Abstract
Mechanisms governing the transcription of p16INK4a, one of the master regulators of cellular senescence, have been extensively studied. However, little is known about chromatin dynamics taking place at its promoter and distal enhancer. Here, we report that Forkhead box A1 protein (FOXA1) is significantly upregulated in both replicative and oncogene-induced senescence, and in turn activates transcription of p16INK4a through multiple mechanisms. In addition to acting as a classic sequence-specific transcriptional activator, FOXA1 binding leads to a decrease in nucleosome density at the p16INK4a promoter in senescent fibroblasts. Moreover, FOXA1, itself a direct target of Polycomb-mediated repression, antagonizes Polycomb function at the p16INK4a locus. Finally, a systematic survey of putative FOXA1 binding sites in the p16INK4a genomic region revealed an ∼150 kb distal element that could loop back to the promoter and potentiate p16INK4a expression. Overall, our findings establish several mechanisms by which FOXA1 controls p16INK4a expression during cellular senescence.
Keywords: FOXA1, p16INK4a, polycomb complex, senescence
Introduction
Senescence is a permanent state of halted cellular proliferation, triggered by replicative exhaustion or elicited as homeostatic defence mechanisms against DNA damage, oncogenic activation, oxidative stresses and other detrimental growth conditions (Hayflick and Moorhead, 1961; Hayflick, 1965; Kuilman et al, 2010). Induction of senescence would not only establish a barrier against malignant progression of tumour cells, but also impose potential disruptions on tissue structure and function with ageing (Rohme, 1981; Campisi, 2001a, 2001b, 2005; Janzen et al, 2006; Krishnamurthy et al, 2006; Molofsky et al, 2006; Baker et al, 2011). In support of this notion, investigations on senescent haematopoietic and epidermal stem cells evidenced the inverse correlation between tissue regeneration capacity and senescence progression (Youn et al, 2004; Wang et al, 2006; Beltrami et al, 2012). Moreover, suppression of senescence network would improve the reprogramming efficiency for induced pluripotent cell (iPS) generation dramatically, underscoring potential deterioration of body functions by senescence in vivo (Banito et al, 2009; Banito and Gil, 2010).
Cellular senescence is characterized by arrays of distinctive phenotypes and biomarkers, such as large and flattened morphology, increased senescence-associated β-galactosidase (SA-β-gal) activity, permanent G1-phase arrest and accumulation of senescence-associated heterochromatic foci (SAHF) (Campisi, 1997; Ly et al, 2000; Narita et al, 2003; Kuilman et al, 2010). Among them, upregulation of the p16INK4a cyclin-dependent kinase inhibitor is one of the pivotal nodes of activated tumour suppressor network during senescence, as inactivation of CDK4 by p16INK4a disables the sensitivity of senescent cells for mitogen signals permanently (Alcorta et al, 1996; Kuilman et al, 2010). In view of the dominant role of p16INK4a in both senescence and tumour suppression, tremendous investigations were applied to uncover the genetic or epigenetic mechanisms driving its activation in ageing or its inactivation in tumorigenesis (Brookes et al, 2004). In this regard, oncogenic factors including RAS (Serrano et al, 1997), BRAF (Michaloglou et al, 2005), MYC (Guney et al, 2006), transcriptional factors (TFs) such as AP1 (Passegue and Wagner, 2000), ETS2 (Ohtani et al, 2001), and telomeric repeat-binding protein TRF2 (Smogorzewska and de Lange 2002) were implicated in p16INK4a transcription activation; helix–loop–helix(HLH) ID protein ID1 (Alani et al, 2001), basic helix–loop–helix (bHLH) protein TAL1 (Hansson et al, 2003), T-box proteins TBX2 (Jacobs et al, 2000) and DNA replication origin binding protein CDC6 of INK4 locus were identified as its repressors (Gonzalez et al, 2006). More recently, opposing functional histone methylation, catalysed by MLL1 and DDB1-CUL4 complex for H3 lysine 4 trimethylation (H3K4me3) and Polycomb complex for H3 lysine 27 trimethylation (H3K27me3), was reported to mark transcriptional activation or repression of p16INK4a, respectively (Itahana et al, 2003; Bracken et al, 2007; Dietrich et al, 2007; Agherbi et al, 2009). In addition, histone positioning at p16INK4a promoter was demonstrated to be differentially associated with its transcriptional outcome (Fatemi et al, 2005; Hinshelwood et al, 2009), suggesting that chromatin remodelling would actively participate in senescence progress through de-condensation of p16INK4a promoter.
Mammalian FOXA1, as a member of TF family characterized by ∼100 amino acid Forkhead DNA-binding domain (DBD), is implicated in endodermic and reproductive organogenesis of liver, pancreas, lung, prostate, and mammary gland (Zaret, 1999; Carlsson and Mahlapuu, 2002; Katoh, 2004; Lee et al, 2005; Wan et al, 2005; Gao et al, 2008; Augello et al, 2011). FOXA family TFs were designated as ‘pioneer factor’, because they could occupy distal regulatory enhancers and create chromatin competency for subsequent recruitment of collaborating TFs, instead of promoting immediate transcription activation (McPherson et al, 1993; Cirillo et al, 1998, 2002; Cirillo and Zaret, 1999; Carroll et al, 2005; Wang et al, 2009, 2011; Zaret and Carroll, 2011). This specialized capacity for ATP-independent chromatin remodelling is believed to be derived from the structural similarity between its Forkhead domain and linker histone H1 (Cirillo et al, 2002; Zaret and Carroll, 2011). Recent progress on lineage-specific occupancy of FOXA1 at discrete genomic regions revealed the importance of active chromatin context, including histone modification marks H3 lysine 4 mono- and di-methylation (H3K4me1/2) and DNA hypomethylation on selective activation of FOXA1-dependent cis-regulatory elements (Lupien et al, 2008; Serandour et al, 2011). Of particular interest, C. elegans PHA-4, which is homologous to human Forkhead box A family of transcription factors and required for embryonic development of the worm pharynx, was recently demonstrated to be essential for adult-specific regulation of diet restriction (DR)-mediated longevity (Horner et al, 1998; Kalb et al, 1998; Panowski et al, 2007).
In this manuscript, we demonstrated that FOXA1 was upregulated in both replicative and oncogene-induced senescence (OIS), and consequently drove p16INK4a activation through sequence-dependent transcriptionally activating and chromatin-dependent remodelling mechanisms. We also identified the epigenetic signatures facilitating FOXA1 recruitment on senescent chromatin and delineated the feedforward loop between dismissal of Polycomb complex and FOXA1 binding for p16INK4a activation. Finally, an ∼150 kb away distal regulatory element was evidenced to communicate with p16INK4a promoter and act as functional enhancer in a FOXA1-dependent fashion.
Results
FOXA1 is upregulated with senescence
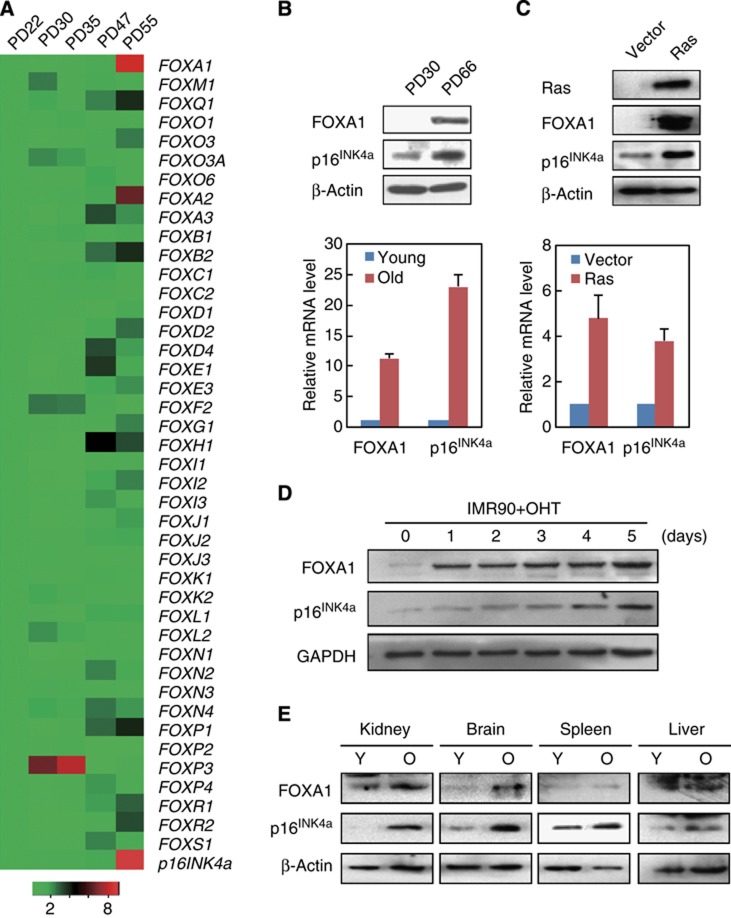
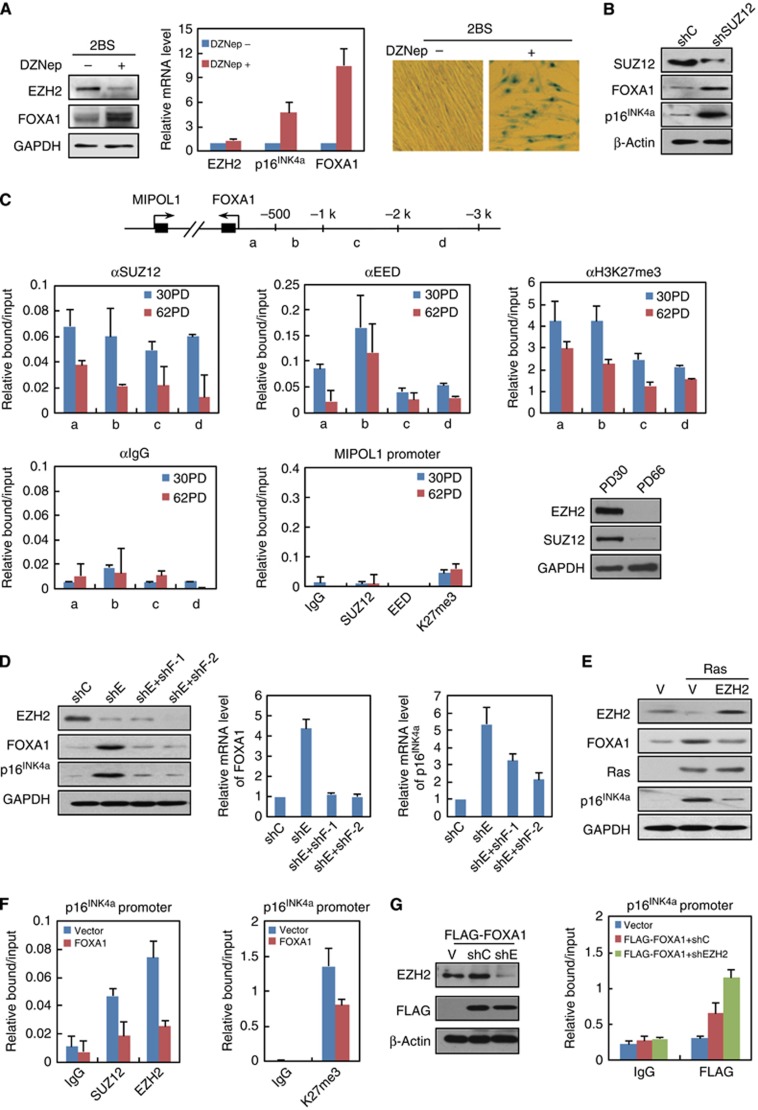
To further characterize transcription factors that contribute to the activation of tumour suppressor network in senescence, we performed real-time reverse-transcriptase PCR (RT-qPCR) to screen the Forkhead transcription factors with significant expression changes in human 2BS diploid fibroblast cells with increasing numbers of population doublings (PDs). The result indicated FOXA subfamily members FOXA1 and FOXA2 were most upregulated and their mRNA levels correlated with increased p16INK4a (Figures 1A and B). Western blotting results further confirmed robust induction of FOXA1 protein in senescent cells, compared with its barely detected level in young cells (Figure 1B). In addition, increased mRNA and protein level of FOXA1 could be observed in the oncogenic Ras-induced premature 2BS cells (Figure 1C). To consolidate this observation, we assessed its expression in human IMR90 diploid fibroblast cells that were derived from a different ethnic origin and stably integrated with a tamoxifen-regulated form of activated Ras (Tarutani et al, 2003). Using the protein lysates from IMR90 cells over a 4-hydroxytamoxifen (OHT) stimulation time course, we observed marked elevation of FOXA1 accompanied by increased p16INK4a activation, in accordance with the results from 2BS cells (Figure 1D).
