Abstract
Replication Licensing Factor (RLF) was originally defined as an essential initiation factor whose regulation prevents re-replication of DNA in a single cell cycle [1, reviewed in 2]. It is required for the initiation of DNA replication, binds to chromatin early in the cell cycle, is removed from chromatin as DNA replicates and is unable to re-bind replicated chromatin until the following mitosis. Chromatography of replication licensing factor (RLF) from Xenopus extracts has shown that it consist of two components termed RLF-B and RLF-M [3]. RLF-M consists of complexes of all 6 Xenopus MCM/P1 proteins (XMcm2,3,4,5,6 and 7), which bind to chromatin in late mitosis and are removed as replication occurs [3-7]. The identity of RLF-B is currently unknown. At least two factors must be present on chromatin before licensing can occur: the Xenopus Origin Recognition Complex (XORC) [8, 9] and Xenopus Cdc6 (XCdc6) [10]. XORC saturates Xenopus sperm chromatin at approximately 1 copy per replication origin and is required for licensing by RLF-B and RLF-M to occur [9-11]. XCdc6 binds to chromatin only if XORC is bound first, and the binding of MCM/P1 proteins to chromatin is dependent on XCdc6 [10]. Although XORC has been shown to be a distinct activity from RLF-B [9], the relationship between XCdc6 and RLF-B is currently unclear. Here we show that active XCdc6 is loaded onto chromatin in RLF-defective extracts, and show that both RLF-M and RLF-B are still required for the licensing of XCdc6-containing chromatin. Further, RLF-B can be separated from XCdc6 by immunoprecipitation and standard chromatography. These experiments demonstrate that RLF-B is both functionally and physically distinct from XCdc6, and that XCdc6 is loaded onto chromatin before RLF-B function is executed.
Keywords: DNA Replication, Licensing Factor, ORC, Cdc6, RLF-B
Results and Discussion
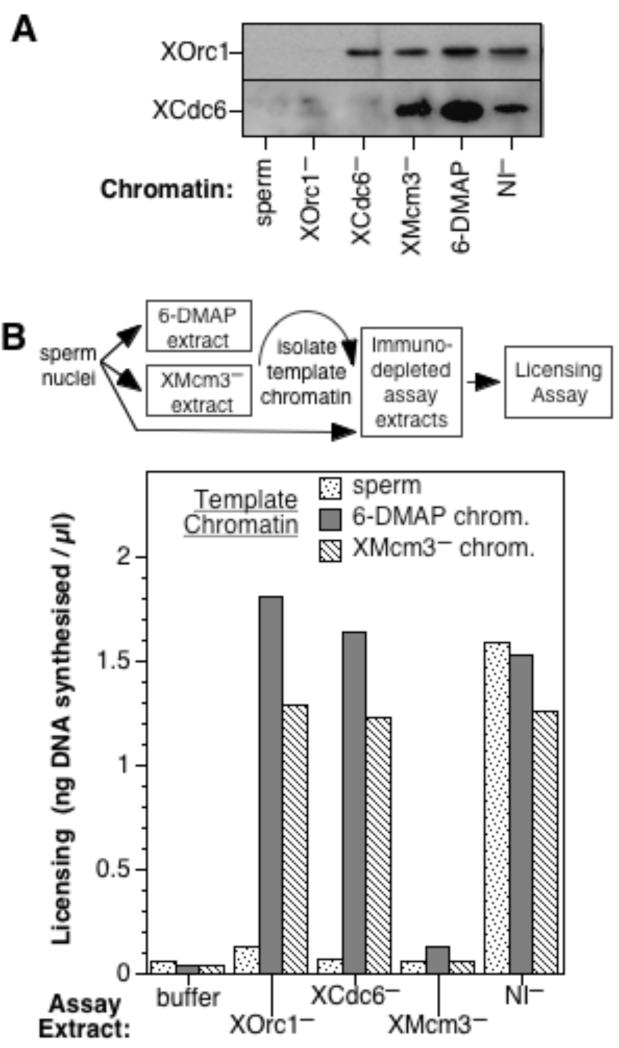
In order to explore different activities required for licensing, we immunodepleted Xenopus egg extracts with antibodies specific for XOrc1 (to remove XORC), XCdc6, or XMcm3 (to remove RLF-M) or with non-immune serum (as a negative control). Extracts were also treated with the kinase inhibitor 6-DMAP as described [12] to generate “6-DMAP extract” which is functionally devoid of RLF activity. Xenopus sperm nuclei were incubated briefly in these extracts to allow proteins to load onto the DNA, and