ABSTRACT
Cysteine peptidases (CPs) of Entamoeba histolytica are considered to be important pathogenicity factors. Previous studies have found that under standard axenic culture conditions, only four (ehcp-a1, ehcp-a2, ehcp-a5, and ehcp-a7) out of 35 papain-like ehcp genes present in the E. histolytica genome are expressed at high levels. Little is known about the expression of CPs in E. histolytica during amoebic liver abscess (ALA) formation. In the current study, a quantitative real-time PCR assay was developed to determine the expression of the various ehcp genes during ALA formation in animal models. Increased expression of four ehcp genes (ehcp-a3, -a4, -a10, and -c13) was detected in the gerbil and mouse models. Increased expression of another three ehcp genes (ehcp-a5, -a6, and -a7) was detected in the mouse model only, and two other ehcp genes (ehcp-b8 and -b9) showed increased expression in the gerbil model only. Trophozoites of the nonpathogenic E. histolytica HM-1:IMSS clone A1, which was unable to induce ALAs, were transfected with vectors enabling overexpression of those CPs that are expressed at high levels under culture conditions or during ALA formation. Interestingly, overexpression of ehcp-b8, -b9, and -c13 restored the pathogenic phenotype of the nonpathogenic clone A1 whereas overexpression of various other peptidase genes had no effect on the pathogenicity of this clone.
IMPORTANCE
Entamoeba histolytica is a widespread and clinically important protozoan parasite. It normally exists in the human intestine without causing clinical symptoms but can invade the intestinal mucosa, which causes serious intestinal (amoebic colitis) and extraintestinal (amoebic liver abscess [ALA]) diseases. The identification of factors responsible for the invasion of the parasite and disease formation is a major topic in the field. Here, we investigate the roles of different papain-like cysteine peptidases (CPs) as pathogenicity factors. We show that the expression of some of the peptidases that are normally expressed at low levels increases during ALA formation. Furthermore, nonpathogenic amoebae can be transformed to pathogenic amoebae, simply by specific overexpression of some of these CPs. Our findings reinforce the importance of CPs as pathogenicity factors of E. histolytica.
Introduction
Entamoeba histolytica is an important pathogen responsible for millions of cases of invasive amoebiasis each year (1). This protozoan parasite passes through a simple life cycle consisting of an infectious cyst that can survive outside the host and a vegetative trophozoite that proliferates in the human gut. The trophozoites of E. histolytica can persist in the gut for months or even years, causing asymptomatic luminal gut infections. However, sometimes E. histolytica penetrates the intestinal mucosa and destroys host tissues. Parasite penetration leads to ulcerative colitis or invasion of other organs, most commonly the liver, and may result in abscess formation.
Identification of factors that are responsible for the pathogenicity of E. histolytica has been a major topic in the field. During recent years, numerous in vitro and in vivo studies have suggested that cysteine peptidases (CPs) play an important role in the pathogenicity of E. histolytica (2–9). The most convincing are from infections of laboratory animals, indicating that E. histolytica trophozoites with reduced CP activity are much less able to induce amoebic disease (9, 10). In addition, overexpression of CPs leads to an increase in amoebic liver abscess (ALA) formation in gerbils (11). To date, only four of the 35 papain-like ehcp genes present in the E. histolytica genome (ehcp-a1, -a2, -a5, and -a7) have been shown to be highly expressed in amoeba trophozoites under standard axenic culture conditions (12–14). Of these, EhCP-A5 is thought to be a major pathogenicity factor, with possible involvement in colon invasion, induction of host inflammatory responses, and ALA formation (10, 11, 15, 16).
However, little is known about the function and regulation of the majority of CPs that are expressed at low levels in axenic culture or about their involvement in excystation and encystation, colon invasion, and most importantly, ALA formation. Several CPs exhibit cyst-specific expression (ehcp-a3, -a4, -a8, -b1, -b3, -b8, -b9, and -b10), with the majority showing low levels of expression in the trophozoite stage in axenic culture (17, 18). In addition, intestinal invasion resulted in a change in the expression of ehcp genes in E. histolytica isolated from mouse ceca, with four ehcp genes (ehcp-a1, -a4, -a6, and -a8) showing increased expression and one (ehcp-a7) showing decreased expression (19). In a proteomic approach analyzing the composition of E. histolytica uropods, the CPs EhCP-A1, -A2, -A4, -C4, -C5, -C6, and -C13 were identified (20). Interestingly, this was the first time that proteins belonging to the EhCP-C family were detected in E. histolytica. In contrast, very little information about CP expression during ALA formation is available. Freitas and colleagues compared the expression levels of ehcp-a5 in ALA samples and axenic cultivated strains and found a higher level of ehcp-a5 mRNA in amoebae derived directly from ALAs (21). Nevertheless, information is lacking on the expression profiles of the entire set of CPs during abscess formation.
In our laboratory, two E. histolytica cell lines (and their derived clones), which differ substantially in their pathogenic properties, are available. Cell line B and its derived clone B2 are highly pathogenic, as evidenced by their abilities to produce considerable ALAs, whereas cell line A and its derived clone A1 are unable to induce ALAs (22) (see Fig. S1 in the supplemental material). We used the pathogenic cell line B and clone B2 to analyze the expression profiles of CPs during ALA formation. For this, we developed a quantitative real-time PCR (qRT-PCR) assay to detect ehcp transcripts directly in ALA material. In total, nine different CP genes showed increased expression during abscess formation in both or either of the two different animal models used, mouse and gerbil. Interestingly, overexpression of three of these CPs in the nonpathogenic clone A1 was sufficient to confer pathogenicity.
RESULTS
Expression of E. histolytica ehcp genes directly derived from ALAs.
To date, the expression of ehcp genes in E. histolytica during ALA formation has been little investigated. Therefore, E. histolytica trophozoites from the pathogenic cell line B and the pathogenic clone B2 were inoculated into gerbil and mice livers, respectively (22). To ensure that no mixture of different cell types was used, cell line B was cloned. All further experiments were performed with clone B2. Using a qRT-PCR approach, ehcp mRNA levels in axenic cultivated trophozoites were compared with those in ALAs in mice and gerbils at different time points after infection. For mice, ehcp gene expression was determined 24, 48, and 72 h postinfection (hpi), whereas for gerbils, ehcp expression was determined at one time point, seven days postinfection (dpi).
Under axenic culture conditions, the ehcp expression profiles were very similar in cell line B and clone B2. The expression of only a few of the analyzed CPs was slightly higher in cell line B than in clone B2 (Table 1). In both, the major expressed ehcp genes were ehcp-a1, -a2, -a5, and -a7. All other ehcp genes were expressed at moderate or low levels, with ehcp-b8 showing the lowest expression levels.
TABLE 1 .
Expression of ehcp genes in trophozoites of cell line B, clone B2, and clone A1 cultivated under axenic conditions
| ehcp gene | GenBank accession no. | ΔCTa |
||
|---|---|---|---|---|
| Cell line B | Clone B2 | Clone A1 | ||
| ehcp-a1 | XM_645064 | −0.91 ± 0.68 | 0.36 ± 0.74 | 0.20 ± 0.24 |
| ehcp-a2 | XM_645550 | −0.92 ± 0.68 | 0.37 ± 0.52 | 0.58 ± 0.30 |
| ehcp-a3 | XM_648162 | 10.73 ± 1.85 | 12.84 ± 0.45 | 11.25 ± 0.26 |
| ehcp-a4 | XM_651510 | 4.74 ± 0.80 | 5.69 ± 0.58 | 5.01 ± 1.07 |
| ehcp-a5 | XM_645845 | 2.06 ± 0.46 | 2.29 ± 0.70 | 1.84 ± 0.39 |
| ehcp-a6 | XM_652272 | 7.37 ± 0.29 | 7.54 ± 0.92 | 5.18 ± 0.42 |
| ehcp-a7 | XM_643904 | 1.75 ± 0.88 | 2.45 ± 0.81 | 3.36 ± 0.52 |
| ehcp-a8 | XM_652354 | 8.21 ± 1.36 | 8.77 ± 0.33 | 8.5 ± 0.13 |
| ehcp-a9 | XM_650583 | ND | 11.18 ± 0.49 | 10.32 ± 0.17 |
| ehcp-a10 | XM_646055 | 8.27 ± 0.15 | 9.51 ± 0.85 | 8.76 ± 0.22 |
| ehcp-a11 | XM_646598 | 4.67 ± 0.59 | 5.52 ± 0.79 | 4.93 ± 0.04 |
| ehcp-a12 | XM_648731 | 9.66 ± 0.23 | 9.61 ± 0.65 | 8.97 ± 0.11 |
| ehcp-a13 | Not annotated | 6.45 ± 0.30 | 7.44 ± 0.95 | 6.58 ± 0.12 |
| ehcp-b1 | XM_646489 | 14.17 ± 2.44 | 14.38 ± 0.67 | 11.4 ± 0.46 |
| ehcp-b2 | XM_001914414 | ND | 14.56 ± 1.20 | 9.3 ± 0.38 |
| ehcp-b3 | XM_651655 | 10.52 ± 2.10 | 12.53 ± 0.55 | 11.76 ± 0.4 |
| ehcp-b4 | XM_643409 | 5.66 ± 0.30 | 7.17 ± 0.72 | 6.2 ± 0.1 |
| ehcp-b5 | XM_647579 | 10.13 ± 0.29 | 10.44 ± 0.89 | 9.46 ± 0.14 |
| ehcp-b7 | XM_645308 | 8.02 ± 0.84 | 8.86 ± 0.65 | 10.68 ± 0.16 |
| ehcp-b8 | XM_645957 | 18.54 ± 0.52 | 17.65 ± 0.47 | 18.31 ± 0.53 |
| ehcp-b9 | XM_647901 | 10.06 ± 0.06 | 10.37 ± 0.40 | 10.3 ± 0.21 |
| ehcp-b10 | XM_643214 | ND | 12.97 ± 4.09 | 14.95 ± 0.92 |
| ehcp-c1 | XM_649361 | 9.93 ± 0.92 | 9.47 ± 0.61 | 8.3 ± 0.23 |
| ehcp-c2 | XM_651540 | ND | 9.93 ± 2.36 | 6.91 ± 0.57 |
| ehcp-c3 | XM_650036 | ND | 7.82 ± 1.05 | 6.62 ± 0.91 |
| ehcp-c4 | XM_650708 | 4.79 ± 0.87 | 4.63 ± 0.74 | 4.48 ± 0.02 |
| ehcp-c5 | XM_649708 | 4.99 ± 0.47 | 5.98 ± 1.44 | 5.77 ± 0.28 |
| ehcp-c6 | XM_646461 | ND | 12.94 ± 3.3 | 7.5 ± 0.02 |
| ehcp-c9 | XM_649919 | ND | 7.40 ± 0.91 | 5.85 ± 0.02 |
| ehcp-c11 | XM_642991 | ND | 13.94 ± 3.37 | 9.79 ± 0.66 |
| ehcp-c12 | XM_645737 | ND | 10.14 ± 1.31 | 8.22 ± 1.1 |
| ehcp-c13 | XM_651464 | 7.19 ± 0.31 | 7.93 ± 0.54 | 6.16 ± 0.01 |
| ehicp1 | XM_648163 | ND | 0.92 ± 0.06 | 0.89 ± 0.22 |
| ehicp2 | XM_644271 | ND | 2.2 ± 0.09 | 1.89 ± 0.13 |
Concentration relative to ehactin, which was used as a normalizer. ND, not determined; ΔCT, threshold cycle. At least two biological replicates were analyzed in duplicate.
