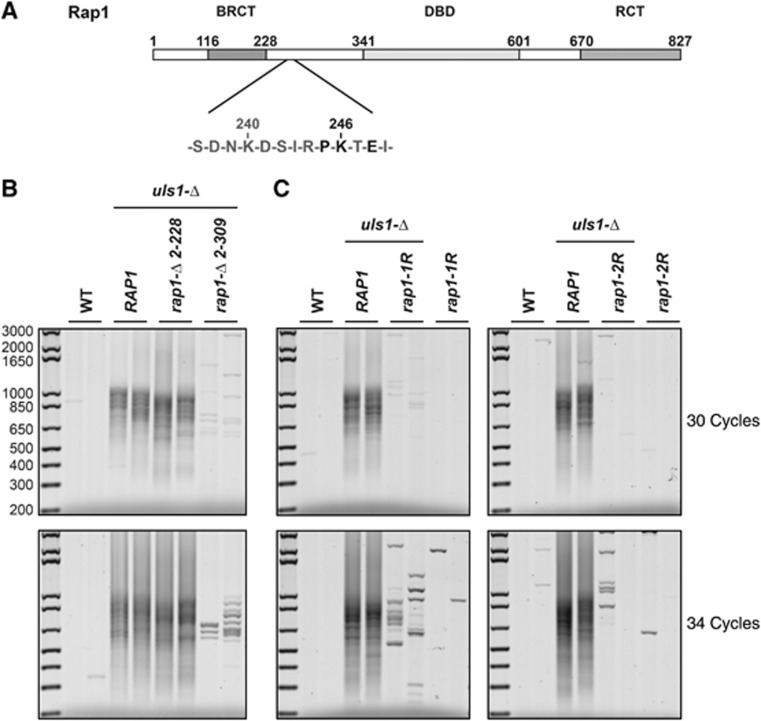
If the poly-SUMOylation of Rap1 is responsible of NHEJ inhibition failure at telomeres in cells lacking Uls1, then a non-SUMOylatable allele of rap1 should suppress this defect. Rap1 displays three distinct and conserved domains: a BRCT domain in its amino-terminal region, a central DNA-binding domain (DBD) and a carboxy-terminal domain called RCT that is unique to Rap1 and its orthologues in other species (Figure 4A) (Chen et al, 2011; Matot et al, 2012). Only Rap1 DBD and RCT domain are required for NHEJ inhibition at telomeres (Marcand et al, 2008). The function of its amino-terminal region, which includes the BRCT domain, remains to be identified. We first tested whether a deletion of this non-essential part of Rap1 could suppress Uls1 loss. Two truncation mutants were introduced in uls1-Δ cells: rap1-Δ2-228 and rap1-Δ2-309. Position 228 is the edge of the BRCT domain and position 309 is the last lysine residue before the essential DBD. In uls1-Δ cells where the first 228 amino acids of Rap1 are missing, telomere fusions still occur but extension of the deletion to amino acid 309 strongly reduces the frequency of telomere fusions (Figure 4B). The average telomere length is shortened by about 50 bp in these rap1 mutant cells (data not shown).
Figure 4.
Suppression of uls1-Δ by rap1 alleles. (A) Schematic representation of S. cerevisiae Rap1. The sequence surrounding lysine 246 is indicated. (B) Suppression of uls1-Δ by rap1-Δ2-309. Strains 196-11c and 196-13b (WT), 196-5a and 196-6a (uls1-Δ), 198-16a and 198-19a (uls1-Δ rap1-Δ2-228) and 196-1a and 196-7c (uls1-Δ rap1-Δ2-309) were grown to stationary phase. Telomere fusions were amplified with 30 and 34 cycles. (C) Suppression of uls1-Δ by mutation of Rap1 lysine 246. rap1-1R is rap1-K246R and rap1-2R is rap1-K240R, K246R. Strains 210-2d and 210-3d (WT), 210-5b and 210-4c (uls1-Δ), 210-10b and 210-5d (uls1-Δ rap1-1R), 210-1b and 210-7b (rap1-1R), 212-2d and 212-10b (WT), 212-1a and 212-8a (uls1-Δ), 212-5b and 212-17d (uls1-Δ rap1-2R), 212-3b and 212-4c (rap1-2R) were grown to stationary phase. Telomere fusions were amplified with 30 and 34 cycles.