the chromatin was then isolated and blotted for XOrc1 and XCdc6 (Fig 1A). Consistent with previous reports [9-11], both XOrc1 and XCdc6 were assembled onto chromatin in control extract (NI−) or in RLF-defective extracts (XMcm3-depleted or 6-DMAP extract), but XCdc6 was not loaded onto chromatin lacking XORC. In parallel, we measured the ability of the different extracts to license sperm nuclei (Fig 1B, stippled bars). Although efficient licensing occurred in extracts mock-depleted with non-immune antibodies, licensing did not occur in 6-DMAP extract or in extracts immunodepleted of either XOrc1, XCdc6 or XMcm3. This demonstrates that, consistent with previous reports [3, 4, 9-11], XORC, XCdc6 and XMcm3 are all independently required for licensing naive sperm nuclei. We next determined whether the XORC and XCdc6 assembled onto chromatin in XMcm3-depleted extract (“XMcm3− chromatin”) or in 6-DMAP extract (“6-DMAP chromatin”) was functional. Chromatin templates were assembled in XMcm3-depleted or 6-DMAP extract, and were then isolated and incubated for a second time in either buffer or in another immunodepleted extract, before being subjected to a functional licensing assay (Fig 1B). Both “XMcm3− chromatin” and “6-DMAP chromatin” were efficiently licensed on subsequent incubation in XOrc1-depleted or XCdc6-depleted extract, but not on incubation in buffer or XMcm3-depleted extract (Fig 1B). This demonstrates that despite being unlicensed, chromatin assembled in either XMcm3-depleted extract or 6-DMAP extract contains active XORC and XCdc6.
Figure 1.
Unlicensed chromatin contains active XORC and XCdc6. A. Xenopus sperm nuclei were incubated for 15 mins in interphase Xenopus extracts previously immunodepleted with antibodies against XOrc1, XCdc6, XMcm3 or with non-immune antibodies (NI−), or in 6-DMAP extract. Chromatin was isolated and subjected to SDS PAGE and immunoblotted with antibodies against XOrc1 and XCdc6. B. Untreated sperm chromatin (“sperm”) or sperm chromatin previously assembled in either 6-DMAP extract (“6-DMAP chrom.”) or in XMcm3-depleted extract (“XMcm3−chrom.”) was isolated and incubated in a second aliquot of extract previously immunodepleted with antibodies against XOrc1, XCdc6 or XMcm3, before being transferred to 6-DMAP treated extract and incubated for a further 90 mins with [α-32P]dATP, to assess the degree of licensing (expressed as ng DNA synthesised/μl).