During ALA formation, the expression of only a few ehcp genes was up- or downregulated relative to their expression in axenic culture. In trophozoites derived from ALAs in gerbils, the expression of six (ehcp-a3, -a4, -a10, -b8, -b9, and -c13) of the 23 ehcp genes analyzed increased significantly by 4- to 56-fold (Table 2). Similarly, the expression of four of the same genes, ehcp-a3, -a4, -a10, and -c13, increased by 3- to 345-fold in trophozoites derived from ALAs in mice. In addition, the expression of other ehcp genes, ehcp-a5 (3- to 8-fold), ehcp-a6 (3- to 12-fold), and ehcp-a7 (4- to 86-fold), was also increased in mice. In detail, for ehcp-a3 and ehcp-a7, the increase was detected at 24 hpi, the first time point analyzed. For ehcp-a5 and ehcp-a6, the increase was first detected at 48 hpi. For ehcp-a10, the increase was detected at 24 hpi in one biological sample and at 48 hpi in the other biological sample (Table 2). For ehcp-c13, for which increased expression was detected in amoebae derived from ALAs in gerbils at seven dpi, an approximately 3-fold increase was detected in amoebae derived from ALAs in mice at 72 hpi. For eight of the 32 ehcp genes analyzed in ALAs of mice, no qRT-PCR product was detected, presumably because there was a low proportion of amoebae relative to hepatic cells within the abscess material as well as low levels of expression of these genes.
TABLE 2 .
Expression of ehcp genes in trophozoites derived from ALAs of gerbils and mice analyzed by qRT-PCR
| ehcp gene | GenBank accession no. | ΔΔCTa |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cell line B (axenic culture) calibrator 7 dpi |
Cell line B gerbil ALA 7 dpib |
Clone B2 (axenic culture) calibrator 7 dpi |
Clone B2 mouse ALA |
|||||||
| 24 hpi |
48 hpi |
72 hpi |
||||||||
| I | II | I | II | I | II | |||||
| ehcp-a1 | XM_645064 | 1 | 2.35 ±0.91 | 1 | 1.53 | 0.84 | 1.08 | 0.55 | 1.69 | 2.67 |
| ehcp-a2 | XM_645550 | 1 | 0.47 ±0.10 | 1 | 0.90 | 0.64 | 0.55 | 1.12 | 0.32 | 0.31 |
| ehcp-a3 | XM_648162 | 1 | 55.59 ±33.42* | 1 | 8.56 | 345.2 | 8.92 | 28.8 | 10.89 | 8.05 |
| ehcp-a4 | XM_651510 | 1 | 16.01 ± 11.94* | 1 | 0.79 | 1.74 | 1.93 | 1.09 | 7.98 | 5.84 |
| ehcp-a5 | XM_645845 | 1 | 1.48 ± 0.99 | 1 | 2.52 | 1.11 | 3.18 | 5.04 | 4.06 | 8.04 |
| ehcp-a6 | XM_652272 | 1 | 2.18 ± 0.32 | 1 | 1.60 | 1.30 | 5.73 | 3.36 | 3.2 | 12.49 |
| ehcp-a7 | XM_643904 | 1 | 2.15 ± 0.52 | 1 | 9.78 | 86.23 | 4.11 | 13.51 | 13.39 | 25.0 |
| ehcp-a8 | XM_652354 | 1 | 1.33 ± 0.47 | 1 | 1.26 | 0.42 | 2.50 | 1.64 | NA | 0.66 |
| ehcp-a9 | XM_650583 | ND | ND | 1 | 1.68 | 1.89 | 2.73 | 2.81 | 2.56 | 1.55 |
| ehcp-a10 | XM_646055 | 1 | 18.45 ± 2.35* | 1 | 2.09 | 20.22 | 5.91 | 12.42 | NA | NA |
| ehcp-a11 | XM_646598 | 1 | 0.61 ± 0.40 | 1 | 0.29 | 0.54 | 1.45 | 1.19 | 1.33 | 0.53 |
| ehcp-a12 | XM_648731 | 1 | 0.83 | 1 | 0.32 | NA | 1.05 | 0.38 | NA | 2.16 |
| ehcp-a13 | Not annotated | 1 | 1.17 ± 0.62 | 1 | 1.07 | NA | 1.4 | 0.77 | NA | NA |
| ehcp-b1 | XM_646489 | 1 | NA | 1 | NA | NA | NA | NA | NA | NA |
| ehcp-b2 | XM_001914414 | ND | ND | 1 | NA | NA | NA | NA | NA | NA |
| ehcp-b3 | XM_651655 | 1 | 2.5 ± 0.33 | 1 | 1.43 | 1.36 | NA | NA | NA | NA |
| ehcp-b4 | XM_643409 | 1 | 0.52 ± 0.33 | 1 | 1.65 | NA | 1.12 | 0.61 | 1.52 | 2.06 |
| ehcp-b5 | XM_647579 | 1 | 1.61 ± 0.83 | 1 | 0.87 | 0.82 | 1.98 | 2.33 | NA | 2.30 |
| ehcp-b7 | XM_645308 | 1 | 0.6 ± 0.42 | 1 | 1.51 | 0.27 | 0.86 | 0.87 | 0.67 | 0.81 |
| ehcp-b8 | XM_645957 | 1 | 116.6 ± 77.37* | 1 | NA | NA | NA | NA | NA | NA |
| ehcp-b9 | XM_647901 | 1 | 11.08 ± 4.0* | 1 | 1.09 | 0.69 | 0.61 | 1.34 | 2.49 | 1.45 |
| ehcp-b10 | XM_643214 | ND | ND | 1 | NA | NA | NA | NA | NA | NA |
| ehcp-c1 | XM_649361 | 1 | 0.8 ± 0.5 | 1 | 0.63 | 0.8 | NA | NA | NA | 2.02 |
| ehcp-c2 | XM_651540 | ND | ND | 1 | NA | NA | NA | NA | NA | NA |
| ehcp-c3 | XM_650036 | ND | ND | 1 | NA | NA | NA | NA | NA | NA |
| ehcp-c4 | XM_650708 | 1 | 2.49 ± 0.95 | 1 | 1.50 | 1.20 | 3.14 | 2.32 | 1.92 | 2.59 |
| ehcp-c5 | XM_649708 | 1 | 0.63 ± 0.32 | 1 | 0.51 | 0.32 | 0.5 | 0.38 | 0.23 | 0.35 |
| ehcp-c6 | XM_646461 | ND | ND | 1 | NA | 2.76 | NA | NA | NA | NA |
| ehcp-c9 | XM_649919 | ND | ND | 1 | 1.27 | 1.14 | 1.28 | 1.33 | NA | 1.42 |
| ehcp-c11 | XM_642991 | ND | ND | 1 | NA | NA | NA | NA | NA | NA |
| ehcp-c12 | XM_645737 | ND | ND | 1 | NA | 1.1 | NA | NA | 2.34 | 1.26 |
| ehcp-c13 | XM_651464 | 1 | 4.25 ± 1.88* | 1 | 2.75 | 1.1 | 2.49 | 2.04 | 3.04 | 2.93 |
Relative differences in gene expression; ehactin was used as a normalizer. Bold indicates differentially expressed genes. NA, no amplificate in respective qRT-PCR; ALA, amoebic liver abscess; ND, not determined; I and II indicate two different animals.
The threshold for defining genes as being differentially expressed was set at 3.0. The expression of the various ehcp genes of amoebae derived from gerbil livers was analyzed at least two times in duplicate. P values of differentially expressed genes are expressed as follows: *, P < 0.05.
Generation of transgenic E. histolytica overexpressing various ehcp genes.
To determine if the ehcp genes that are upregulated during ALA formation are involved in pathogenicity, transfectants of the nonpathogenic E. histolytica clone A1 that specifically overexpressed various peptidase genes were generated. In an earlier study, it was shown that cell line B has a total CP activity approximately ten times higher than that of cell line A (110 and 15 mU/mg, respectively) (22). This correlates well with the CP activity measured for the cell line B-derived clone B2 (123 ± 60 mU/mg) and the cell line A-derived clone A1 (14 ± 10 mU/mg). Surprisingly, the ehcp expression profiles of the two clones were very similar, and strong differences in total CP activity were not reflected at the mRNA level (Table 1). The observed discrepancy cannot be attributed to a differential expression of the genes coding for the CP inhibitors (ehicp1 and ehicp2) (Table 1).
