Abstract
Physiological studies require sensitive tools to directly quantify transport kinetics in the cell/tissue spatial domain under physiological conditions. Although biosensors are capable of measuring concentration, their applications in physiological studies are limited due to the relatively low sensitivity, excessive drift/noise, and inability to quantify analyte transport. Nanomaterials significantly improve the electrochemical transduction of microelectrodes, and make the construction of highly sensitive microbiosensors possible. Furthermore, a novel biosensor modality, self-referencing (SR), enables direct measurement of real-time flux and drift/noise subtraction. SR microbiosensors based on nanomaterials have been used to measure the real-time analyte transport in several cell/tissue studies coupled with various stimulators/inhibitors. These studies include: glucose uptake in pancreatic β cells, cancer cells, muscle tissues, intestinal tissues and P. Aeruginosa biofilms; glutamate flux near neuronal cells; and endogenous indole-3-acetic acid flux near the surface of Zea mays roots. Results from the SR studies provide important insights into cancer, diabetes, nutrition, neurophysiology, environmental and plant physiology studies under dynamic physiological conditions, demonstrating that the SR microbiosensors are an extremely valuable tool for physiology research.
Keywords: Biosensor, Self-referencing, Flux, Enzyme, Nanomaterial
1. Introduction
Electrochemical biosensors are transducers that convert biological information (such as analyte concentration) into electrical signals (such as current or voltage). Electrochemical biosensors are more efficient than conventional measurement techniques (including radioisotope tracing (Guillam et al., 2000; Hellman et al., 1974; Sweet et al., 1996; Zawalich and Matschinsky, 1977), NMR spectroscopy (Weiss et al., 1989), and microfluorometry assays (Moley et al., 1998; Passonneau and Lowry, 1993)) due to the high sensitivity, real-time monitoring capabilities and low cost, while the conventional techniques are complex, expensive and severely limited in terms of spatial and temporal resolution. Most electrochemical biosensors have two major components: the biorecognition element and the transduction element. Biorecognition elements include enzymes (for amperometric sensors (Gouveia-Caridade et al., 2008; Hrapovic et al., 2004; Kang et al., 2007; Salimi et al., 2004; Yao and Shiu, 2007; Zou et al., 2008)) and ionophores (for potentiometric sensors (McLamore and Porterfield, 2011; McLamore et al., 2009; Porterfield, 2007; Porterfield et al., 2009), and the transduction elements include the electrode and nanomaterials (Shi and Porterfield, 2011). Most biosensors function based on a two-step scheme: biorecognition and transduction (Shi and Porterfield, 2011). In biorecognition, the biorecognition element recognizes and binds to the target compound. The specificity associated with the binding ensures the selectivity of the biosensor. In transduction, a series of electrochemical reactions take place in the proximity of the transduction element(s), and sometimes the reactions are driven by an externally applied potential (working potential). The final outcome is an electrical signal (current or voltage) which is proportional to compound concentration.
2. Amperometric biosensor
Amperometric biosensors are used to measure the electroactive molecules such as H2O2 (Marc et al., 1997), NADH (Santos et al., 2006; Tsai et al., 2007) and indole-3-acetic acid (McLamore et al., 2010a). The target molecule is oxidized or reduced by the working potential, and a current proportional to concentration is generated. When coupled with enzymes (biorecognition element of amperometric sensor), amperometric biosensors can measure non-electroactive molecules such as glucose (Shi et al., 2011a), glutamate (McLamore et al., 2010b) and ethanol (Azevedo et al., 2005), because enzymes convert the target compound into an electro-active intermediate such as H2O2. Enzymes can be immobilized on the biosensor via a covalent linker such as glutaraldehyde (Shi et al., 2011b), or via adsorption by polymers (Shi et al., 2011b). Oxidase and dehydrogenase are the most commonly used enzymes. Biosensors based on oxidase rely on H2O2 as the electroactive intermediate, while those based on dehydrogenase rely on NADH as the intermediate. Take ethanol biosensors as an example. Alcohol dehydrogenase (ADH) (Santos et al., 2006; Tsai et al., 2007) and alcohol oxidase (AOx) (Gouveia-Caridade et al., 2008; Yildiz and Toppare, 2006) have been used for ethanol biosensing. ADH converts ethanol into acetaldehyde and NADH in the presence of NAD+ as a cofactor. NADH is then oxidized:
AOx converts ethanol into acetaldehyde and hydrogen peroxide. Hydrogen peroxide is then oxidized:
The use of ADH or AOx has both advantages and disadvantages. Biosensors based on ADH require external NAD+ for the production of NADH. The inefficient diffusion of NAD+ towards the ADH immobilized on the biosensor complicates the measurement process and affects the biosensor sensitivity, detection limit and linear detection range (Azevedo et al., 2005). Biosensors based on AOx require O2 to oxidize ethanol. When the ethanol concentration in the analyte is low, the O2 from ambient air is adequate for the reactions to occur. However, when ethanol concentration increases, O2 is gradually depleted and the oxidation of ethanol is constrained (although the oxidation of H2O2 also generates O2). Therefore, the oxidation of ethanol becomes inadequate. This is reflected as a gradually attenuated current response when ethanol concentration increases. In other words, the current-concentration curve in the high concentration region is non-linear, where it is theoretically linear based on biosensing mechanism.
