Abstract
The Helicobacter pylori (Hp) Asp-tRNAAsn/Glu-tRNAGln amidotransferase (AdT) plays important roles in indirect aminoacylation and translational fidelity. AdT has two active sites, in two separate subunits. Kinetic studies have suggested that interdomain communication occurs between these subunits, however this mechanism is not well understood. To explore domain-domain communication in AdT, an assay was adapted and optimized to kinetically characterize the kinase activity of Hp AdT. This assay was applied to the analysis of a series of point mutations at conserved positions throughout the putative AdT ammonia tunnel that connects the two active sites. Several mutations were identified that caused significant decreases in AdT’s kinase activity (reduced by 55–75 %). Mutations at Thr149 (37 Å distal to the GatB kinase active site) and Lys89 (located at the interface of GatA and GatB) were detrimental to AdT’s kinase activity, suggesting that these mutations have disrupted interdomain communication between the two active sites. Models of wild-type AdT, a valine mutation at Thr149, and an arginine mutation at Lys89 were subjected to molecular dynamics simulations. The comparison of wild-type, T149V and K89R AdT simulation results unmask 59 common residues that are likely involved in connecting the two active sites.
The discovery of the first molecular tunnel, in tryptophan synthase, challenged the conventional mechanism of multiple active site enzymes.1 In contrast to ligand-induced conformational changes that bring multiple active sites into close proximity, some enzymes have evolved molecular tunnels to connect isolated catalytic centers.2–4 These tunnels can enhance the stability of reactive intermediates via sequestration and allow the delivery of intermediates to the next active site. Furthermore, tunnels can serve as a method of interdomain communication, in which the binding of the substrate or the generation of an intermediate at one active site triggers conformational changes at another, distal active site.2–5
Most glutamine-dependent amidotransferases (GATs) contain molecular tunnels. These enzymes play important roles in diverse biosynthetic pathways as they deliver ammonia from the side chain of glutamine to various electrophilic acceptors during the biosynthesis of cofactors,6–8 amino acids,9–11 amino sugars,12 purines,13 and pyrimidines,14 amongst others. The tunnels serve to sequester and deliver ammonia (generated via glutamine hydrolysis in one active site) to the downstream active site. Crystal structures are available for many of these enzymes and reveal that the molecular tunnels are typically hydrophobic or only partially occupied by water;1, 15–20 they range in length from 6 Å (e. g. glucosamine 6-phosphate synthase19) to 45 Å (e. g. carbamoyl phosphate synthetase20).
H. pylori Asp-tRNAAsn/Glu-tRNAGln amidotransferase (AdT) is a heterotrimeric GAT made up of the GatC, GatA, and GatB subunits.21 Like many bacteria, H. pylori is missing both glutaminyl- and asparaginyl-tRNA synthetase (GlnRS and AsnRS, respectively).22, 23 In these species, AdT is essential for protein translation and fidelity because it converts the misacylated tRNAs Glu-tRNAGln and Asp-tRNAAsn into Gln-tRNAGln and Asn-tRNAAsn, respectively.21, 24, 25 AdT catalyzes three distinct reactions. The GatA subunit contains a classic amidase signature sequence and catalyzes the hydrolysis of glutamine into glutamate and ammonia (reaction 1).21 The GatB active site catalyzes phosphorylation and transamidation of both Glu-tRNAGln and Asp-tRNAAsn (reactions 2 and 3, respectively, shown for Glu-tRNAGln only).26 GatC is apparently not involved in catalysis.