Figure 1.
Expression of FOXA1 increases with senescence. (A) Expression changes of Forkhead family genes and p16INK4a in 2BS cells during replicative senescence. The mRNA levels at indicated passages were first normalized with GAPDH and then divided by its level in PD22 cells. The heat map was then produced based on these normalized results. The colour code indicative of fold changes was shown at bottom. (B) FOXA1 was upregulated in replicative senescent 2BS cells. Total protein and RNA were extracted from 2BS cells with PD30 or 66, and analysed for expression of FOXA1 and p16INK4a by western blotting (upper panel) and RT-qPCR (lower panel). mRNA levels of indicated gene were normalized with GAPDH. Each bar represents the mean+s.d. for triplicate experiments. (C) FOXA1 was upregulated in Ras-induced senescent 2BS cells. Total proteins and RNAs were extracted from 2BS cells retrovirally transduced with control vector or H-Ras, and analysed for expression of FOXA1 and p16INK4a by western blotting (upper panel) and RT-qPCR (lower panel). mRNA levels of indicated gene were normalized with GAPDH. Each bar represents the mean+s.d. for triplicate experiments. (D) Induction of FOXA1 in IMR90 cells with tamoxifen-inducible Ras expression. Protein levels of FOXA1 and p16INK4a at different times (0, 1, 2, 3, 4, and 5 days) following OHT treatment were determined by western blotting. GAPDH was used as loading control. (E) Protein levels of FOXA1 and p16INK4a in indicated tissues of young (3 months) and old (18 months) BALB/c mice were determined with western blotting. β-Actin serves as loading control.
Source data for this figure is available on the online supplementary information page.
In order to examine the expression change of FOXA1 with ageing in vivo, we compared its protein level in tissues from young adult BALB/c mice (3 months of age) to those from older ones (18 months). The results suggested that there was significant increment of FOXA1 with age in kidney and brain, and milder in the spleen, compared to the little changes in the liver (Figure 1E).
FOXA1 promotes senescence and activates expression of p16INK4a
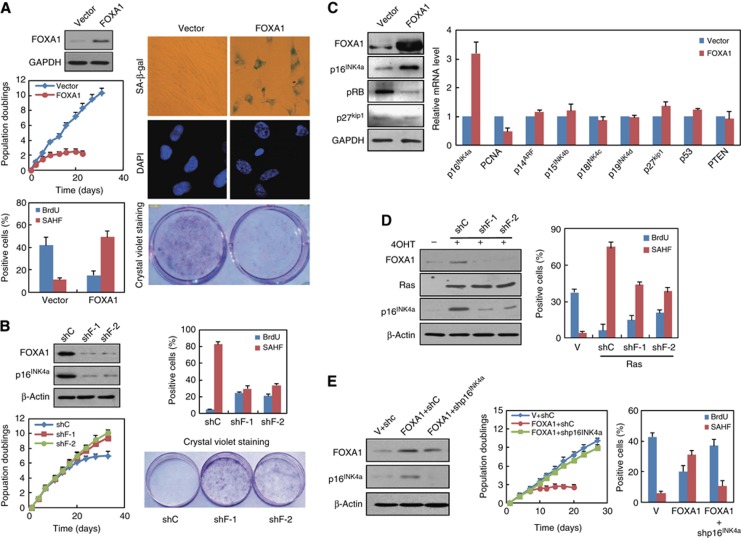
Increased FOXA1 expression in aged cells and mice implied its potential involvement in senescence programs. To determine the functional role of FOXA1 in these processes, we monitored phenotypic changes in 2BS cells with ectopic FOXA1 expression. Growth curve and crystal violet staining assay indicated that 2BS cells transduced with retroviral FOXA1 displayed lower proliferation rate, earlier growth arrest and limited colony formation compared with cells transduced with empty vector (Figure 2A). Meanwhile, ectopic expression of FOXA1 induced morphological features of senescence, including enlarged and flattened cell size, accumulation of granular cytoplasmic inclusions and elevated SA-β-galactosidase activity (Figure 2A). Immunostaining assays further support the pro-senescence action of FOXA1 as reduced BrdU incorporation and the formation of SAHF in ectopic FOXA1-expressed cells (Figure 2A).
Figure 2.
FOXA1 promotes senescence and activates expression of p16INK4a. (A) Ectopic expression of FOXA1 causes premature senescence. 2BS cells were infected with retrovirus expressing FOXA1 or control at early passage and expression of FOXA1 was determined by western blotting (first panel on the left). The infected 2BS cells were selected and cultured for ∼40 days with their growth curve monitored (second panel on the left). PDs for each time point are the mean value of triplicate experiments. Percentage of the indicated cells positive for BrdU and senescence-associated heterochromatin foci (SAHF) was shown (third panel on the left). Data represent the mean+s.d. for triplicate experiments. Representative images of indicated cells stained for SA-β-gal activity (first panel on the right) and DAPI (second panel on the right) were shown. 2BS cells were infected with indicated retroviruses, and seeded per 6-well plates for 10–15 days followed by fixation and staining with crystal violet (third panel on the right). (B) Knockdown of FOXA1 impaired p16INK4a activation and senescence progression. 2BS cells were transduced with two independent lentiviral shRNAs against FOXA1 (shF-1 and shF-2) or a scramble control vector (shC) at middle passage. About 1 month post infection, western blotting was performed for analysis of FOXA1 and p16INK4a expression (upper panel in the left). Growth curve monitoring, cell staining for BrdU and SAHF, and colony formation assay with crystal violet staining were performed as in (A). Data represent the mean+s.d. for triplicate experiments. (C) Overexpression of FOXA1 upregulated p16INK4a. 2BS cells were infected with retrovirus expressing FOXA1 or control as in (A). Total proteins extracted from infected 2BS cells were subjected to western blotting analysis with indicated antibodies (left panel). mRNA analysis of indicated senescence-associated genes was determined by RT-qPCR (right panel). Data represent the mean+s.d. for triplicate experiments. (D) Tow independent shRNAs against FOXA1 (shF-1, shF-2) or a control shRNA(shC) were infected in IMR90 cells with tamoxifen-inducible Ras expression. Seven days following OHT treatment, western blotting analysis was performed with indicated antibodies (left panel). Percentage of the indicated cells positive for BrdU and senescence-associated heterochromatin foci (SAHF) was shown in the right panel. Data represent the mean+s.d. for triplicate experiments. (E) p16INK4a was indispensible for FOXA1 overexpression-induced senescence. 2BS cells were transduced with a combination of retroviral FOXA1 or empty vector (V) and lentiviral shRNA against p16INK4a (shp16INK4a) or control shRNA (shC). The expression of FOXA1 and p16INK4a was determined by western blotting (left panel). Phenotypic differences in these cells were monitored through population doubling analysis (middle panel), BrdU incorporation assay and SAHF formation assay (right panel) as done in (A). Data represent the mean+s.d. for triplicate experiments.
Source data for this figure is available on the online supplementary information page.
Next, we took a short hairpin RNAi (shRNA)-based knockdown approach to examine the requirement of FOXA1 for senescence progression. Middle-aged 2BS cells (PD40) were transduced with two independent lentiviral shRNA against FOXA1 or a non-silence control and the efficiency of FOXA1 depletion was validated by western blotting (Figure 2B). In concert with the effect of ectopic FOXA1, its removal resulted in reduced senescence and partial restoration of growth arrest as indicated by elevated BrdU incorporation, augmented colony formation, decreased presence of SAHF, and continued cell proliferation (Figure 2B).
Senescence is characterized by activation of a distinctive tumour suppressor network; we therefore examined expression changes of several key cell-cycle regulators and senescence-associated genes when FOXA1 was overexpressed in 2BS cells. We found that p16INK4a, but not other genes such as p27kip1, p14ARF, p15INK4b, p18INK4c, p19INK4d, p53, or PTEN, increased markedly upon ectopic FOXA1 expression (Figure 2C). This point was further supported by the downregulation of phosphorylated Rb (pRb) (Figure 2C, left panel), which could be negatively modulated by p16INK4a (Serrano et al, 1993). Besides, downregulation of PCNA, a reflection of Rb-dependent repression of cell-cycle progression, was also observed (Figure 2C).
To determine whether FOXA1 was required for p16INK4a activation in replicative exhaustion and OIS, we knocked down FOXA1 using lentiviral shRNA and examined the resultant expression change of p16INK4a. As expected, induction of p16INK4a protein at late passage was impaired in 2BS cells which integrated with lentiviral shRNA for FOXA1 in middle-aged stage (Figure 2B). In parallel, when we silenced FOXA1 in Ras or BRAF V600E-induced 2BS cells, a similar decline of p16INK4a could be detected by RT–qPCR or western blotting (Figure 2D; Supplementary Figure 1B). Phenotypic changes as elevated BrdU incorporation and less presence of SAHF were also observed in OIS with FOXA1 ablation (Figure 2D, right panel). In agreement with the removal of senescence barrier following FOXA1 knockdown, overexpression of oncogenic Ras and EZH2 accompanied by FOXA1 depletion greatly promoted anchorage-independent growth in soft agar relative to control RNAi (unpublished data). This point was further consolidated as cancer gene expression profile analysis revealed the inverse association of FOXA1 with neoplastic progression in several cancer types, such as gastric adenocarcinoma, desmoplastic medulloblastoma, pancreatic ductal adenocarcinoma, and colon adenocarcinoma (Supplementary Figure 7).
