Abstract
Background
The cell renews, adapts, or expands its mitochondrial population during episodes of cell damage or periods of intensified energy demand by the induction of mitochondrial biogenesis. This bi-genomic program is modulated by redox-sensitive signals that respond to physiological nitric oxide (NO), carbon monoxide (CO), and mitochondrial reactive oxygen species (ROS) production.
Scope of Review
This review summarizes our current ideas about the pathways involved in the activation of mitochondrial biogenesis by the physiological gases leading to changes in the redox milieu of the cell with an emphasis on the responses to oxidative stress and inflammation.
Major Conclusions
The cell’s energy supply is protected from conditions that damage mitochondria by an inducible transcriptional program of mitochondrial biogenesis that operates in large part through redox signals involving the nitric oxide synthase and the heme oxygenase-1/CO systems. These redox events stimulate the coordinated activities of several multifunctional transcription factors and co-activators also involved in the elimination of defective mitochondria and the expression of counter-inflammatory and anti-oxidant genes, such as IL10 and Sod2, as part of a unified damage-control network.
General Significance
The redox-regulated mechanisms of mitochondrial biogenesis schematically outlined in the graphical abstract link mitochondrial quality control to an enhanced capacity to support the cell’s metabolic needs while improving its resistance to metabolic failure and avoidance of cell death during periods of oxidative stress.
Keywords: Carbon monoxide, cell metabolism, heme oxygenase, inflammation, mitochondria, nitric oxide, oxidative stress, reactive oxygen species
Introduction
Oxidative stress and other cellular injuries that damage mitochondria may impair the cell’s capacity to generate sufficient adenosine triphosphate (ATP) for homeostasis, ultimately leading to apoptosis or necrosis [1]. The protection of mitochondrial integrity and quality is the task of cellular programs that monitor and replace dysfunctional mitochondria with organelles better suited to energy prevision under the conditions prevailing in the tissue microenvironment [2]. This process of mitochondrial quality control involves a bi-genomic program of nuclear- and mitochondrial-encoded gene regulation that rapidly adjust mitochondrial mass, distribution, and phenotype [3, 4]. The program requires the transcription and replication of mitochondrial DNA (mtDNA), mitochondrial protein synthesis, and distinct structural events in the cytoplasm, including mitochondrial fusion/fission, mitochondrial autophagy (mitophagy), and mitochondrial proliferation, as well as specific reorganization of the cytoskeleton.
Cells do not generate mitochondria de novo, but instead identify and dispose of defective mitochondria while stimulating healthy mitochondria to proliferate through mitochondrial biogenesis [5]. In the process, highly functional mitochondrial subpopulations are segregated from poorly functional mitochondria, which are targeted for degradation. The elucidation of this high-level capability is an area of emphasis in mitochondrial research, and this review summarizes those aspects of redox-regulation of mammalian mitochondrial biogenesis known to be involved in maintaining and restoring mitochondrial function after oxidative damage. The emphasis here is on the roles of nitric oxide and carbon monoxide, and excludes hydrogen sulfide, about which far less is known.
Mitochondrial Biogenesis
Normally, mitochondrial biogenesis is activated by changes in physiological state that require increases in the rates of ATP utilization approaching the existing capacity of the cells to produce it. Such events include, but are not limited to thermogenesis, exercise, calorie restriction, and several others listed in Table 1 [4, 6, 7]. The focus here is the activation of mitochondrial biogenesis by mitochondrial damage from oxidative stress and pathological inflammation [3, 4, 6-18]. Under such conditions, mitochondrial biogenesis indispensably supports energy-dependent cell processes, including those involved in the repair of cell and tissue damage.
Table 1.
Major Initiators of Mitochondrial Biogenesis
|
Mitochondrial biogenesis is regulated mainly at the level of transcription, and numerous nuclear-encoded mitochondrial genes must be expressed in synchrony with the 13 mitochondrial-encoded genes. This coordinated bi-genomic program includes nuclear-encoded mitochondrial proteins that control mtDNA transcription and replication, and requires the induction of mitochondrial DNA polymerase (gamma), mitochondrial transcription factor A (TFAM), and TFB2M [3, 4]. These nuclear control mechanisms also lead to the induction of tissue- and signal-specific subsets of genes that serve specialized functions. Much of the mitochondrial proteome is dedicated to such lineage-specific proteins [19], and hence, the transcriptional program matches the cell’s mitochondrial mass and phenotype to the physiological energy needs and the functions of each tissue.
Many nuclear-encoded genes for mitochondrial proteins, for instance, electron transport and oxidative phosphorylation proteins contain conserved binding motifs for nuclear respiratory factors-1 (NRF-1) and NRF-2 (also called GA-binding protein A or GABPA). The peroxisome proliferator-activated receptor gamma (PPARγ) co-activator 1- protein (PGC-1α) has been identified as a central transcriptional co-activator for NRF-1, GABPA, and the PPARs, and it is involved in the physiological integration of mitochondrial biogenesis with oxidative metabolism [8]. Two related co-activators (PGC-1β and the PGC-1-related co-activator, PRC) provide overlapping and reinforcing regulation of many nuclear-encoded mitochondrial genes [20]. All three family members activate genes encoding for proteins for mtDNA transcription and replication and mitochondrial protein importation [4, 8, 20-22].
The physiological induction of mitochondrial biogenesis in working tissues such as contracting skeletal and cardiac muscle involves intracellular calcium (Ca2+) signaling [23–29]. Energy depletion is marked by the accumulation of adenosine monophosphate (AMP) an impending energy crisis activates the serine/threonine AMP-activated protein kinase (AMPK) [30–36]. AMPK interrupts ATP-consuming reactions and activates ATP-generating pathways [32]. AMPK also promotes mitochondrial biogenesis [30, 31, 34] and blocks the growth regulating mTOR pathway [37, 38] by phosphorylation of the TSC2 tumor suppressor, which co-operates with TSC1 (tuberin) [37]. Activation of mTOR signaling by Akt/PKB involves the phosphorylation and inactivation of TSC2 [39]. Akt/PKB also promotes mitochondrial biogenesis through the phosphorylation of NRF-1 and cyclic AMP response element binding protein (CREB1), thereby enabling their nuclear translocation and target gene activation[40]. This process also increases mitochondrial hexokinase (HK) activity, committing glucose to glucose-6-phosphate using ATP generated by mitochondria and coupling glycolysis to oxidative phosphorylation [41].
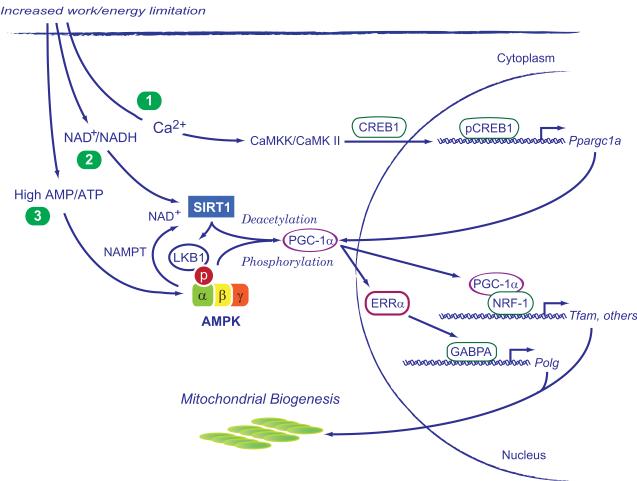
Energy homeostasis in the face of changes in nutrient availability is regulated in part by the NAD-dependent deacetylase, sirtuin-1 (SIRT1) [35, 42-45]. During nutrient depletion, SIRT1 increases the NAD+/NADH ratioand deacetylates PGC-1α, allowing the co-activator to facilitate target gene transcription [46]. SIRT1 positively regulates lipid homeostasis in the liver during fasting and starvation through nuclear PPAR, which cooperates in PGC-1 activation [43, 47]. A diagram of these physiological aspects of mitochondrial biogenesis is shown in Figure 1.
Figure 1.
Three major pathways in the activation of mitochondrial biogenesis during aerobic work and/or energy limitation. These are: 1) intracellular calcium release, particularly in contractile tissues, which activates calcium/calmodulin kinase II (CaMKII) and cAMP-responsive element binding protein 1 (CREB1); 2) the NAD+/NADH ratio, which activates sirtuin-1 (SIRT1) causing deacetylation of peroxisome proliferator-activated receptor gamma co-activator 1- PGC-1 , and 3) a high AMP/ATP ratio, which activates AMP-activated kinase (AMPK), allowing phosphorylation of PGC-1 . PGC-1 co-activates nuclear respiratory factor-1 (NRF-1) and other transcription factors involved in mitochondrial biogenesis. Other abbreviations are NAMPT, nicotinamide phosphoribosyltransferase; CaMKK, calcium/calmodulin-dependent protein kinase kinase 2; ERR , estrogen-related receptor alpha; GABPA, GA binding protein alpha; Polg, DNA-directed polymerase gamma; Tfam, mitochondrial transcription factor A.
Other transcription factors are also involved in mitochondrial biogenesis [48]. Aerobic tissues express nuclear estrogen-related receptors (ERRs), particularly the orphan nuclear receptor ERRα, a PGC-1-partner in the expression of the genes necessary for fatty acid β-oxidation [49]. In addition to CREB1, the YY1 initiator binding factor contributes to the constitutive expression of respiratory and other genes of energy metabolism [50–55].
Other genes important for mitochondrial biogenesis include the nuclear-encoded proto-oncogene c-Myc, an activator of PGC-1, and the myocyte-specific enhancer factor 2A (MEF2A) [56, 57], a critical regulator of oxidative capacity in skeletal and cardiac muscle activated by NRF-1 [58–60]. MEF2A also activates growth factor and stress-induced genes and promotes cell growth and cell survival [60]. It is noteworthy that the p53 tumor suppressor, a well-known pro-apoptotic protein, also protects mitochondria through activation of genes encoding for respiratory and mtDNA maintenance proteins [61, 62]. In the mitochondrial matrix, p53 is bound to the hsp70 and hsp60 mitochondrial import proteins and interacts with mitochondrial DNA polymerase gamma (Pol γ) to enhance its fidelity [63]. Since the accuracy of Pol γ influences mtDNA integrity, p53 may assure mitochondrial genomic stability, and the loss of p53 indeed increases the sensitivity of mtDNA to genotoxic stress [64]. Putative p53-recognition sequences are found within mtDNA in the Complex I genes and the 12S and 16S rDNA regions [65]. The mitochondrial 16S rDNA sequence in human cancer cells appears to be p53-responsive, but it is not clear whether p53 directly binds mtDNA or indirectly affects transcription. In any case, the implication is that p53 plays integral roles in mitochondrial maintenance and ROS control.
Nitric Oxide
NO synthesis from L-arginine and O2 by the nitric oxide synthases (NOS) occurs in almost all mammalian cells [66]. Of the three recognized NOS enzyme isoforms, two, the endothelial (eNOS; NOS3) and the neuronal (nNOS; NOS1), are regulated by calcium, while the third (iNOS; NOS2) is calcium-independent and inducible by inflammation. Moreover, all three NOS isoforms can be expressed constitutively or selectively up-regulated by specific transcriptional and post-transcriptional events in a number of tissues [67].
A potent physiological vasodilator, NO also acts on mitochondria in important ways. Clearly, NOS3-dependent blood flow regulation to tissue helps supply carbon substrates to mitochondria, but NO is also released allosterically from red blood cells and thus directly regulates the O2 supply to mitochondria [68]. In the cell, nitric oxide acts on the respiratory chain by binding to subunits of Complex I and Complex IV [69]. Two chemical interactions, S-nitrosation and heme-metal binding, inhibit OXPHOS, but the inhibition of cytochrome c oxidase by the latter is especially active at low O2 concentrations [70]. Thus, for certain cells and tissues, reversible NO/cytochrome c oxidase binding is seen as an acute O2 sensing system [71]. Some mitochondria also have an associated NO synthase (mitochondrial NOS, mtNOS) probably involved in the direct regulation of respiratory function [72, 73].