Trophozoites of the nonpathogenic clone A1 were transfected with either an empty expression vector (pNC; control) or one of 11 different expression vectors containing the open reading frame for ehcp-a1, -a2, -a3, -a4, -a5, -a6, -a7, -a10, -b8, -b9, or -c13 (Fig. 1). After transfection, the trophozoites were cloned and at least four clones per vector were analyzed for overexpression of the relevant ehcp gene by qRT-PCR. For each transfectant, the clone with the highest ehcp expression was used for all further experiments (Table 3). After transfection of clone A1 with the control vector, pNC, the CP activity dropped from approximately 14 mU/mg to 0.5 mU/mg, as measured using an enzymatic CP assay with Z-Arg-Arg as a substrate (Table 3). It may be that selection of the transfectants with the protein synthesis inhibitor G418 was responsible for this. With two exceptions (pNC-CPA3 and pNC-CPA6 transfectants), the overexpression of particular ehcp genes led to a significantly increased CP activity for all transfectants in vitro (Table 3). Three transfectants (pNC-CPA10, -CPB9, and -CPC13) showed a 4- to 8-fold increase in activity compared to the control, another three transfectants (pNC-CPA4, -CPA7, and -CPB8) showed a 10- to 15-fold increase, and three further transfectants (pNC-CPA1, -CPA2, and -CPA5) showed a 50- to 140-fold increase in activity.
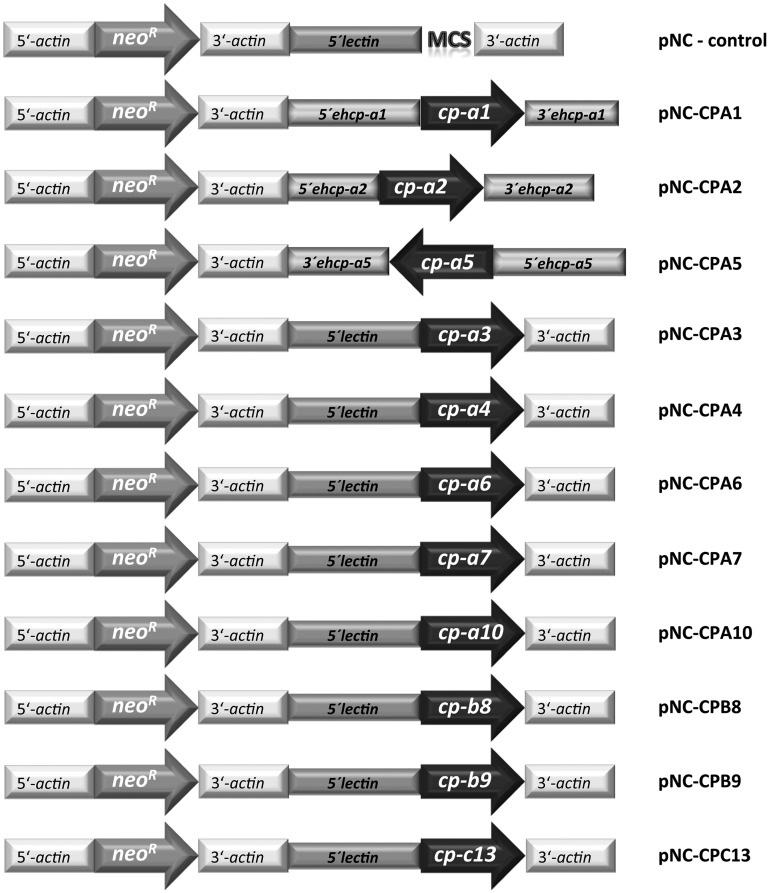
FIG 1.
Schematic of plasmid vectors used for stable episomal transfection of E. histolytica clone A1 trophozoites. Neomycin phosphotransferase (neoR) flanked by the 5′- and 3′-untranslated sequence of an E. histolytica actin gene was used as a selectable marker. Ehcp-a1, -a2, and -a5 are flanked by the respective gene-specific 3′- and 5′-untranslated regions (11). For all other plasmids, the coding sequences of the ehcp genes are flanked by the 5′-untranslated sequence of an E. histolytica lectin gene and by a 3′-untranslated sequence of an E. histolytica actin gene. MCS, multiple cloning site.
TABLE 3 .
Expression and CP activity of clone A1 transfectants overexpressing various ehcp genes
| Clone A1 transfectant | Relative expression (ΔΔCT method)a | CP activity |
|
|---|---|---|---|
| mU/mg | Fold activity to control (P value) | ||
| pNC | 1 | 0.49 ± 0.47 | 1 |
| pNC-CPA1 | 14.1 ± 13 | 24.3 ± 1.1 | 49.6 (<0.0001)*** |
| pNC-CPA2 | 6.1 ± 1.7 | 69.5 ± 19.4 | 141.8 (0.0012)** |
| pNC-CPA3 | 22.20 ± 20.35 | 1.2 ± 0.38 | 2.4 (NS) |
| pNC-CPA4 | 387.5 ± 94.5 | 7.7 ± 2.9 | 15.7 (0.0007)*** |
| pNC-CPA5 | 3.16 ± 2.01 | 30.6 ± 18.3 | 62.4 (0.0011)** |
| pNC-CPA6 | 119.2 ± 21.7 | 0.9 ± 0.6 | 1.8 (NS) |
| pNC-CPA7 | 7 ± 4.4 | 5.2 ± 4.5 | 10.6 (0.0080)** |
| pNC-CPA10 | 642.4 ± 54.1 | 2.9 ± 2.4 | 5.9(0.0043)** |
| pNC-CPB8 | 9,370 ± 13,028 | 3.6 ± 2.9 | 7.3(0.043)* |
| pNC-CPB9 | 589 ± 1,097 | 3.8 ± 3.6 | 7.8(0.0173)* |
| pNC-CPC13 | 19.83 ± 12.17 | 2.1 ± 0.9 | 4.3(0.0043)** |
Relative expression of the ehcp gene that should be overexpressed in the respective transfectant was analyzed. Ehactin was used as a normalizer. NS, not significant; CP, cysteine peptidases.
In addition, for none of the transfectants could activity against any of the four additionally used chromogenic substrates (Z-Ala-Ala, Z-Phe-Ala, Z-Gly-Ala, Z-Ala-Pro) be detected.
Furthermore, the CPs of the transfectants were analyzed using substrate gel electrophoresis. For the nontransfected pathogenic clone B2, the major activity bands were easily assigned to EhCP-A1, EhCP-A2, and EhCP-A5 (Fig. 2, far left lane). For the nonpathogenic clone A1, all of the activity bands were much weaker (Fig. 2, lane second from left). No activity bands were detected for the control transfectant, pNC (Fig. 2, third lane from the left), a finding which is in good agreement with its low CP activity (Table 3). This was also the case for the transfectants pNC-CPA3, -CPA6, -CPA10, -CPB8, -CPB9, and -CPC13, all of which had specific peptidolytic activities below 4 mU/mg (Fig. 2; Table 3). In contrast, an increase in the intensity of the band representing the transfected gene was observed for the transfectants pNC-CPA1, -CPA2, -CPA4, and -CPA7 (Fig. 2), and their specific peptidolytic activities were between 5 and 70 mU/mg (Table 3). For pNC-CPA1 transfectants, an increase in the intensity of the band representing EhCP-A2 was also observed. As already described in a previous study, transfection of E. histolytica with pNC-CPA5 leads to an increase in intensity of several peptidase activity bands (EhCP-A1, -A2, -A4, -A5, and -A7) (11).
FIG 2.
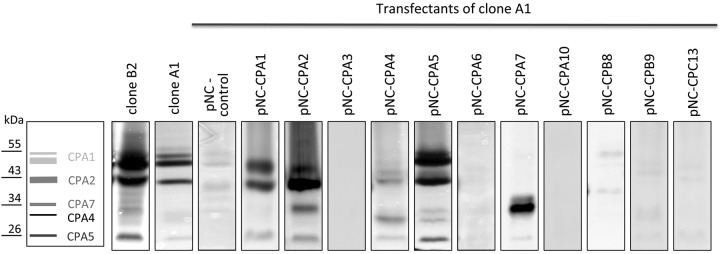
Substrate gel electrophoresis of E. histolytica clone B2, clone A1, and transgenic clone A1 trophozoites overexpressing various ehcp genes. Trophozoites of E. histolytica clone A1 were transfected with episomal plasmids as indicated and selected with G418. Subsequently, lysates (4 µg protein/ml) of the cells were separated on SDS-PAGE gels copolymerized with gelatin. To visualize the CP activity of proteins, gels were stained with Coomassie blue and the picture was inverted. Standards are indicated in kDa on the left.