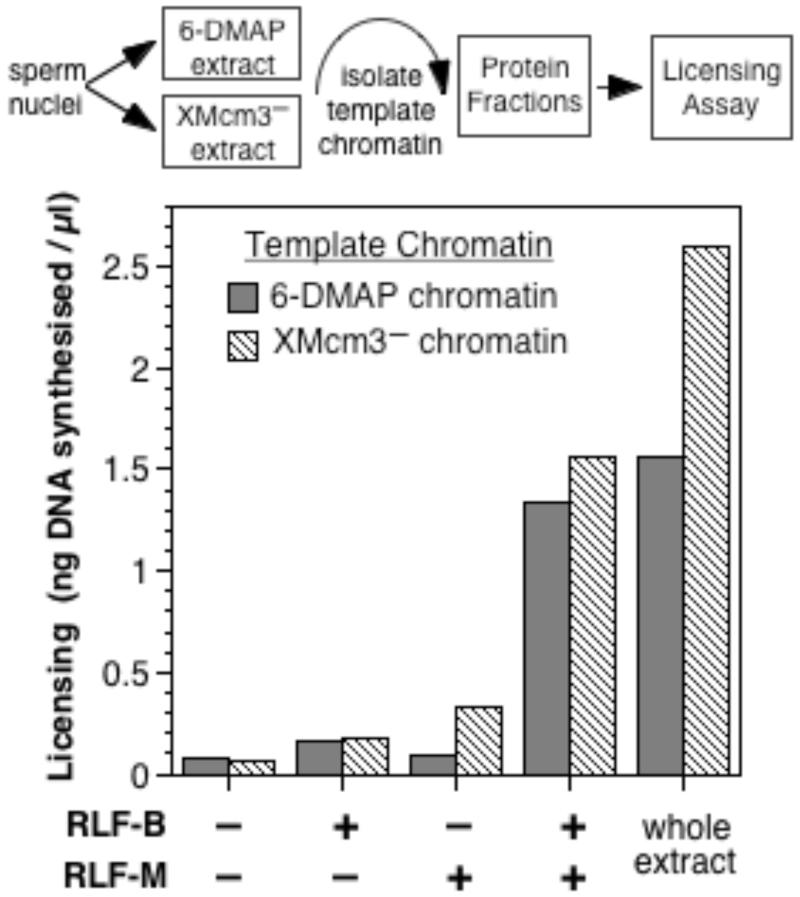
We have previously shown that the two essential components of RLF, RLF-M (consisting of complexes of the MCM/P1 proteins [3, 4, 6, 7]) and the currently unpurified RLF-B, can be separated by differential precipitation with polyethylene glycol (PEG) [3, 13]. We therefore prepared unlicensed chromatin by isolating Xenopus sperm chromatin after incubation in either XMcm3-depleted extract or in 6-DMAP-treated extract. We next determined which RLF components are required for this “XMcm3-chromatin” and “6-DMAP chromatin” to become licensed. Fig 2 shows that only when these chromatin templates were incubated with both RLF-B and RLF-M fractions did they become functionally licensed. Since both “XMcm3-chromatin” and “6-DMAP chromatin” contain functional XORC and XCdc6 and could be licensed in XOrc1-depleted or XCdc6-depleted extract (Fig 1), this demonstrates that RLF-B is an essential component of the licensing system that is functionally distinct from either XORC or XCdc6. The requirement for RLF-B in the licensing of chromatin assembled in XMcm3-depleted extract also demonstrates that RLF-B is not simply required to reverse an abnormal effect that occurs in 6-DMAP-treated extract, but is likely to represent a physiological activity required for the licensing of chromatin in a normal cell cycle.
Figure 2.
6-DMAP and XMcm3− chromatin require both RLF-B and RLF-M for licensing. Sperm chromatin previously assembled in either 6-DMAP extract (“6-DMAP chromatin”) or in XMcm3-depleted extract (“XMcm3− chromatin”) was isolated and incubated in combinations of PEG-cut fractions containing RLF-B or RLF-M or in untreated extract, before being transferred to 6-DMAP treated extract and incubated for a further 90 mins with [α-32P]dATP, to assess the degree of licensing (expressed as ng DNA synthesised/μl).