3. Nanomaterials and biosensing
In order to apply biosensors to physiological research, miniaturization of sensors is required for a high spatial resolution, because the sensors are often operated at cell or tissue level. Miniaturization increases the resistance of the sensor, which significantly decreases the maximum attainable sensitivity (Bard and Faulkner, 2000). The sensitivity issue affects not only the limit of detection, but also the capability of measuring very small changes in concentration over time (Shi et al., 2011a), while the small changes can be key to exploring important physiological phenomena, such as β cell glucose consumption during insulin secretion (Jung et al., 2000). One effective way to solve the low sensitivity problem is to enhance electrochemical transduction via incorporating nanomaterials. Carbon nanotube (CNT) (Claussen et al., 2011; McLamore et al., 2010a; McLamore et al., 2010b; McLamore et al., 2011; Shi et al., 2011a; Shi et al., 2011b; Shi et al., 2011c), graphene (Kang et al., 2009; Shao et al., 2010; Zhang et al., 2011), graphene oxide (Shi et al., in press) and metal nanomaterials (Claussen et al., 2011; McLamore et al., 2010a; McLamore et al., 2010b; McLamore et al., 2011; Shi et al., 2011a; Shi et al., 2011b; Shi et al., 2011c) are the most commonly used nanomaterials for biosensing enhancement. Carbon nanomaterials increase electrochemical transduction partially due to the unique structure and the different local density of states, which increase the electronic interaction width and decrease the activation energy for redox reactions (Nugent et al., 2001), and partially due to the defect sites which facilitate the chemisorption of molecules and lower the activation energy (Chakrapani et al., 2003). Metal nanomaterials possess electrocatalytic activities due to the multiple oxidation states, enabling reactants such as H2O2 to form intermediates at the surface, and lowering the activation energy of reactions such as H2O2 oxidation (Li et al., 2005).
The combination of metal and carbon nanomaterials for biosensor enhancement has proved to be feasible and more effective than using a single material (Hrapovic et al., 2004; Kang et al., 2007, 2008; McLamore et al., 2010a; McLamore et al., 2010b; McLamore et al., 2011; Shi et al., 2011a; Shi et al., 2011c; Shi and Porterfield, 2011; Zou et al., 2008). For example, CNT can act as the molecular template for Pt black electrodeposition, and the resultant hybrid nanocomposite is more effective in enhancing biosensor performance compared with either CNT or Pt black used (Shi et al., 2011a).
4. Self-referencing
While most biosensors only measure the concentration of the target compound, there has been a need to measure the real-time transport (flux) of the compound, as flux characterizes the dynamic transmembrane transport activities, which is important for studying transport related physiological processes. Self-referencing (SR) technique has been developed for this purpose (Porterfield, 2007). SR, based on Fick’s first law of diffusion (eq.1), operates within a concentration gradient produced by cells or tissues (Porterfield, 2007):
| (1) |
where J is the flux between two points termed the “near pole” and the “far pole” at the surface of the tissues or cells, D is the diffusion coefficient of the target compound, ΔC is the concentration difference of the target compound between the near pole and the far pole, and ΔX is the distance between the near pole and the far pole (10–50 μm). From eq. 1, J is proportional to ΔC, thus, the dynamic flux can be derived from the static differential concentration. SR microbiosensor was first used by Zisman to measure the contact potential difference in metals (Zisman, 1932), while Jeffe et al. significantly refined this technique by using a lock-in amplifier (Jaffe and Nuccitelli, 1974).
A typical SR system consists of a microscope and a head-stage sensor amplifier driven by a translational motion control system. A microbiosensor is connected to the amplifier. During measurements, the microbiosensor oscillates between the near pole and the far pole, and takes measurements at both positions (move-wait-measure). The position and the movement of the microbiosensor are controlled by the computer driven motion control system (Porterfield, 2007) and the frequency of the translational movement is usually 0.2–0.5 Hz (Kuhtreiber and Jaffe, 1990). SR technique has been combined with amperometric (McLamore et al., 2010a; McLamore et al., 2010b; McLamore et al., 2011; Shi et al., 2011c), potentiometric (Smith et al., 1999) and optical (Porterfield et al., 2006) microbiosensors with a detection limit of as low as 1 fmol cm−2 s−1. One advantage of the SR technique is that a static concentration sensor is converted into a dynamic biophysical flux sensor, which provides information on both magnitude and direction of transport. Another advantage is the high signal-to-noise ratio because the drift and noise patterns are common to the output signals at both the near pole and the far pole. By calculating ΔC, the interferences are eliminated from the differential signal (phase discrimination) (Porterfield, 2007). A schematic of the SR system is shown in Fig. S1.
5. Physiological applications of SR microbiosensors based on nanomaterials
A previous paper reviewed the applications of SR ion, NO, ascorbate, H2O2 and O2 sensors for physiological studies (Porterfield, 2007). Recently, the development of enzyme based SR microbiosensors enabled the flux measurement for many non-electroactive species (e.g., glucose and glutamate), while the incorporation of nanomaterials into sensor construction significantly increased the sensitivity and detection limit. These improvements have significantly broadened the applications of the SR technique (McLamore et al., 2010a; McLamore et al., 2010b; McLamore et al., 2011; Shi et al., 2011c).