Between positions 228 and 309 of Rap1, a sequence close to the most common SUMOylation consensus ΨKXE/D (Ψ: bulky hydrophobic residue) is present around lysine 246 (Figure 4A). The position of this lysine suggests that it could be a SUMOylation site whose loss in the rap1-Δ2-309 mutant contributes to uls1-Δ suppression. To test this hypothesis, lysine 246 was mutated to a non-SUMOylatable arginine in full-length Rap1. This rap1-1R allele strongly reduces the frequency of telomere fusions caused by Uls1 loss (Figure 4C). A few telomere fusions are still detected in uls1-Δ rap1-1R cells. The additional mutation of nearby lysine 240 to arginine (rap1-2R) further reduces this very low level of fusions (Figure 4C; data not shown), suggesting that SUMOylation at lysine 240 might contribute to the weak NHEJ inhibition defect observed in uls1-Δ rap1-1R cells. The average telomere length remains unchanged in these mutant cells (data not shown). Although these results do not rule out SUMOylation at other lysines in Rap1 or that SUMOylation of other proteins contributes to the uls1-Δ phenotype, they indicate that NHEJ inhibition failure in the absence of Uls1 is in part due to Rap1 SUMOylation at lysines 240 and 246.
Increased level of poly-SUMOylated Rap1 in uls1-Δ cells
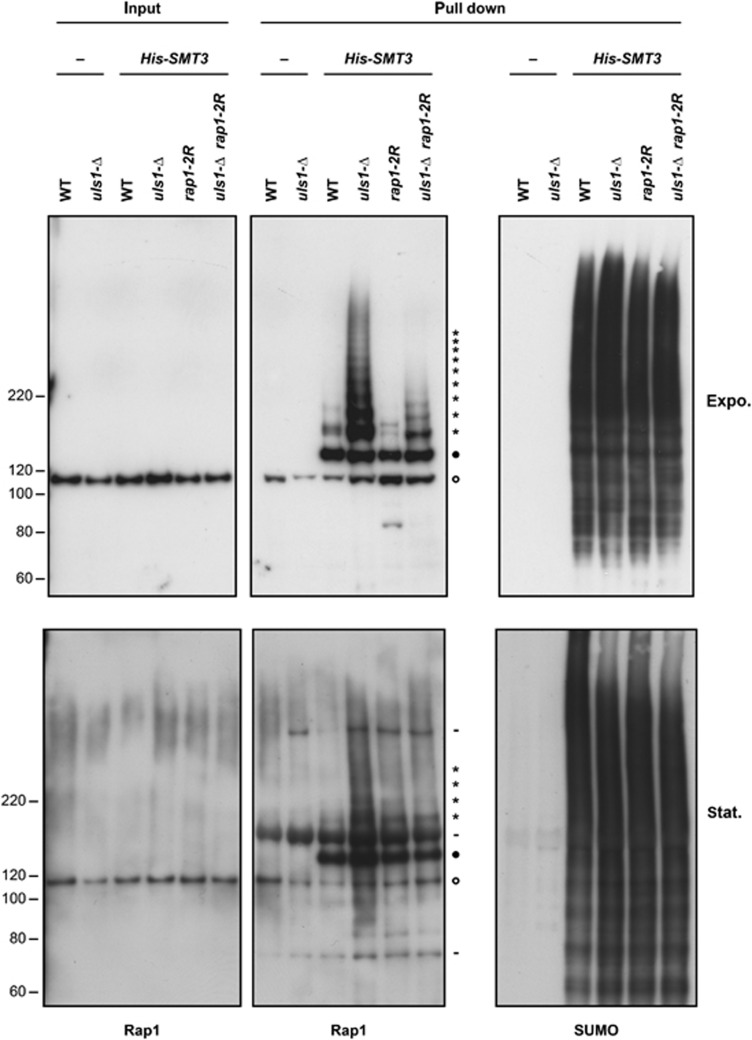
A prediction of the previous results is that SUMOylated Rap1 molecules accumulate in uls1-Δ cells. SUMOylated proteins are rare and their detection often requires an enrichment step. To detect SUMOylated Rap1, we used a method described by Ulrich and Davies (2009). In short, His-tagged SUMO expressed in vivo allows the bulk purification of SUMOylated proteins under denaturing conditions by Ni-NTA pull-down. The presence of a particular SUMO conjugate can then be detected by western blot analysis. Rap1 SUMOylation was tested in exponentially growing cells (Figure 5, upper panel) and in stationary cells (lower panel). As previously observed (Hang et al, 2011), Rap1 is SUMOylated in wild-type cells, mostly as mono-SUMO conjugates (Figure 5; filled circle). In cells lacking Uls1, the level of SUMOylated Rap1 increases, mostly as poly-SUMO conjugates (stars), indicating that Uls1 is required to limit the accumulation of poly-SUMOylated Rap1 molecules. In rap1-2R cells, poly-SUMOylated Rap1 molecules are less frequent, indicating that a large fraction of the Rap1 poly-SUMO conjugates are linked to lysines 240 and 246. The persistence of mono- and some poly-SUMO conjugates in rap1-2R cells reveals that, in addition to lysines 240 and 246, other Rap1 lysines can be SUMOylated. Together, these observations reinforce the notion that Uls1 addresses the consequences of poly-SUMOylated Rap1 molecules by eliminating these conjugates.
Figure 5.