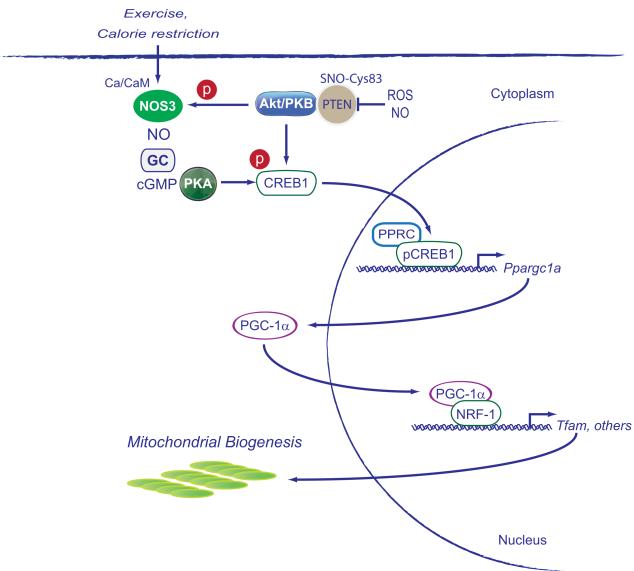
The effects of NO on the regulation of the electron transport chain also lead to increased mitochondrial superoxide anion production [74, 75]. Mitochondria also consume NO, and moderate increases in NO stimulate mitochondrial biogenesis [15, 76] mainly through cGMP-dependent gene expression and activation of regulatory proteins such as peroxisome proliferator-activated receptor γ (PPARγ) co-activator 1α(PGC1-α), nuclear respiratory factor 1 (NRF-1), and mitochondrial transcription factor A (Tfam) [3, 77]. A diagram of the original pathway for mitochondrial biogenesis proposed for NOS3 is given in Figure 2.
Figure 2.
The induction of mitochondrial biogenesis by nitric oxide synthase 3 (NOS3). Calcium/calmodulin (Ca/CM)-dependent NOS activates guanylate cyclase (GC) to generate cyclic GMP, which activates protein kinase A (PKA). PKA phosphorylates CREB1 at Ser 133 enabling its nuclear translocation and activation of the PGC-1 gene (Ppargc1a). PGC-1 protein is a co-activator for NRF-1 and other transcription factors for mitochondrial biogenesis. Other abbreviations are Akt, protein kinase B; PPRC, peroxisome proliferator-activated receptor gamma, co-activator-related 1, and PTEN, the phosphatase and tensin homolog. Other abbreviations are as in Figure 1.
In contractile tissues, calcium transients that activate calcineurin A and serine/threonine calcium/calmodulin-dependent protein kinases (CaMK), particularly CaMK II, can activate PGC-1α gene transcription [78]. The calcineurin A-mediated activation of PGC-1α gene transcription depends on MEF2 response elements, whereas CaMK-mediated regulation requires CREB1. In HeLa cells, mitochondrial biogenesis increases after transfection with NOS3, while inhibition of NOS abolishes the effect [15]. Furthermore, NOS3-deficient mice exhibit decreased levels of mtDNA, Complex IV and cytochrome c in brain, liver and heart tissue, indicating that loss of NOS3 is sufficient to reduce mitochondrial biogenesis even though the mice still express the other NOS isoforms [79].
Also, NOS3 expression and guanylate cyclase (GC) activation, but not NOS1 or NOS2 expression is induced in mice by calorie restriction (CR), which stimulates SIRT1 expression and mitochondrial biogenesis [17]. During CR, other mitochondrial proteins increase including NRF-1, Tfam, mitofusin 1 (Mfn1), mitofusin 2 (Mfn2), cytochrome c and cytochrome c oxidase. The NOS3 knockout mouse does not show the effect of CR on mitochondrial biogenesis and the mice have low SIRT1 levels and decreased longevity consistent with inability to generate new mitochondrial proteins [17].
Two other aspects of NO in mitochondria affect gene expression. One is the diversion of O2 to the cytoplasm, which causes the down-regulation of hypoxia-inducible factor 1 (HIF-1α) and anaerobic glycolysis, and the other is the release of mitochondrial calcium resulting in activation of the ATF6 transcription factor. In contrast, HIF-1α stabilization in tumor cells inhibits mitochondrial biogenesis through suppression of c-Myc transcriptional activity by two mechanisms. First, HIF-1α activates the Mxi1 gene, which encodes for a c-Myc transcriptional repressor, Max-interacting protein-1. Second, HIF-1α promotes proteasome-dependent degradation of c-Myc protein, which interferes with gene transcription for NRF-1 and PGC-1β [80].
Other transcription factors also contribute to NO’s impact on mitochondrial quality control, particularly those responsive to redox signals, such as the NF-E2 related factor (Nfe2l2 or Nrf2) [81], which physiologically regulates anti-oxidant and cytoprotective genes [82, 83]. In endothelial cells, nitric oxide facilitates the nuclear translocation of Nrf2 by a mechanism blocked by antioxidants and by inhibitors of mitogen-activated protein kinases (MAPK) [84]. And increases in NOS3 activation (22) and expression (23) triggered by laminar flow also induces the expression of certain Nrf2 target genes [85, 86].
One of the key transcriptional regulators activated by NO is CREB1. CREB1 is widely expressed and is involved along with NRF-1 in the growth-regulated expression of cytochrome c [21, 87]. NRF-1 regulates many nuclear-encoded genes required for the expression of the mitochondrial electron transport chain, mtDNA transcription and replication [10, 88], genes for heme biosynthesis [89, 90] and for mitochondrial protein importation and assembly [91]. NRF-1 regulates MEF2A expression in muscle, which controls muscle-specific cytochrome c oxidase subunits and other muscle-specific MEF2 target genes [60]. NRF-1 is also transcriptionally regulated by Nrf2 and NRF-1 induction by Nrf2 is followed by the induction of Tfam and mitochondrial transcription [92]. In addition, a link between NRF-1 and the LPS-induced inflammatory response is demonstrated by the identification of NF-kB-responsive elements in the NRF-1 promoter region [93]. This response is facilitated by CREB1 binding to the promoter and a functional interaction between NF-kB and CREB1 in controlling NRF-1-dependent mitochondrial biogenesis during the resolution of inflammation.
The PGC-1 co-activator genes are regulated by pathways that modulate growth, differentiation, and metabolism [4, 94] including those that converge on the CREB1-dependent induction of PGC-1α in response to nutrient deprivation, cold exposure, and exercise [29, 87, 95]. As mentioned earlier, PGC-1 activation and expression of mitochondrial fatty acid oxidation enzymes also occurs by post-translational protein modification by SIRT1, which deacetylates multiple lysine residues in PGC-1α [96]. In muscle, energy depletion triggers AMPK activation, which phosphorylates PGC-1α [36]. The subsequent mitochondrial gene expression is consistent with the idea that AMPK mediates its major effects through PGC-1α [97]. Also as indicated, SIRT1 and AMPK cooperate in promoting calcium-dependent mitochondrial biogenesis in myocytes [35].
PGC-1α is also a link between calorie restriction and mitochondrial oxidant production through SIRT3 expression. PGC-1α co-activates SIRT3 gene expression through ERRα, which binds to the proximal SIRT3 promoter [98, 99]. The SIRT3 protein is imported into mitochondria where it optimizes several key enzymatic activities of metabolic function [99]. It also opposes the effects of oxidant stress by triggering a series of reactions beginning with the activation of isocitrate dehydrogenase 2 and culminating in ROS detoxification by glutathione peroxidase [100]. It deacetylates mitochondrial superoxide dismutase (SOD2) leading to increased activity and enhanced oxidant scavenging [101, 102]. SIRT3 also deacetylates specific subunits in the respiratory complexes, and cells devoid of the SIRT3 gene show impaired energy homeostasis [103]. These SIRT3-dependent mechanisms have been implicated in mediating antioxidant effects associated with increased longevity [104].
Although mitochondrial ROS serve important redox signaling functions, excessive superoxide and H2O2 are certainly well-known disruptors of mitochondrial and cellular function. H2O2 is a potential trigger for intrinsic apoptosis, particularly in connection with the calcium-dependent mitochondrial permeability transition (MPT). For instance, one of the two mitochondrial-targeted isoforms of the gene for SHC-transforming protein 1, p66Shc, after phosphorylation at Ser36, accumulates in the intermembrane space [105]. Although initially implicated in cell growth, subsequent studies have indicated that p66Shc increases mitochondrial H2O2 production [106, 107] and predisposition to the MPT. In mice, p66Shc deletion confers resistance to oxidative damage and prolongs the lifespan [106, 107]. Genetic p66Shc−/−mice are also protected from certain diabetes complications, such as nephropathy and cardiomyopathy, while in diabetic patients, elevated p66Shc levels correlate with oxidative stress and poor glycemic control [108].
Mitochondrial biogenesis and mtDNA replication is not the sole mechanism by which nitric oxide supports bioenergetic function. Mitochondrial biogenesis is coordinated with mitochondrial division (fission),, and although the process of synchronization is incompletely understood, NO does alter intracellular mitochondrial structural networks. Mitochondria exist as a dynamic network, the nature of which is regulated by several factors, including fusion and fission and interactions with the cytoskeleton [109]. It is thought that mitochondrial fission functionally responds to O2 and metabolic fluctuations, but whether ROS promote mitochondrial fission or fission promotes mitochondrial ROS generation is not settled. However, during myogenic differentiation, NO-induced mitochondrial elongation (fusion) is a necessary for the repression of the pro-fission GTPase dynamin-related protein-1 (Drp1) [110]. Blocking these NO-mediated pathways in differentiating myoblasts negatively impacts mitochondrial function and oxidative phosphorylation. Drp1 is also a target for S-nitrosation, which promotes the toxic effects of β-amyloid in Alzheimer disease due to NO-mediated enhancement of Drp1-dependent fission and mitochondrial fragmentation [111]. A later study on the role of Drp1 in Alzheimer’s disease, has suggested instead that NO promotes Drp1activation by phosphorylation rather than – SNO modification because abundant SNO-Drp1 is present in normal brains in addition to those from Parkinson’s and Alzheimer’s diseases [112]. In other words, the extent to which S-nitrosation, cGMP signaling or both modulate fission is still being worked out. Nonetheless, the stimulation of mitochondrial biogenesis by NO promotes matching of cell metabolism to tissue blood flow and oxygenation in relation to the physiological demand for energy.
The HO-1/CO System
The heme oxygenase enzymes (HO-1 and -2) control the degradation of heme, which if left unattended, causes oxidative stress [113]. Free heme is toxic to the cell, and both HO isoforms, in removing it, generate physiological carbon monoxide (CO), release iron, and produce biliverdin, which is promptly converted to bilirubin [114]. The inducible HO-1 gene (Hmox1) is particularly protective, safeguarding the cell against many types of stress including heat, oxidants, inflammation, heavy metals, and hypoxia [83, 115]. Although biliverdin and bilirubin have anti-oxidant properties, CO and iron are typically pro-oxidant, and CO is a relatively weak activator of GC [116]. HO-1 induction also leads to augmentation of the cell’s iron-handling defenses, and most cells respond to CO by up-regulating anti-oxidant enzymes such as SOD2 [117]. Despite numerous publications demonstrating the protective effects of this enzyme and its products against a wide variety of tissue injuries, a unifying biological explanation has been elusive.
