Summary
Small RNAs regulate genetic networks through a ribonucleo-protein complex called the RNA-induced silencing complex (RISC), which, in mammals, contains at its center one of four Argonaute proteins (Ago1–Ago4) (reviewed in [1–4]). A key regulatory event in the RNA interference (RNAi) and microRNA (miRNA) pathways is Ago loading, wherein double-stranded small-RNA duplexes are incorporated into RISC (pre-RISC) and then become single-stranded (mature RISC), a process that is not well understood [5, 6]. The Agos contain an evolutionarily conserved PAZ (Piwi/Argonaute/Zwille) domain [7, 8] whose primary function is to bind the 3′ end of small RNAs [9–13]. We created multiple PAZ-domain-disrupted mutant Ago proteins and studied their biochemical properties and biological functionality in cells. We found that the PAZ domain is dispensable for Ago loading of slicing-competent RISC. In contrast, in the absence of slicer activity or slicer-substrate duplex RNAs, PAZ-disrupted Agos bound duplex small interfering RNAs, but were unable to unwind or eject the passenger strand and form functional RISC complexes. We have discovered that the highly conserved PAZ domain plays an important role in RISC activation, providing new mechanistic insights into how miRNAs regulate genes, as well as new insights for future design of miRNA- and RNAi-based therapeutics.
Results and Discussion
Ago2 Lacking the PAZ Domain Retains the Ability to Associate Small RNAs and Elicit Noncleavage Gene Repression
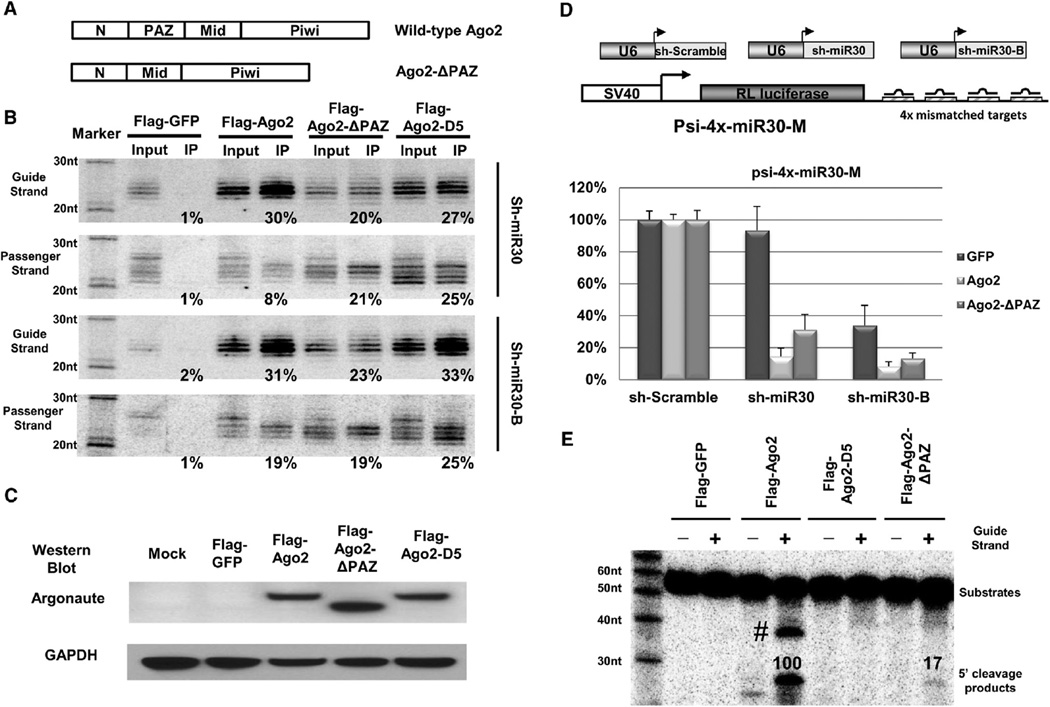
We investigated the role of the PAZ (Piwi/Argonaute/Zwille) domain by creating a truncated Ago2 with a complete PAZ-domain deletion (Ago2-ΔPAZ) (Figure 1A). We measured the association between the truncated Ago2-ΔPAZ and small RNAs processed from small hairpin RNAs (shRNAs) using immunoprecipitation (IP) assays. Specifically, two U6-driven shRNAs (sh-miR30 and sh-miR30-B), designed to generate the same guide strand as that derived from the endogenous hsa-miR30a-3p microRNA (miRNA) but having differing stem structures and thermodynamic stabilities [14], were coexpressed with various Flag-tagged Ago2 variants or a control Flag-green fluorescent protein (GFP) expression cassette in Ago2 knockout (KO) mouse embryonic fibroblast (MEF) cells [15] (Figure 1B). Western blot experiments established that the Flag-tagged version of Ago2 and the truncated mutant (Ago2-ΔPAZ) were expressed at nearly equivalent amounts in MEF cells (Figure 1C). Quantification of the sh-miR30 or sh-miR30-B RNAs bound to each of the Ago2 variants was determined 24 hr posttransfection by anti-Flag IP and northern blotting. The Ago2 variant pull-down results were confirmed to be Ago-specific because of the lack of signal in the GFP pull-down control (Figure 1B). Interestingly, deletion of the PAZ domain did not abolish the interaction between Ago2 and small RNA, as IP experiments showed that Ago2-ΔPAZ was able to bind the guide-strand RNA (derived from sh-miR30 and sh-miR30-B), albeit at a reduced level compared to the wild-type (Figure 1B).