Accumulation of SUMOylated Rap1 molecules in uls1-Δ cells. Strains 210-2d (WT) and 210-5b (uls1-Δ) transformed with empty vector pRS316 and strains 210-2d (WT), 210-5b (uls1-Δ), 210-3b (rap1-2R) and 210-13c (uls1-Δ rap1-2R) transformed with YEp195-CUP-His-Smt3 were grown exponentially (upper panel) or allowed to reach stationary phase (lower panel). Protein extraction and pull-down were done under denaturing conditions. Input sample is 1/2000th of the total extract subjected to the pull down. Rap1 was detected with a polyclonal antibody (left and central panels). The membrane was then rehybridized with a second polyclonal antibody against SUMO (right panel; the antibody sensitivity did not allow us to detect SUMO conjugates in the input samples (data not shown)). Position of mono-SUMOylated Rap1 is marked with a filled circle. Positions of poly-SUMOylated Rap1 are marked with stars. A background of unmodified Rap1 is detected in all pull-down samples and its position is marked with an empty circle. Total protein extraction from stationary cells was less efficient. Non-specific signals are marked with minus signs.
Synergy between Uls1 and Sir4
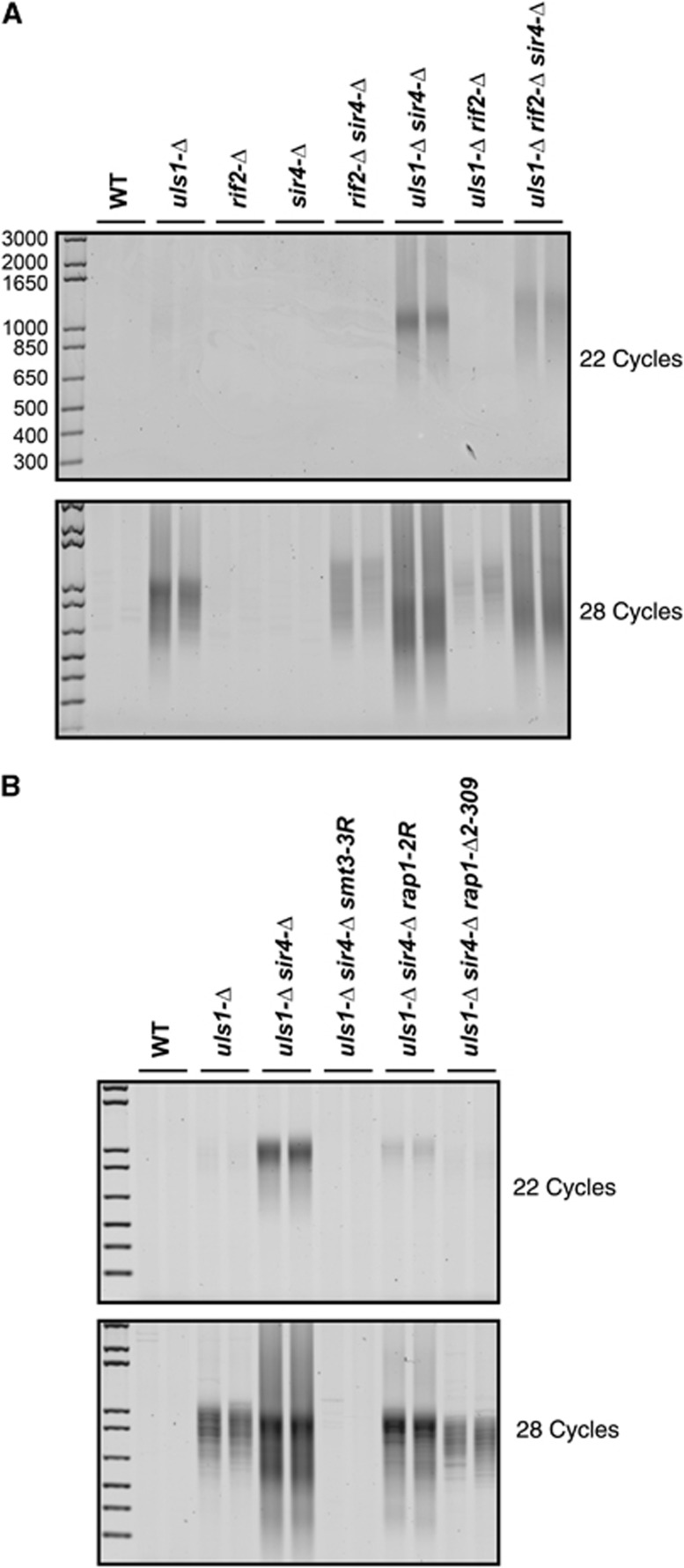
Several pathways synergize to inhibit NHEJ at telomeres. The Rap1 C-terminal domain establishes two parallel inhibitory pathways through the proteins Rif2 and Sir4. In addition, the essential central part of Rap1 inhibits NHEJ independently of Rif2 and Sir4 (Marcand et al, 2008). As shown previously, fusions are detected in rif2-Δ sir4-Δ double mutant cells (Figure 6A, lower panel). They are detected as longer PCR products than in uls1-Δ cells, a consequence of the telomere elongation caused by Rif2 loss (Supplementary Figure S2). To know whether Uls1 is linked to these pathways, we tested the genetic interactions between deletions of ULS1, RIF2 and SIR4. As shown in Figure 6A, in uls1-Δ sir4-Δ cells and in usl1-Δ sir4-Δ rif2-Δ cells, telomeres fuse more frequently than in uls1-Δ cells. By contrast, fusions occur slightly less frequently in uls1-Δ rif2-Δ cells. Thus in the absence of Uls1, NHEJ inhibition at telomeres still relies on Sir4 but inhibition by Rif2 is lost. Since a rif2 deletion alone is insufficient to cause a detectable level of fusions and since fusions are more frequent in usl1-Δ sir4-Δ rif2-Δ cells than in sir4-Δ rif2-Δ cells, the pathway independent of Rif2 and Sir4 is also at least partially defective in the absence of Uls1. The slight decrease in fusion frequency observed in uls1-Δ rif2-Δ cells relative to uls1-Δ cells might be a consequence of telomere elongation and increased Sir4 recruitment caused by Rif2 loss (Wotton and Shore, 1997; Luo et al, 2002; Feeser and Wolberger, 2008; Marcand et al, 2008; Chen et al, 2011).