Like many other anti-oxidant enzyme genes, Hmox1 expression is also rapidly up-regulated by the Nrf2 transcription factor [118]. The Nrf2 protein, docked in the cytoplasm to Keap1, a redox-sensitive cytosolic adaptor protein, is targeted by a cullin-based ubiquitin ligase for proteasomal degradation, which normally prevents Nrf2 target gene expression [83]. In the nucleus, Nrf2 binds to antioxidant response elements (ARE) in the promoter regions of xenobiotic- and anti-oxidant genes, including Hmox1 [119]. Most electrophiles and oxidants, including the pro-oxidant actions of CO, activate Nrf2, leading to the transcription of ARE-containing genes, including Nrf2 itself [120].
A mitochondrial Keap1-binding protein, PGAM5, a phosphoglycerate mutase without PGM enzyme activity is targeted to the outer mitochondrial membrane by an N-terminal localization sequence. PGAM5 forms a ternary complex with Keap1 and Nrf2 in which Keap1 dimer simultaneously binds PGAM5 and Nrf2 through conserved E (S/T) GE motifs [121]. Loss of Keap1 or PGAM5 activates Nrf2-dependent genes.
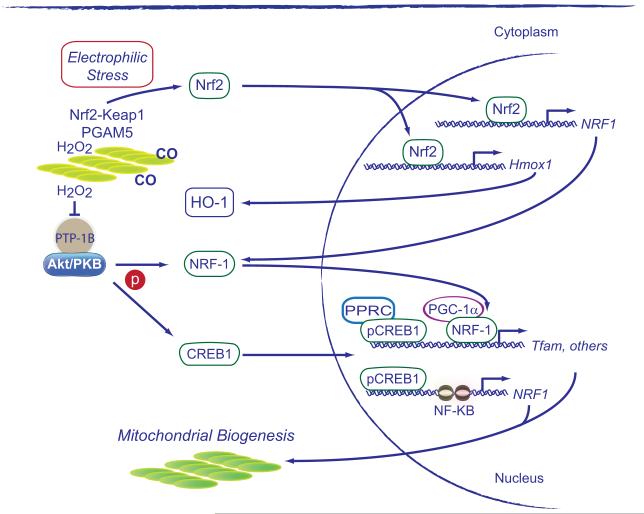
The role of HO-1 and endogenous CO production in mitochondrial biogenesis was discovered in 2007 [122]. An increase in endogenous CO level stimulates SOD2 expression and mitochondrial H2O2 production at Complex III, which leads to activation of Akt/PKB and deactivation of glycogen synthase kinase-3 , permitting Nrf2 nuclear translocation and occupancy of AREs in the NRF-1 promoter [92]. Activation of this pathway leads to strong induction of mitochondrial biogenesis as illustrated by Figure 3.
Figure 3.
The induction of mitochondrial biogenesis by the HO-1/CO system. The diagram illustrates how endogenous CO and mitochondrial H2O2 release contribute to the redox activation of Nrf2-Keap1. The oxidation of Keap1 in the vicinity of mitochondria stabilizes the protein complex and allows free cytoplasmic Nrf2 to translocate to the nucleus where it activates NRF-1 and HO-1. This creates a feed-forward cycle for the initiation and maintenance of mitochondrial biogenesis following oxidative and inflammatory stress. PTP-1B, Tyrosine-protein phosphatase non-receptor type 1 (PTPN1); PGAM5, mitochondrial serine/threonine-protein phosphatase, Other abbreviations as in Figures 1 and 2.
This transcriptional mechanism also provided the first link to the expansion of mitochondrial mass with the induction of the xenobiotic and antioxidant defenses. In the mouse heart, HO-1 stimulation of mitochondrial biogenesis protects against the induction of cardiomyopathy by the anthracycline chemotherapeutic, doxorubicin [123], an agent which disrupts mitochondrial biogenesis and causes intrinsic apoptosis, necrosis, inflammation, and myocardial fibrosis. But the administration of low level CO or the expression of active HO-1 mitigates all of these effects [123].
The induction of HO-1 during inflammation not only triggers mitochondrial biogenesis, but leads to activation of counter-inflammatory genes such as IL-10 [124], and anti-apoptotic proteins such as BclXL [125]. These responses encompass the expression of the IL-1 receptor antagonist (IL-1Ra) and the suppressor of cytokine synthesis-3 (SOCS3) [126]. Mechanistically, CO release by HO-1 promotes binding of the Nrf2, GABPA, and MEF2 transcription factors to the IL10 promoter and the NRF-1 and MEF2 transcription factors to the IL1Ra promoter. In liver cells and in macrophages, Nrf2 or Hmox1 gene silencing blocks IL-10 and IL-1Ra up-regulation, and hepatic Hmox1 induction fails in response to bacterial-induced inflammation in Nrf2-/- mice.
HO-1 induction by mitochondrial ROS leads to a cycle of Nrf2 activation and further HO-1 induction, which is necessary to sustain NRF-1 and Tfam expression during mitochondrial biogenesis and for the induction of counter-inflammatory genes. The integration of this work with that mentioned above indicating that PPARs, AMPK, SIRT1, and PCG-1 family proteins produce anti-inflammatory effects implies that the program for mitochondrial biogenesis is part of an integrated transcriptional network linking cell metabolism to the cell’s anti-oxidant, anti-apoptotic, and counter-inflammatory defenses.
Oxidative Damage and Inflammation
Mitochondria are vulnerable to oxidative and nitrosative damage by the effects of proinflammatory cytokines, such as tumor necrosis factor alpha (TNF- ) and interleukins generated during innate immune activation [127]. Defective mitochondria generate high ROS levels and release calcium and intrinsic apoptosis proteins during inflammation [128-130]. Excess ROS and NO production via immune effector pathways also compromises mitochondrial structure and function through direct oxidation of proteins and lipids or by peroxynitrite generation [131]. This may cause sufficient oxidative mitochondrial damage to cause cell death by apoptosis or necrosis.
During infections, higher eukaryotes recognize pathogens as “non-self” by means of evolutionarily-conserved pattern-recognition receptors (PRRs) [132-134].” The PRR detects pathogen-associated molecular patterns or PAMPs— entire molecules such as LPS or conserved motifs recognized as pathogenic. Plasma membranes or endosomes, for instance in macrophages, express toll-like receptors (TLR) that initiate the immune response to PAMPs from bacteria, fungi, parasites, and viruses. Specific adaptor proteins, MyD88 and TRIF, activate the NF-kB transcription factor and initiate signals for the synthesis of inflammatory cytokines, chemokines, antimicrobial peptides, and type I interferon [134]. TLR4 signaling through MyD88 is critical to the rapid synthesis of TNF-interleukin-1 beta (IL-1 ) and NOS2 in response to LPS [133].
Many cytokines and chemokines promote pathogen elimination by recruiting myeloid cells to sites of infection and stimulating phagocytosis, antigen presentation or antibody production [135]. Although NO and ROS production are characteristic of the mammalian innate immune response, these species in excess are pro-inflammatory [136, 137]. NF-kB activation by ROS in inflammatory and vascular cells enhances TNF- release, which further increases ROS production [138, 139]. TNF- is also a catabolic agent, and it inhibits respiration, mitochondrial biogenesis, and oxidative fiber phenotype in skeletal muscle [140]. In cardiomyocytes, TNF- stimulates binding of the NF-kB p65 subunit to PGC-1 , which may interfere with how one or both molecules effect downstream gene activation [141].
In addition to the up-regulation of anti-oxidant defenses, inflammation increases NRF-1 and PGC-1 expression [40, 142]. PGC-1 contributes to the induction of ROS-detoxifying enzymes, for instance SOD2 and glutathione peroxidase-1 (GPx1), and in neuronal cells after oxidant stress, CREB1 activates PGC-1 [142]. PGC-1 also promotes myocardial SOD2 and thioredoxin (Trx2) expression and protects the mouse heart against oxidative stress, hypertrophy, and contractile dysfunction [143]. PGC-1 over-expression in macrophages inhibits NF-kB-dependent cytokine production, which opposes inflammatory stress [144]. And NF-kB activates anti-oxidant enzyme genes, including SOD2 [145], while NF-kB inhibition increases TNF-mediated apoptosis in part by allowing the FAS-associating death domain-containing protein (FADD) to join a death-inducing signaling complex (DISC) that triggers caspase-8 activation [146]. The cell can avert this mode of death by shedding TNF-R1 [147].
TNF-R1 activation also increases cytosolic Ca2+ release and mitochondrial Ca2+ uptake [148], thus stimulating respiration and mitochondrial ROS production [149]. In lung vascular cells, high SOD2 activity increases the peroxide leak rate at Complex III, activating the TNF-α converting enzyme (TACE), a metalloproteinase that cleaves TNF-R1, causing receptor shedding and dampening of the TNF- response [150].
In muscle-specific PGC-1 knockout mice, high TNF- and IL-6 levels cause fiber damage and muscle wasting during exercise [151-154]. By comparison, muscle-specific PGC-1 over-expression in mice opposes aging-related declines in mitochondrial function and muscle integrity [155]. High muscle PGC-1 levels prevent muscle wasting by reducing apoptosis, autophagy, and proteasomal activity. This muscle protection also slows the decline in bone density and chronic inflammation of aging. In smooth muscle and endothelial cells, Ppargc1a over-expression suppresses mitochondrial ROS production induced by NF-kB and TNF- [139].
In the liver, TLR2/4 activation by bacterial products is accompanied by mtDNA depletion and impaired mitochondrial transcription [12] followed by a wave of mitochondrial proliferation [12, 156]. Histological observations provide evidence of accelerated mitochondrial turnover involving mitochondrial biogenesis and mitochondrial autophagy (mitophagy), which restores mitochondrial mass and mitigates cell death [12, 157-159]. TLR signaling post-inoculation with Staphylococcal aureus also leads to prompt Ppargc1a and Ppargc1b expression, but these genes are deregulated in TLR2-/- mice (fail to increase) and in TLR4-/- mice (highly increased) [160].
The initial depletion of mtDNA is related to NF-kB activation and TNF- and NO over-production. In LPS-mediated infections, TLR4 null mice show less mtDNA depletion, but restoration of mtDNA copy number is delayed [161]. The mtDNA damage induces the base excision repair glycosylase OGG1 [162] and is abrogated by increasing SOD2 levels or by limiting reactive nitrogen species production [163]. Redox- mechanisms for NRF-1 induction and including phosphorylation of the protein by Akt enable it to translocate to the nucleus and activate the genes for Tfam and other mitochondrial transcriptome proteins, allowing restoration of mtDNA copy number [164].
The TLR system also fosters the early up-regulation of mitochondrial biogenesis [160, 161] through NF-kB [93, 165], CREB1 [93], Nrf2 [126], and interferon response factors (IRF-3,IRF-7) [166]. The early-phase host defense enhances mitochondrial H2O2 production and initiation of mitochondrial biogenesis. AMPK also promotes NO production [167, 168], regulates autophagy [169] and inhibits NF-κB–dependent cytokine expression [170-177]. Conversely, low AMPK activity leads to enhanced inflammation. The NF-kB inhibition may involve PGC-1α, Forkhead box O transcription factors, and/or SIRT1 [20, 35, 178, 179]. SIRT1 is also anti-inflammatory [180] and SIRT1 deletion interferes with PPAR and -oxidation, whereas SIRT1 over-expression induces PPAR target genes. Liver-specific SIRT1−/− mice develop steatosis, ER stress, and hepatic inflammation. In peritoneal macrophages SIRT1 gene silencing stimulates the inflammatory response to LPS while SIRT1 activation inhibits it [181].