Figure 1. Truncated Ago2 without the PAZ Domain Can Associate Small RNAs, Elicit Noncleavage Gene Repression, and Slice Target RNA.
(A) Schematic representation of human Ago2 and its Flag-tagged truncated version lacking the PAZ domain.
(B) Perfect-stem sh-miR30 or bulged-stem shRNA-miR30-B coexpressed with Flag-GFP (negative control), Flag-Ago2, or its mutants in Ago2 KO MEF cells. 24 hr posttransfection, IP experiments with anti-Flag antibody were performed on cell lysates. RNA extracted from either 20% of input or IP were run on 15% polyacrylamide 7 M urea denaturing gels. Guide- and passenger-strand RNAs from each sample were identified by sequential northern blotting. The intensity of the bands was determined by phosphoimaging. The pull-down efficiency listed in the figure was calculated by dividing the IP signal with total input.
(C) Cell lysates were also subjected to a western blot using Ago2 antibody. Wild-type Ago2, Ago2-ΔPAZ, and Ago2-D5 were expressed at similar levels. The signal from endogenous GAPDH served as the loading control.
(D) PsiCHECK vectors with four tandem target sites in the 3′ UTR, which were mismatched to the guide strand of sh-miR30 and sh-miR30-B, were cotransfected with various shRNAs in Ago2 KO MEF cells. Dual-luciferase assays were performed 24 hr posttransfection. RL-luciferase activities were normalized with Firefly (FF)-luciferase, and the percentage of relative enzyme activity compared to the negative control (treated with sh-scramble) was plotted. Error bars represent the SD from two independent experiments, each performed in triplicate transfections.
(E) The cleavage activities of the wild-type human Ago2 and the mutant derivatives were measured with an in vitro cleavage assay. Target RNA was incubated with 32P-labeled sh-miR30 guide-strand RNA and IP-derived Argonaute proteins at 26°C for 90 min. Products of the cleavage reaction were purified, separated, and detected by autoradiography. The expected size of 5′ cleavage products is 27 nucleotides (nt) as indicated in the figure. The intensity of the bands was determined by phosphoimaging as shown in the figure (the Ago2-treated sample was assigned a value of 100). A nonspecific band is labeled with # and was not observed in a repeat experiment. See also Figure S1.
In Ago2 KO cells, cleavage-based gene silencing is abolished, but residual small interfering RNA (siRNA)- or shRNA-induced gene silencing through noncleavage repression is detectable due to the presence of Ago1, Ago3, and Ago4 [14–16]. Nevertheless, the noncleavage repression specifically mediated by the Ago2 mutants can be determined via complementation experiments in these cells. To do this, we utilized well-characterized reporter systems in which four tandem target sites were inserted into the 3′ untranslated region (UTR) of the Renilla (RL)-luciferase reporter gene (Figure 1D). Sh-miR30 and sh-miR30-B generated the same guide strands that were mismatched to reporter psi-4×-miR30-M, providing a means to measure the noncleavage repression. Concordant with the small-RNA association results, Ago2-ΔPAZ demonstrated a substantial (p < 0.0001) albeit reduced amount of gene repression compared to wild-type Ago2 (Figure 1D), indicating that the truncated Ago2 without a PAZ domain retained its ability to associate with small RNAs and to silence genes. This conclusion was confirmed with a second shRNA sequence based on Drosophila bantam miRNA (Figure S1 available online).
HsAgo2 Slicer Activity Is Retained with Deletion of the PAZ Domain
Although the Ago2-ΔPAZ retained the ability to silence mismatched targets through noncleavage repression, we wanted to investigate whether the PAZ deletion affected the slicer activity. To directly measure the slicer activity of Ago2-ΔPAZ, we performed an in vitro cleavage assay using a radioactive-labeled RNA target perfectly matched to the guide strand of sh-miR30 and sh-miR30-B. As shown in Figure 1E, a substantially lesser amount of cleavage product was detected in the sample treated with Ago2-ΔPAZ compared to that of wild-type Ago2, demonstrating that Ago2-ΔPAZ had weak slicer activity. The remaining cleavage activity was confirmed to be Ago2-ΔPAZ-specific because of the total absence of signal in the Ago2-D5 control (Figure 1E). At this point, it was unclear whether the reduction in cleavage activity was due to impairment of inherent slicer activity, a result of less-efficient binding between Ago2-ΔPAZ and guide-strand RNA, or both. Nonetheless, the cleavage product of Ago2-ΔPAZ was the same size as that of wild-type Ago2 (Figure 1E), establishing that the cleavage process mediated by Ago2-ΔPAZ was still precise, which implies that deletion of the PAZ domain did not disrupt the ternary complex of the protein, guide strand, and target RNA [17].