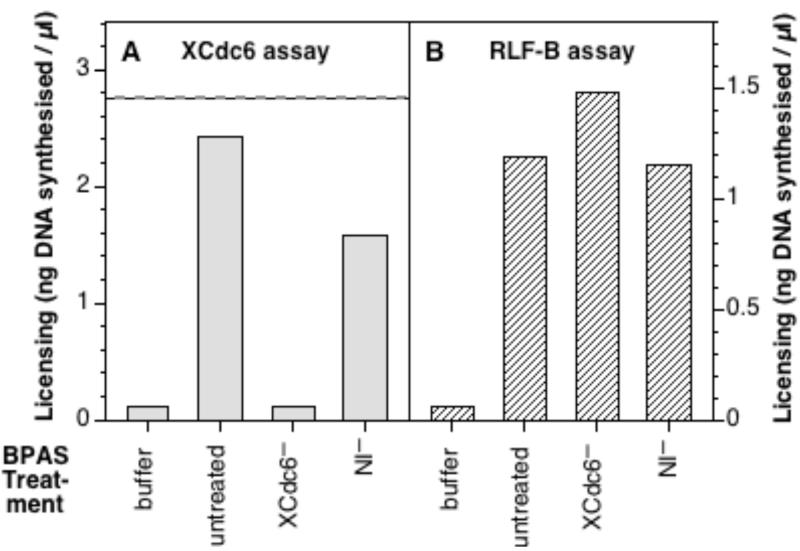
We next investigated whether the different RLF-B and XCdc6 activities are physically distinct, and whether they can be separated from one another. The fractionation of RLF-B over three sequential chromatographic steps (PEG precipitation, phosphocellulose chromatography and ammonium sulphate precipitation) has been described previously [9, 13]. The resultant fraction (called “BPAS”) contains both XORC and XCdc6 in addition to RLF-B ([9] and below). The BPAS fraction was immunodepleted using either antibodies to XCdc6 (XCdc6−) or non-immune antibodies (NI−) and the resultant material was then assayed for XCdc6 activity (Fig 3A) and RLF-B activity (Fig 3B). This showed that the BPAS fraction had both XCdc6 and RLF-B activity (Figs 3A and B, “untreated”), and that the XCdc6 activity was completely removed by depletion with anti-XCdc6 antibodies (Fig 3A, “XCdc6−”). In contrast, the RLF-B activity of BPAS was not reduced by the depletion of XCdc6 (Fig 3B, “XCdc6−”), showing that RLF-B had been separated from XCdc6.
Figure 3.
Immunodepletion of XCdc6 from partially-purified RLF-B. The BPAS fraction was prepared and then immunodepleted with anti-XCdc6 antibodies (XCdc6−) or with an equivalent quantity of non-immune antibody (NI−). The resultant material was then compared with buffer alone or untreated BPAS in assays for XCdc6 activity (A) or RLF-B activity (B). A. Aliquots were added directly to whole interphase extract previously immunodepleted of XCdc6. Sperm nuclei were then incubated in each aliquot for 15 mins, before being transferred to 6-DMAP treated extract and incubated for a further 90 mins with [α-32P]dATP, as a measure of the rescue of XCdc6 activity. The dashed line shows the amount of licensing observed in whole interphase extract depleted with non-immune antibodies. B. Aliquots were incubated with partially-purified RLF-M and 6-DMAP chromatin for 15 mins, before being transferred to 6-DMAP treated extract and incubated for a further 90 mins with [α-32P]dATP to measure the RLF-B activity.