| Reaction 1 |
| Reaction 2 |
| Reaction 3 |
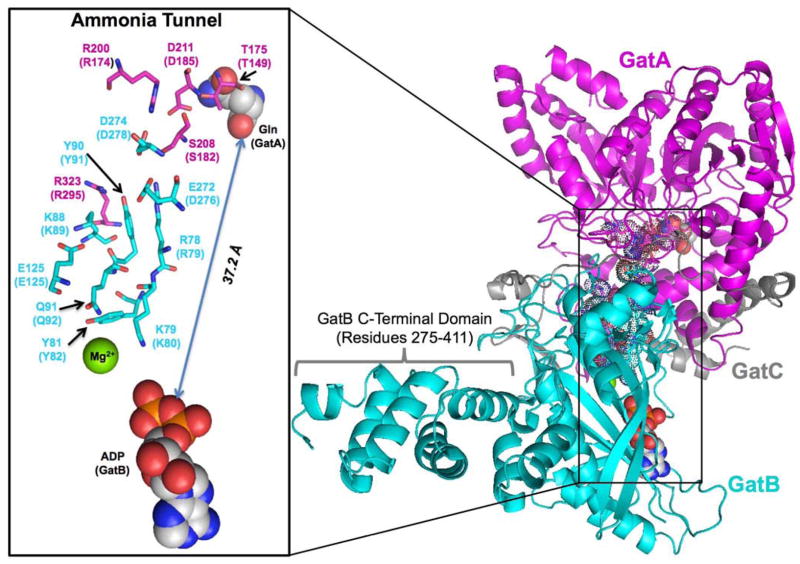
The crystal structure of S. aureus AdT revealed that the GatA and GatB active sites are separated by 37 Å, and a putative ammonia tunnel was identified that connects the two catalytic centers (Figure 1).26 This tunnel is unique, compared to other known ammonia tunnels,2 because it is highly populated with ordered water molecules and lined with conserved polar and ionic amino acids.26 Tanaka and colleagues proposed that ammonia delivery could occur through a series of protonation and deprotonation events.26 To our knowledge, such a mechanism would be unprecedented amongst enzymes that utilize ammonia tunnels.
Figure 1.
The structure of the S. aureus AdT and its putative ammonia tunnel. This image was built from the S. aureus Mu50 AdT crystal structure using Pymol and PDB files 2F2A and 2G5I. GatA is shown in magenta, GatB in cyan, and GatC in grey. Conserved residues along the proposed tunnel are highlighted in sticks and dots.43 The glutamine in the GatA active site (from PDB file 2F2A) and the ADP in the GatB active site (from PDB file 2G5I) are shown as spaced filled molecules. The GatB C-terminal flexible domain (residues 275–411) is indicated; see text for details. The inset shows a close-up of the conserved residues lining the ammonia tunnel. The residues are numbered according to S. aureus Mu50 AdT, with parenthetical notations indicating the corresponding positions in H. pylori AdT.
In many respects, AdT is well characterized for its role in indirect tRNA aminoacylation, translational fidelity, catalysis, specificity for both Asp-tRNAAsn and Glu-tRNAGln, and the formation of a transamidosome complex with a non-discriminating aspartyl-tRNA synthetase (ND-AspRS).25, 27, 28 In contrast, little is known about whether the GatA and GatB subunits communicate with each other, except that the addition of ATP and Glu-tRNAGln stimulates the glutaminase activity of GatA.25, 29
AdT’s kinase and glutaminase reactions must both occur before transamidation. We hypothesized that GatB’s kinase activity might serve as a probe to examine domain-domain connectivity between the GatA and GatB active sites. We optimized and applied a UV-based colorimetric method30, 31 to the quantification of the kinase activity of AdT. This assay was used to kinetically characterize a series of mutations at conserved positions throughout the putative AdT tunnel. Effects ranged from negligible to an approximately 75 % reduction in kinase activity. Mutations at T149 and K89 caught our attention because even conservative mutations at these positions caused substantial decreases in kinase activity in GatB. This effect was essentially the same whether rates were measured in the presence or absence of glutamine. Additionally, T149 is located at the entrance of the ammonia tunnel in GatA distal to the GatB kinase active site, and K89 lies at the interface of GatA and GatB. Our kinetic analyses of these mutations suggest that they participate in directing communication between the glutaminase and kinase active sites of AdT.
To investigate this domain-domain communication, we carried out molecular dynamics (MD) simulations on wild-type, T149V and K89R AdT, all with and without glutamine bound to the glutaminase active site. Root mean square deviation and residue-wise correlation analyses indicate a significant difference in structural mobility for certain regions in the mutant structures compared to the wild-type enzyme. This effect is more pronounced when Gln is bound in the glutaminase active site of the T149V AdT in contrast to the observed steady-state kinetic measurements. Analyses of the simulation results point to 59 AdT residues that show an elevated level of mobility in both T149V and K89R AdT variants. These residues cluster mainly along the tunnel or close to the tRNA binding site. Taken together with our kinetic analyses, these results strongly suggest a pathway for domain-domain communication in AdT.