The PAZ Domain Is Required for HsAgo1, HsAgo3, and HsAgo4 to Unwind Small-RNA Duplexes during RISC Activation
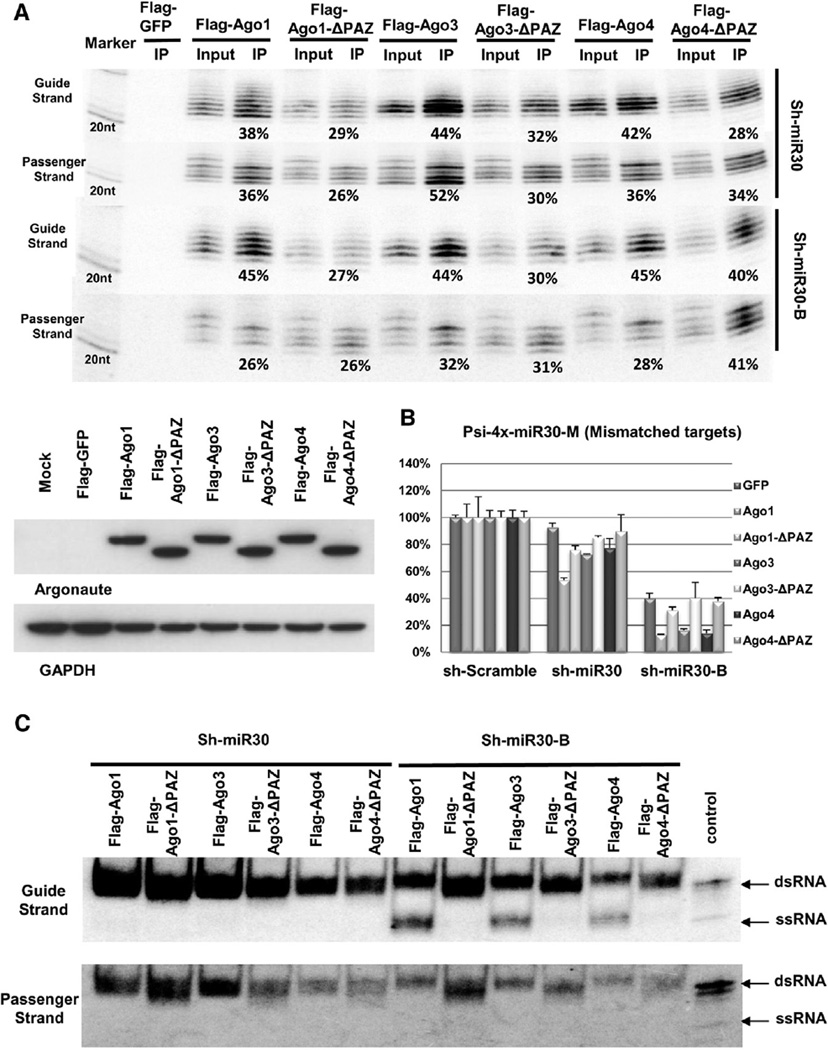
Consistent with the role of the PAZ domain in small-RNA association, fewer guide and passenger strands processed from sh-miR30 and sh-miR30-B were pulled down with truncated, noncleaving Agos (Ago1-ΔPAZ, Ago3-ΔPAZ, and Ago4-ΔPAZ [Ago1/3/4-ΔPAZ]) than were obtained with their wild-type counterparts (Figure 2A).
Figure 2. The PAZ Domain Is Required for HsAgo1, HsAgo3, and HsAgo4 to Form Mature RISC.
(A) Sh-miR30 or shRNA-miR30-B coexpressed with Flag-GFP or various Flag-Agos and their mutants in Ago2 KO MEF cells. IP and northern blotting experiments were performed as described in Figure 1. The expression levels of the proteins used in the assays were assessed by western blotting with anti-Flag antibody and are shown in the lower panel.
(B) A psiCHECK vector with four tandem target sites in the 3′ UTR, which were mismatched to the guide strand of sh-miR30 and sh-miR30-B, was co-transfected with various shRNAs and Argonaute-expressing plasmids in Ago2 KO MEF cells. Dual-luciferase assays were performed and analyzed as described in Figure 1. Error bars represent the SD from two independent experiments, each performed in triplicate transfections.
(C) IP samples in (A) were separated on 15% polyacrylamide native gels. 20 fmol synthetic si-miR30 duplex was also loaded on the gel as a control. Guide- and passenger-strand RNAs were identified by sequential northern blot. Notably, the si-miR30 control ran as a mixture of dsRNA and a trace amount of ssRNA. See also Figure S2.