Figure 6.
Synergy between Uls1 and Sir4. (A) Two independent cultures of strains Lev346 (WT), 183-30d (rif2-Δ), 183-10c (uls1-Δ sir4-Δ) and 183-2d (uls1-Δ rif2-Δ), and strains 184-48c and 184-9d (uls1-Δ), Lev575 and Lev576 (sir4-Δ), 184-14c and Lev601 (rif2-Δ sir4-Δ) and 183-25a and 184-49a (uls1-Δ rif2-Δ sir4-Δ) were grown to stationary phase. Telomere fusions were amplified with 22 and 28 cycles. (B) Two independent cultures of strains 205-9a (WT), 205-14c (uls1-Δ), 205-16a (uls1-Δ sir4-Δ smt3-3R) and 207-14d (uls1-Δ sir4-Δ rap1-Δ2-309) and strains 212-3c and 212-4b (uls1-Δ sir4-Δ), 212-1b and 212-2a (uls1-Δ sir4-Δ rap1-2R) were grown to stationary phase. Telomere fusions were amplified with 22 and 28 cycles.
Next, we asked whether telomere fusions occurring in uls1-Δ sir4-Δ cells are still in part linked to Rap1 poly-SUMOylation. The smt3-3R, rap1-2R and rap1-Δ2-309 alleles were combined with the uls1-Δ sir4-Δ double mutant. As shown in Figure 6B, all three alleles suppress the telomere fusions caused by Uls1 loss in the absence of Sir4, indicating that the bulk of telomere fusions remains a consequence of Rap1 poly-SUMOylation. However, suppression by rap1-2R and rap1-Δ2-309 is only partial. It is possible that the loss of one NHEJ inhibition pathway sensitizes telomeres to SUMOylation at other sites than Rap1 lysines 240 and 246. Sir4 loss may also expose other SUMOylation sites within Rap1 but this remains to be addressed.
Discussion
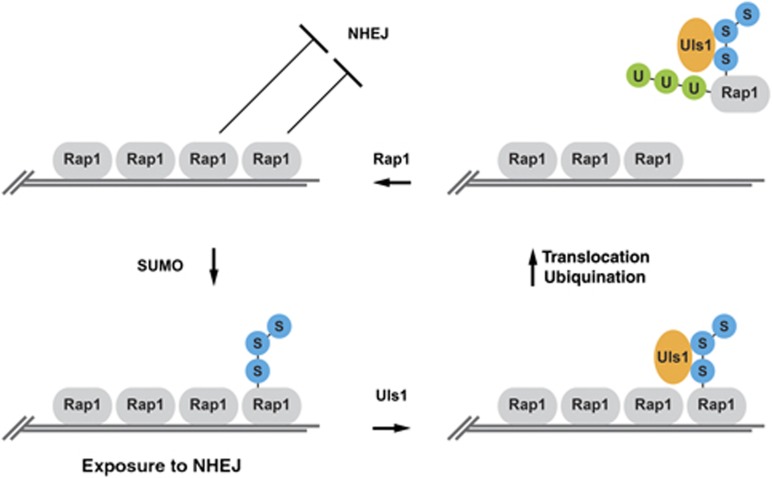
In this study, we observed that the SUMO-dependent ubiquitin ligase and translocase Uls1 is required for efficient NHEJ inhibition at telomeres. This requirement is bypassed by the absence of poly-SUMO chains and by Rap1 mutants lacking SUMOylation sites. In addition, Uls1 is required to avoid the accumulation of poly-SUMOylated Rap1 molecules. Together, these results show that the failure to inhibit NHEJ at telomeres in the absence of Uls1 is at least in part the consequence of the accumulation of poly-SUMOylated Rap1 that are normally eliminated by Uls1. Rap1 poly-SUMOylation seems to cause a loss of Rap1 function. A simple model is that when Rap1 poly-SUMOylation occurs on one or several Rap1 molecule(s) bound to DNA at a telomere, it cripples its ability to inhibit NHEJ (Figure 7). Uls1 recognizes poly-SUMO chains conjugated to Rap1. Its translocase activity dissociates these molecules from DNA and ubiquitinylation targets them for degradation by the proteasome (Uzunova et al, 2007). Unmodified Rap1 from the pool of free Rap1 molecules rapidly replace the freed binding sites, re-establishing full protection against NHEJ.