Danger signals lead to the full development of tissue inflammation through the assembly of intracellular inflammasomes. The NLRP3 inflammasome and caspase-1 (and caspase-11 in mice) generate IL-1 by cleavage of its pro-form. Mitochondria are a main source of ROS for NLRP3 activation and the NLRP3 inflammasome also influences glycolysis and lipogenesis [182-184]. In macrophages, inflammasome activation is impaired by inactivation of the outer membrane voltage dependent anion channel, VDAC, or by mtDNA depletion [184]. Complex I and Complex III inhibition also activate the NLRP3 inflammasome [183]. Dysfunctional ROS-generating mitochondria and counter-regulation the NLRP3 inflammasome occurs in part through mitochondrial autophagy or mitophagy [185]. A loss of mitophagy allows persistence of ROS-generating mitochondria and NLRP3 activation [183, 184].
Another common metabolic disorder, Type 2 diabetes, is associated with ROS production, inflammation, and alterations in mitochondrial density and function [186, 187]. Analysis of the mitochondrial transcriptome in the skeletal muscle of patients with type 2 diabetes has indicated impaired expression of OXPHOS genes [188, 189]. Hyperglycemia also increases intracellular ROS levels by several mechanisms. One that operates in neuronal tissue involves hyperpolarization of the inner mitochondrial membrane, which increases ROS leakage out of the mitochondrion [190]. In hyperglycemic, hyperlipidemic mice, muscle ROS production is associated with mitochondrial swelling and a decrease in mRNA expression for PGC1-α and its target genes. In hyperglycemia-associated oxidative stress, low molecular weight anti-oxidant administration can improve mitochondrial density and structure [191]. This implies that hyperglycemia and hyperlipidemia-induced mitochondrial dysfunction in skeletal muscle is due at least in part to increased mitochondrial ROS production and impaired redox signaling. In animal models of insulin resistance and high fat diets, AMPK activity is also down-regulated [192].
These pro-survival pathways also impinge on the regulation of apoptosis. The pro-apoptotic BAD, a BH3-only domain pro-apoptotic member of the Bcl-2 family is phosphorylated by survival kinases resulting in cytoplasmic localization and inactivation [193]. In mitochondria, BAD is part of the large protein complex that through glucokinase (HK4) catalyzes the first step of glycolysis. The loss of BAD restricts respiration in response to glucose, while glucose deprivation leads to BAD dephosphorylation and to apoptosis [194]. Although normally pro-survival, the disproportionate induction of mitochondrial biogenesis may indicate persistent mitochondrial dysfunction [195]. Interference with mitochondrial biogenesis during periods of oxidative stress exacerbates inflammation and promotes apoptosis, but rapid mitochondrial turnover under the stimulus of continuous mitochondrial damage also increases the likelihood of immune suppression by counter-inflammatory mediator over-production [196-200].
Summary and Conclusions
Mitochondrial biogenesis can be activated by NO, CO, and mitochondrial ROS by means of retrograde signaling from damaged mitochondria to the nucleus. These redox signals give rise to nuclear transcriptional responses that lead to mitochondrial protein synthesis. The response is structured to compensate for mitochondrial dysfunction and to avoid compromise of cell metabolism, triggered for example, by oxidative stress during a hyperactive host immune response. Immune effectors elaborated by both structural and immune cells, such as TNF- and IL-1 promote oxidative stress, calcium deregulation, and NO production, which not only contributes to mitochondrial damage, but to activation of mitochondrial biogenesis. These same effectors may also interfere sufficiently with these redox signals to obfuscate such adaptive responses, ultimately leading to loss of homeostasis. There are also interactions, although not yet fully defined, among the NO, CO and calcium-mediated control mechanisms for mitochondrial biogenesis. Still, it is evident that this multifaceted transcriptional network is coordinated with the removal of damaged mitochondria and that together these processes re-establish an anti-oxidant counter-inflammatory milieu that protects against energy failure and cell death.
Highlights.
The physiological gases NO and CO are redox regulators of mitochondrial biogenesis
NO activates mitochondrial biogenesis mainly through cGMP and CREB1
CO activates mitochondrial biogenesis mainly through ROS and Nrf2-dependent genes
Adaptive mitochondrial biogenesis opposes oxidative stress and prevents energy failure
Acknowledgements
NIH HL090679 and GM084116
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Singer M. Mitochondrial function in sepsis: acute phase versus multiple organ failure. Crit Care Med. 2007;35:S441–448. doi: 10.1097/01.CCM.0000278049.48333.78. [DOI] [PubMed] [Google Scholar]
- [2].Okamoto K, Kondo-Okamoto N. Mitochondria and autophagy: Critical interplay between the two homeostats. Biochimica et biophysica acta. 2011 doi: 10.1016/j.bbagen.2011.08.001. [DOI] [PubMed] [Google Scholar]
- [3].Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- [4].Scarpulla RC. Transcriptional paradigms in Mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- [5].Kowald A, Kirkwood TB. Evolution of the mitochondrial fusion-fission cycle and its role in aging. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:10237–10242. doi: 10.1073/pnas.1101604108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Scarpulla RC. Transcriptional activators and coactivators in the nuclear control of mitochondrial function in mammalian cells. Gene. 2002;286:81–89. doi: 10.1016/s0378-1119(01)00809-5. [DOI] [PubMed] [Google Scholar]
- [7].Carraway MS, Suliman HB, Jones WS, Chen CW, Babiker A, Piantadosi CA. Erythropoietin activates mitochondrial biogenesis and couples red cell mass to mitochondrial mass in the heart. Circ Res. 2010;106:1722–1730. doi: 10.1161/CIRCRESAHA.109.214353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- [9].Xia Y, Buja LM, Scarpulla RC, McMillin JB. Electrical stimulation of neonatal cardiomyocytes results in the sequential activation of nuclear genes governing mitochondrial proliferation and differentiation. Proc Natl Acad Sci U S A. 1997;94:11399–11404. doi: 10.1073/pnas.94.21.11399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Scarpulla RC. Nuclear activators and coactivators in mammalian mitochondrial biogenesis. Biochim Biophys Acta. 2002;1576:1–14. doi: 10.1016/s0167-4781(02)00343-3. [DOI] [PubMed] [Google Scholar]
- [11].Guidot DM. Endotoxin pretreatment in vivo increases the mitochondrial respiratory capacity in rat hepatocytes. Arch Biochem Biophys. 1998;354:9–17. doi: 10.1006/abbi.1998.0699. [DOI] [PubMed] [Google Scholar]
- [12].Suliman HB, Carraway MS, Piantadosi CA. Postlipopolysaccharide oxidative damage of mitochondrial DNA. Am J Respir Crit Care Med. 2003;167:570–579. doi: 10.1164/rccm.200206-518OC. [DOI] [PubMed] [Google Scholar]
- [13].Himms-Hagen J. Cellular thermogenesis. Annu Rev Physiol. 1976;38:315–351. doi: 10.1146/annurev.ph.38.030176.001531. [DOI] [PubMed] [Google Scholar]
- [14].Mutvei A, Husman B, Andersson G, Nelson BD. Thyroid hormone and not growth hormone is the principle regulator of mammalian mitochondrial biogenesis. Acta Endocrinol (Copenh) 1989;121:223–228. doi: 10.1530/acta.0.1210223. [DOI] [PubMed] [Google Scholar]
- [15].Nisoli E, Clementi E, Paolucci C, Cozzi V, Tonello C, Sciorati C, Bracale R, Valerio A, Francolini M, Moncada S, Carruba MO. Mitochondrial biogenesis in mammals: the role of endogenous nitric oxide. Science. 2003;299:896–899. doi: 10.1126/science.1079368. [DOI] [PubMed] [Google Scholar]
- [16].Lopez-Lluch G, Hunt N, Jones B, Zhu M, Jamieson H, Hilmer S, Cascajo MV, Allard J, Ingram DK, Navas P, de Cabo R. Calorie restriction induces mitochondrial biogenesis and bioenergetic efficiency. Proc Natl Acad Sci U S A. 2006;103:1768–1773. doi: 10.1073/pnas.0510452103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Nisoli E, Tonello C, Cardile A, Cozzi V, Bracale R, Tedesco L, Falcone S, Valerio A, Cantoni O, Clementi E, Moncada S, Carruba MO. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science. 2005;310:314–317. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- [18].Lee HC, Yin PH, Chi CW, Wei YH. Increase in mitochondrial mass in human fibroblasts under oxidative stress and during replicative cell senescence. J Biomed Sci. 2002;9:517–526. doi: 10.1007/BF02254978. [DOI] [PubMed] [Google Scholar]
- [19].Mootha VK, Bunkenborg J, Olsen JV, Hjerrild M, Wisniewski JR, Stahl E, Bolouri MS, Ray HN, Sihag S, Kamal M, Patterson N, Lander ES, Mann M. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115:629–640. doi: 10.1016/s0092-8674(03)00926-7. [DOI] [PubMed] [Google Scholar]
- [20].Scarpulla RC. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochimica et biophysica acta. 2011;1813:1269–1278. doi: 10.1016/j.bbamcr.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Vercauteren K, Pasko RA, Gleyzer N, Marino VM, Scarpulla RC. PGC-1-related coactivator: immediate early expression and characterization of a CREB/NRF-1 binding domain associated with cytochrome c promoter occupancy and respiratory growth. Mol Cell Biol. 2006;26:7409–7419. doi: 10.1128/MCB.00585-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wu Z, Huang X, Feng Y, Handschin C, Feng Y, Gullicksen PS, Bare O, Labow M, Spiegelman B, Stevenson SC. Transducer of regulated CREB-binding proteins (TORCs) induce PGC-1alpha transcription and mitochondrial biogenesis in muscle cells. Proc Natl Acad Sci U S A. 2006;103:14379–14384. doi: 10.1073/pnas.0606714103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Freyssenet D, Di Carlo M, Hood DA. Calcium-dependent regulation of cytochrome c gene expression in skeletal muscle cells. Identification of a protein kinase c-dependent pathway. J Biol Chem. 1999;274:9305–9311. doi: 10.1074/jbc.274.14.9305. [DOI] [PubMed] [Google Scholar]
- [24].Freyssenet D, Irrcher I, Connor MK, Di Carlo M, Hood DA. Calcium-regulated changes in mitochondrial phenotype in skeletal muscle cells. Am J Physiol Cell Physiol. 2004;286:C1053–1061. doi: 10.1152/ajpcell.00418.2003. [DOI] [PubMed] [Google Scholar]
- [25].Guerfali I, Manissolle C, Durieux AC, Bonnefoy R, Bartegi A, Freyssenet D. Calcineurin A and CaMKIV transactivate PGC-1alpha promoter, but differentially regulate cytochrome c promoter in rat skeletal muscle. Pflugers Arch. 2007;454:297–305. doi: 10.1007/s00424-007-0206-6. [DOI] [PubMed] [Google Scholar]