Interestingly, Ago1/3/4-ΔPAZ mutants were almost completely unable to induce transgene silencing in the functional assay, despite the fact that substantial amounts of guide strands from sh-miR30 and sh-miR30-B were detected in the respective Ago pull-down experiments (Figures 2A and 2B). Specifically, overexpression of Flag-tagged Ago1/3/4-ΔPAZ caused no increase in the repression activity of both sh-miR30 and sh-miR30-B (Figure 2B). Similar results were observed with sh-Bantam (Figure S2).
We established previously that noncleavage Agos (Ago1/3/4) failed to generate sufficient amounts of active RISC with stable sh-miR30 because of the inefficiency of the unwinding step [14]. Thus we hypothesized that Ago1/3/4-ΔPAZ were incapable of unwinding an associated duplex RNA. To directly test this idea, we analyzed the same Ago-IP samples using a native gel that provides a means to distinguish between Ago-associated single-stranded and duplexed RNAs [14]. The stable, less active sh-miR30 RNAs associated with noncleaving Agos (Ago1/3/4) were almost exclusively found as duplexes, whereas the less stable but more active sh-miR30-B RNAs associated with the noncleaving Agos were more prominently found as single-stranded guide-strand RNAs (Figure 2C). In contrast to the differences between sh-miR30 and sh-miR30-B, virtually all of the guide and passenger strands from both shRNAs found to be associated with the PAZ-deletion mutants were present as duplex-RNA forms (Figure 2C). These results strongly suggest that the RISC activation is dependent on the PAZ domain; in the absence of the PAZ domain, RISC is unable to unwind even thermodynamically unstable small-RNA duplexes. Thus, Ago1/3/4-ΔPAZ maintained their ability to physically associate with small-RNA duplexes, but not the ability to unwind and form active RISC.
HsAgo2-DPAZ RISC Achieved Activation by Eliminating the Passenger Strand through its Residual Slicer Activity
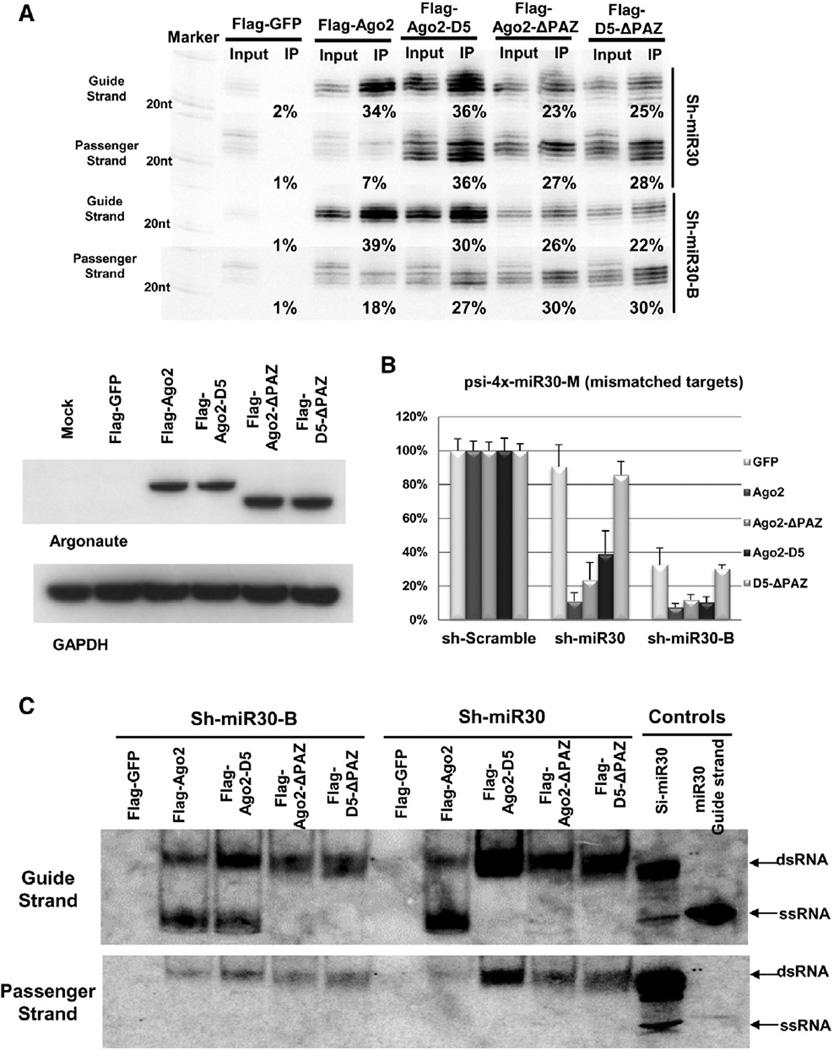
Because the PAZ domain was critical for the Agos’ unwinding of the guide-passenger duplex RNAs, we wanted to establish whether the generation of active Ago2-ΔPAZ-RISC complexes (Figure 1D) was solely dependent on its residual slicer activity. To test this idea, we generated a double mutation of Ago2 deficient in both slicer activity and the PAZ domain (ΔPAZ-D5).