5.1. SR glucose sensor
Glucose is the key molecule in many biochemical pathways, such as glycolysis. Glucose biosensors measure glucose via two steps:
Step 1. The biorecognition element glucose oxidase (GOx) converts glucose and O2 into gluconic acid and H2O2,
Step 2. H2O2 is electrochemically oxidized at +500 mV vs. Ag/AgCl,
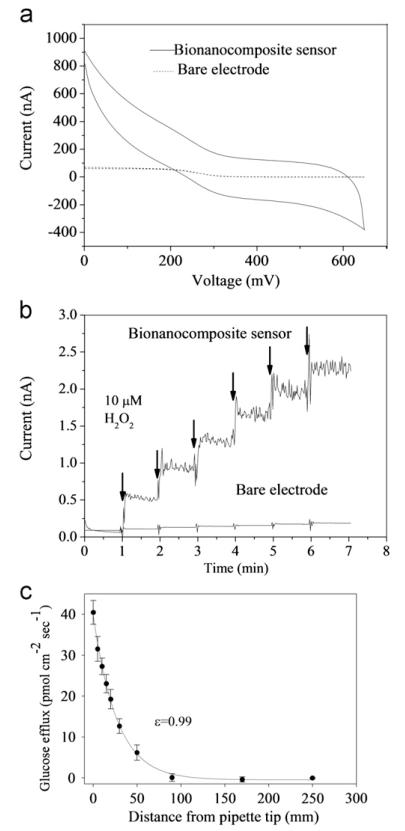
Recent works incorporated multi-walled carbon nanotubes (MWCNT) and Pt black in glucose microbiosensor construction, and the microbiosensor was combined with the SR platform for the measurement of glucose flux (McLamore et al., 2011; Shi et al., 2011c). In this scheme, Pt black was first electrodeposited on a Pt/Ir microelectrode to increase electrocatalytic transduction, and MWCNT was subsequently immobilized using Nafion. MWCNT/Nafion was used for adsorbing enzymes (Wang et al., 2003). A sigmoid CV curve with steady state diffusion-limited current was observed for unmodified microelectrodes (Fig. 1a), which was characteristic of microelectrodes (Heinze, 1993). For the bionanocomposite sensor, non-steady state diffusion-limited characteristics was observed (Fig. 1a), which was likely due to the high film resistance of Nafion (Hrapovic et al., 2004). According to the following equation (Heinze, 1993),
where ilim is the diffusion-limited current, K is a geometric constant (K=2π for hemispherical diffusion model), n is the number of electrons transferred during the redox of , F is the faradic constant, D is the diffusion coefficient for potassium ferricyanide (6.70±0.02 × 10−6 cm2/s), C is the concentration of potassium ferricyanide (4 mM) and r is the microelectrode tip radius, the area of the CV curve reflects the effective surface area. Thus, the surface area for bionanocomposite modified microelectrodes has been significantly increased compared with unmodified microelectrodes. The electrocatalytic activities in terms of H2O2 sensitivity have been shown to be increased as well (Fig.1b) (McLamore et al., 2011; Shi et al., 2011c).
Fig. 1.
(a) CV in 4 mM Fe(CN)63-/1 M KNO3 for a bare micro electrode and a bionanocomposite sensor at a scan rate 20 mV/s. (b) Representative current response to H2O2 for a bionanocomposite sensor and a bare electrode. (c) Abiotic step back experiment from pulled micropipette containing 3 mM glucose and 0.5% agar in PBS at 37 °C. Correlation coefficient (ε) between measured (●) and predicted flux (solid line) was 0.99. All error bars represent the standard error of the arithmetic mean. (Reprinted with permission from (Shi et al. 2011c)).
The capability of SR glucose sensor for measuring flux was calibrated through “step-back” experiment that involved using gradients created from artificial sources (e.g., pipette filled with agar and concentrated glucose solution) (Porterfield, 2007). The process was mainly used to characterize the impact of moving the sensor within the concentration gradient, because the movement of the sensor may cause mechanical convection to flux measurement. The flux values versus the distance to the source were compared to the mathematically modeled flux. The coefficient of correlation between the measured and modeled flux represented the dynamic efficiency of the sensor. As shown in Fig. 1c, a efficiency of 0.99 indicated that the SR glucose microbiosensor based on Pt black/MWCNT/Nafion could be readily used for glucose flux measurement (Shi et al., 2011c).
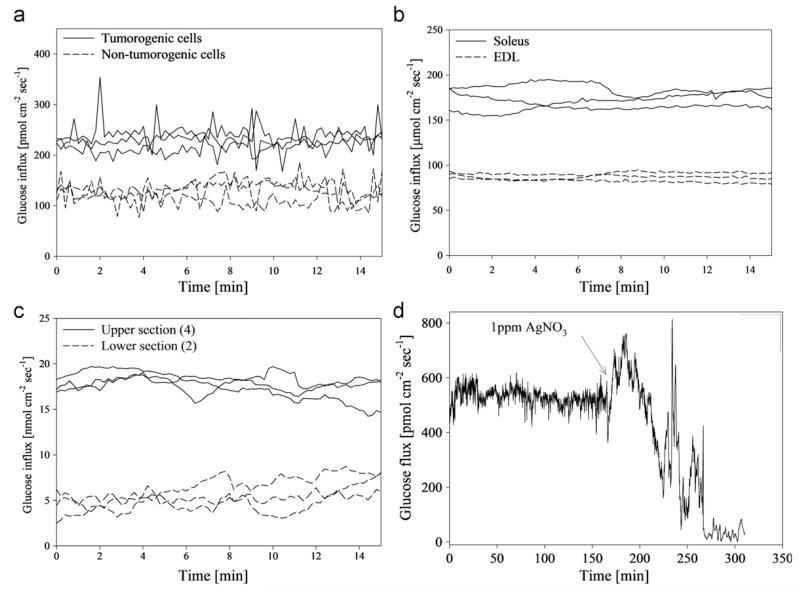
The SR glucose sensor was used to measure glucose flux in various cells/tissues (Fig. 2). With the SR sensor, we reported a higher glucose uptake in tumor cells than non-tumor cells (Fig. 2a), because tumor cells produce energy at a much higher rate than normal cells due to a higher rate of aerobic glycolysis, while normal cells produce most ATP in oxidative phosphorylation (Warburg effect) (Lu et al., 2002; Matoba et al., 2006; Pelicano et al., 2006; Xu et al., 2005). The soleus muscle tissue showed a significantly higher glucose uptake than the EDL tissue because soleus contained more slow-contracting type fibers, significantly more mitochondria, and typically more transmembrane glucose transport proteins (Fig. 2b) (Balon and Nadler, 1997; Holloszy and Hansen, 1996; Lagord et al., 1998). For intestine tissues, glucose uptake in upper jejunum tissue was significantly higher than the average flux in lower jejunum tissue due to higher sucrase activity in the upper section (Fig. 2c) (Goda and Koldovsk, 1985).
Fig. 2.