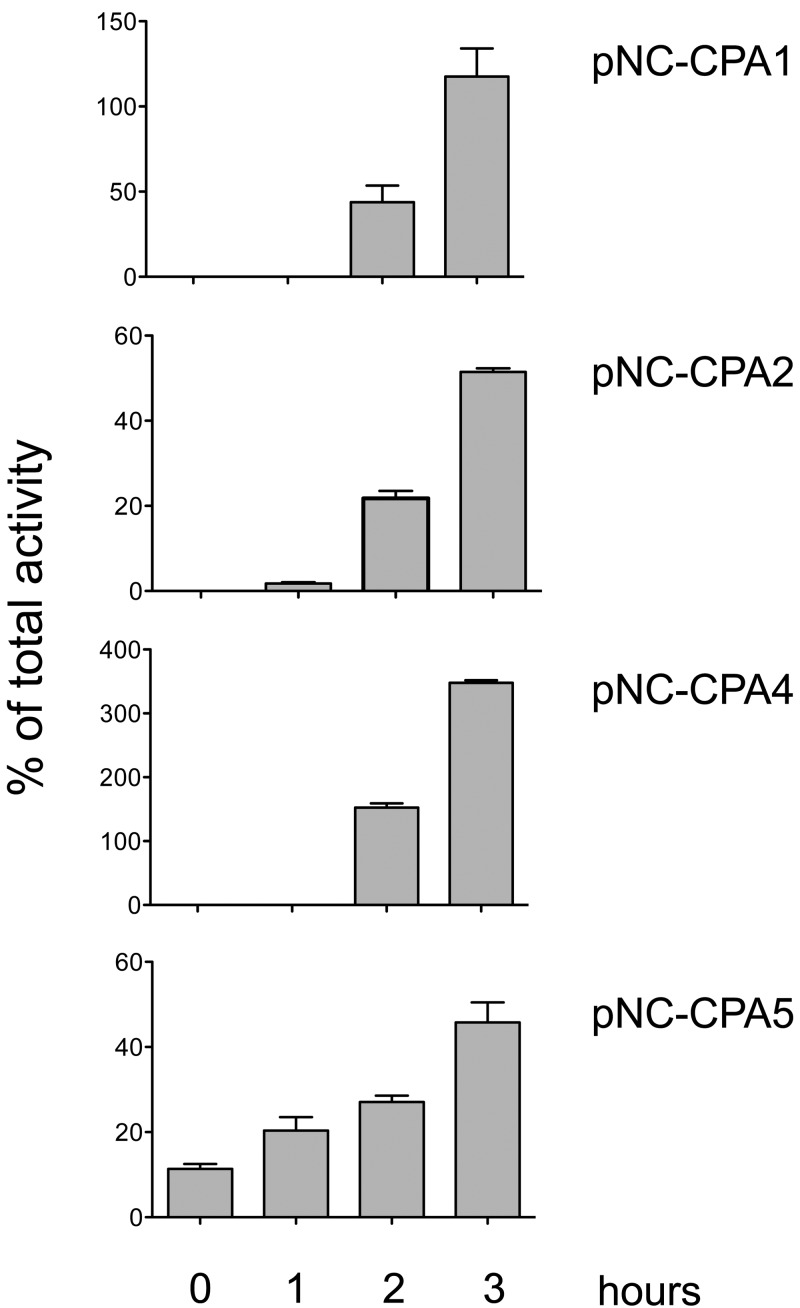
A secretion of CPs over time was able to be verified for the transfectants pNC-CPA1, -CPA2, -CPA4, and -CPA5 (Fig. 3). These four are the transfectants with the highest CP activities (15- to 140-fold higher than that of the control). Moreover, the respective overexpressed cysteine peptidases have already been described as being secreted (23–25). A peptidase release was also described for EhCP-B9 (24). Thus, it cannot be excluded that the lack of detectable secretion of some of the transfectants is due to a low total CP activity.
FIG 3.
To determine the secretion of CPs, 1 × 106 trophozoites were suspended in 500 µl TYI_secretion medium. After 0 min, 1 h, 2 h, and 3 h of incubation, the amoebae were sedimented and the supernatant was removed to measure CP activity. To determine the total CP activity, soluble extracts of 1 × 106 trophozoites/500 µl were generated. Secretion was outlined as the percentage of total activity. Each time point was measured at least two times in duplicate.
ALA formation in mice using transfectants of the nonpathogenic clone A1 overexpressing different ehcp genes.
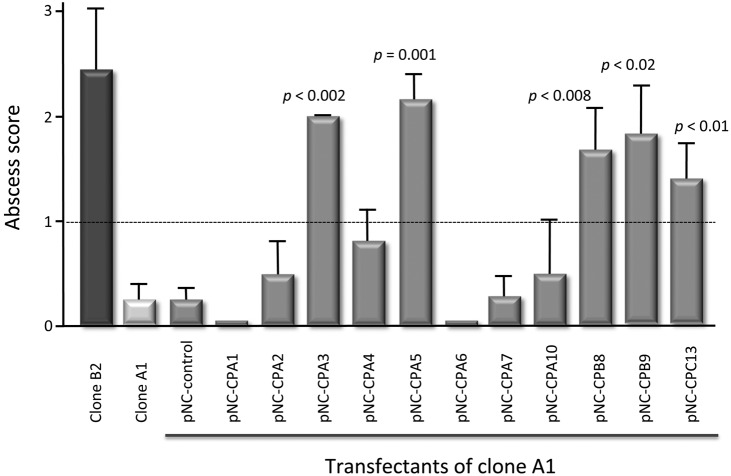
It was previously shown that cell line B forms abscesses in both the gerbil model and the immune-competent mouse model whereas cell line A is nonpathogenic (22, 26). Similar results were obtained for clone B2 and clone A1 in the mouse model in the current study. Infections with trophozoites of clone B2 induced ALAs between 3 and 4 mm in diameter (Fig. 4) (abscess score, 2.4 ± 1.3), whereas no ALA formation was observed seven days postinfection using clone A1. Transfection of clone A1 with the control vector pNC did not induce any abscess formation (abscess score, 0.2 ± 0.4). This was also true for the transfectants overexpressing ehcp-a1, -a2, -a4, -a6, -a7, and -a10 (all had abscess scores of ≤1). In contrast, transfection with the plasmids pNC-CPA3, -CPA5, -CPB8, -CPB9, and -CPC13 made clone A1 pathogenic. For all these transfectants, ALA formation was significantly higher (abscess scores of between 1.4 and 2.1) than ALA formation in the pNC transfectant (Fig. 4).
FIG 4.
Abscess formation in mouse livers following infection with E. histolytica clone B2 or clone A1 trophozoites as well as various transgenic trophozoites overexpressing various ehcp genes. Data are shown in the bar graphs as means ± standard errors of the means (SEM). For each transfectant, ALA formation in 5 to 15 animals was analyzed (clone B2, pNC-CPA3 and -CPA10, n = 5; pNC-CPA1, n = 6; pNC-CPA2, n = 8; pNC-CPA4, -CPA6, and -CPB8, n = 9; pNC-CPC13, n = 10; clone A1, pNC-CPA7, n = 11; pNC, n = 14, pNC-CPA5, n = 15). P values were calculated using the Mann-Whitney test. Abscess score: 0, no abscess; 1, <1 mm; 2, 1 to 5 mm; 3, >5 mm. Seven days postinfection.
ehcp expression profiles of E. histolytica clone A1 transfectants with conferred pathogenic phenotypes.
The ehcp expression profiles of the clone A1 transfectants that conferred a pathogenic phenotype were analyzed in detail. For pNC-CPB8, -CPB9, and -CPC13 transfectants, the overexpression was highly specific, with a significant overexpression of the relevant ehcp gene and weak overexpression of only a few other ehcp genes (Table 4). For example, in pNC-CPB8 transfectants, the expression of two peptidase genes (ehcp-b2 and ehcp-c2) decreased and the expression of one (ehcp-b10) slightly increased (Table 4). Despite the strong increase in expression of the transfected peptidase gene, as demonstrated by the qRT-PCR assay, this was not reflected at the protein level, with substrate gel electrophoresis showing no increase in intensity of the relevant protein band (Fig. 2).
TABLE 4 .
Expression of ehcp genes in clone A1 transfectants showing an increased ability to induce ALAs in mice
| ehcp gene (target) | ΔCT for clone A1_pNC (axenic culture)a |
Relative expression (ΔΔCTb method) of each transfectant |
|||||
|---|---|---|---|---|---|---|---|
| Clone A1_pNC (axenic culture) calibrator |
pNC-CPA3 | pNC-CPA5 | pNC-CPB8 | pNC-CPB9 | pNC-CPC13 | ||
| ehcp-a1 | 0.47 ± 0.65 | 1 | 9.41 ± 5.19*c | 1.46 ± 0.39 | 0.47 ± 0.2 | 0.5 ± 0.24 | 0.81 ± 0.22 |
| ehcp-a2 | 0.65 ± 0.92 | 1 | 10.24 ± 8.07* | 4.9 ± 0.9* | 0.62 ± 0.05 | 0.78 ± 0.18 | 0.8 ± 0.24 |
| ehcp-a3 | 12.29 ± 0.9 | 1 | 22.20 ± 20.35** | 1.71 ± 1.04 | 1.7 ± 0.33 | 0.82 ± 0.55 | 1.03 ± 0.35 |
| ehcp-a4 | 5.37 ± 0.8 | 1 | 6.3 ± 1.35* | 2.89 ± 1.48 | 0.71 ± 0.58 | 0.73 ± 0.2 | 1.14 ± 0.76 |
| ehcp-a5 | 1.68 ± 0.86 | 1 | 8.65 ± 1.22* | 3.16 ± 2.01** | 0.85 ± 0.16 | 0.31 ± 0.05* | 0.68 ± 0.11 |
| ehcp-a6 | 5.25 ± 0.59 | 1 | 9.35 ± 2.09* | 0.48 ± 0.041 | 1.01 ± 0.76 | 0.44 ± 0.15 | 0.81 ± 0.14 |
| ehcp-a7 | 3.94 ± 0.59 | 1 | 467.2 ± 501.9* | 2.48 ± 1.09 | 0.83 ± 0.23 | 13.49 ± 6.3* | 1.28 ± 0.43 |
| ehcp-a8 | 8.47 ± 0.42 | 1 | 13.56 ± 6.54* | 0.43 ± 0.1 | 0.65 ± 0.27 | 0.62 ± 0.084 | 1.18 ± 0.17 |
| ehcp-a9 | 10.94 ± 0.4 | 1 | 27.30 ± 21.23* | 0.83 ± 0.0 | 1.06 ± 0.35 | 2.9 ± 1.24 | 1.57 ± 0.33 |
| ehcp-a10 | 8.57 ± 2.19 | 1 | 3.44 ± 1.75* | 0.86 ± 0.56 | 1.18 ± 0.53 | 2.91 ± 3.1 | 0.78 ± 0.44 |
| ehcp-a11 | 4.72 ± 0.59 | 1 | 30.29 ± 24.91* | 0.53 ± 0.05 | 1.03 ± 0.45 | 0.64 ± 0.09 | 1.1 ± 0.13 |
| ehcp-a12 | 9.73 ± 0.54 | 1 | 6.27 ± 5.06* | 0.96 ± 0.06 | 0.84 ± 0.35 | 0.65 ± 0.12 | 1 ± 0.29 |
| ehcp-a13 | 7.46 ± 0.69 | 1 | 6.02 ± 3.03* | 0.76 ± 0.31 | 0.78 ± 0.24 | 0.4 ± 0.11 | 1.43 ± 0.47 |
| ehcp-b1 | 13.17 ± 0.65 | 1 | 7.1 ± 5.27* | 1.7 ± 0.53 | 1.32 ± 0.33 | 0.76 ± 0.45 | 0.72 ± 0.21 |
| ehcp-b2 | 10.05 ± 0.49 | 1 | 0.41 ± 0.01 | 0.08 ± 0.04* | 0.12 ± 0.05* | 0.97 ± 0.33 | 0.37 ± 0.14 |
| ehcp-b3 | 11.38 ± 0.76 | 1 | 31.28 ± 33.97* | 1.86 ± 1.49 | 1.63 ± 0.51 | 0.87 ± 0.32 | 0.9 ± 0.23 |
| ehcp-b4 | 6.7 ± 0.6 | 1 | 2.61 ± 1.33 | 0.54 ± 0.28 | 0.91 ± 0.08 | 0.71 ± 0.14 | 1.07 ± 0.35 |
| ehcp-b5 | 9.94 ± 0.47 | 1 | 0.79 ± 0.39 | 0.74 ± 0.46 | 0.74 ± 0.44 | 0.51 ± 0.28 | 0.81 ± 0.48 |
| ehcp-b7 | 9.8 ± 0.18 | 1 | 1.81 ± 0.72 | 2.16 ± 1.24 | 0.69 ± 0.1 | 0.6 ± 0.43 | 0.98 ± 0.53 |
| ehcp-b8 | 15.42 ± 0.41 | 1 | NA | 2.98 ± 1.24 | 9,370 ± 13,028** | NA | NA |
| ehcp-b9 | 10.49 ± 0.59 | 1 | 2.03 ± 0.01 | 1.08 ± 0.42 | 1.76 ± 1.35 | 589 ± 1,097** | 1.43 ± 0.43 |
| ehcp-b10 | 14.48 ± 0.89 | 1 | 4.57 ± 1.81* | 0.84 ± 0.1 | 4.23 ± 1.4* | 1.35 ± 1.19 | 2.49 ± 2.8 |
| ehcp-c1 | 10.20 ± 0.89 | 1 | 42.56 ± 47.02* | 0.7 ± 0.23 | 1.27 ± 0.31 | 0.52 ± 0.09 | 0.74 ± 0.35 |
| ehcp-c2 | 7.73 ± 0.78 | 1 | NA | 0.42 ± 0.06 | 0.3 ± 0.03* | 2.17 ± 2.24 | 0.55 ± 0.64 |
| ehcp-c3 | 9.15 ± 2.52 | 1 | 6.67 ± 0.59* | 0.48 ± 0.01 | 0.65 ± 0.27 | 2.43 ± 3.28 | 0.35 ± 0.01 |
| ehcp-c4 | 4.46 ± 0.35 | 1 | 6.58 ± 3.23* | 0.76 ± 0.22 | 0.73 ± 0.1 | 0.74 ± 0.22 | 2.19 ± 2.17 |
| ehcp-c5 | 5.39 ± 0.53 | 1 | 1.54 ± 0.59 | 0.65 ± 0.07 | 0.71 ± 0.18 | 1.1 ± 0.76 | 1.1 ± 0.25 |
| ehcp-c6 | 14.28 ± 0.26 | 1 | NA | NA | 0.62 ± 0.33 | 1.77 ± 1.36 | 1.22 ± 0.75 |
| ehcp-c9 | 5.87 ± 0.53 | 1 | 4.4 ± 4.34NS | 0.75 ± 0.13 | 0.98 ± 0.24 | 1.51 ± 1.08 | 0.94 ± 0.41 |
| ehcp-c11 | 11.76 ± 1.13 | 1 | 3.45 ± 2.14NS | 0.31 ± 0.21* | 0.92 ± 0.65 | 0.35 ± 0.2 | 1.03 ± 0.24 |