MATERIALS AND METHODS
Materials
Oligonucleotides were purchased from Invitrogen and used without further purification. Pfu polymerase was purchased from Stratagene. Taq polymerase was from New England Biolabs. All buffers were filtered through a 0.22 μm filter prior to use. When appropriate, solutions were autoclaved. Unless otherwise stated, reagents were used without further purification.
Cloning of Hp AdT
The H. pylori gatCAB operon was originally amplified from H. pylori strain 26695. The genes coding for the three subunits of AdT were sub-cloned into two compatible plasmids (Drs. Stéphane Skouloubris and Pitak Chuawong, unpublished results). Plasmid pPTC032 is derived from pCDF-1b (Novagen) and codes for GatC and GatA in a single operon with an N-terminal six-histidine tag appended onto GatC. Plasmid pSS003 (derived from pET-28a) encodes for GatB with an N-terminal six-histidine (His6) tag (pPTC032).
Site–directed mutagenesis
Individual point mutations were introduced into either the gatA or gatB gene (encoded in plasmids pPTC032 and pSS003, respectively) by QuikChange© mutagenesis according to the directions provided by Stratagene. Oligonucleotide sequences and plasmids are listed in the Supporting Information in Table S1. All gene constructs were verified by DNA sequencing of the entire gene prior to use.
Overexpression and purification of Hp AdT
For wild-type AdT, E. coli strain BL21(DE3) calcium-chloride competent cells were transformed with both pPTC032 and pSS003. For expression of each mutant AdT, BL21(DE3) was first transformed with the plasmid encoding for the wild-type subunit(s) and then individually transformed with each mutated plasmid. For example, to produce the T149A AdT mutant, cells were transformed with pSS003, encoding for wild-type His6-GatB, and then with plasmid pSF005, encoding for His6-GatC and T149A GatA in an operon. In all cases, colonies containing both plasmids were selected on agar plates supplemented with kanamycin (50 μg/mL) and streptomycin (50 μg/mL) and incubated at 37 °C overnight. A 5 mL Luria Broth (LB) culture, containing the same two antibiotics, was inoculated with a single colony. The overnight culture was scaled up to 500 mL in the same medium. When the absorbance at 600 nm was between 0.4 and 0.6, IPTG was added to a final concentration of 1 mM to induce overexpression. Cells were harvested by centrifugation at 5,000 rpm for 5 minutes at 4 °C after a 4-hour induction, and AdT or its mutants were purified by HisPur™ Cobalt resin (Pierce) as per the manufacturer’s instruction. With the exception of R174A AdT, all mutations were readily purified to homogeneity (See Supporting Information, Figure S1). Protein concentrations were determined by UV/Vis spectroscopy with the extinction coefficients for each protein determined using the ExPASy Proteomics server (http://ca.expasy.org/tools/protparam.html).32
In vivo transcription of H. pylori tRNAGln and tRNA purification
H. pylori tRNAGln was in vivo transcribed in the E. coli strain MV1184 and the tRNA was purified and quantified as previously described.33
Preparation of Glu-tRNAGln
H. pylori tRNAGln was aminoacylated with glutamate using H. pylori GluRS233 as previously described.34 The resultant product was extracted with phenol/chloroform (1/1); excess ATP and ADP were removed as previously described.35 The Glu-tRNAGln yield was quantified by running a parallel reaction that contained 3H-labeled glutamic acid.