(a) Average glucose flux at the surface of tumorogenic and non-tumorogeinc human breast endothelial cells in αMEM (n=3). Glucose flux in cancerous cells was significantly higher than non-tumorogenic cells. (b) Average glucose flux at the surface of soleus and gastrocnemius muscle tissue from a mouse model (n=3). Glucose uptake in soleus tissue was significantly higher than gastrocnemius tissue for all preps. (c) Average glucose flux at the surface of upper jejunum and lower jejunum of excised mouse small intestinal tissue in RPMI media (n=3). Glucose flux in upper intestinal tissue was significantly higher than the lower intestine tissue. (d) Glucose flux into a mature (30 day old) Pseudomonas aeruginosa biofilm grown on a hollow fiber silicone membrane. (Reprinted with permission from (McLamore et al. 2011)).
P. aeruginosa is an opportunistic pathogen in water that may cause infectious disease in cystic fibrosis patients (Costerton et al., 1999). The measurement of glucose flux in P. aeruginosa and other bacteria was combined with antimicrobial studies, such as the effects of ionic silver on P. aeruginosa glucose metabolism (Hwang et al., 2007). Results showed that after AgNO3 treatment, glucose uptake was inhibited (Fig. 2d), indicating a sharp reduction in metabolism due to the anti-microbial activities of silver. This experiment has provided useful information on the physiology of biofilm, and the efficiency of antimicrobials in disinfecting drinking water.
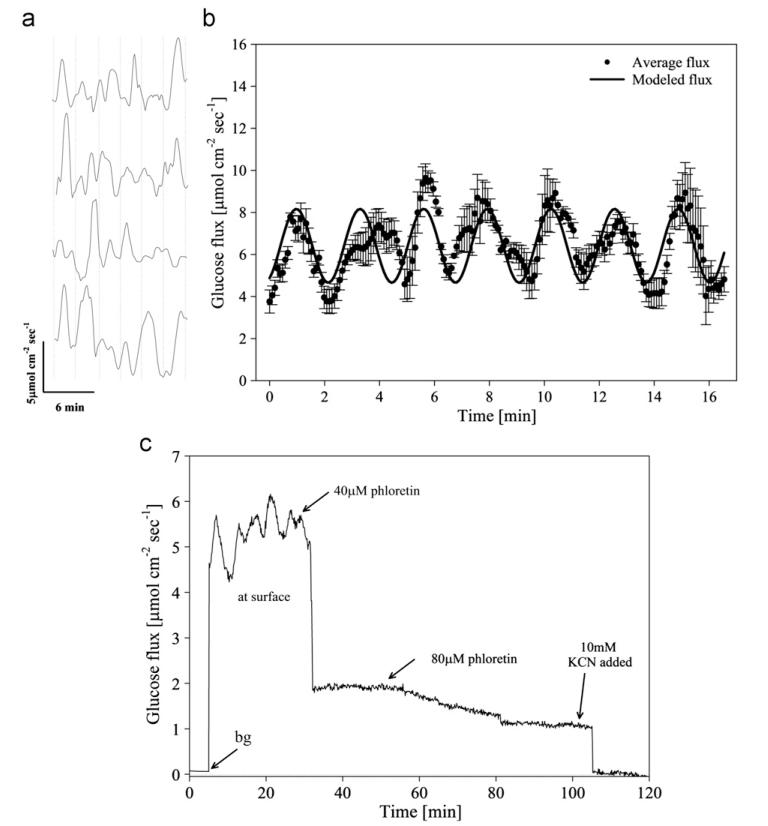
β cells are a type of cell in the mammalian pancreas that secrete insulin in a stimulus-secretion oscillatory manner (Bergstrom et al., 1989; Chou and Ipp, 1990; Lefebvre et al., 1987; Longo et al., 1991; Weigle, 1987a). The SR glucose sensor was used to measure glucose uptake around β cells and observed an oscillatory manner with a period around 3 min for the uptake (Fig. 3a & b) (Shi et al., 2011c), which was close to the period of oxygen influx, cytosolic Ca2+, ATP/ADP ratio, and insulin secretion (Barbosa et al., 1996; Kennedy et al., 2002; Longo et al., 1991; Nilsson et al., 1996; Porterfield et al., 2000). Since oscillations correlate with insulin secretion (Tornheim, 1997), and the oscillatory secretion of insulin is lost in both individuals with type 2 diabetes and their near relatives due to defective temporal coordination of β cell function (O’Rahilly et al., 1988; Polonsky et al., 1988; Weigle, 1987b), the measurement of flux oscillation in pancreatic β cells would contribute to the understanding of β cell physiology and diabetes. Subsequently, the kinetics of glucose transporters, and the effects of inhibitors (Fig. 3c) (e.g., phloretin (GLUT2 inhibitor) and KCN (oxidative phosphorylation inhibitor)) on glucose metabolism were studied using this technique, demonstrating the application of the SR sensor as a useful tool for pharmacology research.
Fig. 3.
(a) Representative oscillatory glucose flux in four replicate confluent INS 1 clusters out of 12 total. Average oscillation period (indicated by a vertical dashed line) was 2.9±0.6 min (n=12). (b) Average glucose flux (dots, n=12 replicate confluent INS 1 cells) and harmonic oscillator model (solid line): J=J0+a* sin(2π/T) *t(a=1.75 μmol cm−2 s−1, T=2.9 min, J0=6.41 μmol cm−2 s−1). Average basal glucose influx in cultured β cell (INS 1) clusters was 5.9±1.4 μmol cm−2 s−1 (n=12). Correlation coefficient between modeled and measured data was 0.78. (c) Representative glucose flux measured at the surface of cultured INS 1 β cells during inhibition by external addition of phloretin. A minimum of three successive oscillations were measured under basal conditions (average period was 2.9±0.4 min), and glucose transport was then inhibited by addition of 40 μM phloretin and 80 μM phloretin (total concentration noted). Glucose transport was subsequently abolished via addition of 10 mM KCN. (Reprinted with permission from (Shi et al. 2011)).