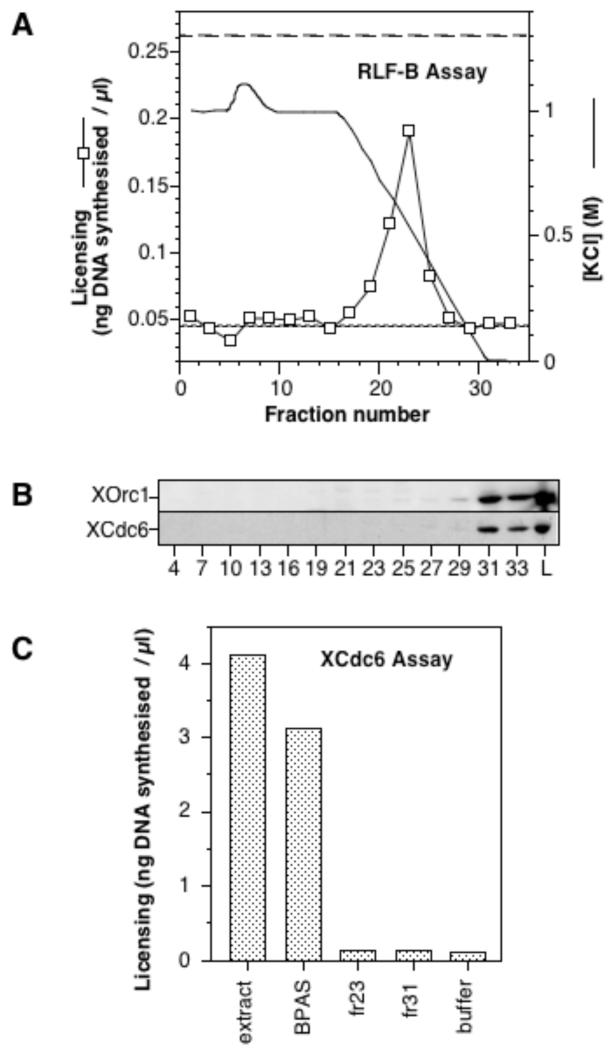
As an alternative method of separating RLF-B and XCdc6, we chromatographed the BPAS fraction on phenyl Sepharose. A peak of RLF-B activity eluted at ~500 mM KCl (Fig 4A, fractions 21 - 25) representing >90% of the activity loaded onto the column. In contrast, XOrc1 and XCdc6 were not eluted until much lower salt concentrations, with the majority of XOrc1 and XCdc6 remaining bound to the column under these elution conditions (Fig 4B and data not shown). No XCdc6 could be detected in the fractions containing RLF-B activity: note that this is a very long exposure of the blot, and that 25× less material is loaded in the BPAS lane L than in the individual fractions. Although the BPAS loaded onto the phenyl Sepharose column could restore activity to a XCdc6-depleted extract (Fig 3C, “BPAS”), there was insufficient XCdc6 in either the RLF-B peak (fraction 23) or even in the last fraction of the elution (fraction 31) to provide any XCdc6 activity (Fig 4C, “fr 23” and “fr 31”). This demonstrates a clear physical separation of RLF-B and XCdc6 by standard chromatography.
Figure 4.
Separation of RLF-B and XCdc6 by hydrophobic chromatography. The BPAS fraction, containing both RLF-B and XCdc6, was fractionated on phenyl Sepharose. A. Chromatography profile, showing RLF-B activity of individual fractions (squares), plus the KCl elution gradient. The RLF-B assay was performed by incubating diluted column fractions with partially-purified RLF-M and 6-DMAP chromatin for 15 mins, before being transferred to 6-DMAP treated extract and incubated for a further 90 mins with [α-32P]dATP. The fine dashed line shows the background activity of buffer alone, whilst the coarse dashed line shows the activity of the starting BPAS fraction at a comparable dilution. B. 50% of each fraction was DOC-TCA precipitated, electrophoresed and then immunoblotted with antibodies against XOrc1 and XCdc6. Fraction L is 2% of the BPAS sample loaded onto the phenyl Sepharose column. C. Aliquots of either crude extract (“Extr.”), the BPAS load to the phenyl Sepharose column (“BPAS”), or fractions 23 or 31 were added directly to extract immunodepleted of XCdc6. Sperm nuclei were then incubated in each aliquot for 15 mins, before being transferred to 6-DMAP treated extract and incubated for a further 90 mins with [α-32P]dATP, as a measure of the rescue of XCdc6 activity.
The experiments shown here demonstrate a clear functional and physical separation of RLF-B from XCdc6 using a variety of immunological and chromatographic techniques. We have also observed that recombinant XCdc6 protein can restore licensing activity to XCdc6-depleted extract, but provides no RLF-B activity (data not shown; see supplementary material). We are currently attempting to complete the RLF-B purification in order to identify it and understand its function. Despite it being unidentified, the results shown here strongly suggest that RLF-B acts in concert with RLF-M on chromatin that already contains XORC and XCdc6. Fig 2 shows that both RLF-M and RLF-B are required for the licensing of chromatin previously assembled in an XMcm3-depleted extract. Since XMcm3-depleted extract contains active RLF-B [3], this demonstrates that RLF-B cannot execute its essential function before RLF-M loading. Given that XCdc6 must be loaded onto chromatin prior to RLF-M [10 and this paper, Fig 1], these experiments also show that the essential function of RLF-B in licensing can only be executed after XCdc6 has bound to chromatin.