As expected, introducing a single-point mutation to abolish the slicer activity did not change the binding affinity of Ago2-ΔPAZ. Guide strands of sh-miR30 and sh-miR30-B were pulled down by ΔPAZ-D5 with a similar efficiency to that obtained by Ago2-ΔPAZ (Figure 3A). However, ΔPAZ-D5 was unable to enhance gene repression in a complementation assay (Figures 3B and S3), suggesting that ΔPAZ-D5-RISC was functionally inactive even though it maintained its ability to bind small duplex RNAs.
Figure 3. Ago2-ΔPAZ-Forming Mature RISC through a Slicer-Dependent Pathway.
(A) Sh-miR30 or shRNA-miR30-B coexpressed with Flag-GFP or various Flag-Ago2 mutants in Ago2 KO MEF cells. IP and northern blotting experiments were performed as described in Figure 1. The expression levels of the proteins used in the assays were assessed by western blotting with Ago2 antibody. Signals from GAPDH served as transfection and loading controls.
(B) A psiCHECK vector with four tandem target sites in the 3′ UTR, which were mismatched to the guide strand of sh-miR30 and sh-miR30-B, was cotransfected with various shRNAs and Argonaute-expressing plasmids in Ago2 KO MEF cells. Dual-luciferase assays were performed and analyzed as described in Figure 1. Error bars represent the SD from two independent experiments, each performed in triplicate transfections.
(C) IP samples in (A) were separated on 15% polyacrylamide native gels. 20 fmol synthetic RNAs (si-miR30 duplex and single-stranded guide-strand RNA of miR-30) were also loaded on the gel as controls. Guide- and passenger-strand RNAs were identified by sequential northern blot. Notably, the si-miR30 control ran as a mixture of dsRNA and a trace amount of ssRNA. See also Figure S3.
To test whether the failure of ΔPAZ-D5 to mediate transgene repression was indeed due to its inability to unwind RNA duplexes, we analyzed the Ago-IP samples with a native gel and directly assessed the ratio between single-stranded RNA (ssRNA)-associated RISC and double-stranded RNA (dsRNA)-associated RISC. Consistent with our hypothesis, guide strands processed from sh-miR30 and sh-miR30-B were mainly associated with ΔPAZ-D5 as duplex RNAs (Figure 3C). Interestingly, extremely few, if any, ssRNA-associated Ago2-ΔPAZ was detected, indicating that the activation of Ago2-ΔPAZ into an active RISC was rather inefficient (Figure 3C). This was consistent with the idea that the only pathway available for Ago2-ΔPAZ to achieve activation was cleavage of the passenger strand through its inefficient slicer activity.
Notably, whereas ssRNA-associated mature Ago2-ΔPAZRISC was almost undetectable in the native gel (Figure 3C), the level of noncleavage repression mediated by Ago2- ΔPAZ-RISC was similar and comparable to Ago2-D5-RISC and Ago2-ΔPAZ-RISC (Figure 3B). Similar results were found when we tested shRNAs with a different sequence (sh-Bantam) (Figure S3). Our results suggest that, once successfully loaded with a guide-strand ssRNA, the repressive ability of an individual Ago2-ΔPAZ was actually greater than its wild-type counterpart. At this point, the mechanism of the Ago2-ΔPAZ hyperactivity is unclear. One possible scenario is that the 3′ portion of the guide strand is able to precipitate and strengthen the guide-target hybridization when freed from PAZ-domain binding. Therefore, the enhanced ability of Ago2-ΔPAZ-RISC to mediate noncleavage repression is the result of a more stable interaction between RISC and target messenger RNAs.