AdT kinase assay
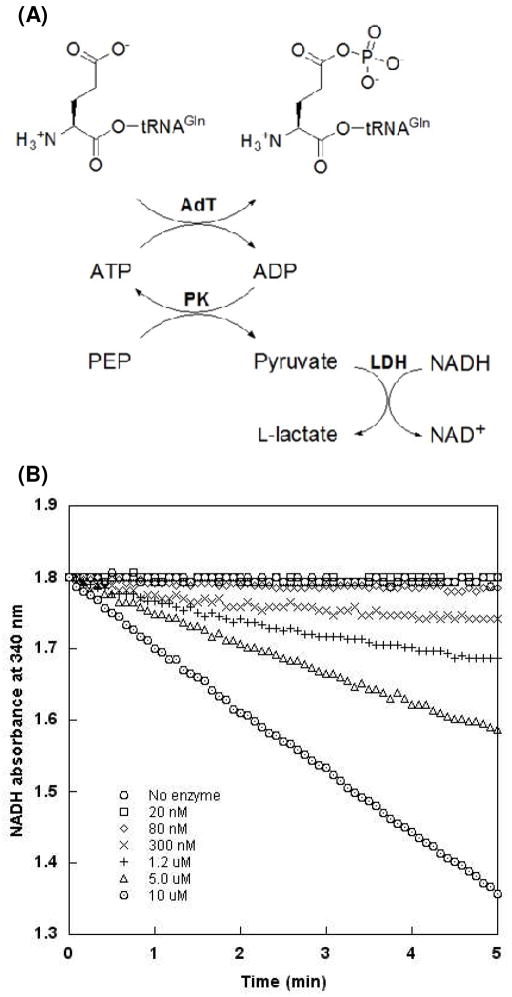
The kinase activity of AdT was monitored via adaptation of a coupled enzyme assay (see Figure. 2A).30, 31 Unless otherwise stated, assays were performed at room temperature in buffer containing 20 mM HEPES-KOH (pH 7.5), 5 mM MgCl2, 0.1 mM DTT, 2 mM phosphoenolpyruvate (PEP), 2 mM ATP, 20 units/mL pyruvate kinase (PK), 20 units/mL L-lactate dehydrogenase (LDH) and NADH to give an absorbance of 0.3 to 0.4 at 340 nm (between 50 and 70 μM, based on standards of known concentration). For initial rate determinations, saturating concentrations of Glu-tRNAGln (10 μM) were used so that the observed initial rates would approximate kcat for each residue. This approach allowed us to focus our analysis on catalytic defects that were conveyed across distances through the enzyme. The possibility that KM is altered for one or more mutations cannot be ruled out but was not expected to impact data analysis; these values may be determined at a later date. For Michaelis-Menten analyses, the concentration of Glu-tRNAGln was varied from 0 to 8 μM and AdT was held constant at 300 nM. All other assays were initiated with 200 nM AdT enzyme. Reaction progress was monitored on a Beckman DU 800 spectrometer at 340 nm. Full Michaelis-Menten values were only determined for wild-type AdT.
Figure 2.
Coupled enzyme assay to measure AdT’s phosphorylation activity with Glu-tRNAGln. (A) Assay schematic. The hydrolysis of ATP is coupled to the oxidation of NADH; the resultant decrease in NADH concentration is monitored by UV at 340 nm. (B) Assay is responsive over a wide range of AdT concentrations (○ no enzyme; □20 nM; ◇ 80 nM; × 300 nM; + 1.2 μM; △ 5.0 μM; ⊙ 10 μM).
As part of the optimization of this assay, wild-type AdT concentrations were varied from 0 to 10 μM to confirm that the coupling enzyme conditions were reporting kinase activity (rather than the activities of PK or LDH). Additionally, the NADH concentration was optimized (reduced from 200 μM to < 70 μM) to produce the largest observable change in absorbance without impacting the actual rate of phosphorylation; this optimization led to more accurate and reproducible results.
Molecular dynamics simulations of AdT and the two mutants
All molecular dynamics (MD) simulations were performed with the PMEMD program in the AMBER11 software suite with the amber99SB force field.36 The SHAKE algorithm37 was applied on all bonds between heavy atoms and hydrogens. All the simulations were done under periodic boundary conditions and the smooth particle mesh Ewald method was used to account for long-range interactions.38, 39
The crystal structure for AdT from Staphylococcus aureus was taken from the Protein Data Bank (PDB: 2F2A26). This crystal structure was chosen since there are no reported crystal structures for H. pylori AdT and it is highly homologous to S. aureus AdT. The crystal structure was checked and protonated with MolProbity.40 Protein termini for all subunits and the glutamine substrate bound to GatA (when present) were capped using acetyl (ACE) and N-methyl acetamide (NME) groups.