Figure 7.
A schematic model for Uls1 function at telomeres. SUMO is depicted in blue and ubiquitin in green. The proposed scenario is described in the main text.
Once bound to DNA, Rap1 detaches slowly in vitro (Williams et al, 2010). In vivo Rap1 residence time on DNA is estimated to be in the 30–60 min range at telomeres (Lickwar et al, 2012). If this slow turnover was the only way to lose SUMOylated Rap1 molecules, a telomere could be exposed to NHEJ long enough to create a reasonable probability that another telomere is simultaneously exposed at some point, allowing telomere fusions. Uls1 ATP-dependent translocation can prevent it by accelerating the dislodging of these molecules from DNA. Their subsequent ubiquitination and degradation prevent these molecules from rebinding DNA at telomeres or at other sites. In addition, because telomere fusions occurring in non-dividing cells are cumulative events, even low steady levels of SUMOylated Rap1 and relatively infrequent and transient telomere exposure to NHEJ could result in a significant accumulation of fusions over time.
How poly-SUMOylation perturbs Rap1 function is unknown but the requirement for Uls1 translocase activity indicates that Rap1 DNA binding ability must remain significant. Rap1 poly-SUMOylation antagonizes NHEJ inhibition by Rif2 and by the pathway independent of Rif2 and Sir4 but not by the Sir4 pathway, which remains proficient in the absence of Uls1. One possibility is that once at telomeres Sir4 can remain anchored through the silent chromatin and thereby be partially insensitive to a transient Rap1 loss of function. The Sir4-Rap1 complex or the mechanism of NHEJ inhibition by Sir4 may also be specifically insensitive to Rap1 SUMOylation. In all scenarios, the independence between Sir4 and Uls1 adds a level of redundancy that further protects telomeres from NHEJ. The lack of telomere elongation in uls1-Δ cells despite a loss of Rif2 function in NHEJ inhibition is surprising. Whether Rap1 SUMOylation can perturb Rif2 function in telomere length homeostasis during replication remains unknown. Perhaps in the absence of Uls1 poly-SUMOylated Rap1 molecules are too rare to influence most telomeres at each replication cycle, thus allowing telomere length homeostasis by functional Rap1 molecules to buffer for the drift of a few telomeres stochastically affected by Rap1 SUMOylation. By contrast, the static nature of telomere fusions may allow the dominance of Rap1 SUMOylation over Rif2 function to manifest itself by the progressive accumulation of telomere fusions. It would be interesting to test this model.
A different issue is whether Rap1 mono- and poly-SUMO conjugates have a positive role in cells. rap1-1R and rap1-2R mutants display normal growth, normal telomere length and no detectable telomere fusion (Figure 4B; data not shown). This indicates that SUMOylation at lysines 240 and 246 is not essential for the core Rap1 functions. It does not rule out a positive function for Rap1 SUMOylation that would be redundant with another mechanism or only relevant in contexts that do not often occur in normal growth conditions, for instance situations where Rap1 poly-SUMOylation could rapidly and transiently inactivate Rap1 function. Interestingly, DNA damage induces the hyper-SUMOylation of many proteins including Rap1 (Cremona et al, 2011; Hang et al, 2011). Maybe the modulation of Rap1 function at telomeres or at promoters helps the cell to cope with DNA damage-induced stress. An alternative possibility is that Rap1 poly-SUMOylation has no biological function and is part of a background of SUMOylation in the cell. In this model, Uls1 role would be to keep low the steady level of unselected poly-SUMO conjugates, Rap1 being one target among others.
Uls1 is the only known STUbL with a translocase activity. It may have a large number of targets and therefore can be in principle the general molecular sweeper for the clearance of poly-SUMOylated proteins on DNA in eukaryotes. Uls1 has previously been implicated in homologous recombination at breaks and at replication forks, functions for which its targets remain to be identified (Zhang and Buchman, 1997; Shah et al, 2010; Cal-Bakowska et al, 2011; Chi et al, 2011). In addition, the new and yeast-specific REPULS domain we identified in Uls1 and whose functions remain unknown suggests that additional modes of Uls1 regulation or mobilization are likely to exist.