HsAgo2 with Point Mutations in PAZ Is Defective in Slicer-Independent RISC Unwinding
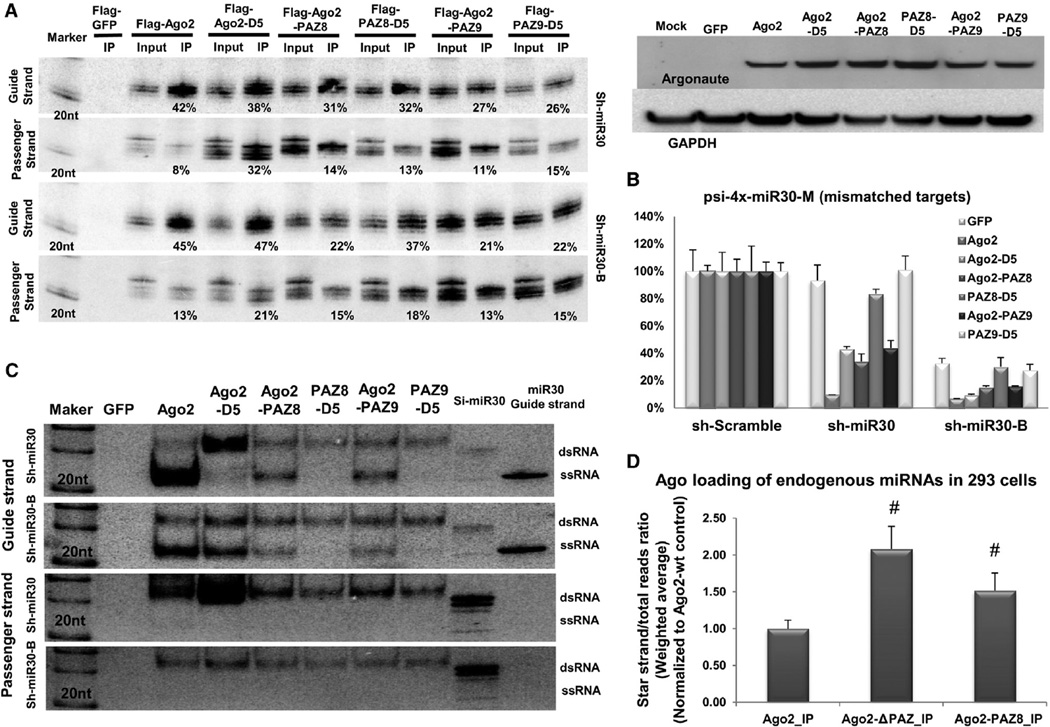
To confirm that the deficiency in RISC activation was specific to the function of the PAZ domain, but not secondary to gross structural changes caused by the deletion of the whole domain, we created Ago2 variants with a series of point mutations in the PAZ domain. PAZ8 and PAZ9 contain eight or nine point mutations, respectively, which are involved in PAZ-siRNA association [9, 12, 18]. Similar to what we found using the PAZ-deleted Ago2 truncations, both Ago2-PAZ8 and Ago2-PAZ9 and their slicer-deficient counterparts (PAZ8-D5 and PAZ9-D5) were able to associate, albeit less efficiently than wild-type Ago2, with small RNAs processed from sh-miR30 and sh-miR30-B (Figure 4A). However, only Ago2-PAZ8 and Ago2-PAZ9, but not PAZ8-D5 and PAZ9-D5, were capable of inducing transgene silencing in the reporter assay (Figures 4B and S4). The native-gel analysis further confirmed that the failure of PAZ8-D5 and PAZ-D5 to mediate non-cleavage repression was due to their inability to unwind RNA duplexes (Figure 4C). Taken together, these results strongly suggest that the PAZ-small RNA association is critical for RISC maturation.
Figure 4. Ago2 with Point Mutations in PAZ Is Defective in Slicer-Independent RISC Unwinding.
(A) Sh-miR30 or shRNA-miR30-B coexpressed with Flag-GFP or various Flag-Ago2 mutants in Ago2 KO MEF cells. IP and northern blotting experiments were performed as described in Figure 1. The expression levels of the proteins used in the assays were assessed by western blotting with Ago2 antibody. The signal from GAPDH served as the transfection and loading controls.
(B) A psiCHECK vector with four tandem target sites in the 3′ UTR, which were mismatched to the guide strand of sh-miR30 and sh-miR30-B, was cotransfected with various shRNAs and Argonaute-expressing plasmids in Ago2 KO MEF cells. Dual-luciferase assays were performed and analyzed as described in Figure 1. Error bars represent the SD from two independent experiments, each performed in triplicate transfections.
(C) IP samples in (A) were separated on 15% polyacrylamide native gels. 20 fmol synthetic RNAs (si-miR30 duplex and single-stranded guide-strand RNA of miR-30) were also loaded on the gel as controls. Guide- and passenger-strand RNAs were identified by sequential northern blot. Notably, the si-miR30 control ran as a mixture of dsRNA and a trace amount of ssRNA.
(D) The amount of miRNAs and their corresponding star strands were measured in various Ago-IP samples by deep sequencing. For each miRNA, the percentage of its star strand to total reads was determined. The weighted average and SD were calculated by using the total reads of each miRNA as the weight. Number sign (#) indicates statistical significance (p < 0.05, unpaired t test, using Ago2-IP as a control group). See also Figure S4 and Table S1.
HsAgo2 with PAZ Domain Mutations Is Deficient in Unwinding of Endogenous miRNAs
To study the RISC unwinding of endogenous miRNAs, we deep sequenced the Ago-associated small RNAs (IP samples) and measured the amount of each detectable miRNA and its corresponding star strands in HEK293 cells. Consistent with our previous results using shRNA expression sequences, the average percentage of endogenous star strands was significantly higher (p < 0.05) in PAZ-domain mutations (Ago2-ΔPAZ and Ago2-PAZ8) than in wild-type Ago2 controls (Figure 4D), further supporting their inability to unwind duplex RNAs. Interestingly, the slicer-defective PAZ mutants (ΔPAZ-D5 and PAZ8-D5) had an even higher average percentage of star strands, suggesting a role of slicer activity in Ago2 loading of miRNAs (Figure S4C). On average, the miRNAs behaved similarly to the shRNAs, whereas some of the individual miRNAs did not. For example, ΔPAZ-D5 blocked the unwinding of hsa-miR-17 and hsa-miR-424 but had marginal effects on hsa-miR-15b and hsa-miR-9 (Table 1), suggesting the presence of additional RISC-assembling pathway(s) specific to miRNA subgroups.