The simulated systems included wild-type, T149V and K89R AdT in the presence or absence of glutamine. Based on the protein sequence alignment of H. pylori and S. aureus AdT (see Figure S4 in the Supporting Information), residue T175 in S. aureus, homologous to T149 in H. pylori, was replaced by valine in the mutant structures; and residue K88 in S. aureus, homologous to K89 in H. pylori, was replaced by arginine in the mutant structures. All structures were created using the UCSF Chimera software suite.41
The protonated structures were minimized using the sander program in AMBER with 50 steps of steepest descent and 450 steps of conjugate gradient with constraints on the protein (a force constant of 500 kcal•mol−1Å−2 on the protein). The minimized structures were solvated using the TIP3P water model inside rectangular boxes of 125 × 96 × 142 Å. Charges were neutralized in all four structures by adding Na+ ions using the xleap program in AMBER.
The added water molecules were minimized again using the PMEMD program in AMBER with 50 steps of steepest descent and 450 steps of conjugate gradient. During this minimization the protein and substrates were restrained with a force constant of 500 kcal•mol−1Å−2.
Following minimization, the density of all the systems was increased to 1 g•cm−3 by NPT simulations restraining all protein and substrate atoms with a force constant of 500 kcal•mol−1Å−2. Pressure was kept constant by anisotropic pressure.36 Subsequently, the systems were gradually heated up to 300 K in 10 steps (10 ps each) at constant volume with constraints on the protein (500 kcal•mol−1Å−2 force constant on the protein). For the wild type without glutamine bound to the GatA active site, the warm-up was performed in 10 steps of 20 ps each. Once the systems achieved the target temperature, the constraints were gradually removed over 80 ps until no constraints were left. The temperature was kept constant using a Berendsen thermostat.42 Subsequently, simulations were carried out for an additional 20 ns for each system with snapshots taken every 1 ps.
RESULTS
Production of H. pylori AdT and its Mutants
Initially, the gatCAB genes were assembled into a single operon in the pET-28a vector; a plasmid (pSS003) encoding only for His6-GatB (also in pET-28a) was also constructed (Dr. Stéphane Skouloubris, unpublished results). To facilitate purification, and building from a previous report,28 the gatCA operon was transplanted into the pCDF-1b vector (compatible with pET-28a) with concomitant addition of an N-terminal His6 tag onto GatC (His6-GatC, pPTC032). The two resultant plasmids were co-transformed into E. coli and wild-type AdT was readily purified to homogeneity, essentially as previously reported (see Supporting Information Figure S1).28 Following purification, the ratio of GatA and GatB was examined to insure that AdT concentrations were not inaccurate due to dramatic excess of one subunit over the other.
Each mutant was purified using the same approach optimized for wild-type AdT (see Supporting Material Figure S1). For GatA mutations, plasmid pSS003 was separately transformed with each mutant variant of pPTC032. For GatB mutations, plasmid pPTC032 was separately co-transformed with each mutant variant of pSS003 (see Supporting Information, Table S1, for list of mutant plasmids).
Development of a Coupled-Enzyme Assay for AdT-Catalyzed Phosphorylation of Glu-tRNAGln
Conversion of ATP into ADP can be observed indirectly via a coupled, spectroscopic enzyme assay that monitors the oxidation of NADH to NAD+ (Figure 2A).31 In this assay, AdT-catalyzed phosphorylation of Glu-tRNAGln causes concomitant production of ADP. ATP is regenerated in the presence of excess phosphoenolpyruvate (PEP) and pyruvate kinase (PK), to produce pyruvate. Lactate dehydrogenase (LDH) reduces the resultant pyruvate to L-lactate, via conversion of NADH to NAD+. This reaction can be monitored spectrophotometrically because NADH has an absorbance maximum at 340 nm and NAD+ does not absorb UV light at this wavelength.
Coupled-enzyme assays carry the risk that one is inadvertently quantifying the activity of the downstream enzyme(s) (LDH and/or PK in this case), rather than the enzyme of interest (AdT). To rule out this concern, several control experiments were conducted before progressing with quantitative assays of AdT and its mutants. The previously reported buffer conditions were adapted to those for AdT.25 Different AdT enzyme concentrations were also examined (Figure 2B). As the concentration of AdT was increased (ranging from 20 nM to 10 μM), the rate of ATP hydrolysis also increased, confirming that the assay was directly reporting on AdT activity. Previously reported UV-based assays typically used NADH concentrations of 200 μM;30 with AdT, this concentration led to an untenable signal to noise ratio (S/N, See supporting information, Figure S2A). To address this issue, the concentration of NADH was decreased to between 50 and 70 μM. This reduction led to a much more acceptable S/N while producing the same rate of ATP hydrolysis (again, indicating that AdT activity, and not PK or LDH, is being reported, see Figure S2B in the online Supporting Information). An NADH calibration curve (Supporting Information, Figure S3) allowed us to accurately correlate UV absorbance with concentration.