To determine the dependence of FOXA1’s pro-senescence function on p16INK4a, we performed an epistasis analysis by co-expression of FOXA1 with a shRNA against p16INK4a or a control vector in 2BS cells and measured DNA synthesis and SAHF formation using BrdU incorporation and DAPI staining, respectively. As shown in Figure 2E, growth arrest and SAHF formation in FOXA1-overexpressed 2BS cells was greatly reduced when p16INK4a was removed, confirming that FOXA1-regulated senescence was imparted by p16INK4a.
p16INK4a is a direct transcriptional target gene of FOXA1
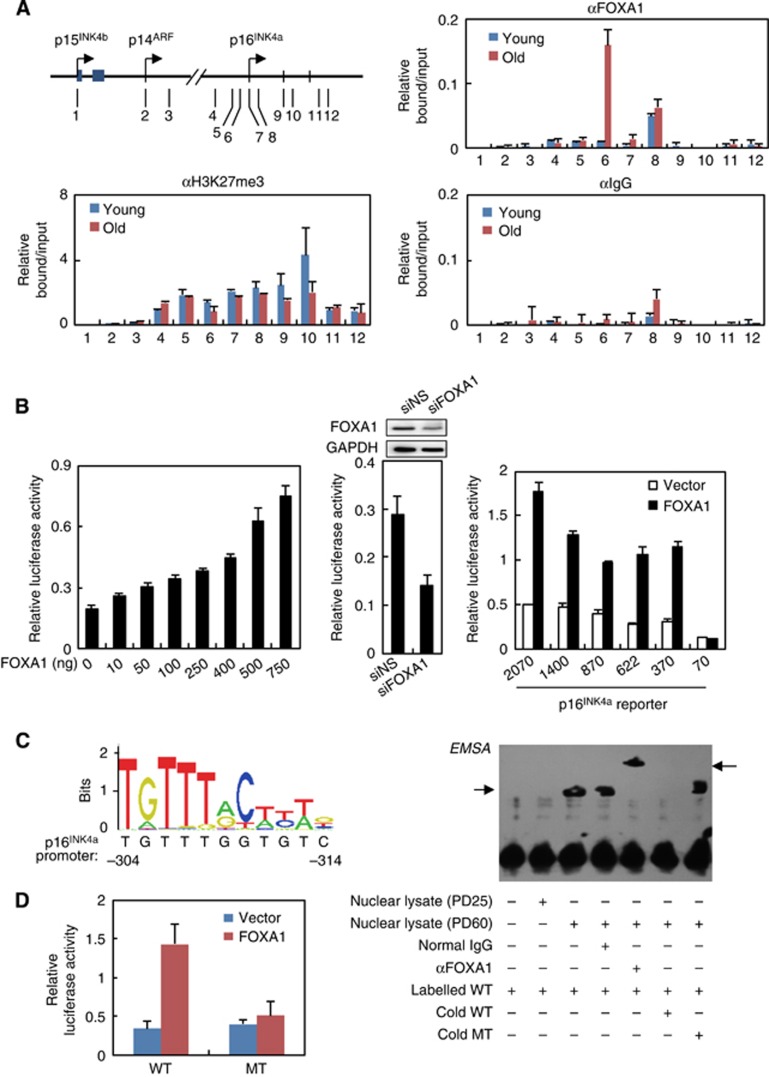
We next sought to explore the molecular mechanism underlying FOXA1-mediated transcriptional activation of p16INK4a. To test the possibility that FOXA1 directly bound to its cognate cis-regulatory element proximal to p16INK4a, we first performed chromatin immunoprecipitation assay followed by quantitative PCR (ChIP-qPCR) to determine FOXA1’s binding pattern within the INK4 locus in young and aged 2BS cells. The results indicated significant enrichment of FOXA1 in senescent cells surrounding p16INK4a transcription start site (TSS), in contrast to its barely detected binding in young cells (Figure 3A). Notably, such association was only observed at the p16INK4a promoter rather than the remaining 11 regulatory regions throughout the INK4 locus, in concert with specific transcriptional activation of p16INK4a but no other CDK inhibitors by FOXA1 (Figures 2C and 3A). In addition, decreased H3K27me3 modification was also observed at this region as previously described (Bracken et al, 2007; Agherbi et al, 2009), implying an active chromatin context accompanying FOXA1 recruitment during senescence.
Figure 3.
FOXA1 activates p16INK4a through directly binding on its promoter. (A) ChIP analysis of FOXA1 occupancy on the INK4 loci in young and replicative senescent 2BS cells. The precipitated DNA was amplified by qPCR using primers illustrated in the left panel. Error bars represent standard deviation from triplicate experiments. (B) FOXA1 enhanced the transactivation of p16INK4a promoter. Increasing amounts of FOXA1 together with a luciferase construct harbouring 2070-bp fragment of p16INK4a promoter were transfected in HeLa cells, and 48 h after transfection cells were harvested for luciferase activity assay (left panel). MCF-7 cells were co-transfected with siRNA against FOXA1 (siFOXA1) or non-sense control (siNS), along with p16INK4a promoter reporter. Depletion of endogenous FOXA1 was confirmed by western blotting (top panel, middle). Luciferase activity was shown below (bottom panel, middle). A series of luciferase reporters with various spanning coverage of p16INK4a promoter (2070, 1400, 870, 622, 370, and 70 base pairs in length) were constructed. These constructs were co-transfected with FOXA1 or empty vector in HeLa cells, and 48 h after transfection the relative luciferase activity was measured (right panel). Data represent the mean+s.d. for triplicate experiments. (C) The sequence logo for FOXA1 binding motif was retrieved from JASPAR database (http://jaspar.genereg.net). Predication of FOXA1 binding site on p16INK4a promoter was performed using JASPAR analysis tools with the default settings (left panel). EMSA was performed using biotin-labelled probes containing the predicated FOXA1 binding motif at p16INK4a promoter together with nuclear lysate from young (PD25) or old 2BS cells (PD60) (right panel). Inclusion of normal IgG or anti-FOXA1 antibody, and exceeded unlabelled wild-type probe (cold WT) or unlabelled mutant probe (cold MT) was indicated below. (D) A luciferase reporter was constructed with substitution of the four most conserved thymine bases for cytosine in the predicted FOXA1 motif. This construct (MT) and its parent (WT) were transfected in HeLa cells, and their responsiveness to FOXA1 was measured. Data represent the mean+s.d. for triplicate experiments.
Source data for this figure is available on the online supplementary information page.
The association between FOXA1 recruitment and activation of p16INK4a promoted us to test its trans-activating activity on the promoter of p16INK4a. For this purpose, we co-transfected HeLa cells with a p16INK4a promoter-driven luciferase reporter harbouring a 2070-bp fragment upstream of p16INK4a TSS and FOXA1 expression construct. As shown in Figure 3B, overexpression of FOXA1 was able to activate p16INK4a promoter in a dose-dependent manner (Figure 3B, left panel). Furthermore, a weakened p16INK4a promoter activity was observed when FOXA1 was knocked down in MCF-7 cells, a breast cell line with relatively high level of endogenous FOXA1 (Figure 3B, middle panel), indicating the requirement of FOXA1 for maintenance of p16INK4a promoter activity. In addition, luciferase activity analysis with domain mutagenesis of FOXA1 indicated that DBD was essential for p16INK4a promoter activation (Supplementary Figure 2B), reinforcing the importance of sequence-specific association between FOXA1 and p16INK4a promoter.
Luciferase reporter assay using a series of constructs with various spanning coverage of p16INK4a promoter indicated that enforced expression of FOXA1 could activate the reporters with 1400-, 870-, 622- and 370-bp fragments comparable to the 2070-bp construct, in contrast to the disappeared responsiveness of the 70-bp construct, suggesting a putative binding site of FOXA1 might locate between 370 and 70 bp upstream of p16INK4a TSS (Figure 3B, right panel). Bioinformatics analysis on this region revealed an 11 base pair sequence ranging from −314 to −304 bp closely resembled a putative FOXA1 binding motif (Figure 3C, left panel). Coordinately, substitution of the four most conserved thymine bases for cytosine in the predicted motif completely abolished its responsiveness to FOXA1 in the luciferase report assay (Figure 3D). Electrophoretic mobility shift assay (EMSA) experiments confirmed that recombinant FOXA1 protein was able to specifically bind to the p16INK4a promoter probes containing this element, since such occupancy was dominated by unlabelled wild-type competitor probes, but not by unlabelled probes with a mutation in this element (Supplementary Figure 2A). EMSA with young and aged nuclear lysates instead of purified FOXA1 protein further consolidated this point, as old 2BS cell lysate but not young ones could shift the labelled probe and incubation with antibody against FOXA1 resulted in a super-shift of FOXA1-DNA probe complex (Figure 3C, right panel). The binding specificity was also confirmed with unlabelled competitors.
FOXA1 promotes nucleosome loss at p16INK4a promoter
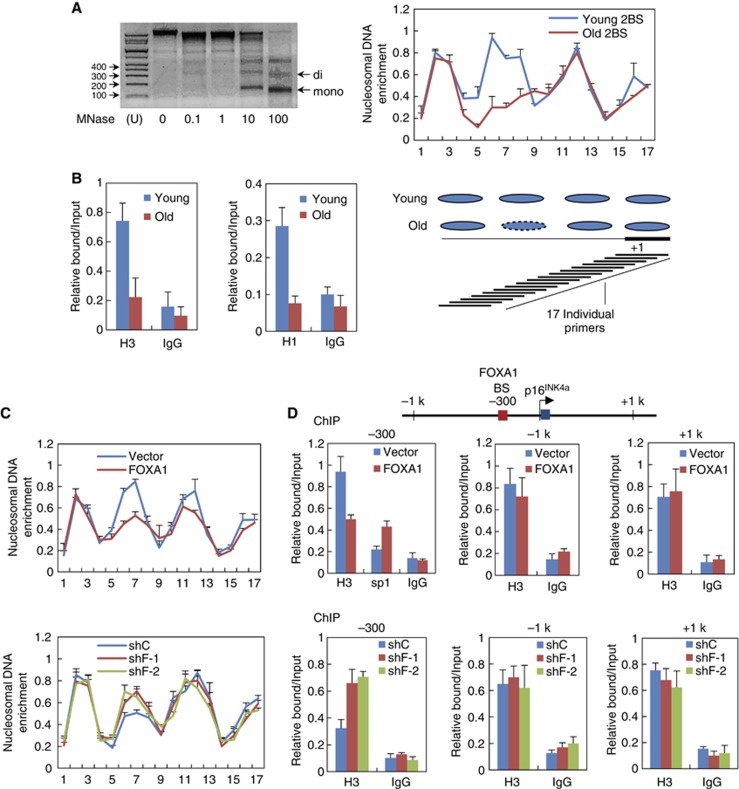
In addition to the physical association and functional link between p16INK4a and FOXA1 on histone-free DNA template in vitro, we sought to get further insight into the chromatin dynamics underlying FOXA1-dependent p16INK4a activation at its cis-regulatory elements packed with nucleosomes, given the structural similarity between its DBD domain and repressive linker histones H1 (Clark et al, 1993; Cirillo et al, 1998; Cirillo and Zaret, 2007). For this purpose, we employed a previously developed high-resolution MNase mapping assay to analyse in vivo nucleosome positioning pattern at p16INK4a promoter along with replicative senescence (Sekinger et al, 2005; Petesch and Lis, 2008). In this assay, chromatin from young and senescent 2BS fibroblast cells was isolated and further digested with micrococcal nuclease (MNase), and the resulting mono-nucleosome sized DNA fragments were visualized and extracted through gel electrophoresis (Figure 4A, left panel). The purified DNA samples were subjected to qPCR with overlapping primer pairs spanning ∼600 bp DNA stretch totally at p16INK4a promoter. We measured nucleosome density at a given region by the relative ratio of the amount of digested DNA to the undigested control, as the ability of a primer pair to amplify depends on the amount of contiguous DNA between the primers that remains after MNase digestion (Petesch and Lis, 2008).