- [26].Ojuka EO, Jones TE, Han DH, Chen M, Wamhoff BR, Sturek M, Holloszy JO. Intermittent increases in cytosolic Ca2+ stimulate mitochondrial biogenesis in muscle cells. Am J Physiol Endocrinol Metab. 2002;283:E1040–1045. doi: 10.1152/ajpendo.00242.2002. [DOI] [PubMed] [Google Scholar]
- [27].Ojuka EO, Jones TE, Nolte LA, Chen M, Wamhoff BR, Sturek M, Holloszy JO. Regulation of GLUT4 biogenesis in muscle: evidence for involvement of AMPK and Ca(2+) Am J Physiol Endocrinol Metab. 2002;282:E1008–1013. doi: 10.1152/ajpendo.00512.2001. [DOI] [PubMed] [Google Scholar]
- [28].Tavi P, Westerblad H. The role of in vivo Ca(2) signals acting on Ca(2)-calmodulin-dependent proteins for skeletal muscle plasticity. The Journal of physiology. 2011;589:5021–5031. doi: 10.1113/jphysiol.2011.212860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wu H, Kanatous SB, Thurmond FA, Gallardo T, Isotani E, Bassel-Duby R, Williams RS. Regulation of mitochondrial biogenesis in skeletal muscle by CaMK. Science. 2002;296:349–352. doi: 10.1126/science.1071163. [DOI] [PubMed] [Google Scholar]
- [30].Bergeron R, Ren JM, Cadman KS, Moore IK, Perret P, Pypaert M, Young LH, Semenkovich CF, Shulman GI. Chronic activation of AMP kinase results in NRF-1 activation and mitochondrial biogenesis. Am J Physiol Endocrinol Metab. 2001;281:E1340–1346. doi: 10.1152/ajpendo.2001.281.6.E1340. [DOI] [PubMed] [Google Scholar]
- [31].Zong H, Ren JM, Young LH, Pypaert M, Mu J, Birnbaum MJ, Shulman GI. AMP kinase is required for mitochondrial biogenesis in skeletal muscle in response to chronic energy deprivation. Proc Natl Acad Sci U S A. 2002;99:15983–15987. doi: 10.1073/pnas.252625599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hardie DG. The AMP-activated protein kinase pathway--new players upstream and downstream. J Cell Sci. 2004;117:5479–5487. doi: 10.1242/jcs.01540. [DOI] [PubMed] [Google Scholar]
- [33].Atherton PJ, Babraj J, Smith K, Singh J, Rennie MJ, Wackerhage H. Selective activation of AMPK-PGC-1alpha or PKB-TSC2-mTOR signaling can explain specific adaptive responses to endurance or resistance training-like electrical muscle stimulation. Faseb J. 2005;19:786–788. doi: 10.1096/fj.04-2179fje. [DOI] [PubMed] [Google Scholar]
- [34].Reznick RM, Shulman GI. The role of AMP-activated protein kinase in mitochondrial biogenesis. J Physiol. 2006;574:33–39. doi: 10.1113/jphysiol.2006.109512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Steinberg GR, Kemp BE. AMPK in Health and Disease. Physiol Rev. 2009;89:1025–1078. doi: 10.1152/physrev.00011.2008. [DOI] [PubMed] [Google Scholar]
- [37].Li Y, Corradetti MN, Inoki K, Guan KL. TSC2: filling the GAP in the mTOR signaling pathway. Trends in biochemical sciences. 2004;29:32–38. doi: 10.1016/j.tibs.2003.11.007. [DOI] [PubMed] [Google Scholar]
- [38].Fingar DC, Blenis J. Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene. 2004;23:3151–3171. doi: 10.1038/sj.onc.1207542. [DOI] [PubMed] [Google Scholar]
- [39].Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Molecular cell. 2002;10:151–162. doi: 10.1016/s1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- [40].Piantadosi CA, Suliman HB. Mitochondrial transcription factor A induction by redox activation of nuclear respiratory factor 1. J Biol Chem. 2006;281:324–333. doi: 10.1074/jbc.M508805200. [DOI] [PubMed] [Google Scholar]
- [41].Gottlob K, Majewski N, Kennedy S, Kandel E, Robey RB, Hay N. Inhibition of early apoptotic events by Akt/PKB is dependent on the first committed step of glycolysis and mitochondrial hexokinase. Genes & development. 2001;15:1406–1418. doi: 10.1101/gad.889901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha} J Biol Chem. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- [43].Rodgers JT, Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc Natl Acad Sci U S A. 2007;104:12861–12866. doi: 10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- [45].Civitarese AE, Carling S, Heilbronn LK, Hulver MH, Ukropcova B, Deutsch WA, Smith SR, Ravussin E. Calorie Restriction Increases Muscle Mitochondrial Biogenesis in Healthy Humans. PLoS Med. 2007;4:e76. doi: 10.1371/journal.pmed.0040076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Dominy JE, Jr., Lee Y, Gerhart-Hines Z, Puigserver P. Nutrient-dependent regulation of PGC-1alpha’s acetylation state and metabolic function through the enzymatic activities of Sirt1/GCN5. Biochimica et biophysica acta. 2010;1804:1676–1683. doi: 10.1016/j.bbapap.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell metabolism. 2009;9:327–338. doi: 10.1016/j.cmet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Handschin C, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr Rev. 2006;27:728–735. doi: 10.1210/er.2006-0037. [DOI] [PubMed] [Google Scholar]
- [49].Schreiber SN, Emter R, Hock MB, Knutti D, Cardenas J, Podvinec M, Oakeley EJ, Kralli A. The estrogen-related receptor alpha (ERRalpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha)-induced mitochondrial biogenesis. Proc Natl Acad Sci U S A. 2004;101:6472–6477. doi: 10.1073/pnas.0308686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Spiegelman BM, Puigserver P, Wu Z. Regulation of adipogenesis and energy balance by PPARgamma and PGC-1. Int J Obes Relat Metab Disord. 2000;24(Suppl 4):S8–10. doi: 10.1038/sj.ijo.0801492. [DOI] [PubMed] [Google Scholar]
- [51].Wang YX, Zhang CL, Yu RT, Cho HK, Nelson MC, Bayuga-Ocampo CR, Ham J, Kang H, Evans RM. Regulation of muscle fiber type and running endurance by PPARdelta. PLoS Biol. 2004;2:e294. doi: 10.1371/journal.pbio.0020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Baar K. Involvement of PPAR gamma co-activator-1, nuclear respiratory factors 1 and 2, and PPAR alpha in the adaptive response to endurance exercise. Proc Nutr Soc. 2004;63:269–273. doi: 10.1079/PNS2004334. [DOI] [PubMed] [Google Scholar]
- [53].Andersen G, Wegner L, Yanagisawa K, Rose CS, Lin J, Glumer C, Drivsholm T, Borch-Johnsen K, Jorgensen T, Hansen T, Spiegelman BM, Pedersen O. Evidence of an association between genetic variation of the coactivator PGC-1beta and obesity. J Med Genet. 2005;42:402–407. doi: 10.1136/jmg.2004.026278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Barish GD, Narkar VA, Evans RM. PPAR delta: a dagger in the heart of the metabolic syndrome. J Clin Invest. 2006;116:590–597. doi: 10.1172/JCI27955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Madrazo JA, Kelly DP. The PPAR trio: regulators of myocardial energy metabolism in health and disease. J Mol Cell Cardiol. 2008;44:968–975. doi: 10.1016/j.yjmcc.2008.03.021. [DOI] [PubMed] [Google Scholar]
- [56].Morrish F, Giedt C, Hockenbery D. c-MYC apoptotic function is mediated by NRF-1 target genes. Genes Dev. 2003;17:240–255. doi: 10.1101/gad.1032503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Dang CV, Li F, Lee LA. Could MYC induction of mitochondrial biogenesis be linked to ROS production and genomic instability? Cell Cycle. 2005;4:1465–1466. doi: 10.4161/cc.4.11.2121. [DOI] [PubMed] [Google Scholar]
- [58].Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, Lowell BB, Bassel-Duby R, Spiegelman BM. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- [59].Czubryt MP, McAnally J, Fishman GI, Olson EN. Regulation of peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PGC-1 alpha ) and mitochondrial function by MEF2 and HDAC5. Proc Natl Acad Sci U S A. 2003;100:1711–1716. doi: 10.1073/pnas.0337639100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Ramachandran B, Yu G, Gulick T. Nuclear respiratory factor 1 controls myocyte enhancer factor 2A transcription to provide a mechanism for coordinate expression of respiratory chain subunits. J Biol Chem. 2008;283:11935–11946. doi: 10.1074/jbc.M707389200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Saleem A, Adhihetty PJ, Hood DA. Role of p53 in mitochondrial biogenesis and apoptosis in skeletal muscle. Physiological genomics. 2009;37:58–66. doi: 10.1152/physiolgenomics.90346.2008. [DOI] [PubMed] [Google Scholar]
- [62].Maddocks OD, Vousden KH. Metabolic regulation by p53. J Mol Med (Berl) 2011;89:237–245. doi: 10.1007/s00109-011-0735-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Laptenko O, Prives C. Transcriptional regulation by p53: one protein, many possibilities. Cell death and differentiation. 2006;13:951–961. doi: 10.1038/sj.cdd.4401916. [DOI] [PubMed] [Google Scholar]
- [64].Achanta G, Sasaki R, Feng L, Carew JS, Lu W, Pelicano H, Keating MJ, Huang P. Novel role of p53 in maintaining mitochondrial genetic stability through interaction with DNA Pol gamma. The EMBO journal. 2005;24:3482–3492. doi: 10.1038/sj.emboj.7600819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Heyne K, Mannebach S, Wuertz E, Knaup KX, Mahyar-Roemer M, Roemer K. Identification of a putative p53 binding sequence within the human mitochondrial genome. FEBS letters. 2004;578:198–202. doi: 10.1016/j.febslet.2004.10.099. [DOI] [PubMed] [Google Scholar]
- [66].Alderton WK, Cooper CE, Knowles RG. Nitric oxide synthases: structure, function and inhibition. The Biochemical journal. 2001;357:593–615. doi: 10.1042/0264-6021:3570593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Stuehr DJ, Santolini J, Wang ZQ, Wei CC, Adak S. Update on mechanism and catalytic regulation in the NO synthases. J Biol Chem. 2004;279:36167–36170. doi: 10.1074/jbc.R400017200. [DOI] [PubMed] [Google Scholar]
- [68].Allen BW, Stamler JS, Piantadosi CA. Hemoglobin, nitric oxide and molecular mechanisms of hypoxic vasodilation. Trends Mol Med. 2009;15:452–460. doi: 10.1016/j.molmed.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Erusalimsky JD, Moncada S. Nitric oxide and mitochondrial signaling: from physiology to pathophysiology. Arterioscler Thromb Vasc Biol. 2007;27:2524–2531. doi: 10.1161/ATVBAHA.107.151167. [DOI] [PubMed] [Google Scholar]
- [70].Brown GC. Nitric oxide and mitochondria. Front Biosci. 2007;12:1024–1033. doi: 10.2741/2122. [DOI] [PubMed] [Google Scholar]
- [71].Clementi E, Nisoli E. Nitric oxide and mitochondrial biogenesis: a key to long-term regulation of cellular metabolism. Comp Biochem Physiol A Mol Integr Physiol. 2005;142:102–110. doi: 10.1016/j.cbpb.2005.04.022. [DOI] [PubMed] [Google Scholar]
- [72].Elfering SL, Sarkela TM, Giulivi C. Biochemistry of mitochondrial nitric-oxide synthase. J Biol Chem. 2002;277:38079–38086. doi: 10.1074/jbc.M205256200. [DOI] [PubMed] [Google Scholar]
- [73].Venkatakrishnan P, Nakayasu ES, Almeida IC, Miller RT. Absence of nitric-oxide synthase in sequentially purified rat liver mitochondria. J Biol Chem. 2009;284:19843–19855. doi: 10.1074/jbc.M109.003301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Borutaite V, Brown GC. Rapid reduction of nitric oxide by mitochondria, and reversible inhibition of mitochondrial respiration by nitric oxide. The Biochemical journal. 1996;315(Pt 1):295–299. doi: 10.1042/bj3150295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Poderoso JJ, Carreras MC, Lisdero C, Riobo N, Schopfer F, Boveris A. Nitric oxide inhibits electron transfer and increases superoxide radical production in rat heart mitochondria and submitochondrial particles. Archives of biochemistry and biophysics. 1996;328:85–92. doi: 10.1006/abbi.1996.0146. [DOI] [PubMed] [Google Scholar]