Table 1.
Ago2 Variants with PAZ-Domain Mutations Are Defective in Unwinding of Endogenous miRNAs in HEK293 Cells
| Ago2 | Ago2-D5 | Ago2-ΔPAZ | ΔPAZ-D5 | Ago2-PAZ8 | PAZ8-D5 | F181A | |
|---|---|---|---|---|---|---|---|
| Hsa-miR-17 | 13.07% | 18.44% (1.4) | 38.03% (2.9) | 49.23% (3.8) | 17.36% (1.3) | 35.13% (2.7) | 15.15% (1.2) |
| Hsa-miR-15b | 0.69% | 0.51% (0.7) | 0.67% (1.0) | 0.68% (1.0) | 0.88% (1.3) | 0.95% (1.4) | 0.95% (1.4) |
| Hsa-miR-221 | 0.12% | 0.13% (1.1) | 0.05% (0.5) | 0.74% (6.2) | 0.39% (3.3) | 1.18% (9.8) | 0.14% (1.2) |
| Hsa-miR-20a | 2.81% | 3.19% (1.1) | 4.49% (1.6) | 8.00% (2.8) | 7.65% (2.7) | 31.88% (11.3) | 2.83% (1.0) |
| Hsa-miR-106b | 26.67% | 24.72% (0.9) | 27.03% (1.0) | 38.46% (1.4) | 58.62% (2.2) | 47.87% (1.8) | 34.48% (1.3) |
| Hsa-miR-15a | 1.82% | 5.566% (3.1) | 16.92% (9.3) | 9.09% (5.0) | 8.33% (4.6) | 5.12% (2.8) | 3.19% (1.8) |
| Hsa-miR-126 | 19.44% | 30.16% (1.6) | 29.29% (1.5) | 25.00% (1.3) | 25.00% (1.3) | 27.16% (1.4) | 17.65% (0.9) |
| Hsa-miR-424 | 8.33% | 10.77% (1.3) | 39.58% (4.8) | 41.67% (5.0) | 7.50% (0.9) | 48.15% (5.8) | 7.46% (0.9) |
| Hsa-miR-9 | 10.53% | 30.51% (2.9) | 18.37% (1.7) | 18.18% (1.7) | 34.37% (3.3) | 19.44% (1.8) | 9.26% (0.9) |
| Hsa-miR-183 | 12.50% | 29.17% (2.3) | 69.23% (5.5) | 66.67% (5.3) | 18.75% (1.5) | 68.18% (5.5) | 15.00% (1.2) |
The abundance of miRNAs and their corresponding star strands were measured in various immunoprecipitated (IP) Ago samples by deep sequencing. For each miRNA, the percentage of its star strands to total reads was determined. Results of ten miRNAs with both guide and passenger strands detected are listed here, and the weighted average of each sample is presented in Figure S4C. The higher the percentage of star strands in the IP sample, the bigger the unwinding defects of Ago2 variants. The relative fold of star-strand percentage to the wild-type Ago2 control is indicated by the number inside the parentheses.
A Proposed Model for Slicer-Independent RISC Unwinding
Our observations raised intriguing questions regarding the mechanism(s) involved in the slicer-independent unwinding pathway and the requirement for the PAZ domain in this process. In light of published results [6, 14, 19–23] and the data presented here, we propose a modified model for slicer-independent unwinding (Figure S5A). In brief, the Agos undergo a conformational change involving the movement of the PAZ domain during the loading of duplex RNAs. During this transition, the PAZ domain will tightly associate with the 3′ end of the guide strand and function as a handle to peel off the duplex (see Supplemental Discussion for details).
A recent report indicated that the N domain was involved in duplex unwinding [24]. To compare the contributions of the PAZ and N domains directly, we tested two of these N-domain mutants, Ago2-ΔWedge (truncation) and Ago2-F181A (point mutation) [24]. Unlike the PAZ mutations, both N-domain mutants and their corresponding slicer-defective versions (ΔWedge-D5 and F181A-D5) were still able to unwind the unstable sh-miR-30-B (Figure S5B). This conclusion was further supported by the observation that all N-domain mutants could induce transgene silencing, albeit some (Ago2-ΔWedge and ΔWedge-D5) were less efficient than wild-type Ago2 (Figures S5C and S5D). The apparent contradiction could be explained by the difference in the method used to measure the RISC activation. Although the N-domain study [24] measured the changes in the unwinding rate in the first 30 min of RISC formation in a reconstitution system, our analysis was performed in living cells under steady-state conditions (24 hr posttransfection) and suggests that this domain is not essential for unwinding. It is possible that the N domain is involved in the initiation of the unwinding process and only affects the efficiency of the process. Consistent with this idea, our deep-sequencing analyses found that the N-domain mutation (Ago2-F181A) had a smaller impact on endogenous-miRNA unwinding than PAZ-domain mutations did (Figure S4C and Table 1). Whether or not this quantitative difference is due to the fact that the N-domain mutants may affect the structural integrity of the PAZ domain is not clear.