5.2. SR glutamate sensor
Glutamate is an important neurotransmitter in the central nervous system. The release and uptake of glutamate plays a vital role in synaptic plasticity, differentiation, apoptosis, and long-term potentiation (McLamore et al., 2010b; Ye et al., 1999). We have constructed a glutamate microbiosensor based on the scheme similar to that for the glucose microbiosensor, which incorporated Pt black and MWCNT. Glutamate oxidase was used as the biorecognition element, and the following reactions took place during measurement:
Step 1. The biorecognition element glutamate oxidase converts glutamate, H2O and O2 into 2-oxoglutarate, NH3 and H2O2,
Step 2. H2O2 is electrochemically oxidized at + 700 mV vs. Ag/AgCl. The oxidation potential used (+ 700 mV) was different from the potential used for SR glucose sensor (+500 mV), because the rate of H2O2 production by step 1 (glutamate oxidase catalyzed reaction) was much slower compared with glucose oxidase catalyzed reaction. To obtain the same level of current response, a higher oxidation potential was used.
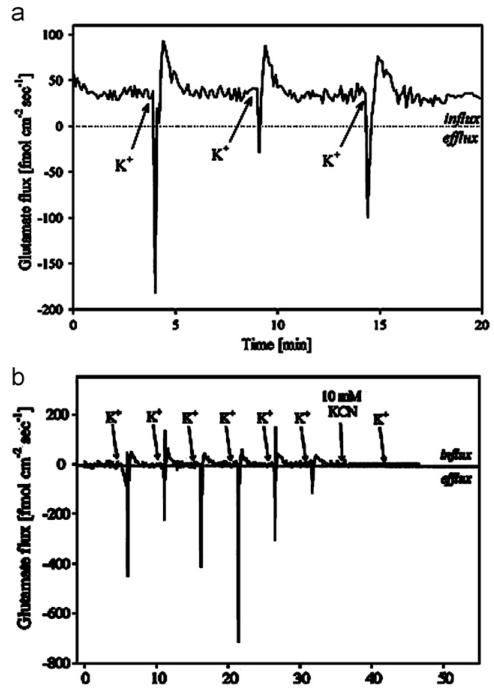
The sensor was operated in the SR modality to measure the physiological glutamate release and uptake in the extrasynaptic space of mixed neural culture (neurons, astrocytes, oligodendrocytes and fibroblasts), and we reported that when cells were electrochemically stimulated with Locke’s buffer containing 53 mM potassium, rapid efflux (release) of glutamate was observed (negative flux), followed by subsequent uptake (positive flux) (Fig. 4a), because potassium stimulated glutamate release via EAAT transporters (Wu and Fujikawa, 2002). To verify this underlying mechanism, we added threo-β-benzyloxyaspartate (TBOA), an effective competitive inhibitor of glutamate uptake by all of the major EAATs (Jabaudon et al., 1999), to the cells prior to stimulation. Although potassium stimulation was observed for TBOA treated cells, the average surface flux was significantly lower than for untreated cells (Fig. 4b), indicating inhibited glutamate transport by TBOA. The ability to quantify the direction and magnitude of glutamate transport will improve our understanding of spatially and temporally dynamic glutamate transport, and provide important insights into neuroscience research.
Fig. 4.
(a) Flux recorded 1 μM above the cells during three successive potassium stimulations with 150 μL of Locke’s buffer containing 53 mM KCl. Increase in Glu concentration and subsequent decrease in concentration were observed following stimulation. Phase sensitive detection provided signal noise filtration required to quantify biophysical glutamate efflux and influx. (b) Potassium stimulation of neural cells following 30 min of exposure to 100 μM threo-β-benzyloxyaspartate (TBOA). (Reprinted with permission from (McLamore et al. 2010b)).
5.3. SR indole-3-acetic acid sensor
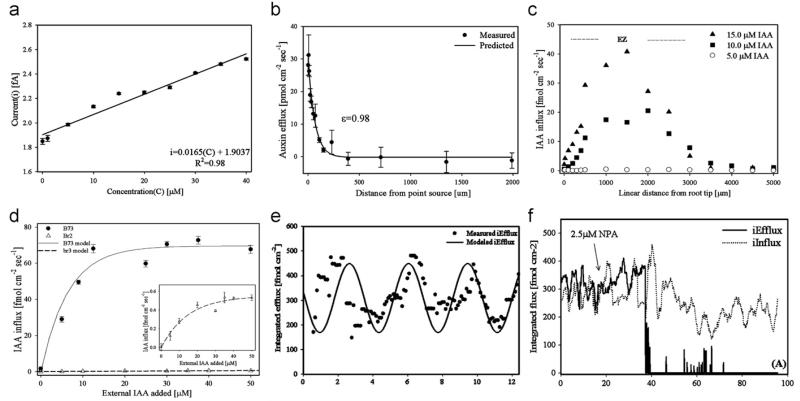
Indole-3-acetic acid (IAA) is a primary phytohormone produced in the cells of apex (bud) and very young leaves of a plant. The distribution and transport of IAA regulates plant growth and development. IAA contains a pyrrole ring, and the nitrogen atom in which could be electro-oxidized by the working potential of the biosensor, generating a current proportional to IAA concentration (Wu et al., 2003). To overcome the low sensitivity issue commonly associated with microbiosensors, we electrodeposited Pt black on the tip of the microelectrode, and then applied MWCNT suspended in dimethyl formamide to the modified electrode (McLamore et al., 2010a). The sensor showed excellent performance towards measuring IAA concentration (Fig. 5a) and flux from an artificial source (Fig. 5b). When operated in the SR modality, we observed induced IAA influx from B73 maize roots (Fig. 5c). We also observed that the IAA uptake in the distal elongation zone (DEZ) in B73 roots was significantly higher than in mutant br2 roots for all external additions of IAA (Fig. 5d), indicating a significant reduced capacity for polar auxin transport in the br2 mutant.
Fig. 5.