- [76].Nisoli E, Falcone S, Tonello C, Cozzi V, Palomba L, Fiorani M, Pisconti A, Brunelli S, Cardile A, Francolini M, Cantoni O, Carruba MO, Moncada S, Clementi E. Mitochondrial biogenesis by NO yields functionally active mitochondria in mammals. Proc Natl Acad Sci U S A. 2004;101:16507–16512. doi: 10.1073/pnas.0405432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Nisoli E, Carruba MO. Nitric oxide and mitochondrial biogenesis. J Cell Sci. 2006;119:2855–2862. doi: 10.1242/jcs.03062. [DOI] [PubMed] [Google Scholar]
- [78].Chin ER. The role of calcium and calcium/calmodulin-dependent kinases in skeletal muscle plasticity and mitochondrial biogenesis. Proc Nutr Soc. 2004;63:279–286. doi: 10.1079/PNS2004335. [DOI] [PubMed] [Google Scholar]
- [79].Gutsaeva DR, Carraway MS, Suliman HB, Demchenko IT, Shitara H, Yonekawa H, Piantadosi CA. Transient hypoxia stimulates mitochondrial biogenesis in brain subcortex by a neuronal nitric oxide synthase-dependent mechanism. J Neurosci. 2008;28:2015–2024. doi: 10.1523/JNEUROSCI.5654-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Zhang H, Gao P, Fukuda R, Kumar G, Krishnamachary B, Zeller KI, Dang CV, Semenza GL. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11:407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- [81].Nguyen T, Sherratt PJ, Pickett CB. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annual review of pharmacology and toxicology. 2003;43:233–260. doi: 10.1146/annurev.pharmtox.43.100901.140229. [DOI] [PubMed] [Google Scholar]
- [82].Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochemical and biophysical research communications. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- [83].Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci U S A. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Buckley BJ, Marshall ZM, Whorton AR. Nitric oxide stimulates Nrf2 nuclear translocation in vascular endothelium. Biochemical and biophysical research communications. 2003;307:973–979. doi: 10.1016/s0006-291x(03)01308-1. [DOI] [PubMed] [Google Scholar]
- [85].Davis ME, Cai H, Drummond GR, Harrison DG. Shear stress regulates endothelial nitric oxide synthase expression through c-Src by divergent signaling pathways. Circulation research. 2001;89:1073–1080. doi: 10.1161/hh2301.100806. [DOI] [PubMed] [Google Scholar]
- [86].Hosoya T, Maruyama A, Kang MI, Kawatani Y, Shibata T, Uchida K, Warabi E, Noguchi N, Itoh K, Yamamoto M. Differential responses of the Nrf2-Keap1 system to laminar and oscillatory shear stresses in endothelial cells. The Journal of biological chemistry. 2005;280:27244–27250. doi: 10.1074/jbc.M502551200. [DOI] [PubMed] [Google Scholar]
- [87].Herzig RP, Scacco S, Scarpulla RC. Sequential serum-dependent activation of CREB and NRF-1 leads to enhanced mitochondrial respiration through the induction of cytochrome c. J Biol Chem. 2000;275:13134–13141. doi: 10.1074/jbc.275.17.13134. [DOI] [PubMed] [Google Scholar]
- [88].Cam H, Balciunaite E, Blais A, Spektor A, Scarpulla RC, Young R, Kluger Y, Dynlacht BD. A common set of gene regulatory networks links metabolism and growth inhibition. Mol Cell. 2004;16:399–411. doi: 10.1016/j.molcel.2004.09.037. [DOI] [PubMed] [Google Scholar]
- [89].Braidotti G, Borthwick IA, May BK. Identification of regulatory sequences in the gene for 5-aminolevulinate synthase from rat. The Journal of biological chemistry. 1993;268:1109–1117. [PubMed] [Google Scholar]
- [90].Aizencang GI, Bishop DF, Forrest D, Astrin KH, Desnick RJ. Uroporphyrinogen III synthase. An alternative promoter controls erythroid-specific expression in the murine gene. The Journal of biological chemistry. 2000;275:2295–2304. doi: 10.1074/jbc.275.4.2295. [DOI] [PubMed] [Google Scholar]
- [91].Blesa JR, Prieto-Ruiz JA, Hernandez JM, Hernandez-Yago J. NRF-2 transcription factor is required for human TOMM20 gene expression. Gene. 2007;391:198–208. doi: 10.1016/j.gene.2006.12.024. [DOI] [PubMed] [Google Scholar]
- [92].Piantadosi CA, Carraway MS, Babiker A, Suliman HB. Heme oxygenase-1 regulates cardiac mitochondrial biogenesis via Nrf2-mediated transcriptional control of nuclear respiratory factor-1. Circ Res. 2008;103:1232–1240. doi: 10.1161/01.RES.0000338597.71702.ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Suliman HB, Sweeney TE, Withers CM, Piantadosi CA. Co-regulation of nuclear respiratory factor-1 by NF{kappa}B and CREB links LPS-induced inflammation to mitochondrial biogenesis. J Cell Sci. 2010;123:2565–2575. doi: 10.1242/jcs.064089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Houten SM, Auwerx J. PGC-1alpha: turbocharging mitochondria. Cell. 2004;119:5–7. doi: 10.1016/j.cell.2004.09.016. [DOI] [PubMed] [Google Scholar]
- [95].Handschin C, Rhee J, Lin J, Tarr PT, Spiegelman BM. An autoregulatory loop controls peroxisome proliferator-activated receptor gamma coactivator 1alpha expression in muscle. Proc Natl Acad Sci U S A. 2003;100:7111–7116. doi: 10.1073/pnas.1232352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. The EMBO journal. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Jager S, Handschin C, St-Pierre J, Spiegelman BM. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Kong X, Wang R, Xue Y, Liu X, Zhang H, Chen Y, Fang F, Chang Y. Sirtuin 3, a new target of PGC-1alpha, plays an important role in the suppression of ROS and mitochondrial biogenesis. PLoS ONE. 2010;5:e11707. doi: 10.1371/journal.pone.0011707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Verdin E, Hirschey MD, Finley LW, Haigis MC. Sirtuin regulation of mitochondria: energy production, apoptosis, and signaling. Trends in biochemical sciences. 2010;35:669–675. doi: 10.1016/j.tibs.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Someya S, Yu W, Hallows WC, Xu J, Vann JM, Leeuwenburgh C, Tanokura M, Denu JM, Prolla TA. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell. 2010;143:802–812. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell metabolism. 2010;12:662–667. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- [102].Tao R, Coleman MC, Pennington JD, Ozden O, Park SH, Jiang H, Kim HS, Flynn CR, Hill S, Hayes McDonald W, Olivier AK, Spitz DR, Gius D. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Molecular cell. 2010;40:893–904. doi: 10.1016/j.molcel.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, Deng CX, Finkel T. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:14447–14452. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Bell EL, Guarente L. The SirT3 divining rod points to oxidative stress. Molecular cell. 2011;42:561–568. doi: 10.1016/j.molcel.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Cosentino F, Francia P, Camici GG, Pelicci PG, Luscher TF, Volpe M. Final common molecular pathways of aging and cardiovascular disease: role of the p66Shc protein. Arteriosclerosis, thrombosis, and vascular biology. 2008;28:622–628. doi: 10.1161/ATVBAHA.107.156059. [DOI] [PubMed] [Google Scholar]
- [106].Giorgio M, Migliaccio E, Orsini F, Paolucci D, Moroni M, Contursi C, Pelliccia G, Luzi L, Minucci S, Marcaccio M, Pinton P, Rizzuto R, Bernardi P, Paolucci F, Pelicci PG. Electron transfer between cytochrome c and p66Shc generates reactive oxygen species that trigger mitochondrial apoptosis. Cell. 2005;122:221–233. doi: 10.1016/j.cell.2005.05.011. [DOI] [PubMed] [Google Scholar]
- [107].Migliaccio E, Giorgio M, Mele S, Pelicci G, Reboldi P, Pandolfi PP, Lanfrancone L, Pelicci PG. The p66shc adaptor protein controls oxidative stress response and life span in mammals. Nature. 1999;402:309–313. doi: 10.1038/46311. [DOI] [PubMed] [Google Scholar]
- [108].Paneni F, Mocharla P, Akhmedov A, Costantino S, Osto E, Volpe M, Luscher TF, Cosentino F. Gene Silencing of the Mitochondrial Adaptor p66Shc Suppresses Vascular Hyperglycemic Memory in Diabetes. Circulation research. 2012;111:278–289. doi: 10.1161/CIRCRESAHA.112.266593. [DOI] [PubMed] [Google Scholar]
- [109].Westermann B. Molecular machinery of mitochondrial fusion and fission. The Journal of biological chemistry. 2008;283:13501–13505. doi: 10.1074/jbc.R800011200. [DOI] [PubMed] [Google Scholar]
- [110].De Palma C, Falcone S, Pisoni S, Cipolat S, Panzeri C, Pambianco S, Pisconti A, Allevi R, Bassi MT, Cossu G, Pozzan T, Moncada S, Scorrano L, Brunelli S, Clementi E. Nitric oxide inhibition of Drp1-mediated mitochondrial fission is critical for myogenic differentiation. Cell death and differentiation. 2010;17:1684–1696. doi: 10.1038/cdd.2010.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Cho DH, Nakamura T, Fang J, Cieplak P, Godzik A, Gu Z, Lipton SA. S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324:102–105. doi: 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Bossy B, Petrilli A, Klinglmayr E, Chen J, Lutz-Meindl U, Knott AB, Masliah E, Schwarzenbacher R, Bossy-Wetzel E. S-Nitrosylation of DRP1 does not affect enzymatic activity and is not specific to Alzheimer’s disease. Journal of Alzheimer’s disease: JAD. 2010;20(Suppl 2):S513–526. doi: 10.3233/JAD-2010-100552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Wagener FA, Volk HD, Willis D, Abraham NG, Soares MP, Adema GJ, Figdor CG. Different faces of the heme-heme oxygenase system in inflammation. Pharmacol Rev. 2003;55:551–571. doi: 10.1124/pr.55.3.5. [DOI] [PubMed] [Google Scholar]
- [114].Maines MD. Heme oxygenase: function, multiplicity, regulatory mechanisms, and clinical applications. Faseb J. 1988;2:2557–2568. [PubMed] [Google Scholar]
- [115].Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the aminoterminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Maines MD. The heme oxygenase system: update 2005. Antioxid Redox Signal. 2005;7:1761–1766. doi: 10.1089/ars.2005.7.1761. [DOI] [PubMed] [Google Scholar]
- [117].Ryter SW, Alam J, Choi AM. Heme oxygenase-1/carbon monoxide: from basic science to therapeutic applications. Physiol Rev. 2006;86:583–650. doi: 10.1152/physrev.00011.2005. [DOI] [PubMed] [Google Scholar]
- [118].Alam J, Igarashi K, Immenschuh S, Shibahara S, Tyrrell RM. Regulation of heme oxygenase-1 gene transcription: recent advances and highlights from the International Conference (Uppsala, 2003) on Heme Oxygenase. Antioxid Redox Signal. 2004;6:924–933. doi: 10.1089/ars.2004.6.924. [DOI] [PubMed] [Google Scholar]
- [119].Alam J, Stewart D, Touchard C, Boinapally S, Choi AM, Cook JL. Nrf2, a Cap’n’Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J Biol Chem. 1999;274:26071–26078. doi: 10.1074/jbc.274.37.26071. [DOI] [PubMed] [Google Scholar]