Figure 4.
FOXA1 promotes nucleosome loss at p16INK4a promoter. (A) Nucleosome positioning patterns in the vicinity of p16INK4a TSS in young versus old 2BS cells measured by MNase (micrococcal nuclease) mapping assay. Chromatin was isolated from 2BS cells followed by digestion with increasing concentration of MNase as indicated (left panel). Mono-nucleosome sized DNAs were recovered from gel and used as PCR template to determine nucleosomal DNA enrichment at the indicated positions of p16INK4a promoter (upper panel on the right). Nucleosome density at a given region was measured as the relative ratio of the amount of digested DNA to the undigested control. Data represent the mean+s.d. for triplicate experiments. Overlapping primer pairs (typically spaced 30 bp apart) producing ∼100 bp products individually and the mapped nucleosomes were shown (bottom panel on the right). (B) ChIP experiments were performed in young and aged 2BS cells with indicated antibodies to measure histone density by using the primer pair 6 as depicted in (A). Data represent the mean+s.d. for triplicate experiments. (C) 2BS cells were transduced with retrovirus expressing FOXA1 or its control vector at early passage (PD25). The resultant chromatin was extracted and subjected to MNase mapping assay as in (A) (upper panel). Middle-aged 2BS at PD40 cells was transduced with two independent lentivirus expressing shRNA against FOXA1 (shF-1 and shF-2) or a non-silencing control. Approximately 30 days post infection, the resultant chromatin was subjected to MNase mapping assay. Data represent the mean+s.d. for triplicate experiments. (D) Overexpression or knockdown of FOXA1 in 2BS cells was conducted with the same procedure in (C). The resultant chromatin was extracted and subjected to ChIP assay with indicated antibodies followed by qPCR (bottom panel). The primer pairs to amplify −1 kb, +1 kb, and −300 bp regions relative to p16INK4a TSS were illustrated in the upper panel. Data represent the mean+s.d. for triplicate experiments. BS, binding site.
As shown in Figure 4A, we observed four nucleosomes positioned well on p16INK4a promoter (namely −3, −2, −1, and +1 relative to the TSS) with no apparent gap in young 2BS cells. Interestingly, a specific decrease in DNA amount was detected at the −2 histone positioning site rather than the other three nucleosomal regions in senescent cells, indicating a specialized chromatin remodelling event occurred at p16INK4a promoter during senescence progression (Figure 4A, right panel). To exclude the possibility that chromatin structure changes measured after MNase processing might be derived from increased accessibility of the DNA for MNase digestion, we performed traditional ChIP assay with antibody against histone H3 and H1 to directly quantify histone residence; and the results supported the occupancy of both histone H3 and H1 decreased at p16INK4a promoter in senescent 2BS cells (Figure 4B).
Of note, −2 histone positioning site contains the FOXA1’s binding motif identified through ChIP, luciferase reporter assay and EMSA as stated above (Figure 3), implying the implication of ATP-independent chromatin remodelling activity of FOXA1 for histone dynamic change at p16INK4a promoter. We therefore ectopically expressed FOXA1 in young 2BS cells, and monitored histone density by MNase mapping experiments. The results revealed a specific loss of −2 positional histone compared with its steady positioning at other sites upon enforced FOXA1 expression, recapitulating chromatin dynamics in natural senescence progression (Figure 4C, upper panel). Independent approach as low-resolution ChIP assay with antibody against histone H3 and H1 further supported that FOXA1 could regulate chromatin remodelling at p16INK4a promoter other than distal regions (Figure 4D, upper panel; Supplementary Figure 3B, upper panel). Consistent with these gain-of-function experiments, depletion of FOXA1 with two independent shRNA in senescent 2BS cells was sufficient to eliminate such remodelling events, evidenced by both MNase mapping (Figure 4C, bottom panel) and ChIP assay (Figure 4D, bottom panel; Supplementary Figure 3B, lower panel).
In view of kinetic chromatin state in replicative senescence, measurement of DNA accessibility in OIS model revealed active de-condensation at p16INK4a promoter following oncogenic stress (Supplementary Figure 3A). The open chromatin context at FOXA1 binding region might be then exploited for subsequent recruitment of collaborating TFs at p16INK4a promoter. This point was proved as Sp1 binding at p16INK4a promoter increased when FOXA1 was introduced in young 2BS fibroblast cells (Figure 4D).
FOXA1 is an important player in the transcriptional feedforward loop for p16INK4a activation induced by PRC2 removal
In order to delineate the upstream signalling molecules responsible for upregulation of FOXA1 in senescence, we challenged 2BS fibroblast cells with established senescence inducers, including oxidative/DNA damage, nutrient overload and manipulation of key genes for senescence program. Among them, elevated expression of FOXA1 was consistently observed when the component of Polycomb Repressive Complex 2 (PRC2) was removed (Figure 5A). In that case, growing 2BS fibroblast cells were treated by 3-Deazaneplanocin A (DZNep), which effectively promotes the depletion of cellular levels of PRC2 components and inhibition of associated histone H3 lysine 27 trimethylation (Fiskus et al, 2009). Similarly to the premature characteristics of fibroblast cells depleted with PRC2 components (Bracken et al, 2007; Wilson et al, 2010), administration of DZNep induced senescence, as illustrated by increased expression of p16INK4a and high β-galactosidase activity (Figure 5A). Importantly, we observed dramatically induced FOXA1 protein along with degradation of EZH2 upon DZNep treatment; RT-qPCR results also supported that the induction was at transcription level (Figure 5A). To exclude the possibility that upregulation of FOXA1 was derived from non-specific pharmaceutical effect of DZNep, we depleted SUZ12 component of PRC2 with RNAi-based approach and observed increment of FOXA1 protein following SUZ12 removal (Figure 5B). In line with the results from loss-of-function experiment, overexpression of EZH2 in Ras-mediated OIS blunted the upregulation of FOXA1 (Figure 5E). These results were reasonable, because FOXA1 as a developmental gene was transcriptionally silenced by PRC2 in human embryonic stem cells (Bracken et al, 2006; Lee et al, 2006; Schuettengruber et al, 2007).
Figure 5.
FOXA1 is an important player in the transcriptional feedforward loop for p16INK4a activation induced by PRC2 removal. (A) FOXA1 was upregulated following DZNep-mediated degradation of EZH2. 2BS cells were treated with 5 μM DZNep for 48 h followed by an extended culture for 2 weeks. The cells were then harvested for western blotting (left panel), RT-qPCR (middle panel), and SA-β-gal activity analysis (right panel). Data represent the mean+s.d. for triplicate experiments. (B) Early-passage 2BS cells were infected with retroviral shRNA against SUZ12 (shSUZ12) or a non-silencing control (shC). Total cell lysates were harvested for western blotting analysis with indicated antibodies. (C) ChIP analysis of SUZ12, EED, and histone H3 lysine 27 trimethylation (H3K27me3) enrichment on FOXA1 promoter in young (PD30) and old (PD62) 2BS cells was performed. The primer pairs used for PCR on FOXA1 promoter (designated as ‘a’, ‘b’, ‘c’, and ‘d’) and MIPOL1 promoter were illustrated at the top panel. Data represent the mean+s.d. for triplicate experiments. The protein level of EZH2 and SUZ12 in young and aged 2BS cells was determined by western blotting assay (bottom panel on the right). (D) FOXA1 was required for EZH2 silence-mediated p16INK4a activation. Young 2BS cells infected with shRNA against EZH2 (shE), or a combination of shE and shRNA against FOXA1 (shF-1 or shF-2) were subjected to western blotting (left panel) and RT-qPCR analysis (right panel). Data represent the mean+s.d. for triplicate experiments. (E) Western blotting analysis was performed in 2BS cells transduced with empty vector (V), Ras or a combination of Ras and EZH2 using indicated antibodies. (F) ChIP analysis of EZH2, SUZ12, and H3K27me3 enrichment on p16INK4a promoter was performed in empty vector- or FOXA1-infected 2BS cells. Data represent the mean+s.d. for triplicate experiments. (G) 2BS cells were transduced with empty vector (V), or a combination of retroviral FLAG-FOXA1 and shRNA against EZH2 (shE) or non-silencing control (shC). Western blotting analysis with indicated antibodies was shown in the left panel. ChIP analysis with anti-FLAG antibody on p16INK4a promoter was shown in the right panel. Data represent the mean+s.d. for triplicate experiments.
Source data for this figure is available on the online supplementary information page.
ChIP-qPCR analyses in 2BS cells revealed prominent binding of EED and SUZ12 components and also the enrichment of histone H3K27me3 modification on FOXA1 promoter in young 2BS cells at all tested regions, compared to the control IgG (Figure 5C). Importantly, in replicative senescent cells almost all the tested regions experienced decreased binding of EED and SUZ12 coupled with a loss of H3K27me3 mark, especially at the most proximal region; and this result was consistent with globally decreased protein levels of PRC2 members in aged cells (Figure 5C, lower panel). The barely detected PRC2 subunits at the promoter region of MIPOL1, a gene located adjacent to FOXA1 on the chromosome, supported the regulation of FOXA1 by PRC2 was specific (Figure 5C, lower panel).
The fact that p16INK4a was tightly controlled by repressive PRC2 (Bracken et al, 2007), promoted us to test the convergence of PRC2-removal mediated de-repression and FOXA1-dependent transcriptional activation at p16INK4a promoter. We therefore co-infected 2BS fibroblast cells with retroviral-shRNA against EZH2 and lentiviral-shRNA against FOXA1; and western blotting assay indicated that the protein level of p16INK4a increased when EZH2 was knocked down, however, such effect was severely impaired after FOXA1 was further depleted (Figure 5D). On the other hand, enforced EZH2 overexpression weakened FOXA1-mediated p16INK4a upregulation in OIS model (Figure 5E), while ectopic expression of FOXA1 promoted the dismissal of PRC2 from p16INK4a promoter (Figure 5F). These results pinpointed the mutual elusive interplay between FOXA1 and Polycomb complex for control of p16INK4a activation; however, co-immunoprecipitation experiment did not support the physical interaction between FOXA1 and PRC1/2 complex (Supplementary Figure 4B), hence their functional exclusion was not likely resulted from a direct effect. In order to determine whether the chromatin context with the depletion of PRC2-deposited repressive epigenetic code could facilitate FOXA1 recruitment beyond the differential FOXA1 expression levels, we co-transduced the 2BS cells with a combination of FLAG-tagged exogenous FOXA1 and shEZH2 or control shRNA. ChIP experiments with antibody against FLAG confirmed the contribution of chromatin context, as the enrichment of FLAG-tagged FOXA1 increased following EZH2 depletion (Figure 5G).