Materials and methods
Yeast strains and plasmids
The strains used in this study are listed in Supplementary Table 1. Mutations within ULS1 and RAP1 were created by overlapping multiple PCRs whose products were assembled to a plasmid by single-strand annealing in yeast. Mutant alleles were recovered, sequenced, cloned into pRS406 and integrated at the endogenous locus by pop-in pop-out selection. Correct integrations were identified by PCR and sequencing. The smt3-3R::TRP1 and rap1-DAmP::KANr alleles were amplified by PCR from strains GBY1 (a gift from Erica Johnson) and YNL216 (a gift from Jesse Platt and Bradley Johnson) respectively and integrated at their endogenous locus in a W303 wild-type strain.
Amplification of the telomere–telomere fusions by PCR
Cells were streaked on rich medium plate (YPD) at 30°C for 24 h and then grown in liquid-rich medium at 30°C to saturation for 6 days (excepted in Figure 1B where exponentially growing cells were from cultures whose OD600 nm was maintained below 1 for 24 h). Genomic DNA was extracted with phenol/chloroform and 800–1200 μm acid-washed glass beads, ethanol-precipitated and resuspended in TE pH 8.0 with RNAse A (∼10 ng/μl).
Fusions between X and Y′ telomeres were amplified with primers X2 5′-TGTGGTGGTGGGATTAGAGTGGTAG-3′ and Y′2 5′-TTAGGGCTATGTAGAAGTGCTG-3′. PCRs (30 μl) contained genomic DNA ∼10 ng, Phusion HF buffer 1 × , DMSO 3%, dNTP 200 μM each, primers 0.5 μM each, HotStart Phusion polymerase 0.6 unit (Finnzymes). Reaction mixes were prepared at room temperature. Amplification conditions were 98°C 30 s, then 22, 26, 30 or 34 cycles of 98°C 10 s, 65°C 20 s, 72°C 1 min, followed by 72°C 5 min, using GeneAmp PCR System 9700 (Applied Biosystems). The products (10 μl) were separated on a 1% agarose gel containing 1 × Gel Red nucleic acid stain (Biotium). Fluorescence was analysed on a Typhoon imager (GE).
Detection of SUMO conjugates
Cells were transformed with plasmids pRS316 or YEp195-CUP-His-Smt3 (URA3) (Ulrich and Davies, 2009). Cells were grown exponentially in synthetic medium lacking uracil followed by 2 h (expo.) or 5 days (stat.) in rich medium (YPD). Extraction and Ni-NTA pull-down of SUMO conjugates were done as described by Ulrich and Davies (2009). For stationary cells, lysis in NaOH/BME on ice was improved with 0.8 mm zirconium beads and four pulses of 30 s vortex at 4°C. Input aliquots and pull-downs were separated in a NuPage Tris-Acetate 3–8% gel (Novex Life Science) and transferred overnight onto a PVDF membrane at 25 V at 4°C in 25 mM Tris, 192 mM Glycine, 10% v/v Ethanol, 0.01% SDS. Anti-Rap1 rabbit polyclonal antibody (1/500) is from Santa Cruz (Y-300; ref sc-20167). Anti-Smt3 rabbit polyclonal antibody (1/500) is from Abcam (ref ab14405).
Supplementary Material
Acknowledgments
We thank Erica Johnson for the smt3-3R allele, Jesse Platt and Bradley Johnson for the rap1-DAmP allele, Helle Ulrich for the YEp195-CUP-His-Smt3 plasmid, Marie Frank for unpublished data, Hannah Klein for very fruitful discussions at early and late stages of this project, Julie Cooper for a key suggestion, as well as Ofer Rog, Serge Gangloff, Karine Dubrana and Nathalie Hardouin for important comments. This work is supported by grants from ARC and ANR (ANR-06-BLAN-076; Blanc-SVSE-8-2011-TELO&DICENs). RL was supported by an ARC postdoctoral fellowship.