- [120].Lee BS, Heo J, Kim YM, Shim SM, Pae HO, Kim YM, Chung HT. Carbon monoxide mediates heme oxygenase 1 induction via Nrf2 activation in hepatoma cells. Biochem Biophys Res Commun. 2006;343:965–972. doi: 10.1016/j.bbrc.2006.03.058. [DOI] [PubMed] [Google Scholar]
- [121].Lo SC, Hannink M. PGAM5 tethers a ternary complex containing Keap1 and Nrf2 to mitochondria. Experimental cell research. 2008;314:1789–1803. doi: 10.1016/j.yexcr.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Suliman HB, Carraway MS, Tatro LG, Piantadosi CA. A new activating role for CO in cardiac mitochondrial biogenesis. J Cell Sci. 2007;120:299–308. doi: 10.1242/jcs.03318. [DOI] [PubMed] [Google Scholar]
- [123].Suliman HB, Carraway MS, Ali AS, Reynolds CM, Welty-Wolf KE, Piantadosi CA. The CO/HO system reverses inhibition of mitochondrial biogenesis and prevents murine doxorubicin cardiomyopathy. J Clin Invest. 2007;117:3730–3741. doi: 10.1172/JCI32967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Lee TS, Chau LY. Heme oxygenase-1 mediates the anti-inflammatory effect of interleukin-10 in mice. Nat Med. 2002;8:240–246. doi: 10.1038/nm0302-240. [DOI] [PubMed] [Google Scholar]
- [125].Kruger AL, Peterson SJ, Schwartzman ML, Fusco H, McClung JA, Weiss M, Shenouda S, Goodman AI, Goligorsky MS, Kappas A, Abraham NG. Up-regulation of heme oxygenase provides vascular protection in an animal model of diabetes through its antioxidant and antiapoptotic effects. The Journal of pharmacology and experimental therapeutics. 2006;319:1144–1152. doi: 10.1124/jpet.106.107482. [DOI] [PubMed] [Google Scholar]
- [126].Piantadosi CA, Withers CM, Bartz RR, Macgarvey NC, Fu P, Sweeney TE, Welty-Wolf KE, Suliman HB. Heme oxygenase-1 couples activation of mitochondrial biogenesis to anti-inflammatory cytokine expression. J Biol Chem. 2011 doi: 10.1074/jbc.M110.207738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Schulze-Osthoff K, Bakker AC, Vanhaesebroeck B, Beyaert R, Jacob WA, Fiers W. Cytotoxic activity of tumor necrosis factor is mediated by early damage of mitochondrial functions. Evidence for the involvement of mitochondrial radical generation. J Biol Chem. 1992;267:5317–5323. [PubMed] [Google Scholar]
- [128].Llesuy S, Evelson P, Gonzalez-Flecha B, Peralta J, Carreras MC, Poderoso JJ, Boveris A. Oxidative stress in muscle and liver of rats with septic syndrome. Free radical biology & medicine. 1994;16:445–451. doi: 10.1016/0891-5849(94)90121-x. [DOI] [PubMed] [Google Scholar]
- [129].Taylor DE, Ghio AJ, Piantadosi CA. Reactive oxygen species produced by liver mitochondria of rats in sepsis. Archives of biochemistry and biophysics. 1995;316:70–76. doi: 10.1006/abbi.1995.1011. [DOI] [PubMed] [Google Scholar]
- [130].Kantrow SP, Taylor DE, Carraway MS, Piantadosi CA. Oxidative metabolism in rat hepatocytes and mitochondria during sepsis. Arch Biochem Biophys. 1997;345:278–288. doi: 10.1006/abbi.1997.0264. [DOI] [PubMed] [Google Scholar]
- [131].Kurose I, Miura S, Fukumura D, Yonei Y, Saito H, Tada S, Suematsu M, Tsuchiya M. Nitric oxide mediates Kupffer cell-induced reduction of mitochondrial energization in hepatoma cells: a comparison with oxidative burst. Cancer research. 1993;53:2676–2682. [PubMed] [Google Scholar]
- [132].Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- [133].Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, Takeda K, Akira S. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity. 1999;11:443–451. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- [134].Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- [135].Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- [136].Schreck R, Rieber P, Baeuerle PA. Reactive oxygen intermediates as apparently widely used messengers in the activation of the NF-kappa B transcription factor and HIV-1. The EMBO journal. 1991;10:2247–2258. doi: 10.1002/j.1460-2075.1991.tb07761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Koay MA, Christman JW, Segal BH, Venkatakrishnan A, Blackwell TR, Holland SM, Blackwell TS. Impaired pulmonary NF-kappaB activation in response to lipopolysaccharide in NADPH oxidase-deficient mice. Infect Immun. 2001;69:5991–5996. doi: 10.1128/IAI.69.10.5991-5996.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Figari IS, Mori NA, Palladino MA., Jr. Regulation of neutrophil migration and superoxide production by recombinant tumor necrosis factors-alpha and -beta: comparison to recombinant interferon-gamma and interleukin-1 alpha. Blood. 1987;70:979–984. [PubMed] [Google Scholar]
- [139].Kim HJ, Park KG, Yoo EK, Kim YH, Kim YN, Kim HS, Kim HT, Park JY, Lee KU, Jang WG, Kim JG, Kim BW, Lee IK. Effects of PGC-1alpha on TNF-alpha-induced MCP-1 and VCAM-1 expression and NF-kappaB activation in human aortic smooth muscle and endothelial cells. Antioxidants & redox signaling. 2007;9:301–307. doi: 10.1089/ars.2006.1456. [DOI] [PubMed] [Google Scholar]
- [140].Remels AH, Gosker HR, Schrauwen P, Hommelberg PP, Sliwinski P, Polkey M, Galdiz J, Wouters EF, Langen RC, Schols AM. TNF-alpha impairs regulation of muscle oxidative phenotype: implications for cachexia? The FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2010;24:5052–5062. doi: 10.1096/fj.09-150714. [DOI] [PubMed] [Google Scholar]
- [141].Alvarez-Guardia D, Palomer X, Coll T, Davidson MM, Chan TO, Feldman AM, Laguna JC, Vazquez-Carrera M. The p65 subunit of NF-kappaB binds to PGC-1alpha, linking inflammation and metabolic disturbances in cardiac cells. Cardiovascular research. 2010;87:449–458. doi: 10.1093/cvr/cvq080. [DOI] [PubMed] [Google Scholar]
- [142].St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jager S, Handschin C, Zheng K, Lin J, Yang W, Simon DK, Bachoo R, Spiegelman BM. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- [143].Lu Z, Xu X, Hu X, Fassett J, Zhu G, Tao Y, Li J, Huang Y, Zhang P, Zhao B, Chen Y. PGC-1 alpha regulates expression of myocardial mitochondrial antioxidants and myocardial oxidative stress after chronic systolic overload. Antioxidants & redox signaling. 2010;13:1011–1022. doi: 10.1089/ars.2009.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Vats D, Mukundan L, Odegaard JI, Zhang L, Smith KL, Morel CR, Wagner RA, Greaves DR, Murray PJ, Chawla A. Oxidative metabolism and PGC-1beta attenuate macrophage-mediated inflammation. Cell Metab. 2006;4:13–24. doi: 10.1016/j.cmet.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Kiningham KK, Xu Y, Daosukho C, Popova B, St Clair DK. Nuclear factor kappaB-dependent mechanisms coordinate the synergistic effect of PMA and cytokines on the induction of superoxide dismutase 2. Biochem J. 2001;353:147–156. [PMC free article] [PubMed] [Google Scholar]
- [146].Chen G, Goeddel DV. TNF-R1 signaling: a beautiful pathway. Science. 2002;296:1634–1635. doi: 10.1126/science.1071924. [DOI] [PubMed] [Google Scholar]
- [147].Ogura H, Tsukumo Y, Sugimoto H, Igarashi M, Nagai K, Kataoka T. Ectodomain shedding of TNF receptor 1 induced by protein synthesis inhibitors regulates TNF-alpha-mediated activation of NF-kappaB and caspase-8. Exp Cell Res. 2008;314:1406–1414. doi: 10.1016/j.yexcr.2008.01.019. [DOI] [PubMed] [Google Scholar]
- [148].Kim BC, Kim HT, Mamura M, Ambudkar IS, Choi KS, Kim SJ. Tumor necrosis factor induces apoptosis in hepatoma cells by increasing Ca(2+) release from the endoplasmic reticulum and suppressing Bcl-2 expression. The Journal of biological chemistry. 2002;277:31381–31389. doi: 10.1074/jbc.M203465200. [DOI] [PubMed] [Google Scholar]
- [149].Balaban RS. The role of Ca(2+) signaling in the coordination of mitochondrial ATP production with cardiac work. Biochim Biophys Acta. 2009;1787:1334–1341. doi: 10.1016/j.bbabio.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Rowlands DJ, Islam MN, Das SR, Huertas A, Quadri SK, Horiuchi K, Inamdar N, Emin MT, Lindert J, Ten VS, Bhattacharya S, Bhattacharya J. Activation of TNFR1 ectodomain shedding by mitochondrial Ca2+ determines the severity of inflammation in mouse lung microvessels. The Journal of clinical investigation. 2011;121:1986–1999. doi: 10.1172/JCI43839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Handschin C, Chin S, Li P, Liu F, Maratos-Flier E, Lebrasseur NK, Yan Z, Spiegelman BM. Skeletal muscle fiber-type switching, exercise intolerance, and myopathy in PGC-1alpha muscle-specific knock-out animals. The Journal of biological chemistry. 2007;282:30014–30021. doi: 10.1074/jbc.M704817200. [DOI] [PubMed] [Google Scholar]
- [152].Handschin C, Choi CS, Chin S, Kim S, Kawamori D, Kurpad AJ, Neubauer N, Hu J, Mootha VK, Kim YB, Kulkarni RN, Shulman GI, Spiegelman BM. Abnormal glucose homeostasis in skeletal muscle-specific PGC-1alpha knockout mice reveals skeletal muscle-pancreatic beta cell crosstalk. The Journal of clinical investigation. 2007;117:3463–3474. doi: 10.1172/JCI31785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Handschin C, Spiegelman BM. The role of exercise and PGC1alpha in inflammation and chronic disease. Nature. 2008;454:463–469. doi: 10.1038/nature07206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Handschin C. Peroxisome proliferator-activated receptor-gamma coactivator-1alpha in muscle links metabolism to inflammation. Clinical and experimental pharmacology & physiology. 2009;36:1139–1143. doi: 10.1111/j.1440-1681.2009.05275.x. [DOI] [PubMed] [Google Scholar]
- [155].Wenz T, Rossi SG, Rotundo RL, Spiegelman BM, Moraes CT. Increased muscle PGC-1alpha expression protects from sarcopenia and metabolic disease during aging. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:20405–20410. doi: 10.1073/pnas.0911570106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [156].Suliman HB, Welty-Wolf KE, Carraway M, Tatro L, Piantadosi CA. Lipopolysaccharide induces oxidative cardiac mitochondrial damage and biogenesis. Cardiovasc Res. 2004;64:279–288. doi: 10.1016/j.cardiores.2004.07.005. [DOI] [PubMed] [Google Scholar]
- [157].Crouser ED, Julian MW, Huff JE, Struck J, Cook CH. Carbamoyl phosphate synthase-1: a marker of mitochondrial damage and depletion in the liver during sepsis. Crit Care Med. 2006;34:2439–2446. doi: 10.1097/01.CCM.0000230240.02216.21. [DOI] [PubMed] [Google Scholar]