These results suggested that FOXA1 did not only respond to depression of PRC2, but also was implicated in reinforcing the active epigenetic remodelling at p16INK4a promoter to orchestrate a feedforward loop governed by PRC2 removal for fully activated p16INK4a transcription.
Identification of a distal FOXA1-bound region ∼150 kb away from TSS as a putative enhancer for p16INK4a
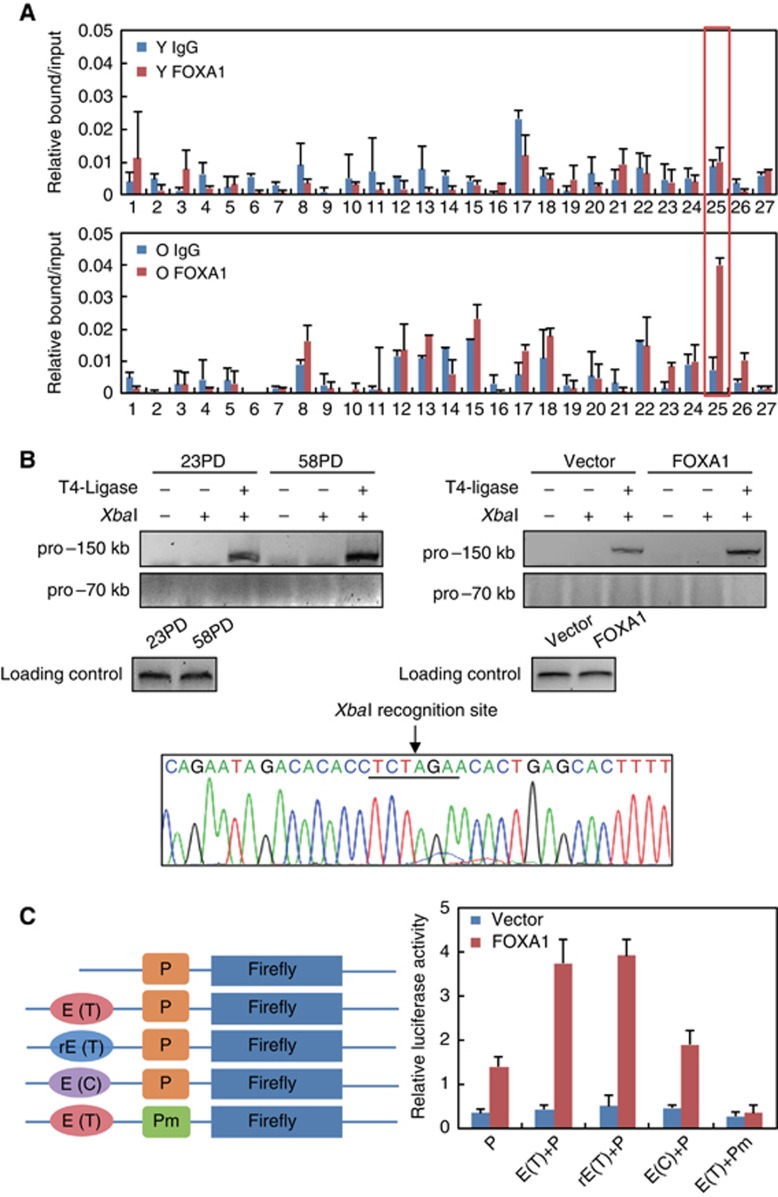
Gene transcription requires collaborations between proximal promoters and distal enhancers. However, to our knowledge, there are few investigations on distal enhancer elements compared to the overwhelming researches on promoter regions for the studies of p16INK4a activation during senescence. In light of the prevalent chromosomal looping mechanism of FOXA1 to promote nuclear receptor-mediated gene transcription (Carroll et al, 2005; Lupien et al, 2008), identification of FOXA1 as a putative activator for p16INK4a afforded us the opportunity to search for the potential p16INK4a enhancer elements. The candidates were chosen based on both predicated FOXA1 motif score (Figure 3C) and histone H3K4me1/H3K4me2 marks indicative of enhancer elements in ENCODE database (Ohtani et al, 2001; Heintzman et al, 2007). As a result, 27 sites scattered within ∼200 kb encompassing p16INK4a TSS were finally picked up for further investigations.
ChIP-qPCR analysis at all the 27 sites in young and replicative senescent 2BS cells revealed a unique site ∼150 kb away from p16INK4a TSS was significantly bound by FOXA1 in aged but not in young cells (Figure 6A). To test whether this ∼150 kb away site could physically communicate with p16INK4a promoter via chromosome looping, we performed chromosome conformation capture (3C) experiments to assess their interaction (Smogorzewska and de Lange, 2002; Carroll et al, 2005). In this experiment, young and replicative senescent 2BS cells were collected, and the fixed chromatin was digested with a specific restriction enzyme XbaI. The resulting DNA samples were diluted and re-ligated by T4 DNA ligase to achieve maximal intermolecular joining. After ligation, the crosslinking was reversed and the recombinant DNA was purified. The interaction frequency between this putative enhancer element and p16INK4a promoter was then measured by PCR with the primer to amplify ligation products bridging these two sites (Zhang et al, 2010). The results in Figure 6B illustrated a prominent band at expected position from senescent 2BS cells indicative of genuine interaction between the −150 kb element and p16INK4a promoter, while such interaction was weaker in young cells. Sequencing result of these PCR products confirmed their derivation, and the XbaI site was also evidenced to be located at the joint (Figure 6B, lower panel). To determine the sufficiency of FOXA1 to drive such proximity, we overexpressed lentiviral FOXA1 in growing 2BS fibroblast cells and performed 3C assay to examine the resultant interacting frequency. As expected, enforced expression of FOXA1 alone could induce strong interaction between these two sites comparable to that in replicative senescent cells (Figure 6B, right upper panel). As a negative control, the interaction between p16INK4a promoter and a −70 kb site was not observed in both replicative senescent and FOXA1-overexpressed cells (Figure 6B, upper panel).
Figure 6.
Identification of a distal FOXA1-bound region ∼150 kb away from TSS as a putative enhancer for p16INK4a. (A) ChIP analysis of FOXA1 enrichment on candidate enhancer regions was performed in young (upper panel) and senescent (lower panel) 2BS cells. The prominent binding of FOXA1 at the region ∼150 kb away from p16INK4a TSS was highlighted by a red box. Data represent the mean+s.d. for triplicate experiments. (B) Chromosome conformation capture (3C) assay confirmed the communication between p16INK4a promoter and the putative enhancer element highlighted in (A). In this experiment, chromatin extracted from young and replicative senescent 2BS cells was subjected to 3C assay with ‘pro--150 kb’ or ‘pro--70 kb’ primers, which were used to amplify the re-ligated products composed of p16INK4a promoter as the ‘constant fragment’ and DNA stretches −150 or −70 kb away from TSS as ‘candidate interacting fragment’ (left panel). Fragments within two adjacent XbaI sites were amplified as a loading control. 3C-PCR product with the ‘pro--150 kb’ primer was sequenced as shown in the bottom panel and the XbaI site linking two fragments was underlined. 3C assay was also performed in retroviral FOXA1- or control-infected 2BS cells (right panel). (C) The indicated reporter constructs were introduced into HeLa cells for testing their responsiveness to FOXA1. Data represent the mean+s.d. for triplicate experiments. Firefly, luciferase gene; P, p16INK4a promoter; Pm, p16INK4a promoter with mutated FOXA1 binding site; E(T), −150 kb enhancer with rs10811661-T (see below); rE(T), reversed E(T); E(C), −150 kb enhancer with rs10811661-C (see below).
Source data for this figure is available on the online supplementary information page.
To further investigate the possibility that this 150 kb element can function as authentic enhancer, we inserted a stretch of genomic sequence centred on this site into the luciferase vector containing the p16INK4a promoter and co-transfected these constructs with FOXA1 expression plasmids into HeLa cells. As shown in Figure 6C, inclusion of the putative enhancer sequences regardless of the direction (‘E(T)’ and ’rE(T)’, see below) resulted in an additional ∼3-fold increment of response to FOXA1 compared with the sole promoter construct. Importantly, mutation of FOXA1 binding site at promoter region completely abolished the induced luciferase activity, underscoring that the function of −150 kb enhancer was dependent on its communication with the promoter bridged by FOXA1 (Figure 6C).
DNA variant at the −150 kb enhancer influences FOXA1-dependent p16INK4a activation
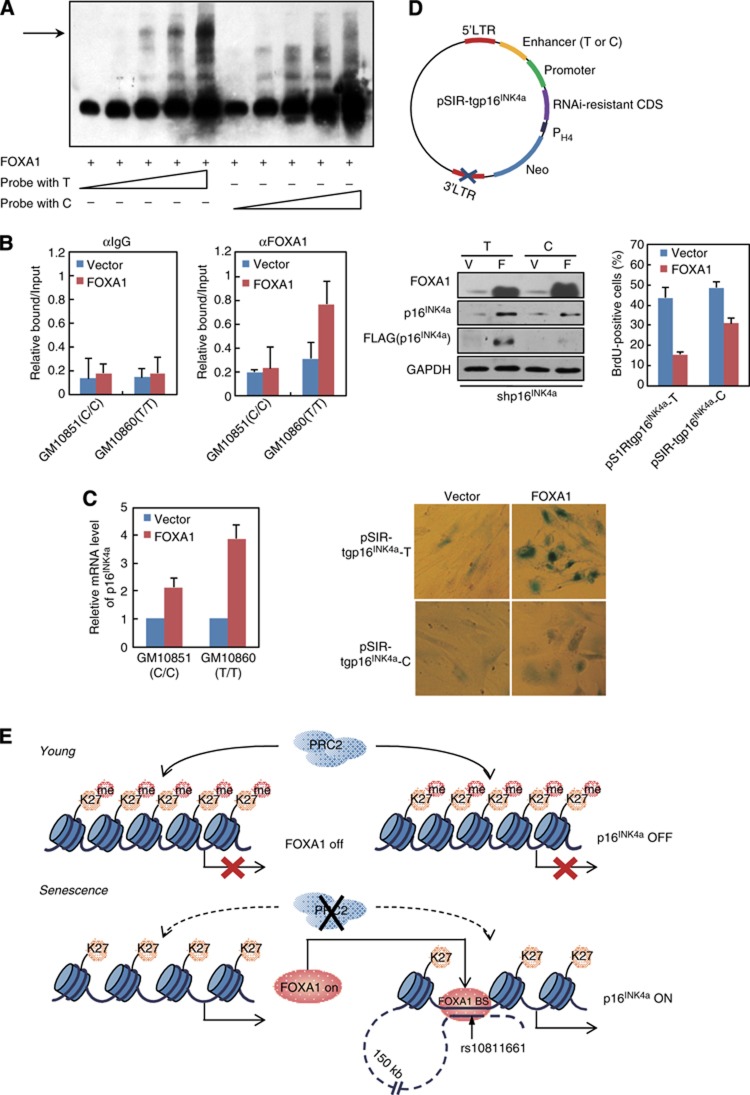
Detailed sequence analysis on the −150-kb enhancer revealed a single-nucleotide polymorphism (SNP) site rs10811661 (C/T) resides in the core region of predicted FOXA1 binding motif and maps to sites of evolutionary conservation among several mammals, implying its biological relevance (Supplementary Figure 5). Interestingly, the risk allele with polymorphism T was independently identified to be associated with type 2 diabetes mellitus (T2D) in several recent genome-wide association studies (GWAS) (Wen et al, 2010; Cugino et al, 2011; Bao et al, 2012). Since the FOXA family proteins play crucial roles in pancreatic development and glucose metabolism, it was reasonable to examine the influence of C/T polymorphism on FOXA1 binding at this −150-kb enhancer element. We thus performed EMSA with in vitro synthesized C or T containing probes to compare their affinity towards FOXA1; and the result indicated that T probe was preferable to be bound by recombinant FOXA1 protein in accordance with predicated binding preference (Figure 7A; Supplementary Figure 6A). To determine whether the similar scenario exists in vivo, two commercially available lymphocytic cell lines possessing T/T or C/C genotype of rs10811661 were utilized. Genotyping by direct DNA sequencing of PCR products confirmed that GM10860 was homozygous for rs10811661 T allele, while GM10851 was homozygous for C allele (Supplementary Figure 6B). We lentivirally overexpressed FOXA1 in these two cell lines and ChIP-qPCR analysis indicated that FOXA1 was more readily to be detected on this −150-kb enhancer region in GM10860 (T/T) cells than that in GM10851 (C/C) cells (Figure 7B), in concert with higher p16INK4a level in the T/T cells responding to enforced FOXA1 expression (Figure 7C). Interestingly, although 2BS fibroblast cells were heterozygous for rs10811661, sequencing results of FOXA1 bound DNA fragments in ChIP experiments (Figure 6A) revealed their inclination for T allele as the ratio of T-allele clones versus C-allele clones was 8:2. Moreover, in agreement with our observation, a recent publication on breast cancer risk-associated SNPs identified an allele with rs4784227-T variant at the same position as rs10811661 (Supplementary Figure 6) in FOXA1 binding motif could favour its recruitment over rs4784227 C allele (Cowper-Sal lari et al, 2012).