(a) Calibration curve of Pt black/CNT-modified IAA sensor between 0 and 40 μM IAA in ¼ MS media. Data are means and standard deviations of five independent experiments, R2=0.98. (b) Abiotic validation of SR IAA microsensor. Validation was achieved by measuring the gradient formed near the tip of a pipette filled with 5 mM IAA and 2% agar immersed in ¼ MS media. Data are means and standard deviations of three independent experiments. Solid line indicates predicted value based on empirical Fickian diffusion model. The correlation (ε) between the predicted and measured IAA efflux was 0.98, validating the use of the SR microsensor in growth media. (c) Induced IAA influx for a 4-day B73 root in ¼ MS media following increasing external addition of IAA. The area where maximum induced influx occurred (i.e. distal elongation zone) was approximately 1.45±0.2 mm from the root tip (linearized to account for root curvature, n=5). (d) Induced IAA influx at the distal elongation zone of B73 and br2 roots. (e) Oscillation of IAA flux at DEZ. Integrated IAA flux at the distal elongation zone showing oscillatory patterns for B73 integrated efflux. (f) Endogenous IAA flux at the DEZ of 4-day B73 roots. After 20 min of continuous measurement, 2.5 μM NPA was added, causing a decrease in IAA iEfflux after approximately 20 min (iInflux trended below basal levels over the next 60 min). (Reprinted with permission from (McLamore et al. 2010a)).
We then measured IAA flux without external IAA stimulation, and observed that integretated flux followed an oscillatory pattern (Fig. 5e), which indicated that the IAA transport at the DEZ was transient. Inhibition studies using the non-competitive IAA uptake inhibitor NPA showed decreased IAA iFlux (Fig. 5 f). Results from the experiments demonstrated that the SR IAA sensor was an important tool for studying the biochemical mechanisms involved in root development and plant physiology.
6. Conclusions and future perspectives
We have reviewed the physiological applications of SR microbiosensors incorporating enzymes and nanomaterials under a wide range of cell/tissue culture conditions. The ability to directly measure transmembrane (or trans-tissue) flux is a major technological improvement over the conventional biosensor techniques which can characterize concentration only. Compared with other analytical techniques such as mass spectrometry and liquid chromatography which require sample extraction, the SR technique is non-invasive and real-time. In addition, SR provides a high signal-to-noise ratio by eliminating the drift and noise associated with physiological measurements. The incorporation of enzymes greatly extends the list of target compounds which can be measured by the SR sensor, and the incorporation of nanomaterials significantly enhances the performance of the sensor by overcoming the low sensitivity issue due to miniaturization. The nanomaterial based SR sensors have demonstrated applications in cancer cell physiology, neurophysiology, diabetes, muscle physiology, nutrition, microbial biofilm physiology and plant physiology studies. This highly sensitive/selective biosensor technique is a powerful tool to study transport related phenomena in biomedical, environmental, and agricultural research. Currently, omics research mainly relies on mass spectrometry and NMR for data collection. The ability of the SR sensor to characterize cellular mechanisms including biophysical transport makes it a potential tool for omics research. In the meanwhile, there has always been a need to characterize sub-cellular transport for physiological research, which constantly motivates researchers to develop more sensitive and selective nanoscale sensing technology.
Supplementary Material
Acknowledgments
The authors are grateful for the financial support provided by ONR N00014-09-1-0110 and NIH 5R21RR026249-02.
Footnotes
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.bios.2012.06.059.
References
- Azevedo AM, Prazeres DMF, Cabral JMS, Fonseca LP. Biosensors and Bioelectronics. 2005;21(2):235–247. doi: 10.1016/j.bios.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Balon TW, Nadler JL. Journal of applied Physiology: Respiratory, Environmental and Exercise Physiology. 1997;82(1):359–363. doi: 10.1152/jappl.1997.82.1.359. [DOI] [PubMed] [Google Scholar]
- Barbosa RM, Silva A, Tome A, Stamford JA, Santos AS, Rosario L. Biochemical and Biophysical Research Communications. 1996;228(1):100–104. doi: 10.1006/bbrc.1996.1622. [DOI] [PubMed] [Google Scholar]
- Bard AJ, Faulkner LR. Electrochemical Methods: Fundamentals and Applications. 2nd ed. Wiley; New York: 2000. [Google Scholar]
- Bergstrom RW, Fujimoto WY, Teller DC, de Haen C. The American Journal of Physiology. 1989;4(257):479–485. doi: 10.1152/ajpendo.1989.257.4.E479. [DOI] [PubMed] [Google Scholar]
- Chakrapani N, Zhang YM, Nayak SK, Moore JA, Carroll DL, Choi YY, Ajayan PM. The Journal of Physical Chemistry B. 2003;107(35):9308–9311. [Google Scholar]
- Chou HF, Ipp E. Diabetes. 1990;39(1):112–117. doi: 10.2337/diacare.39.1.112. [DOI] [PubMed] [Google Scholar]