| ehcp-c12 | 8.56 ± 0.71 | 1 | 2.06 ± 0.68 | 0.73 ± 0.05 | 0.7 ± 0.16 | 0.8 ± 0.05 | 0.92 ± 0.4 |
| ehcp-c13 | 7.27 ± 0.63 | 1 | 1.48 ± 0.29 | 1.02 ± 0.14 | 1.08 ± 0.18 | 2.06 ± 1.2 | 19.83 ± 12.17** |
| ehicp1 | 4.49 ± 0.41 | 1 | 2.03 ± 0.28 | 3.55 ± 1.05 | ND | ND | ND |
| ehicp2 | 5.07 ± 0.46 | 1 | 1.1 ± 0.18 | 2.75 ± 0.16 | ND | ND | ND |
Concentration relative to ehactin, which was used as a normalizer.
Relative differences in gene expression.
Expression of the various ehcp genes was analyzed at least two times in duplicate. Bold indicates differentially expressed genes. The threshold for defining genes as being differentially expressed was set at 3.0. P values of differentially expressed genes are expressed as follows: *, P < 0.05; **, P < 0.01; NS, not significant. ND, not determined; NA, no amplificate in respective qRT-PCR.
In the pNC-CPA5 transfectants, a 3-fold increase in ehcp-a5 expression was observed, but there was also a 5-fold increase in ehcp-a2 expression (Table 4). This result was confirmed by substrate gel electrophoresis, which showed that bands representing EhCP-A5 and EhCP-A2 were more intense than that of the control, pNC (Fig. 2). There was also an increase in the intensity of the band representing EhCP-A1 in pNC-CPA5 relative to pNC transfectants. Therefore, it is unclear whether the increased pathogenicity of the pNC-CPA5 transfectants was the result of the increased expression of ehcp-a5 or other ehcp genes.
Surprisingly, in pNC-CPA3 transfectants, the expression of all ehcp-a genes increased significantly between 3.5- and 470-fold (Table 4). In addition, a significant increase in expression was also observed for three members of the ehcp-b family and three members of the ehcp-c family. So far, we have no explanation for this phenomenon. Even more surprisingly, this increase in expression was not reflected at the protein level, as no significant increase in CP activity with respect to the control was detected (Table 3; Fig. 2).
The discrepancies between mRNA levels and CP activities may be due to increased amounts of cysteine peptidase inhibitors in the transfectants. However, no differences in the amounts of transcripts of the two inhibitors of cysteine peptidase activity, EhICP1 and EhICP2, in pNC-CPA3 transfectants and the pNC control were detected by qRT-PCR (Table 4). This was confirmed by titration experiments in which fresh crude extracts from wild-type clone B2 cells were titrated against boiled trophozoite lysates from either pNC or pNC-CPA3 transfectants; similar inhibitor activities were measured in all heat-inactivated extracts (data not shown).
DISCUSSION
Earlier studies have identified two cell lines derived from the pathogenic E. histolytica isolate HM-1:IMSS that differ substantially in their pathogenic properties (22). Cell line B is highly pathogenic, characterized by its ability to produce considerable ALAs, whereas cell line A is unable to induce ALAs (22). To ensure that these cells were not a mixture of different cell types, both cell lines were cloned. The clones A1 and B2 were very similar in pathogenicity to their parental cell lines, with clone B2 causing ALAs in the mouse and gerbil models and clone A1 not causing ALAs in either animal (Fig. 4; see also Fig. S1 in the supplemental material). Additionally, qRT-PCR analysis of the genes known to be differentially expressed in cell lines A and B (27) revealed similar differential expression in clones A1 and B2 (Table S1). Finally, both clones have CP activities comparable to those of their parental cell lines (22), with the activity of clone B2 being approximately ten times higher than that of clone A1. Surprisingly, the differences in CP activity between clones B2 and A1 were not reflected in the expression profiles of the whole set of ehcp genes, which were very similar for the two clones (Table 1). The reasons for this discrepancy require further investigation.
Several studies have demonstrated that CPs of E. histolytica play a role in abscess formation; therefore, low CP activity may be at least partially responsible for the nonpathogenic phenotype of clone A1. Ankri and colleagues showed that a 90% decrease in CP activity, triggered by antisense inhibition of the expression of ehcp-a5, resulted in a reduction in pathogenicity (10, 28). Nevertheless, the ehcp-a5 antisense mRNA inhibited not only ehcp-a5 expression but also the expression of other CPs. Thus, the observed reduction in pathogenicity of these transfectants may have been due to the general reduction in CP activity rather than to that of a single peptidase.
Little is known about the regulation of ehcp expression during the process of ALA formation, with only a few reports describing increased expression of a few ehcp genes during induction of cecal colitis. For example, ehcp-a4, which is expressed at moderate levels in axenic culture, is upregulated in cecal colitis and following exposure to intestinal cells (19, 25). In addition, Gilchrist and colleagues reported increased expression of three additional peptidase genes (ehcp-a1, -a6, and -a8) in trophozoites isolated from the colon of mice (19). If a peptidase gene is upregulated during ALA formation, then it may play a role in this process. This hypothesis can be tested by determining if overexpressing the candidate gene in nonpathogenic E. histolytica mediates a pathogenic phenotype.
We analyzed and compared ehcp expression during ALA formation in gerbil and mouse models. The gerbil model is routinely used in several laboratories worldwide. Nevertheless, the toolbox that allows state-of-the-art immunological investigations in this animal is not available. Therefore, a mouse model was recently established (26). In both animals, the time course of abscess formation is self-limited and amoebic lesions are cleared within approximately 30 days postinfection. Only a few ehcp genes were found to be expressed at high levels during abscess formation in the current study. Some of them (ehcp-a3, -a4, -a10, and -c13) were upregulated in both the mouse and gerbil models, whereas a few others were upregulated in either the gerbil (ehcp-b8 and -b9) or mouse (ehcp-a5, -a6, and -a7) model only. The expression of a few of these ehcp genes has been reported to be affected by heat stress, with Weber and colleagues reporting an increase in the expression of ehcp-a4 and -a5 (29) and Tillack and colleagues reporting an increase in the expression of ehcp-a5 and -a6 (13). Thus, the enhanced expression of ehcp-a4, -a5, and -a6 reported in the current study may simply be a response to the higher temperatures of the gerbil and mouse livers.
To investigate if the total amount of CPs or the increase in expression of a specific CP in response to ALA formation is related to pathogenicity, transfectants of the nonpathogenic clone A1 overexpressing those ehcp genes that were upregulated during ALA formation in the mouse and gerbil models were generated. If overexpression of a particular ehcp mediates pathogenicity in a nonpathogenic amoeba, it is likely that this gene plays an important role in pathogenicity.