Figure 7.
DNA variant at the −150 kb enhancer influences FOXA1-dependent p16INK4a activation. (A) FOXA1 possessed higher affinity towards T-allele binding site located in the −150 kb enhancer. EMSA was performed with recombinant FOXA1 protein and increasing amount of biotin labelled probe. The probe harbours a DNA variant rs10811661 for T or C, which is enclosed in the predicted FOXA1 binding motif at the −150 kb enhancer. (B) ChIP analysis of FOXA1 enrichment at the −150 kb enhancer was performed in FOXA1- or control vector-infected GM10851 (carrying C allele) and GM10860 (carrying T allele) lymphocytes. Data represent the mean+s.d. for triplicate experiments. (C) GM10851 and GM10860 lymphocytes were transduced with lentiviral FOXA1 or control vector. RT-qPCR analysis for p16INK4a was conducted in these cells. Data represent the mean+s.d. for triplicate experiments. (D) A transgenic construct designated as pSIR-tgp16IN4a hosted on a self-inactivating retrovirus-derived vectors was demonstrated in the top panel. RNAi-resistant CDS, FLAG-tagged full-length p16INK4a coding sequence with synonymous mutations to abolish its pairing with shp16INK4a; promoter, p16INK4a promoter with 370 bp DNA sequence upstream from its TSS; enhancer (T or C), −150 kb enhancer with DNA variant rs10811661 for T or C. PH4, promoter for histone H4. Early-passage 2BS cells with endogenous p16INK4a silenced by its targeting shRNA (shp16INK4a) were co-transduced with pSIR-tgp16INK4a (T or C) and retroviral FOXA1 or control vector. The cells were selected and cultured for 2 weeks. Western blotting analysis was performed with indicated antibodies (middle panel, left). Phenotypic differences in these cells were monitored through BrdU incorporation assay (middle panel, right) and SA-β-gal activity analysis (bottom panel). Data represent the mean+s.d. for triplicate experiments. (E) A model depicts the molecular mechanism underlying FOXA1-mediated p16INK4a activation during senescence.
Source data for this figure is available on the online supplementary information page.
To test the functional significance of rs10811661, we performed site-directed mutagenesis to introduce the variant C allele to the established enhancer luciferase construct carrying T allele. As shown in Figure 6C, the C-allele construct had little enhancer activity responding to enforced FOXA1 expression, correlating with its lower affinity to FOXA1. In order to examine the importance of rs10811661 on FOXA1-regulated senescence, it should be a better choice of pairing cell lines with similar genetic background but harbouring homozygous allele for T or C. However, the available cell lines are generally transformed with pRB inactivated, impeding our examination on cell proliferation and cell-cycle exit. Alternatively, we assembled a transgenic construct hosted on self-inactivating retrovirus-derived vector with defective 3’ long terminal repeat (LTR) element, which was suitable for transcription regulation study due to the absence of enhancer and promoter sequences in both LTRs of the integrated provirus (Yu et al, 1986). On the other hand, the p16INK4a protein derived from the transgenic construct should not be complicated by its endogenous counterpart, we therefore designed a FLAG-tagged and p16INK4a-RNAi resistant open reading frame (ORF) called as resp16INK4a. The final retroviral construct was composed of p16INK4a promoter, resp16INK4a, and −150-kb enhancer element carrying C or T allele, and thus termed as pSIR-tgp16INK4a-C and pSIR-tgp16INK4a-T, respectively (Figure 7D). We co-transduced growing 2BS cells with lentivirual-FOXA1, shp16INK4a, and pSIR-tgp16INK4a-C or pSIR-tgp16INK4a-T; and western blotting results confirmed the successful expression of FOXA1 and depletion of endogenous p16INK4a by shp16INK4a (Figure 7D, middle panel). Importantly, when co-infected with lentiviral-FOXA1, 2BS cells with pSIR-tgp16INK4a-T showed higher induction of FLAG-tagged p16INK4a relative to pSIR-tgp16INK4a-C, consistent with the results from luciferase reporter assays (Figure 7D, middle panel). And consequently, pSIR-tgp16INK4a-T transduced 2BS cells displayed stronger SA-β-galactosidase activity and lower proportion of BrdU-positive cells (Figure 7D, bottom panel).
Discussion
In this study, we initially observed that FOXA1 was the most induced transcription factor of Forkhead family proteins during replicative senescence and also was accumulated in aged mouse (Figure 1). Further biochemical and phenotypic experiments suggested that FOXA1 exerts its pro-senescence function through direct transcriptional activation of p16INK4a (Figure 7E). Interestingly, a previous genome-wide RNAi-based screening for target genes required for oncogenic BRAF-activated senescence barrier identified FOXA1 as an essential component for p16INK4a activation (Wajapeyee et al, 2008).
FOXA2 as a close paralogue of FOXA1 was similarly identified to be upregulated in replicative senescence (Figure 1A); however, enforced expression of FOXA1 but not FOXA2 drove the induction of p16INK4a in 2BS fibroblast cells consistently (Supplementary Figure 1A). Although the molecular basis for this differential regulation was currently unknown and correlation of FOXA2 and p16INK4a in other biological context was not determined in this study, distinct sets of target genes of these paralogues were recently demonstrated in the adult liver through genome-wide location analysis and such functional diversification was proposed to confer the maintenance of both genes during evolution (Bochkis et al, 2012). Intriguingly, while FOXA homologue PHA-4 in C. elegans was reported to be involved in DR-mediated longevity, FOXA1-regulated ageing process in mammals should be more complicated due to the presence of evolved INK4 family proteins, which are lacked in C. elegans (Gil and Peters, 2006). On the other hand, the functional role of FOXA1 in neoplastic transformation should not be restricted to its ‘pioneering’ collaborations with extra oncogenic factors, such as oestrogen or androgen receptors; the p16INK4a-centred tumour suppressor network as a senescence barrier, which might be imposed by FOXA1 activation, should be further examined in the context of FOXA1-associated tumour progression. In support of this notion, although FOXA1 was reported to be involved in initiating breast and prostate cancers, downregulation of FOXA1 was proposed to be an unfavourable prognostic signature in progressive prostate cancer accompanied by aberrant cell cycles (Wang et al, 2009, 2011).
Consistent with previous reports that transcriptional activation of p16INK4a correlated with loss of histone binding at its promoter (Fatemi et al, 2005; Hinshelwood et al, 2009), we observed −2 positioned nucleosome was specifically removed during replicative senescence (Figure 7E). Reduced histone H3 and H1 binding when FOXA1 ectopically expressed and retention of these histones in aged cells when FOXA1 knocked down (Figure 4), suggested that FOXA1 was a crucial regulator for chromatin remodelling at p16INK4a promoter, likely through an similar mechanism in liver development (Cirillo et al, 1998, 2002; Cirillo and Zaret, 1999). Interestingly, the specific loss of −2 positioned nucleosome renders p16INK4a fall into the category of occupied proximal-nucleosome (OPN) genes relative to depleted proximal-nucleosome (DPN) genes (Zheng et al, 2004). The OPN set of genes, which exhibit relatively high nucleosome occupancy close to the TSS (−100 to 0) coupled with relatively low occupancy at a more distal region (−400 to −150), tend to have high transcriptional plasticity and dynamic nucleosome positioning compared with DPN genes. These characteristics are compatible with varied expression level and dynamic regulation of p16INK4a responding to manifold cellular stimuli (Gil and Peters, 2006). Thus, beyond activation of histone-free DNA template of p16INK4a promoter (Figure 3A–D), FOXA1 would augment the responsiveness of endogenous chromatinized cis-regulatory elements to other TFs, such as Sp1 in our case, by sequence-specific chromatin remodelling (Figure 4D, left panel), and this mechanism should be tested in a broader way to comprehensively understand other signalling pathways to invoke the activation of p16INK4a.
Depletion of FOXA1 severely impaired PRC2 removal coupled p16INK4a activation (Figure 5D), indicating that FOXA1 was an essential component of a feedforward regulatory circuit linking PRC2 relayed signalling pathway and senescence. In addition to p16INK4a, FOXA1’s promoter was evidenced to be another arena for PRC-involved cell-fate determinations (Figures 5 and 7E). Although FOXA1 was observed to potentiate p16INK4a activation in the presence of PRC2 (Figure 2A; Supplementary Figure 4A), both transcriptional repression of FOXA1 itself by RPC2 and enhanced FOXA1 recruitment at p16INK4a promoter in favour of chromatin context with PRC2 depletion suggested the dominance of Polycomb complex’s repression over FOXA1’s activation for p16INK4a activation (Figure 5). It is conceivable that such ‘dominance’ strategy might have significant biological meanings when p16INK4a is intended to be protected from aberrant activation under the circumstance of high FOXA1 expression. In line with this, insulin production was demonstrated to require not only FOXA1 for expansion and differentiation of the pancreatic primordial (Gao et al, 2008, 2010), but also PRC2 to maintain normal β cell expansion and regeneration by transcriptional repression of p16INK4a (Huang et al, 2009; Chen et al, 2011).