Materials and Methods
Preparation of egg extracts
Metaphase-arrested Xenopus egg extracts were prepared as described [13]. For replication assays they were supplemented with 100 μg/ml cycloheximide, 25 mM phosphocreatine, 15 μg/ml creatine phosphokinase and [α-32P]dATP, and were then released into interphase with 0.3 mM CaCl2. Licensing factor-defective “6-DMAP extract” was prepared in a similar way, except 3 mM 6-dimethylaminopurine was added prior to CaCl2 release [12, 13]. Immunodepletion of interphase extracts or BPAS with antibodies raised against XOrc1 [9], XCdc6 [10] or XMcm3 [5] or with antibodies from non-immune rabbit serum, was performed as described [13]. Control extracts were diluted with an equivalent amount of buffer. Once immunodepletion was complete, extracts were snap frozen in liquid nitrogen in 10 μl aliquots for future use.
Chromatin Templates
Demembranated Xenopus sperm nuclei were prepared as described [13]. They were assembled into chromatin by incubation for 15 min at 23 °C in 6-DMAP extracts at 20,000 - 40,000 nuclei/μl (50 - 100 ng DNA/μl) or in immunodepleted extracts at 10,000 - 13,000 nuclei/μl (25 - 30 ng DNA/μl). The lower concentration in immunodepleted extracts is to compensate for the ~3-fold dilution that occurs as a consequence of depletion. The extract was then diluted 10-fold in Nuclear Isolation Buffer [13] (NIB: 50 mM KCl, 50 mM Hepes KOH pH 7.6, 5 mM MgCl2, 2 mM DTT, 0.5 mM spermine 3HCl, 0.15 mM spermine 4HCl, 1 μg/ml each aprotinin, leupeptin and pepstatin) supplemented with 2.5 mM Mg-ATP and 0.01% Triton X-100, and then underlayered with the same buffer containing 15% sucrose. Chromatin was then pelleted at 1,500 × g in a swinging bucket centrifuge for 5 min at 4°C. The diluted extract and the top part of the cushion were carefully removed, and the chromatin pellet resuspended in NIB supplemented with 2.5 mM Mg-ATP and 0.01% Triton X-100. Aliquots of pelleted chromatin were either used for replication assays, or were immunoblotted by standard techniques using 10% SDS-PAGE and ECL visualisation (Amersham).
Chromatography
The BPAS fraction was prepared as described [9, 13]. For phenyl Sepharose chromatography, BPAS was made 1 M with respect to KCl in Hepes Buffer (20 mM Hepes KOH pH 8; 10% sucrose; 1 μg/ml each aprotinin, leupeptin and pepstatin) and applied to a 1ml HiTrap phenyl Sepharose column (Pharmacia) equilibrated in the same buffer. The column was developed with a gradient over 3 column volumes to Hepes Buffer without KCl. In order to overcome the inhibitory affect of the high salt on the RLF-B and XCdc6 assays, aliquots were diluted five-fold with Hepes Buffer prior to assay. For blotting, aliquots were DOC-TCA precipitated as follows: 50% of each sample was supplemented with 0.015% deoxycholate, mixed and incubated for 10 mins at room temperature, supplemented with 5% trichloroacetic acid, mixed and microfuged for 5 mins. The supernatant was removed and the pellet was resuspended in 0.01 N NaOH and then supplemented with Laemmli protein sample buffer for SDS-PAGE.