Our results indicate that five of the eleven transfectants analyzed (pNC-CPA3, -CPA5, -CPB8, -CPB9, and -CPC13) can confer a pathogenic phenotype of clone A1 (Fig. 4). Each of these transfectants produced abscesses of a similar size as those produced by the pathogenic clone B2. Detailed analysis of the ehcp expression profiles indicated that for the pNC-CPB8, -CPB9, and -CPC13 transfectants, the relevant ehcp was specifically overexpressed, with only a few other genes showing a slight up- or downregulation. Therefore, it can be concluded that the specific overexpression of any one of these three CPs is sufficient to transform the nonpathogenic phenotype of E. histolytica clone A1 into a pathogenic E. histolytica phenotype. However, it can only be speculated if the specific overexpression of these CPs directly influences ALA formation or if a processing of additional proteins of amoebic or host origin stimulates abscess formation. In addition, it has to be kept in mind that the expression was analyzed only for genes encoding CPs. Therefore, it is not known if the expression of other non-ehcp genes is influenced.
The picture is not so clear for the other two transfectants (pNC-CPA5 and pNC-CPA3) that mediated a pathogenic phenotype of clone A1. For the pNC-CPA5 transfectants, an approximately 60-fold increase in CP activity over that of the control, pNC, was observed (Table 3). This was confirmed using substrate gel electrophoresis, in which an increase in the intensity of the bands corresponding to EhCP-A1, -A2, and -A5 was detected (Fig. 2). Therefore, it is possible that the high overall CP activity or the specific composition of CPs, rather than the overexpression of ehcp-a5 alone, was responsible for the pathogenicity observed. In earlier studies, Tillack and colleagues used the expression vectors pNC-CPA1, pNC-CPA2, and pNC-CPA5 to transfect trophozoites of the pathogenic cell line HM-1:IMSS (11, 30). Overexpression of ehcp-a1 and ehcp-a2 resulted in an increase of EhCP-A1 and EhCP-A2, respectively, with no influence on ALA formation, a finding which is in good agreement with results of the current study. Overexpression of ehcp-a5 led to an increase of EhCP-A1 and -A2 as well as -A5, and the transfectants produced abscesses covering a 3-fold-greater area of tissue than the respective control transfectants, a finding which is again in good agreement with results from the current study.
For the EhCP-A3 transfectants in the current study, the ehcp expression profile indicated an overexpression of all A family members, three B family members, and three C family members. Nevertheless, this was not confirmed at the protein level, as the EhCP-A3 transfectants had an activity of 1.2 mU/mg, which is only slightly higher than that of the control pNC transfectants (Table 2). Further expression and activity studies indicated that the lower-than-expected CP activity was not the result of an increase in the activity of inhibitors of cysteine peptidases. Therefore, it still remains to be determined whether the restoration of pathogenicity in the pNC-CPA3 transfectants was due to the overexpression of a particular peptidase or to some other factor.
In summary, the specific overexpression of any of the three peptidases, ehcp-b8, -b9, and -c13, whose expression is increased during ALA formation, is sufficient to transform a nonpathogenic phenotype into a pathogenic one. It is difficult to hypothesize on the mechanism(s) by which these CPs mediate pathogenicity to the nonpathogenic clone A1, as there is little mechanistic information available for these peptidases. EhCP-B9 (EhCP112) is known to form a complex with an adherence domain protein (EhADH112), and then, as a complex, both proteins bind to target cells and are translocated during phagocytosis from the plasma membrane to phagocytic vacuoles (24, 31). Furthermore, EhCP-B9 has a putative transmembrane domain and contains an RGD (Arg–Gly–Asp) motif at positions 249 to 251 between the active-site cysteine167 and histidine328 (31). This RDG motif is also found in EhCP-B8 at a similar position (positions 245 to 247) as well as in the proregion of EhCP-A5 (positions 92 to 94). EhCP-A5 binds via its RGD motif to αVβ3 integrin on colonic cells and stimulates NFκB-mediated proinflammatory responses in the pathogenesis of intestinal amoebiasis (32). For EhCP-C13, an N-terminal signal anchor is postulated, and Western blot analysis indicated that the protein is present in the membrane fraction of E. histolytica (see Fig. S2 in the supplemental material).
Nevertheless, it remains to be determined why EhCP-B8, -B9, and -C13 have such a profound effect on amoebic pathogenicity. Localization to the plasma membrane and secretion of the CPs are not sufficient to induce ALA formation. Of the three peptidase genes (ehcp-a1, -a2, and -a7) that are highly expressed under axenic culture conditions, EhCP-A2 was described to be membrane associated and, in addition, like EhCP-A1, secreted (Fig. 3) (23, 33, 34). Secretion was also described for EhCP-A4, which was found beside EhCP-A6 and EhCP-A10 in higher levels during ALA formation (Fig. 3) (25). Nevertheless, EhCP-A1, -A2, and A4 apparently do not influence pathogenicity.
MATERIALS AND METHODS
Cultivation of cells.
The E. histolytica cell lines A and B derived from the isolate HM-1:IMSS were cloned by limited dilution (22). Most experiments were performed with the nonpathogenic clone A1 or the pathogenic clone B2. E. histolytica trophozoites were cultured axenically in TYI-S-33 medium in plastic tissue culture flasks (35). For individual experiments, 1 × 106 trophozoites were cultivated for 24 h in 75-ml culture flasks. Then, after chilling on ice for 5 min, the trophozoites were harvested by sedimentation at 430 × g at 4°C for 5 min. The resulting cell pellet was washed twice either in phosphate-buffered saline (6.7 mM NaHPO4, 3.3 mM NaH2PO4, 140 mM NaCl, pH 7.2; PBS) or in TYI-S-33 medium without serum. For preparation of soluble amoeba extracts, cells were lysed by four freeze-thaw cycles in CO2/ethanol and sedimented by centrifugation (15,000 × g at 4°C for 15 min).
ALA formation in gerbils and mice.
Animal infections were performed with eight-week-old male gerbils (Meriones unguiculatus) or with eight- to 10-week-old C57BL/6 male mice. All mice were maintained in a specific-pathogen-free microenvironment and received care in compliance with guidelines outlined in the Guide for the Care and Use of Laboratory Animals. All work was conducted with the approval of the Government for Science and Health, Hamburg, Germany (TVA 23/11).
For the infection of gerbils, 1 × 106 trophozoites of cell line B were suspended in 100 µl PBS and were then injected into the left liver lobe, as previously described (36). For mice infection experiments, 1.25 × 105 trophozoites of clone B2 or clone A1 transfectants were resuspended in 25 µl PBS and subsequently injected into the liver as described previously (26). It is noteworthy to emphasize that neither cell line B nor any of the clones were passaged through the liver of mice or gerbils prior to the infection experiments.
For analyzing the expression profiles of ehcp genes during ALA formation, mice were sacrificed 24, 48, and 72 hpi and gerbils were sacrificed seven days postinfection. For mouse infection experiments, two animals of each time point were analyzed separately in duplicate. For gerbil infection experiments, the expression was analyzed in samples derived from three animals in duplicate. The area of abscess was cut out and directly utilized for RNA purification. For each time point, areas of tissue containing abscesses from two animals were analyzed.
To analyze ALA formation using the various transfectants, mice were sacrificed seven days after infection and sizes of liver abscesses were determined. The diameters of the abscessed areas within the liver were measured, and the results were expressed using the following scores: 0, no abscess; 1, <1 mm (pinhead); 2, <5 mm; 3, >5 mm. For each transfectant, ALA formation in 5 to 18 animals was analyzed. P values were calculated using the Mann-Whitney test.
RNA extraction and qRT-PCR.
E. histolytica trophozoites (1 × 106) were cultivated in 75-ml culture flasks for 24 h. The cells were harvested via chilling on ice for 5 min and sedimented at 200 × g for 5 min at 4°C. The cell pellet was washed twice with PBS. For isolation of total RNA, trophozoites were treated with Trizol reagent (Life Technologies, Darmstadt, Germany) by following the manufacturer’s instructions. Extracted RNA was further purified using the RNeasy minikit (Qiagen, Hilden, Germany) without β-mercaptoethanol, and DNA was digested with DNase (Qiagen).
For extraction of RNA from ALA material, approximately 100 mg abscess/liver tissue was homogenized in the presence of 500 µl TRIzol reagent (Life Technologies). After the addition of another 500 µl TRIzol reagent, the RNA was extracted as described above.
cDNA synthesis was accomplished with the SuperScript III reverse transcriptase system (Life Technologies). In a final volume of 20 µl, 1 µg of RNase-free and DNase-treated total RNA was mixed with 5× first-strand buffer, 1 mM deoxynucleoside triphosphates (dNTPs), 500 nM OdT-T71 (5′-GAG AGA GGA TCC AAG TAC TAA TAC GAC TCA CTA TAG GGA GAT24), 2 mM dithiothreitol (DTT), 0.5 mM MgCl2, 40 U RnaseOut (Life Technologies), and SuperScript III (200 U/µl). cDNA was synthesized for 1 h at 42°C.
For qRT-PCR experiments, sense and antisense primers were designed to amplify approximately 100 to 120 bp of the relevant gene. In total, qRT-PCR assays were able to be developed for 32 of the 35 ehcp genes (see Table S2 in the supplemental material). Quantitative amplification was performed in a Rotor-Gene PCR apparatus (Corbett) using a RealMasterMix SYBR ROX kit (5 Prime). cDNA (1 µl) was mixed with 2.5× RealMasterMix/20× SYBR and 5 pmol/µl of appropriate sense and antisense primers to a final volume of 20 µl. Amplification conditions were as follows: 40 cycles at 95°C for 15 s, 58°C for 20 s, and 68°C for 20 s and an adjacent melting step (67 to 95°C). Relative differences in gene expression between axenic cultivated control trophozoites (calibrator = 1) and ALA-derived trophozoites or transfectants were calculated using the 2−ΔΔCT method provided by the Rotor-Gene software (37). Depending on the experiment, cell line B trophozoites, clone B2 trophozoites, or clone A1 trophozoites transfected with a control plasmid were used as the calibrator and actin was chosen as the housekeeping gene for normalization. Efficiencies of the primer pairs were determined using a template dilution series of genomic DNA (gDNA) from clone B2 at concentrations ranging from 50 ng to 0.00005 ng (38). Mean efficiencies determined with the template dilution series were used for correction of the relative expression data. The threshold for defining genes as being differentially expressed was set at 3.0.