Author contributions: RL, IC and SM designed the experiments. RL, SP and SM performed the yeast genetics experiments. IC performed the in silico studies. RL, SP, IC and SM interpreted the data. RL, IC and SM wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Anbalagan S, Bonetti D, Lucchini G, Longhese MP (2011) Rif1 supports the function of the CST complex in yeast telomere capping. PLoS Genet 7: e1002024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askree SH, Yehuda T, Smolikov S, Gurevich R, Hawk J, Coker C, Krauskopf A, Kupiec M, McEachern MJ (2004) A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc Natl Acad Sci USA 101: 8658–8663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae NS, Baumann P (2007) A RAP1/TRF2 complex inhibits nonhomologous end-joining at human telomeric DNA ends. Mol Cell 26: 323–334 [DOI] [PubMed] [Google Scholar]
- Bonetti D, Clerici M, Anbalagan S, Martina M, Lucchini G, Longhese MP (2010) Shelterin-like proteins and Yku inhibit nucleolytic processing of Saccharomyces cerevisiae telomeres. PLoS Genet 6: e1000966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylebyl GR, Belichenko I, Johnson ES (2003) The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J Biol Chem 278: 44113–44120 [DOI] [PubMed] [Google Scholar]
- Cal-Bakowska M, Litwin I, Bocer T, Wysocki R, Dziadkowiec D (2011) The Swi2-Snf2-like protein Uls1 is involved in replication stress response. Nucleic Acids Res 39: 8765–8777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli GB, de Lange T (2005) DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol 7: 712–718 [DOI] [PubMed] [Google Scholar]
- Chen Y, Rai R, Zhou ZR, Kanoh J, Ribeyre C, Yang Y, Zheng H, Damay P, Wang F, Tsujii H, Hiraoka Y, Shore D, Hu HY, Chang S, Lei M (2011) A conserved motif within RAP1 has diversified roles in telomere protection and regulation in different organisms. Nat Struct Mol Biol 18: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi P, Kwon Y, Visnapuu ML, Lam I, Santa Maria SR, Zheng X, Epshtein A, Greene EC, Sung P, Klein HL (2011) Analyses of the yeast Rad51 recombinase A265V mutant reveal different in vivo roles of Swi2-like factors. Nucleic Acids Res 39: 6511–6522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, Ding H, Xu H, Han J, Ingvarsdottir K, Cheng B, Andrews B, Boone C, Berger SL, Hieter P, Zhang Z et al. (2007) Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446: 806–810 [DOI] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, Prinz J St, Onge RP, VanderSluis B, Makhnevych T, Vizeacoumar FJ, Alizadeh S, Bahr S, Brost RL, Chen Y, Cokol M et al. (2010) The genetic landscape of a cell. Science 327: 425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremona CA, Sarangi P, Yang Y, Hang LE, Rahman S, Zhao X (2011) Extensive DNA damage-induced sumoylation contributes to replication and repair and acts in addition to the mec1 checkpoint. Mol Cell 45: 422–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeser EA, Wolberger C (2008) Structural and functional studies of the Rap1 C-terminus reveal novel separation-of-function mutants. J Mol Biol 380: 520–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira HC, Luke B, Schober H, Kalck V, Lingner J, Gasser SM (2011) The PIAS homologue Siz2 regulates perinuclear telomere position and telomerase activity in budding yeast. Nat Cell Biol 13: 867–874 [DOI] [PubMed] [Google Scholar]
- Fujita I, Tanaka M, Kanoh J (2012) Identification of the functional domains of the telomere protein Rap1 in Schizosaccharomyces pombe. PLoS ONE 7: e49151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmi B, Soutourina J, Esnault C, Werner M (2007) TFIIS elongation factor and Mediator act in conjunction during transcription initiation in vivo. Proc Natl Acad Sci USA 104: 16062–16067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang LE, Liu X, Cheung I, Yang Y, Zhao X (2011) SUMOylation regulates telomere length homeostasis by targeting Cdc13. Nat Struct Mol Biol 18: 920–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain D, Cooper JP (2010) Telomeric strategies: means to an end. Annu Rev Genet 44: 243–269 [DOI] [PubMed] [Google Scholar]
- Lickwar CR, Mueller F, Hanlon SE, McNally JG, Lieb JD (2012) Genome-wide protein-DNA binding dynamics suggest a molecular clutch for transcription factor function. Nature 484: 251–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K, Vega-Palas MA, Grunstein M (2002) Rap1-Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev 16: 1528–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcand S, Pardo B, Gratias A, Cahun S, Callebaut I (2008) Multiple pathways inhibit NHEJ at telomeres. Genes Dev 22: 1153–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matot B, Le Bihan YV, Lescasse R, Perez J, Miron S, David G, Castaing B, Weber P, Raynal B, Zinn-Justin S, Gasparini S, Le Du MH (2012) The orientation of the C-terminal domain of the Saccharomyces cerevisiae Rap1 protein is determined by its binding to DNA. Nucleic Acids Res 40: 3197–3207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee JS, Phillips JA, Chan A, Sabourin M, Paeschke K, Zakian VA (2011) Reduced Rif2 and lack of Mec1 target short telomeres for elongation rather than double-strand break repair. Nat Struct Mol Biol 17: 1438–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieczkowski PA, Mieczkowska JO, Dominska M, Petes TD (2003) Genetic regulation of telomere-telomere fusions