- Claussen JC, Artiles MS, McLamore ES, Mohanty S, Shi J, Rickus JL, Fisher TS, Porterfield DM. Journal Of Materials Chemistry. 2011;21(30):11224–11231. [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP. Science (New York, NY) 1999;284(5418):1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- Goda T, Koldovsk O. The Biochemical Journal. 1985;229(3):751. doi: 10.1042/bj2290751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouveia-Caridade C, Pauliukaite R, Brett CMA. Electrochimica Acta. 2008;53(23):6732–6739. [Google Scholar]
- Guillam MT, Dupraz P, Thorens B. Diabetes. 2000;49(9):1485–1491. doi: 10.2337/diabetes.49.9.1485. [DOI] [PubMed] [Google Scholar]
- Heinze J. Journal of Angewandte Chemie, International Edition English. 1993;32(9):1268–1288. [Google Scholar]
- Hellman B, Idahl LA, Lernmark A, Taljedal IB. Proceedings of the National Academy of Sciences of the United States of America. 1974;71:3405–3409. doi: 10.1073/pnas.71.9.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloszy J, Hansen P. Reviews of Physiology Biochemistry and Pharmacology. 1996;128:99–193. doi: 10.1007/3-540-61343-9_8. [DOI] [PubMed] [Google Scholar]
- Hrapovic S, Liu Y, Male KB, Luong JH. Analytical Chemistry. 2004;76(4):1083–1088. doi: 10.1021/ac035143t. [DOI] [PubMed] [Google Scholar]
- Hwang MG, Katayama H, Ohgaki S. Water Research. 2007;41(18):4097–4104. doi: 10.1016/j.watres.2007.05.052. [DOI] [PubMed] [Google Scholar]
- Jabaudon D, Shimamoto K, Yasuda-Kamatani Y, Scanziani M, Gähwiler BH, Gerber U. Proceedings of the National Academy of Sciences. 1999;96(15):8733. doi: 10.1073/pnas.96.15.8733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe LF, Nuccitelli R. The Journal of Cell Biology. 1974;63(2):614–628. doi: 10.1083/jcb.63.2.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S-K, Kauri LM, Qian W-J, Kennedy RT. The Journal of Biological Chemistry. 2000;275(9):6642–6650. doi: 10.1074/jbc.275.9.6642. [DOI] [PubMed] [Google Scholar]
- Kang X, Mai Z, Zou X, Cai P, Mo J. Analytical Biochemistry. 2007;369(1):71–79. doi: 10.1016/j.ab.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Kang X, Mai Z, Zou X, Cai P, Mo J. Talanta. 2008;74(4):879–886. doi: 10.1016/j.talanta.2007.07.019. [DOI] [PubMed] [Google Scholar]
- Kang X, Wang J, Wu H, Aksay IA, Liu J, Lin Y. Biosensors and Bioelectronics. 2009;25(4):901–905. doi: 10.1016/j.bios.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Kennedy RT, Kauri LM, Dahlgren GM, Jung S-K. Diabetes. 2002;51(suppl 1):S152–S161. doi: 10.2337/diabetes.51.2007.s152. [DOI] [PubMed] [Google Scholar]
- Kuhtreiber WM, Jaffe LF. The Journal of Cell Biology. 1990;110(5):1565–1573. doi: 10.1083/jcb.110.5.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagord C, Soulet L, Bonavaud S, Bassaglia Y, Rey C, Barlovatz-Meimon G, Gautron J, Martelly I. Cell and Tissue Research. 1998;291(3):455–468. doi: 10.1007/s004410051015. [DOI] [PubMed] [Google Scholar]
- Lefebvre PJ, Paolisso G, Scheen AJ, Henquin JC. Diabetologia. 1987;30(7):443–452. doi: 10.1007/BF00279610. [DOI] [PubMed] [Google Scholar]
- Li X, Heryadi D, Gewirth AA. Langmuir: The ACS Journal of Surfaces and Colloids. 2005;21(20):9251–9259. doi: 10.1021/la0508745. [DOI] [PubMed] [Google Scholar]
- Longo EA, Tornheim K, Deeney JT, Varnum BA, Tillotson D, Prentki M, Corkey BE. The Journal of Biological Chemistry. 1991;266(14):9314–9319. [PubMed] [Google Scholar]
- Lu H, Forbes RA, Verma A. The Journal of Biological Chemistry. 2002;277(26):23111–23115. doi: 10.1074/jbc.M202487200. [DOI] [PubMed] [Google Scholar]
- Marc G, Zre S, Guy Q, Jean-Michel K. Electrochemical behavior of H2O2 on gold. Electroanalysis. 1997:1088–1092. [Google Scholar]
- Matoba S, Kang J-G, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM. Science (New York, NY) 2006;312(5780):1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- McLamore ES, Diggs A, Calvo Marzal P, Shi J, Blakeslee JJ, Peer WA, Murphy AS, Porterfield DM. The Plant Journal. 2010a;63(6):1004–1016. doi: 10.1111/j.1365-313X.2010.04300.x. [DOI] [PubMed] [Google Scholar]
- McLamore ES, Mohanty S, Shi J, Claussen J, Jedlicka SS, Rickus JL, Porterfield DM. Journal of Neuroscience Methods. 2010b;189(1):14–22. doi: 10.1016/j.jneumeth.2010.03.001. [DOI] [PubMed] [Google Scholar]
- McLamore ES, Porterfield DM. Chemical Society Reviews. 2011;40:5308–5320. doi: 10.1039/c0cs00173b. [DOI] [PubMed] [Google Scholar]
- McLamore ES, Porterfield DM, Banks MK. Biotechnology and Bioengineering. 2009:791–799. doi: 10.1002/bit.22128. [DOI] [PubMed] [Google Scholar]
- McLamore ES, Shi J, Jaroch D, Claussen JC, Uchida A, Jiang Y, Zhang W, Donkin SS, Banks MK, Buhman KK, Teegarden D, Rickus JL, Porterfield DM. Biosensors and Bioelectronics. 2011;26(5):2237–2245. doi: 10.1016/j.bios.2010.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moley KH, Chi MMY, Mueckler MM. American Journal of Physiology Endocrinology and Metabolism. 1998;275(1):E38–E47. doi: 10.1152/ajpendo.1998.275.1.E38. [DOI] [PubMed] [Google Scholar]
- Nilsson T, Schultz V, Berggren PO, Corkey BE, Tornheim K. The Biochemical Journal. 1996;314:91–94. doi: 10.1042/bj3140091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent JM, Santhanam KSV, Rubio A, Ajayan PM. Nano Letters. 2001;1(2):87–91. [Google Scholar]