Expression constructs.
For overexpression of ehcp-a1, -a2, and -a5, the plasmids described by Tillack and colleagues were used (pNC-CPA1, -CPA2, and -CPA5) (11). For overexpression of ehcp-a3, -a4, -a6, -a7, -a10, -b8, -b9, and -c13, the coding sequences of the ehcp genes were flanked by 485 bp of the 5′-untranslated sequence of an E. histolytica lectin gene and 600 bp of the 3′-untranslated actin gene. For all, neomycin phosphotransferase was used as a selectable marker (30).
Transfection.
Transfections were performed by electroporation as described previously (39). Two days after transfection, cells were selected with 10 µg/ml of G-418 sulfate (PAA, Pasching, Austria) for approximately two weeks. Subsequently, the cells were cloned by limited dilution and cultivated in the presence of 20 µg/ml of G418. Successful overexpression of at least four clones was checked by qRT-PCR. The clone that showed the highest expression was used for all further experiments. The maintenance of ehcp overexpression was confirmed regularly by qRT-PCR.
Enzymatic assays.
Peptidolytic activity was measured using the synthetic peptides Z-Arg-Arg-pNA, Z-Phe-Arg-pNA, Z-Ala-Ala-pNA, Z-Gly-Pro-pNA, and Z-Ala-Pro-pNA (Z = Cbz = benzyloxycarbonyl; pNA = p-nitroanilide; Bachem) as substrates (40). The peptide substrates were prepared as 10 mM stock solutions. The sample to be measured (1 to 40 µl) was added to 0.1 KH2PO4/2 mM EDTA/1 mM DTT, adjusted to pH 7.0 with KOH. The activity of the transfectants pNCEhCPA1, -A3, -A5, -B8, -B9, and -C13 was in addition tested at pH 4, 5, 6, and 8. The substrates were added at a final concentration of 0.1 mM (total volume, 200 µl). The rate of cleavage of the pNA group from the substrate was monitored at 405 nm in 96-well plates at 25°C during 30 min. One unit of enzymatic activity is defined as the amount that catalyzes the reduction of 1 µmol/min of p-nitroaniline. The activity was measured at least four times in duplicate.
To determine the inhibitory activity, amoebic extract was heated for 5 min at 95°C to release the inhibitors of cysteine peptidases from the bound EhCPs and to inhibit the activity of the peptidases, centrifuged at 20,000 × g at 4°C for 15 min, and then incubated with a fresh amoebic extract of clone B2 for 15 min. The peptidolytic activity was assessed as mentioned above using Z-Arg-Arg-pNA as the substrate.
Assay to determine secretion.
To determine the secretion of CPs, 1 × 106 trophozoites were suspended in 500 µl TYI_secretion medium (TYI-S-33 without serum, supplemented with 10 mM HEPES, 0.15 mM CaCl2, and 0.5 mM MgCl2). After 0 min, 1 h, 2 h, and 3 h of incubation at 35°C, the amoebae were centrifuged (430 × g at 4°C for 2 min) and the supernatant was removed to measure CP activity. To determine the total activity, soluble extracts of 1 × 106 trophozoites/500 µl were generated. The secretion was outlined as the percentage of total activity. Each time point was measured at least two times in duplicate. To prove the integrity of the amoebae, the cytoplasmic alcohol dehydrogenase (EhADH) activity was measured. The enzymatic activity was measured at 35°C by following the reduction of NADP+ at 340 nm. The assay mixture contained 2-propanol (20 mM), NADP+ (0.2 mM), and glycine (50 mM) in a total volume of 200 µl. Only those supernatants with no detectable ADH activity were used to determine CP activity.
Substrate gel electrophoresis.
The substrate gel electrophoresis was performed as described previously (41). In brief, 4 or 20 µg of amoebic extract was incubated with Laemmli buffer containing 20 mM DTT for 5 min at 37°C. For the substrate gel, a 12% SDS-polyacrylamide gel was copolymerized with 0.1% gelatin. After separation of the proteins and incubation in solution A (2.5% Triton X-100) for 1 h and solution B (100 mM sodium acetate, pH 4.5, 1% Triton X-100, 20 mM DTT) for 3 h at 37°C, the gel was stained with Coomassie blue.
SUPPLEMENTAL MATERIAL
Expression of genes in clone A1 and clone B2 known to be differentially expressed in cell line A and cell line B.
List of oligonucleotides used for qRT-PCR.
Abscess formation in gerbil livers following infection with E. histolytica cell line A and its derived clone A1 and cell line B and its derived clone B2. Abscesses were analyzed 7 days postinfection. Download
EhCP-C13 was recombinantly expressed in Escherichia coli using the expression plasmid pJC45. Recombinantly produced protein was applied onto a Ni2+-nitrilotriacetic acid (NTA) column and eluted with imidazole. For the generation of antibodies, 100 µg of recombinant EhCP-C13 was injected into a mouse. The first injection was done in combination with Freund’s complete adjuvant, and the following two booster injections were done in combination with incomplete Freund’s adjuvant after 2 weeks. To prepare amoebic extracts, trophozoites were sedimented, alternately flash frozen in liquid nitrogen, thawed at room temperature, and vortexed for five times. Lysates were centrifuged at 40,000 × g for 1 h at 4°C. The supernatant contains the PBS-soluble proteins (S). Pellets (P) were washed two times in ice-cold PBS and solubilized in PBS supplemented with 1% Triton X-100. Extracts (50 µg/lane) were separated under reducing conditions using 12% SDS-PAGE. Western blot analyses were carried out by a wet blotting technique. For Western blot analyses, the first antibodies were used in a 1:500 dilution and the second antibodies (anti-mouse horseradish peroxidase [HRP]; Dako A/S, Glostrup, Denmark) were used in a 1:10,000 dilution, and blots were developed using enhanced chemiluminescence (ECL) (Amersham ECL plus Western blotting detection reagents; GE Healthcare). Download
ACKNOWLEDGMENTS
We thank Thomas Roeder for critically reading the manuscript and Ina Hennings for skillful technical assistance.
This work has been supported by the Deutsche Forschungsgemeinschaft (DFG; BR 1744/11-2).
Footnotes
Citation Matthiesen J, Bär A-K, Bartels A-K, Marien D, Ofori S, Biller L, Tannich E, Lotter H, Bruchhaus I. 2013. Overexpression of specific cysteine peptidases confers pathogenicity to a nonpathogenic Entamoeba histolytica clone. mBio 4(2):e00072-13. doi:10.1128/mBio.00072-13.
REFERENCES
- 1. WHO 1997. WHO/PAHO/UNESCO report. A consultation with experts on amoebiasis. Mexico City, Mexico 28–29 January, 1997. Epidemiol. Bull. 18:13–14 [PubMed] [Google Scholar]
- 2. Gadasi H, Kessler E. 1983. Correlation of virulence and collagenolytic activity in Entamoeba histolytica. Infect. Immun. 39:528–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lushbaugh WB, Hofbauer AF, Pittman FE. 1985. Entamoeba histolytica: purification of cathepsin B. Exp. Parasitol. 59:328–336 [DOI] [PubMed] [Google Scholar]
- 4. Luaces AL, Barrett AJ. 1988. Affinity purification and biochemical characterization of histolysin, the major cysteine proteinase of Entamoeba histolytica. Biochem. J. 250:903–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reed SL, Keene WE, McKerrow JH. 1989. Thiol proteinase expression and pathogenicity of Entamoeba histolytica. J. Clin. Microbiol. 27:2772–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schulte W, Scholze H. 1989. Action of the major protease from Entamoeba histolytica on proteins of the extracellular matrix. J. Protozool. 36:538–543 [DOI] [PubMed] [Google Scholar]
- 7. Keene WE, Hidalgo ME, Orozco E, McKerrow JH. 1990. Entamoeba histolytica: correlation of the cytopathic effect of virulent trophozoites with secretion of a cysteine proteinase. Exp. Parasitol. 71:199–206 [DOI] [PubMed] [Google Scholar]
- 8. Li E, Yang WG, Zhang T, Stanley SL., Jr 1995. Interaction of laminin with Entamoeba histolytica cysteine proteinases and its effect on amebic pathogenesis. Infect. Immun. 63:4150–4153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stanley SL, Jr, Zhang T, Rubin D, Li E. 1995. Role of the Entamoeba histolytica cysteine proteinase in amebic liver abscess formation in severe combined immunodeficient mice. Infect. Immun. 63:1587–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ankri S, Stolarsky T, Bracha R, Padilla-Vaca F, Mirelman D. 1999. Antisense inhibition of expression of cysteine proteinases affects Entamoeba histolytica-induced formation of liver abscess in hamsters. Infect. Immun. 67:421–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tillack M, Nowak N, Lotter H, Bracha R, Mirelman D, Tannich E, Bruchhaus I. 2006. Increased expression of the major cysteine proteinases by stable episomal transfection underlines the important role of EhCP5 for the pathogenicity of Entamoeba histolytica. Mol. Biochem. Parasitol. 149:58–64 [DOI] [PubMed] [Google Scholar]
- 12. Bruchhaus I, Loftus BJ, Hall N, Tannich E. 2003. The intestinal protozoan parasite Entamoeba histolytica contains 20 cysteine protease genes, of which only a small subset is expressed during in vitro cultivation. Eukaryot. Cell 2:501–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tillack M, Biller L, Irmer H, Freitas M, Gomes MA, Tannich E, Bruchhaus I. 2007. The Entamoeba histolytica genome: primary structure and expression of proteolytic enzymes. BMC Genomics 8:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Clark CG, Alsmark UC, Tazreiter M, Saito-Nakano Y, Ali V, Marion S, Weber C, Mukherjee C, Bruchhaus I, Tannich E, Leippe M, Sicheritz-Ponten T, Foster PG, Samuelson J, Noël CJ, Hirt RP, Embley TM, Gilchrist CA, Mann BJ, Singh U, Ackers JP, Bhattacharya S, Bhattacharya A, Lohia A, Guillén N, Duchêne M, Nozaki T, Hall N. 2007. Structure and content of the Entamoeba histolytica genome. Adv. Parasitol. 65:51–190 [DOI] [PubMed] [Google Scholar]