Combined bioinformatics survey, biochemistry analysis, and functional evaluation led us to obtain a FOXA1-bound element ∼150 kb away from p16INK4a promoter communicating with its proximal region in aged fibroblast cell and serving as a FOXA1-dependent enhancer (Figure 6). This newly identified distal element could be further investigated under other cellular defence models confronted with diverse homeostatic challenges, such as DNA damage, oxidative stress, and even induced pluripotent reprogramming. Notably, given the critical role of FOXA family factors in early pancreatic development and islet β cell functionalities, the finding of SNP rs10811661 associated with T2D residing within FOXA1 binding motif at this −150 kb enhancer implied a tempting aetiological interpretation for sub-population of T2D patients with T-containing allele in favour of FOXA1 binding, whose insulin secreting β cells might be subject to premature functional decline by abuse of FOXA1 factor to upregulate p16INK4a transcription with the assistance of distal enhanceosome. Besides the insulin producing organ, FOXA family factors are implicated in glucose sensing and homeostasis in liver (Kaestner et al, 1999; Shih et al, 1999); therefore, it is also plausible that FOXA1-p16INK4a axis might be deregulated to result in aberrant hepatic gluconeogenesis.
Overall, the results of present work revealed pleiotropic mechanisms underlying FOXA1-based regulatory strategy to translate epigenetic signature on cis-regulatory elements of p16INK4a into an actual senescence program (Figure 7E).
Materials and methods
Measurement of senescence-associated phenotypes
For the SA-β-gal staining experiment, cells fixed with formaldehyde were incubated in a freshly prepared SA-β-gal staining solution at 37°C overnight followed by photomicrography as described previously (Dimri et al, 1995). For colony formation assay, 3 × 103 cells were seeded on 6-well plates, selected and followed by fixation with formaldehyde and staining with crystal violet (Sigma). For population-doubling analysis, cell number was counted and ∼4 × 105cells were plated on 10 cm plates every 3–4 days. The cumulative PDs were summed by the ΔPD as in the formula ΔPD=log2 (Ch/Ci), with cells derived from the primary culture considered to be at PD=0, where Ci is the number of cells when inoculated and Ch is the number of cells when harvested per round of split and passage. For BrdU incorporation analysis, cells were incubated at 37°C with the addition of 5-Bromo-2’-deoxyuridine and 5-fluor-2’-deoxyuridine followed by fixation, permeabilization, denaturation, and blocking. Immunostaining was then performed with antibody against BrdU (Pharmingen) overnight at 4°C followed by incubation with FITC-conjugated antibody. For SAHF formation assay, the cells were stained with 1 μg/ml DAPI and then examined by using TCS SP2 confocal microscope (Leica). Quantitative results of BrdU and SAHF assays were presented as mean+s.d. for triplicated experiments.
Chromosome conformation capture assay (3C)
The 3C assay was done essentially as described (Tolhuis et al, 2002) with minor modifications. Briefly, collected cell pellets were washed with PBS buffer and then crosslinked with formaldehyde (Sigma) to achieve a final concentration of 2%. After a 10-min incubation at 37°C, glycine (0.125 M final concentration) was added to stop the reaction. The pellet was then subjected to cold lysis buffer with protease inhibitors (Roche Applied Science), homogenized with a Dounce homogenizer on ice, and centrifuged to pellet the nuclei. To remove non-crosslinked proteins from DNA, SDS was added to a final concentration of 0.3%. For XbaI digestion, Triton X-100 was added to a final concentration of 1% to sequester excess SDS. A 5-μl aliquot of the sample was kept as an undigested genomic DNA control. XbaI (New England Biolabs) restriction enzyme was added to the remaining sample, and it was digested overnight at 37°C. Digestion efficiency was optimized and monitored by PCR using primer pairs designed specifically for each of the XbaI restriction sites in the FOXA1 occupied p16INK4a promoter and enhancer loci. After complete digestion, 1.6% SDS was added for 20 min at 65°C to inactivate XbaI, and a 5-μl aliquot of the sample was set aside as the digested genomic DNA control. The ligation reaction was performed with 400 units of T4 DNA ligase (New England Biolabs) for 4 h at 16°C, followed by incubation for 30 min at room temperature in a total of 5-ml reaction system. Crosslinking was reversed by overnight incubation of the samples with proteinase K at 65°C, followed by phenol-chloroform purification of DNA. Purified DNA was subjected to PCR amplification with site-specific primer pairs, which were designed to amplify the re-ligated products composed of p16INK4a promoter as the ‘constant fragment’ and DNA stretches −150 or −70 kb away from TSS as ‘candidate interacting fragment’ as shown in Figure 6B. The primer sequences used in this assay are listed below:
pro--150 kb: GACGGACTCCATTCTCAAAGTC (forward) and TCCAACAGGATCTGTTCACG (reverse);
pro--70 kb: CCTAGACCAGAAAAAGTGCTCAG (forward) and TCAACCTAACCTCCCTCTTCC (reverse);
Loading control: GGACGGACTCCATTCTCAAA (forward) and TCATATTCCCTTCCCCCTTT (reverse).
Supplementary Material
Acknowledgments
We appreciate Dr Masashi Narita (Cancer Research UK, Cambridge Research Institute) for providing us the retroviral infection system and helpful proposal. We thank Dr Wolter J Mooi (VU University Medical Center) and Dr Daniel S Peeper (Netherlands Cancer Institute) for the pBabe-BRAFV600E vector. We thank Dr Peter D Adams (CR-UK Beatson Institute, Glasgow University) for the retroviral vector pQCXIN. We thank Dr Yongfeng Shang (Peking University Health Science Center) for providing us the bacluviral-FOXA1 for in vitro protein purification. We also thank Dr Yongfeng Shang and Dr Jing Liang (Peking University Health Science Center) for discussion. This work was supported by grants from the National Basic Research Programs of China, Nos. 2013CB530801 and 2012CB911203 and the National Natural Science Foundation of China, No. 81170319.
Author contributions: QL and YZ designed and carried out the experiments. QL and YZ analysed all data and wrote the paper. TJT supervised the project.
Footnotes
The authors declare that they have no conflict of interest.
References
- Agherbi H, Gaussmann-Wenger A, Verthuy C, Chasson L, Serrano M, Djabali M (2009) Polycomb mediated epigenetic silencing and replication timing at the INK4a/ARF locus during senescence. PLoS ONE 4: e5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alani RM, Young AZ, Shifflett CB (2001) Id1 regulation of cellular senescence through transcriptional repression of p16/Ink4a. Proc Natl Acad Sci USA 98: 7812–7816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcorta DA, Xiong Y, Phelps D, Hannon G, Beach D, Barrett JC (1996) Involvement of the cyclin-dependent kinase inhibitor p16 (INK4a) in replicative senescence of normal human fibroblasts. Proc Natl Acad Sci USA 93: 13742–13747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augello MA, Hickey TE, Knudsen KE (2011) FOXA1: master of steroid receptor function in cancer. EMBO J 30: 3885–3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DJ, Wijshake T, Tchkonia T, LeBrasseur NK, Childs BG, van de Sluis B, Kirkland JL, van Deursen JM (2011) Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature 479: 232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banito A, Gil J (2010) Induced pluripotent stem cells and senescence: learning the biology to improve the technology. EMBO Rep 11: 353–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, Geti I, Pinho S, Silva JC, Azuara V, Walsh M, Vallier L, Gil J (2009) Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev 23: 2134–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao XY, Xie C, Yang MS (2012) Association between type 2 diabetes and CDKN2A/B: a meta-analysis study. Mol Biol Rep 39: 1609–1616 [DOI] [PubMed] [Google Scholar]
- Beltrami AP, Cesselli D, Beltrami CA (2012) Stem cell senescence and regenerative paradigms. Clin Pharmacol Ther 91: 21–29 [DOI] [PubMed] [Google Scholar]
- Bochkis IM, Schug J, Ye DZ, Kurinna S, Stratton SA, Barton MC, Kaestner KH (2012) Genome-wide location analysis reveals distinct transcriptional circuitry by paralogous regulators Foxa1 and Foxa2. PLoS Genet 8: e1002770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K (2006) Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev 20: 1123–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, Theilgaard-Monch K, Minucci S, Porse BT, Marine JC, Hansen KH, Helin K (2007) The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev 21: 525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes S, Rowe J, Gutierrez Del Arroyo A, Bond J, Peters G (2004) Contribution of p16(INK4a) to replicative senescence of human fibroblasts. ExpCell Res 298: 549–559 [DOI] [PubMed] [Google Scholar]
- Campisi J (1997) The biology of replicative senescence. Eur J Cancer 33: 703–709 [DOI] [PubMed] [Google Scholar]
- Campisi J (2001a) Cellular senescence as a tumor-suppressor mechanism. Trends Cell Biol 11: S27–S31 [DOI] [PubMed] [Google Scholar]
- Campisi J (2001b) From cells to organisms: can we learn about aging from cells in culture? Exp Gerontol 36: 607–618 [DOI] [PubMed] [Google Scholar]
- Campisi J (2005) Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell 120: 513–522 [DOI] [PubMed] [Google Scholar]
- Carlsson P, Mahlapuu M (2002) Forkhead transcription factors: key players in development and metabolism. Dev Biol 250: 1–23 [DOI] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M (2005) Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122: 33–43 [DOI] [PubMed] [Google Scholar]
- Chen H, Gu X, Liu Y, Wang J, Wirt SE, Bottino R, Schorle H, Sage J, Kim SK (2011) PDGF signalling controls age-dependent proliferation in pancreatic beta-cells. Nature 478: 349–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS (2002) Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell 9: 279–289 [DOI] [PubMed] [Google Scholar]
- Cirillo LA, McPherson CE, Bossard P, Stevens K, Cherian S, Shim EY, Clark KL, Burley SK, Zaret KS (1998) Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. EMBO J 17: 244–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo LA, Zaret KS (1999) An early developmental transcription factor complex that is more stable on nucleosome core particles than on free DNA. Mol Cell 4: 961–969 [DOI] [PubMed] [Google Scholar]
- Cirillo LA, Zaret KS (2007) Specific interactions of the wing domains of FOXA1 transcription factor with DNA. J Mol Biol 366: 720–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KL, Halay ED, Lai E, Burley SK (1993) Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 364: 412–420 [DOI] [PubMed] [Google Scholar]
- Cowper-Sal lari R, Zhang X, Wright JB, Bailey SD, Cole MD, Eeckhoute J, Moore JH, Lupien M (2012) Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat Genet 44: 1191–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cugino D, Gianfagna F, Santimone I, de Gaetano G, Donati MB, Iacoviello L, Di Castelnuovo A (2011) Type 2 diabetes and polymorphisms on chromosome 9p21: a meta-analysis. Nutr Metab Cardiovasc Dis 22: 619–625 [DOI] [PubMed] [Google Scholar]