- O’Rahilly S, Turner RC, Matthews DR. The New England Journal of Medicine. 1988;318(19):1225–1230. doi: 10.1056/NEJM198805123181902. [DOI] [PubMed] [Google Scholar]
- Passonneau JV, Lowry OH. Enzymatic Analysis: A Practical Guide. Humana Pr Inc; New York: 1993. [Google Scholar]
- Pelicano H, Martin DS, Xu RH, Huang P. Oncogene. 2006;25(34):4633–4646. doi: 10.1038/sj.onc.1209597. [DOI] [PubMed] [Google Scholar]
- Polonsky KS, Given BD, Hirsch LJ, Tillil H, Shapiro ET, Beebe C, Frank BH, Galloway JA, Van Cauter E. The New England Journal of Medicine. 1988;318(19):1231–1239. doi: 10.1056/NEJM198805123181903. [DOI] [PubMed] [Google Scholar]
- Porterfield DM. Biosensors and Bioelectronics. 2007;22(7):1186–1196. doi: 10.1016/j.bios.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Porterfield DM, Corkey RF, Sanger RH, Tornheim K, Smith PJ, Corkey BE. Diabetes. 2000;49(9):1511–1516. doi: 10.2337/diabetes.49.9.1511. [DOI] [PubMed] [Google Scholar]
- Porterfield DM, McLamore ES, Banks MK. Sensors and Actuators: B Chemical. 2009;136(2):383–387. [Google Scholar]
- Porterfield DM, Rickus JL, Kopelman R. Proceedings of SPIE. 2006:63800S. [Google Scholar]
- Salimi A, Compton RG, Hallaj R. Analytical Biochemistry. 2004;333(1):49–56. doi: 10.1016/j.ab.2004.06.039. [DOI] [PubMed] [Google Scholar]
- Santos AS, Pereira AC, Duran N, Kubota LT. Electrochimica Acta. 2006;52(1):215–220. [Google Scholar]
- Shao Y, Wang J, Wu H, Liu J, Aksay IA, Lin Y. Electroanalysis. 2010;22(10):1027–1036. [Google Scholar]
- Shi J, Cha T, Claussen J, Diggs A, Choi JH, Porterfield DM. Analyst. 2011a:4916–4924. doi: 10.1039/c1an15179g. [DOI] [PubMed] [Google Scholar]
- Shi J, Claussen J, McLamore ES, Jaroch D, Haque A, Diggs A, Calvo Marzal P, Rickus J, Porterfield DM. Nanotechnology. 2011b;22(35):355502. doi: 10.1088/0957-4484/22/35/355502. [DOI] [PubMed] [Google Scholar]
- Shi J, McLamore E, Jaroch D, Claussen J, Rickus J, Porterfield DM. Analytical Biochemistry. 2011c;411:185–193. doi: 10.1016/j.ab.2010.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Porterfield DM. Surface Modification Approaches for Electrochemical Biosensors. In: Andrea P, editor. Biosensors for Health, Environment and Biosecurity/ Book. Intech; Vienna: 2011. p. 1. [Google Scholar]
- Shi J, Zhang H, Snyder A, Wang M, Xie J, Marshall Porterfield D, Stanciu LA. Biosensors and Bioelectronics. doi: 10.1016/j.bios.2012.06.007. http://dx.doi.org/10.1016/j.bios.2012.06.007, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P, Hammar JS, Marshall K, Sanger PD, Trimarchi J, R R,H. Microscopy research and technique. 1999;46(6):398–417. doi: 10.1002/(SICI)1097-0029(19990915)46:6<398::AID-JEMT8>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Sweet IR, Li G, Najafi H, Berner D, Matschinsky FM. American Journal of Physiology Endocrinology and Metabolism. 1996;271(3):E606–E625. doi: 10.1152/ajpendo.1996.271.3.E606. [DOI] [PubMed] [Google Scholar]
- Tornheim K. Diabetes. 1997;46(9):1375–1380. doi: 10.2337/diab.46.9.1375. [DOI] [PubMed] [Google Scholar]
- Tsai Y-C, Huang J-D, Chiu C-C. Biosensors and Bioelectronics. 2007;22(12):3051–3056. doi: 10.1016/j.bios.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Wang SG, Zhang Q, Wang R, Yoon SF, Ahn J, Yang DJ, Tian JZ, Li JQ, Zhou Q. Electrochemistry Communications. 2003;5(9):800–803. [Google Scholar]
- Weigle DS. Diabetes. 1987a;36(6):764–775. doi: 10.2337/diab.36.6.764. [DOI] [PubMed] [Google Scholar]
- Weigle DS. Diabetes. 1987b;36(6):764. doi: 10.2337/diab.36.6.764. [DOI] [PubMed] [Google Scholar]
- Weiss RG, Chacko VP, Glickson JD, Gerstenblith G. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(16):6426–6430. doi: 10.1073/pnas.86.16.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A, Fujikawa DG. Brain research. 2002;946(1):119–129. doi: 10.1016/s0006-8993(02)02869-x. [DOI] [PubMed] [Google Scholar]
- Wu K, Sun Y, Hu S. Sensors and Actuators B. Chemical. 2003;96(3):658–662. [Google Scholar]
- Xu R, Pelicano H, Zhou Y, Carew JS, Feng L, Bhalla KN, Keating MJ, Huang P. Cancer research. 2005;65(2):613. [PubMed] [Google Scholar]
- Yao Y, Shiu K-K. Analytical and Bioanalytical Chemistry. 2007;387(1):303–309. doi: 10.1007/s00216-006-0924-1. [DOI] [PubMed] [Google Scholar]
- Ye ZC, Rothstein JD, Sontheimer H. The Journal of Neuroscience. 1999;19(24):10767–10777. doi: 10.1523/JNEUROSCI.19-24-10767.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz HB, Toppare L. Biosensors and Bioelectronics. 2006;21(12):2306–2310. doi: 10.1016/j.bios.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Zawalich WS, Matschinsky FM. Endocrinology. 1977;100(1):1. doi: 10.1210/endo-100-1-1. [DOI] [PubMed] [Google Scholar]
- Zhang L, Li Y, Li DW, Karpuzov D, Long YT. International Journal of Electrochemical Science. 2011;6:819–829. [Google Scholar]
- Zisman WA. The Review of Scientific Instruments. 1932;3:367–368. [Google Scholar]
- Zou Y, Xiang C, Sun L-X, Xu F. Biosensors and Bioelectronics. 2008;23(7):1010–1016. doi: 10.1016/j.bios.2007.10.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.