- 15. Bansal D, Ave P, Kerneis S, Frileux P, Boché O, Baglin AC, Dubost G, Leguern AS, Prevost MC, Bracha R, Mirelman D, Guillén N, Labruyère E. 2009. An ex-vivo human intestinal model to study Entamoeba histolytica pathogenesis. PLoS Negl. Trop Dis. 3:e551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thibeaux R, Dufour A, Roux P, Bernier M, Baglin AC, Frileux P, Olivo-Marin JC, Guillén N, Labruyère E. 2012. Newly visualized fibrillar collagen scaffolds dictate Entamoeba histolytica invasion route in the human colon. Cell. Microbiol. 14:609–621 [DOI] [PubMed] [Google Scholar]
- 17. Irmer H, Tillack M, Biller L, Handal G, Leippe M, Roeder T, Tannich E, Bruchhaus I. 2009. Major cysteine peptidases of Entamoeba histolytica are required for aggregation and digestion of erythrocytes but are dispensable for phagocytosis and cytopathogenicity. Mol. Microbiol. 72:658–667 [DOI] [PubMed] [Google Scholar]
- 18. Ehrenkaufer GM, Haque R, Hackney JA, Eichinger DJ, Singh U. 2007. Identification of developmentally regulated genes in Entamoeba histolytica: insights into mechanisms of stage conversion in a protozoan parasite. Cell. Microbiol. 9:1426–1444 [DOI] [PubMed] [Google Scholar]
- 19. Gilchrist CA, Houpt E, Trapaidze N, Fei Z, Crasta O, Asgharpour A, Evans C, Martino-Catt S, Baba DJ, Stroup S, Hamano S, Ehrenkaufer G, Okada M, Singh U, Nozaki T, Mann BJ, Petri WA. 2006. Impact of intestinal colonization and invasion on the Entamoeba histolytica transcriptome. Mol. Biochem. Parasitol. 147:163–176 [DOI] [PubMed] [Google Scholar]
- 20. Marquay Markiewicz J, Syan S, Hon CC, Weber C, Faust D, Guillen N. 2011. A proteomic and cellular analysis of uropods in the pathogen Entamoeba histolytica. PLoS Negl. Trop Dis. 5:e1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Freitas MA, Fernandes HC, Calixto VC, Martins AS, Silva EF, Pesquero JL, Gomes MA. 2009. Entamoeba histolytica: cysteine proteinase activity and virulence. Focus on cysteine proteinase 5 expression levels. Exp. Parasitol. 122:306–309 [DOI] [PubMed] [Google Scholar]
- 22. Biller L, Schmidt H, Krause E, Gelhaus C, Matthiesen J, Handal G, Lotter H, Janssen O, Tannich E, Bruchhaus I. 2009. Comparison of two genetically related Entamoeba histolytica cell lines derived from the same isolate with different pathogenic properties. Proteomics 9:4107–4120 [DOI] [PubMed] [Google Scholar]
- 23. Mitra BN, Saito-Nakano Y, Nakada-Tsukui K, Sato D, Nozaki T. 2007. Rab11B small GTPase regulates secretion of cysteine proteases in the enteric protozoan parasite Entamoeba histolytica. Cell. Microbiol. 9:2112–2125 [DOI] [PubMed] [Google Scholar]
- 24. Ocádiz R, Orozco E, Carrillo E, Quintas LI, Ortega-López J, García-Pérez RM, Sánchez T, Castillo-Juárez BA, García-Rivera G, Rodríguez MA. 2005. EhCP112 is an Entamoeba histolytica secreted cysteine protease that may be involved in the parasite-virulence. Cell. Microbiol. 7:221–232 [DOI] [PubMed] [Google Scholar]
- 25. He C, Nora GP, Schneider EL, Kerr ID, Hansell E, Hirata K, Gonzalez D, Sajid M, Boyd SE, Hruz P, Cobo ER, Le C, Liu WT, Eckmann L, Dorrestein PC, Houpt ER, Brinen LS, Craik CS, Roush WR, McKerrow J, Reed SL. 2010. A novel Entamoeba histolytica cysteine proteinase, EhCP4, is key for invasive amebiasis and a therapeutic target. J. Biol. Chem. 285:18516–18527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lotter H, Jacobs T, Gaworski I, Tannich E. 2006. Sexual dimorphism in the control of amebic liver abscess in a mouse model of disease. Infect. Immun. 74:118–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Biller L, Davis PH, Tillack M, Matthiesen J, Lotter H, Stanley SL, Tannich E, Bruchhaus I. 2010. Differences in the transcriptome signatures of two genetically related Entamoeba histolytica cell lines derived from the same isolate with different pathogenic properties. BMC Genomics 11:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ankri S, Stolarsky T, Mirelman D. 1998. Antisense inhibition of expression of cysteine proteinases does not affect Entamoeba histolytica cytopathic or haemolytic activity but inhibits phagocytosis. Mol. Microbiol. 28:777–785 [DOI] [PubMed] [Google Scholar]
- 29. Weber C, Guigon G, Bouchier C, Frangeul L, Moreira S, Sismeiro O, Gouyette C, Mirelman D, Coppee JY, Guillén N. 2006. Stress by heat shock induces massive down regulation of genes and allows differential allelic expression of the Gal/GalNAc lectin in Entamoeba histolytica. Eukaryot. Cell 5:871–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hellberg A, Nickel R, Lotter H, Tannich E, Bruchhaus I. 2001. Overexpression of cysteine proteinase 2 in Entamoeba histolytica or Entamoeba dispar increases amoeba-induced monolayer destruction in vitro but does not augment amoebic liver abscess formation in gerbils. Cell. Microbiol. 3:13–20 [DOI] [PubMed] [Google Scholar]
- 31. García-Rivera G, Rodríguez MA, Ocádiz R, Martínez-López MC, Arroyo R, González-Robles A, Orozco E. 1999. Entamoeba histolytica: a novel cysteine protease and an adhesin form the 112 kDa surface protein. Mol. Microbiol. 33:556–568 [DOI] [PubMed] [Google Scholar]
- 32. Hou Y, Mortimer L, Chadee K. 2010. Entamoeba histolytica cysteine proteinase 5 binds integrin on colonic cells and stimulates NFkappaB-mediated pro-inflammatory responses. J. Biol. Chem. 285:35497–35504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Que X, Brinen LS, Perkins P, Herdman S, Hirata K, Torian BE, Rubin H, McKerrow JH, Reed SL. 2002. Cysteine proteinases from distinct cellular compartments are recruited to phagocytic vesicles by Entamoeba histolytica. Mol. Biochem. Parasitol. 119:23–32 [DOI] [PubMed] [Google Scholar]
- 34. Melendez-Lopez SG, Herdman S, Hirata K, Choi MH, Choe Y, Craik C, Caffrey CR, Hansell E, Chávez-Munguía B, Chen YT, Roush WR, McKerrow J, Eckmann L, Guo J, Stanley SL, Jr, Reed SL. 2007. Use of recombinant Entamoeba histolytica cysteine proteinase 1 to identify a potent inhibitor of amebic invasion in a human colonic model. Eukaryot. Cell 6:1130–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Diamond LS, Harlow DR, Cunnick CC. 1978. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans. R. Soc. Trop. Med. Hyg. 72:431–432 [DOI] [PubMed] [Google Scholar]
- 36. Lotter H, Zhang T, Seydel KB, Stanley SL, Jr, Tannich E. 1997. Identification of an epitope on the Entamoeba histolytica 170-kD lectin conferring antibody-mediated protection against invasive amebiasis. J. Exp. Med. 185:1793–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 38. Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hamann L, Nickel R, Tannich E. 1995. Transfection and continuous expression of heterologous genes in the protozoan parasite Entamoeba histolytica. Proc. Natl. Acad. Sci. U. S. A. 92:8975–8979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Leippe M, Sievertsen HJ, Tannich E, Horstmann RD. 1995. Spontaneous release of cysteine proteinases but not of pore-forming peptides by viable Entamoeba histolytica. Parasitology 111:569–574 [DOI] [PubMed] [Google Scholar]
- 41. Hellberg A, Leippe M, Bruchhaus I. 2000. Two major “higher molecular mass proteinases” of Entamoeba histolytica are identified as cysteine proteinases 1 and 2. Mol. Biochem. Parasitol. 105:305–309 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of genes in clone A1 and clone B2 known to be differentially expressed in cell line A and cell line B.
List of oligonucleotides used for qRT-PCR.
Abscess formation in gerbil livers following infection with E. histolytica cell line A and its derived clone A1 and cell line B and its derived clone B2. Abscesses were analyzed 7 days postinfection. Download
EhCP-C13 was recombinantly expressed in Escherichia coli using the expression plasmid pJC45. Recombinantly produced protein was applied onto a Ni2+-nitrilotriacetic acid (NTA) column and eluted with imidazole. For the generation of antibodies, 100 µg of recombinant EhCP-C13 was injected into a mouse. The first injection was done in combination with Freund’s complete adjuvant, and the following two booster injections were done in combination with incomplete Freund’s adjuvant after 2 weeks. To prepare amoebic extracts, trophozoites were sedimented, alternately flash frozen in liquid nitrogen, thawed at room temperature, and vortexed for five times. Lysates were centrifuged at 40,000 × g for 1 h at 4°C. The supernatant contains the PBS-soluble proteins (S). Pellets (P) were washed two times in ice-cold PBS and solubilized in PBS supplemented with 1% Triton X-100. Extracts (50 µg/lane) were separated under reducing conditions using 12% SDS-PAGE. Western blot analyses were carried out by a wet blotting technique. For Western blot analyses, the first antibodies were used in a 1:500 dilution and the second antibodies (anti-mouse horseradish peroxidase [HRP]; Dako A/S, Glostrup, Denmark) were used in a 1:10,000 dilution, and blots were developed using enhanced chemiluminescence (ECL) (Amersham ECL plus Western blotting detection reagents; GE Healthcare). Download