Overall, we defined an additional role for the Ago PAZ domain in the mammalian RNAi pathway. Our studies provide working models for how this domain fits into the biology of small-duplex-RNA-mediated gene silencing. Moreover, our findings suggest that siRNAs and shRNAs require the presence of an interaction with the PAZ domain to form mature RISC with noncleaving Agos, offering a way to design si/shRNAs that would preferentially be loaded into cleaving Agos. This provides insights into how to devise potent si/shRNAs with minimal off-target effects for therapeutic applications.
Supplementary Material
Acknowledgments
We thank Michael M. Pan for technical assistance and Greg Hannon for the MEF Ago 2 KO cells. This work was supported by NIH DK078424.
Footnotes
Accession Numbers
The NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) accession number for the sequence reported in this paper is GSE39146.
Supplemental Information
Supplemental Information includes five figures, Supplemental Discussion, and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.cub.2012.06.040.
References
- 1.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siomi H, Siomi MC. On the road to reading the RNA-interference code. Nature. 2009;457:396–404. doi: 10.1038/nature07754. [DOI] [PubMed] [Google Scholar]
- 5.Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nat. Rev. Genet. 2011;12:19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawamata T, Tomari Y. Making RISC. Trends Biochem. Sci. 2010;35:368–376. doi: 10.1016/j.tibs.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 7.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 8.Peters L, Meister G. Argonaute proteins: mediators of RNA silencing. Mol. Cell. 2007;26:611–623. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 9.Ma JB, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lingel A, Simon B, Izaurralde E, Sattler M. Nucleic acid 3′-end recognition by the Argonaute2 PAZ domain. Nat. Struct. Mol. Biol. 2004;11:576–577. doi: 10.1038/nsmb777. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008;456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan KS, Yan S, Farooq A, Han A, Zeng L, Zhou MM. Structure and conserved RNA binding of the PAZ domain. Nature. 2003;426:468–474. doi: 10.1038/nature02129. [DOI] [PubMed] [Google Scholar]
- 13.Song J-J, Liu J, Tolia NH, Schneiderman J, Smith SK, Martienssen RA, Hannon GJ, Joshua-Tor L. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat. Struct. Biol. 2003;10:1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- 14.Gu S, Jin L, Zhang F, Huang Y, Grimm D, Rossi JJ, Kay MA. Thermodynamic stability of small hairpin RNAs highly influences the loading process of different mammalian Argonautes. Proc. Natl. Acad. Sci. USA. 2011;108:9208–9213. doi: 10.1073/pnas.1018023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 16.Su H, Trombly MI, Chen J, Wang X. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes Dev. 2009;23:304–317. doi: 10.1101/gad.1749809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Juranek S, Li H, Sheng G, Tuschl T, Patel DJ. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Juranek S, Li H, Sheng G, Wardle GS, Tuschl T, Patel DJ. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyoshi T, Takeuchi A, Siomi H, Siomi MC. A direct role for Hsp90 in pre-RISC formation in Drosophila. Nat. Struct. Mol. Biol. 2010;17:1024–1026. doi: 10.1038/nsmb.1875. [DOI] [PubMed] [Google Scholar]
- 21.Iwasaki S, Kobayashi M, Yoda M, Sakaguchi Y, Katsuma S, Suzuki T, Tomari Y. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol. Cell. 2010;39:292–299. doi: 10.1016/j.molcel.2010.05.015. [DOI] [PubMed] [Google Scholar]
- 22.Iki T, Yoshikawa M, Nishikiori M, Jaudal MC, Matsumoto-Yokoyama E, Mitsuhara I, Meshi T, Ishikawa M. In vitro assembly of plant RNA-induced silencing complexes facilitated by molecular chaperone HSP90. Mol. Cell. 2010;39:282–291. doi: 10.1016/j.molcel.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 23.Johnston M, Geoffroy MC, Sobala A, Hay R, Hutvagner G. HSP90 protein stabilizes unloaded argonaute complexes and microscopic P-bodies in human cells. Mol. Biol. Cell. 2010;21:1462–1469. doi: 10.1091/mbc.E09-10-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwak PB, Tomari Y. The N domain of Argonaute drives duplex unwinding during RISC assembly. Nat. Struct. Mol. Biol. 2012;19:145–151. doi: 10.1038/nsmb.2232. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.