in the yeast Saccharomyces cerevisae. Proc Natl Acad Sci USA 100: 10854–10859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KM, Ferreira MG, Cooper JP (2005) Taz1, Rap1 and Rif1 act both interdependently and independently to maintain telomeres. EMBO J 24: 3128–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen JR, Brill SJ (2008) Activation of the Slx5-Slx8 ubiquitin ligase by poly-small ubiquitin-like modifier conjugates. J Biol Chem 283: 19912–19921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai S, Dubrana K, Tsai-Pflugfelder M, Davidson MB, Roberts TM, Brown GW, Varela E, Hediger F, Gasser SM, Krogan NJ (2008) Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science 322: 597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini S, Ribaud V, Bianchi A, Shore D (2007) DNA breaks are masked by multiple Rap1 binding in yeast: implications for telomere capping and telomerase regulation. Genes Dev 21: 292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo B, Marcand S (2005) Rap1 prevents telomere fusions by nonhomologous end joining. EMBO J 24: 3117–3127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pobiega S, Marcand S (2010) Dicentric breakage at telomere fusions. Genes Dev 24: 720–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts PR, Yu H (2007) The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat Struct Mol Biol 14: 581–590 [DOI] [PubMed] [Google Scholar]
- Ribeyre C, Shore D (2012) Anticheckpoint pathways at telomeres in yeast. Nat Struct Mol Biol 19: 307–313 [DOI] [PubMed] [Google Scholar]
- Rog O, Miller KM, Ferreira MG, Cooper JP (2009) Sumoylation of RecQ helicase controls the fate of dysfunctional telomeres. Mol Cell 33: 559–569 [DOI] [PubMed] [Google Scholar]
- Sarthy J, Bae NS, Scrafford J, Baumann P (2009) Human RAP1 inhibits non-homologous end joining at telomeres. EMBO J 28: 3390–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, de Lange T (2012) Removal of shelterin reveals the telomere end-protection problem. Science 336: 593–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, Kabir S, van Overbeek M, Celli GB, de Lange T (2010) Loss of Rap1 induces telomere recombination in the absence of NHEJ or a DNA damage signal. Science 327: 1657–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah PP, Zheng X, Epshtein A, Carey JN, Bishop DK, Klein HL (2010) Swi2/Snf2-related translocases prevent accumulation of toxic Rad51 complexes during mitotic growth. Mol Cell 39: 862–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Hunter T (2012) Poly-small ubiquitin-like modifier (PolySUMO)-binding proteins identified through a string search. J Biol Chem 287: 42071–42083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MT, Arneric M, Sperisen P, Lingner J (2004) Telomere length homeostasis is achieved via a switch between telomerase- extendible and -nonextendible states. Cell 117: 323–335 [DOI] [PubMed] [Google Scholar]
- Ulrich HD (2008) The fast-growing business of SUMO chains. Mol Cell 32: 301–305 [DOI] [PubMed] [Google Scholar]
- Ulrich HD, Davies AA (2009) In vivo detection and characterization of sumoylation targets in Saccharomyces cerevisiae. Methods Mol Biol 497: 81–103 [DOI] [PubMed] [Google Scholar]
- Uzunova K, Gottsche K, Miteva M, Weisshaar SR, Glanemann C, Schnellhardt M, Niessen M, Scheel H, Hofmann K, Johnson ES, Praefcke GJ, Dohmen RJ (2007) Ubiquitin-dependent proteolytic control of SUMO conjugates. J Biol Chem 282: 34167–34175 [DOI] [PubMed] [Google Scholar]
- van Steensel B, Smogorzewska A, de Lange T (1998) TRF2 protects human telomeres from end-to-end fusions. Cell 92: 401–413 [DOI] [PubMed] [Google Scholar]
- Vodenicharov MD, Laterreur N, Wellinger RJ (2010) Telomere capping in non-dividing yeast cells requires Yku and Rap1. EMBO J 29: 3007–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt B, Hofmann K (2012) Bioinformatical detection of recognition factors for ubiquitin and SUMO. Methods Mol Biol 832: 249–261 [DOI] [PubMed] [Google Scholar]
- Williams TL, Levy DL, Maki-Yonekura S, Yonekura K, Blackburn EH (2010) Characterization of the yeast telomere nucleoprotein core: Rap1 binds independently to each recognition site. J Biol Chem 285: 35814–35824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wotton D, Shore D (1997) A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev 11: 748–760 [DOI] [PubMed] [Google Scholar]
- Xhemalce B, Riising EM, Baumann P, Dejean A, Arcangioli B, Seeler JS (2007) Role of SUMO in the dynamics of telomere maintenance in fission yeast. Proc Natl Acad Sci USA 104: 893–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Kerscher O, Kroetz MB, McConchie HF, Sung P, Hochstrasser M (2007) The yeast Hex3.Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J Biol Chem 282: 34176–34184 [DOI] [PubMed] [Google Scholar]
- Yan Z, Costanzo M, Heisler LE, Paw J, Kaper F, Andrews BJ, Boone C, Giaever G, Nislow C (2008) Yeast Barcoders: a chemogenomic application of a universal donor-strain collection carrying bar-code identifiers. Nat Methods 5: 719–725 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Buchman AR (1997) Identification of a member of a DNA-dependent ATPase family that causes interference with silencing. Mol Cell Biol 17: 5461–5472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Blobel G (2005) A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc Natl Acad Sci USA 102: 4777–4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.