- Dietrich N, Bracken AP, Trinh E, Schjerling CK, Koseki H, Rappsilber J, Helin K, Hansen KH (2007) Bypass of senescence by the polycomb group protein CBX8 through direct binding to the INK4A-ARF locus. EMBO J 26: 1637–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, Peacocke M, Campisi J (1995) A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA 92: 9363–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi M, Pao MM, Jeong S, Gal-Yam EN, Egger G, Weisenberger DJ, Jones PA (2005) Footprinting of mammalian promoters: use of a CpG DNA methyltransferase revealing nucleosome positions at a single molecule level. Nucleic Acids Res 33: e176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiskus W, Wang Y, Sreekumar A, Buckley KM, Shi H, Jillella A, Ustun C, Rao R, Fernandez P, Chen J, Balusu R, Koul S, Atadja P, Marquez VE, Bhalla KN (2009) Combined epigenetic therapy with the histone methyltransferase EZH2 inhibitor 3-deazaneplanocin A and the histone deacetylase inhibitor panobinostat against human AML cells. Blood 114: 2733–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao N, Le Lay J, Qin W, Doliba N, Schug J, Fox AJ, Smirnova O, Matschinsky FM, Kaestner KH (2010) Foxa1 and Foxa2 maintain the metabolic and secretory features of the mature beta-cell. Mol Endocrinol 24: 1594–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao N, LeLay J, Vatamaniuk MZ, Rieck S, Friedman JR, Kaestner KH (2008) Dynamic regulation of Pdx1 enhancers by Foxa1 and Foxa2 is essential for pancreas development. Genes Dev 22: 3435–3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil J, Peters G (2006) Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol 7: 667–677 [DOI] [PubMed] [Google Scholar]
- Gonzalez S, Klatt P, Delgado S, Conde E, Lopez-Rios F, Sanchez-Cespedes M, Mendez J, Antequera F, Serrano M (2006) Oncogenic activity of Cdc6 through repression of the INK4/ARF locus. Nature 440: 702–706 [DOI] [PubMed] [Google Scholar]
- Guney I, Wu S, Sedivy JM (2006) Reduced c-Myc signaling triggers telomere-independent senescence by regulating Bmi-1 and p16(INK4a). Proc Natl Acad Sci USA 103: 3645–3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson A, Manetopoulos C, Jonsson JI, Axelson H (2003) The basic helix-loop-helix transcription factor TAL1/SCL inhibits the expression of the p16INK4A and pTalpha genes. Biochem Biophys Res Commun 312: 1073–1081 [DOI] [PubMed] [Google Scholar]
- Hayflick L (1965) The limited in vitro lifetime of human diploid cell strains. Exp Cell Res 37: 614–636 [DOI] [PubMed] [Google Scholar]
- Hayflick L, Moorhead PS (1961) The serial cultivation of human diploid cell strains. Exp Cell Res 25: 585–621 [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318 [DOI] [PubMed] [Google Scholar]
- Hinshelwood RA, Melki JR, Huschtscha LI, Paul C, Song JZ, Stirzaker C, Reddel RR, Clark SJ (2009) Aberrant de novo methylation of the p16INK4A CpG island is initiated post gene silencing in association with chromatin remodelling and mimics nucleosome positioning. Human Mol Genet 18: 3098–3109 [DOI] [PubMed] [Google Scholar]
- Horner MA, Quintin S, Domeier ME, Kimble J, Labouesse M, Mango SE (1998) pha-4, an HNF-3 homolog, specifies pharyngeal organ identity in Caenorhabditis elegans. Genes Dev 12: 1947–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Huen MS, Kim H, Leung CC, Glover JN, Yu X, Chen J (2009) RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat Cell Biol 11: 592–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itahana K, Zou Y, Itahana Y, Martinez JL, Beausejour C, Jacobs JJ, Van Lohuizen M, Band V, Campisi J, Dimri GP (2003) Control of the replicative life span of human fibroblasts by p16 and the polycomb protein Bmi-1. Mol Cell Biol 23: 389–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs JJ, Keblusek P, Robanus-Maandag E, Kristel P, Lingbeek M, Nederlof PM, van Welsem T, van de Vijver MJ, Koh EY, Daley GQ, van Lohuizen M (2000) Senescence bypass screen identifies TBX2, which represses Cdkn2a (p19(ARF)) and is amplified in a subset of human breast cancers. Nat Genet 26: 291–299 [DOI] [PubMed] [Google Scholar]
- Janzen V, Forkert R, Fleming HE, Saito Y, Waring MT, Dombkowski DM, Cheng T, DePinho RA, Sharpless NE, Scadden DT (2006) Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature 443: 421–426 [DOI] [PubMed] [Google Scholar]
- Kaestner KH, Katz J, Liu Y, Drucker DJ, Schutz G (1999) Inactivation of the winged helix transcription factor HNF3alpha affects glucose homeostasis and islet glucagon gene expression in vivo. Genes Dev 13: 495–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalb JM, Lau KK, Goszczynski B, Fukushige T, Moons D, Okkema PG, McGhee JD (1998) pha-4 is Ce-fkh-1, a fork head/HNF-3alpha,beta,gamma homolog that functions in organogenesis of the C. elegans pharynx. Development 125: 2171–2180 [DOI] [PubMed] [Google Scholar]
- Katoh M (2004) Human FOX gene family (Review). Int J Oncol 25: 1495–1500 [PubMed] [Google Scholar]
- Krishnamurthy J, Ramsey MR, Ligon KL, Torrice C, Koh A, Bonner-Weir S, Sharpless NE (2006) p16INK4a induces an age-dependent decline in islet regenerative potential. Nature 443: 453–457 [DOI] [PubMed] [Google Scholar]
- Kuilman T, Michaloglou C, Mooi WJ, Peeper DS (2010) The essence of senescence. Genes Dev 24: 2463–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CS, Friedman JR, Fulmer JT, Kaestner KH (2005) The initiation of liver development is dependent on Foxa transcription factors. Nature 435: 944–947 [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K et al. (2006) Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125: 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M (2008) FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132: 958–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly DH, Lockhart DJ, Lerner RA, Schultz PG (2000) Mitotic misregulation and human aging. Science 287: 2486–2492 [DOI] [PubMed] [Google Scholar]
- McPherson CE, Shim EY, Friedman DS, Zaret KS (1993) An active tissue-specific enhancer and bound transcription factors existing in a precisely positioned nucleosomal array. Cell 75: 387–398 [DOI] [PubMed] [Google Scholar]
- Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, Majoor DM, Shay JW, Mooi WJ, Peeper DS (2005) BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature 436: 720–724 [DOI] [PubMed] [Google Scholar]
- Molofsky AV, Slutsky SG, Joseph NM, He S, Pardal R, Krishnamurthy J, Sharpless NE, Morrison SJ (2006) Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature 443: 448–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita M, Nunez S, Heard E, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW (2003) Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113: 703–716 [DOI] [PubMed] [Google Scholar]
- Ohtani N, Zebedee Z, Huot TJ, Stinson JA, Sugimoto M, Ohashi Y, Sharrocks AD, Peters G, Hara E (2001) Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature 409: 1067–1070 [DOI] [PubMed] [Google Scholar]
- Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A (2007) PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature 447: 550–555 [DOI] [PubMed] [Google Scholar]
- Passegue E, Wagner EF (2000) JunB suppresses cell proliferation by transcriptional activation of p16(INK4a) expression. EMBO J 19: 2969–2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petesch SJ, Lis JT (2008) Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell 134: 74–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohme D (1981) Evidence for a relationship between longevity of mammalian species and life spans of normal fibroblasts in vitro and erythrocytes in vivo. Proc Natl Acad Sci USA 78: 5009–5013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G (2007) Genome regulation by polycomb and trithorax proteins. Cell 128: 735–745 [DOI] [PubMed] [Google Scholar]
- Sekinger EA, Moqtaderi Z, Struhl K (2005) Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol Cell 18: 735–748 [DOI] [PubMed] [Google Scholar]
- Serandour AA, Avner S, Percevault F, Demay F, Bizot M, Lucchetti-Miganeh C, Barloy-Hubler F, Brown M, Lupien M, Metivier R, Salbert G, Eeckhoute J (2011) Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers. Genome Res 21: 555–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M, Hannon GJ, Beach D (1993) A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature 366: 704–707 [DOI] [PubMed] [Google Scholar]
- Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW (1997) Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88: 593–602 [DOI] [PubMed] [Google Scholar]
- Shih DQ, Navas MA, Kuwajima S, Duncan SA, Stoffel M (1999) Impaired glucose homeostasis and neonatal mortality in hepatocyte nuclear factor 3alpha-deficient mice. Proc Natl Acad Sci USA 96: 10152–10157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, de Lange T (2002) Different telomere damage signaling pathways in human and mouse cells. EMBO J 21: 4338–4348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarutani M, Cai T, Dajee M, Khavari PA (2003) Inducible activation of Ras and Raf in adult epidermis. Cancer Res 63: 319–323 [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W (2002) Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell 10: 1453–1465 [DOI] [PubMed] [Google Scholar]
- Wajapeyee N, Serra RW, Zhu X, Mahalingam M, Green MR (2008) Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell 132: 363–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H, Dingle S, Xu Y, Besnard V, Kaestner KH, Ang SL, Wert S, Stahlman MT, Whitsett JA (2005) Compensatory roles of Foxa1 and Foxa2 during lung morphogenesis. J Biol Chem 280: 13809–13816 [DOI] [PubMed] [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, Glass CK, Rosenfeld MG, Fu XD (2011) Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 474: 390–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Li W, Zhang Y, Yuan X, Xu K, Yu J, Chen Z, Beroukhim R, Wang H, Lupien M, Wu T, Regan MM, Meyer CA, Carroll JS, Manrai AK, Janne OA, Balk SP, Mehra R, Han B, Chinnaiyan AM et al. (2009) Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell 138: 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Schulte BA, Zhou D (2006) Hematopoietic stem cell senescence and long-term bone marrow injury. Cell Cycle 5: 35–38 [DOI] [PubMed] [Google Scholar]
- Wen J, Ronn T, Olsson A, Yang Z, Lu B, Du Y, Groop L, Ling C, Hu R (2010) Investigation of type 2 diabetes risk alleles support CDKN2A/B, CDKAL1, and TCF7L2 as susceptibility genes in a Han Chinese cohort. PLoS ONE 5: e9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BG, Wang X, Shen X, McKenna ES, Lemieux ME, Cho YJ, Koellhoffer EC, Pomeroy SL, Orkin SH, Roberts CW (2010) Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell 18: 316–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn SW, Kim DS, Cho HJ, Jeon SE, Bae IH, Yoon HJ, Park KC (2004) Cellular senescence induced loss of stem cell proportion in the skin in vitro. J Dermatol Sci 35: 113–123 [DOI] [PubMed] [Google Scholar]
- Yu SF, von Rüden T, Kantoff PW, Garber C, Seiberg M, Rüther U, Anderson WF, Wagner EF, Gilboa E (1986) Self-inactivating retroviral vectors designed for transfer of whole genes into mammalian cells. Proc Natl Acad Sci 83: 3194–3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret K (1999) Developmental competence of the gut endoderm: genetic potentiation by GATA and HNF3/fork head proteins. Dev Biol 209: 1–10 [DOI] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS (2011) Pioneer transcription factors: establishing competence for gene expression. Genes Dev 25: 2227–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liang J, Li Y, Xuan C, Wang F, Wang D, Shi L, Zhang D, Shang Y (2010) CCCTC-binding factor acts upstream of FOXA1 and demarcates the genomic response to estrogen. J Biol Chem 285: 28604–28613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Wang H, Xue L, Zhang Z, Tong T (2004) Regulation of cellular senescence and p16(INK4a) expression by Id1 and E47 proteins in human diploid fibroblast. J Biol Chem 279: 31524–31532 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.