Replication Assays
DNA synthesis was measured by incorporation of [α-32P]dATP into acid insoluble material as described [13], assuming a dATP pool in the extract of 50 μM [14]. Licensing assays were performed by measuring DNA synthesis in 6-DMAP-treated extract as described [3, 13]. Briefly, 0.3 μl chromatin templates (either demembranated Xenopus sperm or sperm that had been pre-assembled in an RLF-defective extract) were incubated with 2 μl assay fractions for 15 mins. The reaction was then supplemented with 5.7 μl 6-DMAP extract containing [α-32P]dATP and the incubation continued for a further 90 mins, after which the total amount of DNA synthesis was measured. All incubations were performed at 23°C.
Supplementary Material
Acknowledgements
We thank Anne Donaldson for comments on the manuscript, and Alan Prescott for help in preparation of the figures. This work is supported by the Cancer Research Campaign (SP2385).
References
- 1.Blow JJ, Laskey RA. A role for the nuclear envelope in controlling DNA replication within the cell cycle. Nature. 1988;332:546–548. doi: 10.1038/332546a0. [DOI] [PubMed] [Google Scholar]
- 2.Tada S, Blow J. The replication licensing system. Biological Chemistry. 1998;379:941–949. [PMC free article] [PubMed] [Google Scholar]
- 3.Chong JPJ, Mahbubani MH, Khoo C-Y, Blow JJ. Purification of an Mcm-containing complex as a component of the DNA replication licensing system. Nature. 1995;375:418–421. doi: 10.1038/375418a0. [DOI] [PubMed] [Google Scholar]
- 4.Kubota Y, Mimura S, Nishimoto S, Takisawa H, Nojima H. Identification of the yeast MCM3-related protein as a component of Xenopus DNA Replication Licensing Factor. Cell. 1995;81:601–609. doi: 10.1016/0092-8674(95)90081-0. [DOI] [PubMed] [Google Scholar]
- 5.Madine MA, Khoo C-Y, Mills AD, Laskey RA. MCM3 complex required for cell cycle regulation of DNA replication in vertebrate cells. Nature. 1995;375:421–424. doi: 10.1038/375421a0. [DOI] [PubMed] [Google Scholar]
- 6.Thömmes P, Kubota Y, Takisawa H, Blow JJ. The RLF-M component of the replication licensing system forms complexes containing all six MCM/P1 polypeptides. EMBO J. 1997;16:3312–3319. doi: 10.1093/emboj/16.11.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kubota Y, Mimura S, Nishimoto S, Masuda T, Nojima H, Takisawa H. Licensing of DNA replication by a multi-protein complex of MCM/P1 proteins in Xenopus eggs. EMBO J. 1997;16:3320–3331. doi: 10.1093/emboj/16.11.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carpenter PB, Mueller PR, Dunphy WG. Role for a Xenopus Orc2-related protein in controlling DNA replication. Nature. 1996;379:357–360. doi: 10.1038/379357a0. [DOI] [PubMed] [Google Scholar]
- 9.Rowles A, Chong JP, Brown L, Howell M, Evan GI, Blow JJ. Interaction between the origin recognition complex and the replication licensing system in Xenopus. Cell. 1996;87:287–296. doi: 10.1016/s0092-8674(00)81346-x. [DOI] [PubMed] [Google Scholar]
- 10.Coleman TR, Carpenter PB, Dunphy WG. The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- 11.Romanowski P, Madine MA, Rowles A, Blow JJ, Laskey RA. The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr Biol. 1996;6:1416–1425. doi: 10.1016/s0960-9822(96)00746-4. [DOI] [PubMed] [Google Scholar]
- 12.Blow JJ. Preventing re-replication of DNA in a single cell cycle: evidence for a replication licensing factor. J Cell Biol. 1993;122:993–1002. doi: 10.1083/jcb.122.5.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chong JP, Thömmes P, Rowles A, Mahbubani HM, Blow JJ. Characterization of the Xenopus replication licensing system. Methods Enzymol. 1997;283:549–564. doi: 10.1016/s0076-6879(97)83043-1. [DOI] [PubMed] [Google Scholar]
- 14.Blow JJ, Laskey RA. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell. 1986;47:577–587. doi: 10.1016/0092-8674(86)90622-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.