- [158].Watanabe E, Muenzer JT, Hawkins WG, Davis CG, Dixon DJ, McDunn JE, Brackett DJ, Lerner MR, Swanson PE, Hotchkiss RS. Sepsis induces extensive autophagic vacuolization in hepatocytes: a clinical and laboratory-based study. Lab Invest. 2009;89:549–561. doi: 10.1038/labinvest.2009.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Haden DW, Suliman HB, Carraway MS, Welty-Wolf KE, Ali AS, Shitara H, Yonekawa H, Piantadosi CA. Mitochondrial biogenesis restores oxidative metabolism during Staphylococcus aureus sepsis. Am J Respir Crit Care Med. 2007;176:768–777. doi: 10.1164/rccm.200701-161OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Sweeney TE, Suliman HB, Hollingsworth JW, Piantadosi CA. Differential regulation of the PGC family of genes in a mouse model of Staphylococcus aureus sepsis. PLoS One. 2010;5:e11606. doi: 10.1371/journal.pone.0011606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Suliman HB, Welty-Wolf KE, Carraway MS, Schwartz DA, Hollingsworth JW, Piantadosi CA. Toll-like receptor 4 mediates mitochondrial DNA damage and biogenic responses after heat-inactivated E. coli. Faseb J. 2005;19:1531–1533. doi: 10.1096/fj.04-3500fje. [DOI] [PubMed] [Google Scholar]
- [162].Bartz RR, Suliman HB, Fu P, Welty-Wolf K, Carraway MS, Macgarvey NC, Withers CM, Sweeney TE, Piantadosi CA. Staphylococcus Aureus Sepsis and Mitochondrial Accrual of OGG1 DNA Repair Enzyme in Mice. Am J Respir Crit Care Med. 2010 doi: 10.1164/rccm.200911-1709OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Choumar A, Tarhuni A, Letteron P, Reyl-Desmars F, Dauhoo N, Damasse J, Vadrot N, Nahon P, Moreau R, Pessayre D, Mansouri A. Lipopolysaccharide-Induced Mitochondrial DNA Depletion. Antioxidants & redox signaling. 2011 doi: 10.1089/ars.2010.3713. [DOI] [PubMed] [Google Scholar]
- [164].Suliman HB, Carraway MS, Welty-Wolf KE, Whorton AR, Piantadosi CA. Lipopolysaccharide stimulates mitochondrial biogenesis via activation of nuclear respiratory factor-1. J Biol Chem. 2003;278:41510–41518. doi: 10.1074/jbc.M304719200. [DOI] [PubMed] [Google Scholar]
- [165].Bakkar N, Wang J, Ladner KJ, Wang H, Dahlman JM, Carathers M, Acharyya S, Rudnicki MA, Hollenbach AD, Guttridge DC. IKK/NF-kappaB regulates skeletal myogenesis via a signaling switch to inhibit differentiation and promote mitochondrial biogenesis. J Cell Biol. 2008;180:787–802. doi: 10.1083/jcb.200707179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Sweeney TE, Suliman HB, Hollingsworth JW, Welty-Wolf KE, Piantadosi CA. A toll-like receptor 2 pathway regulates the Ppargc1a/b metabolic co-activators in mice with Staphylococcal aureus sepsis. PLoS ONE. 2011;6:e25249. doi: 10.1371/journal.pone.0025249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Chen ZP, Mitchelhill KI, Michell BJ, Stapleton D, Rodriguez-Crespo I, Witters LA, Power DA, Ortiz de Montellano PR, Kemp BE. AMP-activated protein kinase phosphorylation of endothelial NO synthase. FEBS Lett. 1999;443:285–289. doi: 10.1016/s0014-5793(98)01705-0. [DOI] [PubMed] [Google Scholar]
- [168].Morrow VA, Foufelle F, Connell JM, Petrie JR, Gould GW, Salt IP. Direct activation of AMP-activated protein kinase stimulates nitric-oxide synthesis in human aortic endothelial cells. J Biol Chem. 2003;278:31629–31639. doi: 10.1074/jbc.M212831200. [DOI] [PubMed] [Google Scholar]
- [169].Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Giri S, Nath N, Smith B, Viollet B, Singh AK, Singh I. 5-aminoimidazole-4- carboxamide-1-beta-4-ribofuranoside inhibits proinflammatory response in glial cells: a possible role of AMP-activated protein kinase. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2004;24:479–487. doi: 10.1523/JNEUROSCI.4288-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Cacicedo JM, Yagihashi N, Keaney JF, Jr., Ruderman NB, Ido Y. AMPK inhibits fatty acid-induced increases in NF-kappaB transactivation in cultured human umbilical vein endothelial cells. Biochemical and biophysical research communications. 2004;324:1204–1209. doi: 10.1016/j.bbrc.2004.09.177. [DOI] [PubMed] [Google Scholar]
- [172].Hallows KR, Fitch AC, Richardson CA, Reynolds PR, Clancy JP, Dagher PC, Witters LA, Kolls JK, Pilewski JM. Up-regulation of AMP-activated kinase by dysfunctional cystic fibrosis transmembrane conductance regulator in cystic fibrosis airway epithelial cells mitigates excessive inflammation. The Journal of biological chemistry. 2006;281:4231–4241. doi: 10.1074/jbc.M511029200. [DOI] [PubMed] [Google Scholar]
- [173].Hattori Y, Suzuki K, Hattori S, Kasai K. Metformin inhibits cytokine-induced nuclear factor kappaB activation via AMP-activated protein kinase activation in vascular endothelial cells. Hypertension. 2006;47:1183–1188. doi: 10.1161/01.HYP.0000221429.94591.72. [DOI] [PubMed] [Google Scholar]
- [174].Zhao X, Zmijewski JW, Lorne E, Liu G, Park YJ, Tsuruta Y, Abraham E. Activation of AMPK attenuates neutrophil proinflammatory activity and decreases the severity of acute lung injury. Am J Physiol Lung Cell Mol Physiol. 2008;295:L497–504. doi: 10.1152/ajplung.90210.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Okayasu T, Tomizawa A, Suzuki K, Manaka K, Hattori Y. PPARalpha activators upregulate eNOS activity and inhibit cytokine-induced NF-kappaB activation through AMP-activated protein kinase activation. Life Sci. 2008;82:884–891. doi: 10.1016/j.lfs.2008.02.002. [DOI] [PubMed] [Google Scholar]
- [176].Ko HJ, Zhang Z, Jung DY, Jun JY, Ma Z, Jones KE, Chan SY, Kim JK. Nutrient stress activates inflammation and reduces glucose metabolism by suppressing AMP-activated protein kinase in the heart. Diabetes. 2009;58:2536–2546. doi: 10.2337/db08-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Bai A, Ma AG, Yong M, Weiss CR, Ma Y, Guan Q, Bernstein CN, Peng Z. AMPK agonist downregulates innate and adaptive immune responses in TNBS-induced murine acute and relapsing colitis. Biochem Pharmacol. 2010;80:1708–1717. doi: 10.1016/j.bcp.2010.08.009. [DOI] [PubMed] [Google Scholar]
- [178].Salminen A, Hyttinen JM, Kaarniranta K. AMP-activated protein kinase inhibits NF-kappaB signaling and inflammation: impact on healthspan and lifespan. J Mol Med (Berl) 2011;89:667–676. doi: 10.1007/s00109-011-0748-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Barroso E, Eyre E, Palomer X, Vazquez-Carrera M. The peroxisome proliferator-activated receptor beta/delta (PPARbeta/delta) agonist GW501516 prevents TNF-alpha-induced NF-kappaB activation in human HaCaT cells by reducing p65 acetylation through AMPK and SIRT1. Biochemical pharmacology. 2011;81:534–543. doi: 10.1016/j.bcp.2010.12.004. [DOI] [PubMed] [Google Scholar]
- [180].Gillum MP, Erion DM, Shulman GI. Sirtuin-1 regulation of mammalian metabolism. Trends in molecular medicine. 2010 doi: 10.1016/j.molmed.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Yoshizaki T, Schenk S, Imamura T, Babendure JL, Sonoda N, Bae EJ, Oh DY, Lu M, Milne JC, Westphal C, Bandyopadhyay G, Olefsky JM. SIRT1 inhibits inflammatory pathways in macrophages and modulates insulin sensitivity. American journal of physiology. Endocrinology and metabolism. 2010;298:E419–428. doi: 10.1152/ajpendo.00417.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- [183].Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- [184].Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, Fitzgerald KA, Ryter SW, Choi AM. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nature immunology. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Zhang J, Ney PA. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009;16:939–946. doi: 10.1038/cdd.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Lowell BB, Shulman GI. Mitochondrial dysfunction and type 2 diabetes. Science. 2005;307:384–387. doi: 10.1126/science.1104343. [DOI] [PubMed] [Google Scholar]
- [187].Rains JL, Jain SK. Oxidative stress, insulin signaling, and diabetes. Free radical biology & medicine. 2011;50:567–575. doi: 10.1016/j.freeradbiomed.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature genetics. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- [189].Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, Kashyap S, Miyazaki Y, Kohane I, Costello M, Saccone R, Landaker EJ, Goldfine AB, Mun E, DeFronzo R, Finlayson J, Kahn CR, Mandarino LJ. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Starkov AA, Fiskum G, Chinopoulos C, Lorenzo BJ, Browne SE, Patel MS, Beal MF. Mitochondrial alpha-ketoglutarate dehydrogenase complex generates reactive oxygen species. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2004;24:7779–7788. doi: 10.1523/JNEUROSCI.1899-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Bonnard C, Durand A, Peyrol S, Chanseaume E, Chauvin MA, Morio B, Vidal H, Rieusset J. Mitochondrial dysfunction results from oxidative stress in the skeletal muscle of diet-induced insulin-resistant mice. The Journal of clinical investigation. 2008;118:789–800. doi: 10.1172/JCI32601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Ruderman NB, Saha AK. Metabolic syndrome: adenosine monophosphate-activated protein kinase and malonyl coenzyme A. Obesity (Silver Spring) 2006;14(Suppl 1):25S–33S. doi: 10.1038/oby.2006.279. [DOI] [PubMed] [Google Scholar]
- [193].Cory S, Huang DC, Adams JM. The Bcl-2 family: roles in cell survival and oncogenesis. Oncogene. 2003;22:8590–8607. doi: 10.1038/sj.onc.1207102. [DOI] [PubMed] [Google Scholar]
- [194].Danial NN, Gramm CF, Scorrano L, Zhang CY, Krauss S, Ranger AM, Datta SR, Greenberg ME, Licklider LJ, Lowell BB, Gygi SP, Korsmeyer SJ. BAD and glucokinase reside in a mitochondrial complex that integrates glycolysis and apoptosis. Nature. 2003;424:952–956. doi: 10.1038/nature01825. [DOI] [PubMed] [Google Scholar]
- [195].Russell LK, Mansfield CM, Lehman JJ, Kovacs A, Courtois M, Saffitz JE, Medeiros DM, Valencik ML, McDonald JA, Kelly DP. Cardiac-specific induction of the transcriptional coactivator peroxisome proliferator-activated receptor gamma coactivator-1alpha promotes mitochondrial biogenesis and reversible cardiomyopathy in a developmental stage-dependent manner. Circ Res. 2004;94:525–533. doi: 10.1161/01.RES.0000117088.36577.EB. [DOI] [PubMed] [Google Scholar]
- [196].Brandtzaeg P, Osnes L, Ovstebo R, Joo GB, Westvik AB, Kierulf P. Net inflammatory capacity of human septic shock plasma evaluated by a monocyte-based target cell assay: identification of interleukin-10 as a major functional deactivator of human monocytes. J Exp Med. 1996;184:51–60. doi: 10.1084/jem.184.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Lin CY, Tsai IF, Ho YP, Huang CT, Lin YC, Lin CJ, Tseng SC, Lin WP, Chen WT, Sheen IS. Endotoxemia contributes to the immune paralysis in patients with cirrhosis. J Hepatol. 2007;46:816–826. doi: 10.1016/j.jhep.2006.12.018. [DOI] [PubMed] [Google Scholar]
- [198].Wang TS, Deng JC. Molecular and cellular aspects of sepsis-induced immunosuppression. J Mol Med (Berl) 2008;86:495–506. doi: 10.1007/s00109-007-0300-4. [DOI] [PubMed] [Google Scholar]
- [199].Biswas SK, Lopez-Collazo E. Endotoxin tolerance: new mechanisms, molecules and clinical significance. Trends in immunology. 2009;30:475–487. doi: 10.1016/j.it.2009.07.009. [DOI] [PubMed] [Google Scholar]
- [200].Reddy RC, Standiford TJ. Effects of sepsis on neutrophil chemotaxis. Curr Opin Hematol. 2010;17:18–24. doi: 10.1097/MOH.0b013e32833338f3. [DOI] [PubMed] [Google Scholar]