Michaelis-Menten Kinetics
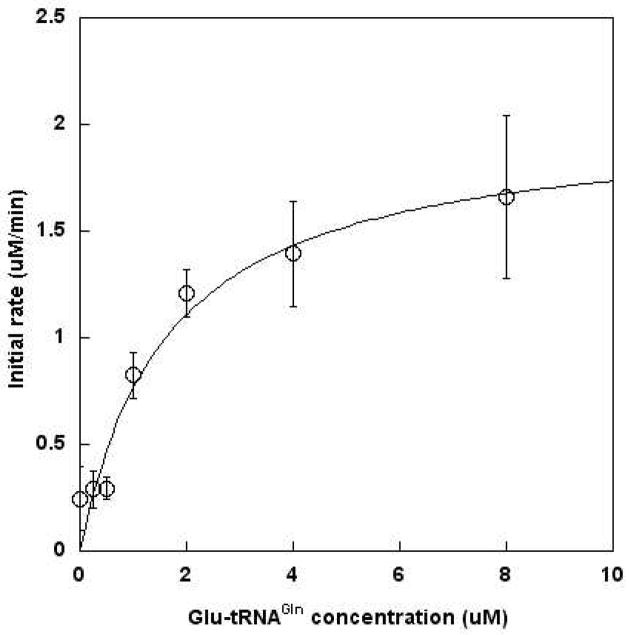
The KM of ATP is approximately 200 μM, when measuring the net reaction of H. pylori AdT (transamidation).43 Thus, the concentration of ATP was set at 2 mM for all assays. Kinetic parameters were determined for Glu-tRNAGln as a substrate for AdT’s kinase activity (Figure 3). The KM for this misacylated tRNA is 1.6 ± 0.5 μM and is consistent with values previously reported (1.18 μM, determined by measuring AdT’s transamidase activity43). The calculated kcat is 0.12 ± 0.01 sec−1, leading to a specificity constant (kcat/KM) of 7.5 × 104 s−1 M−1 for the phosphorylation of Glu-tRNAGln in the absence of glutamine.
Figure 3.
Michaelis-Menten analysis of Glu-tRNAGln as a substrate for AdT-catalyzed phosphorylation. Each reaction was initiated with 300 nM AdT. Data was fit directly to the Michaelis-Menten equation using Kaleidagraph V4.0 (Synergy Software). Error bars represent standard deviation and are from duplicate measurements. Standard deviations reported in Table 1 are from the results of the curve fit analysis.
Design and Characterization of Mutations in the Putative AdT Tunnel
It has been proposed that ammonia is transferred from GatA to GatB through a protonation-deprotonation relay mechanism; this hypothesis is based on the observation that the putative ammonia tunnel contains many hydrophilic and ionic residues and ordered water molecules.26 The H. pylori and S. aureus AdT protein sequences were aligned (see Figure S4 in Supporting Information). A protein sequence alignment between the H. pylori and S. aureus AdT subunits showed that the two GatA subunits share 45 % identity and 75% similarity and the two GatB subunits are 53% identical and 89% similar. A more detailed alignment identified 13 highly conserved residues along the putative ammonia tunnel (Table 1 and Figure 1). 26 Most of these amino acids were separately mutated to alanine and to one or more conservative amino acids to determine which, if any, of these positions is important for phosphorylation of Glu-tRNAGln.
Table 1.
AdT Ammonia Tunnel Conserved Residues
| GatA Mutagenesis
| ||
|---|---|---|
| S. aureus | H. pylori | H. pylori Mutations |
| T175 | T149 | T149A, V, Sa |
| R200 | R174 | R174Ab, K |
| S208 | S182 | S182A, T |
| D211 | D185 | D185A, N |
| R323 | R295 | R295A, K |
| GatB Mutagenesis
| ||
|---|---|---|
| S. aureus | H. pylori | H. pylori Mutations |
| R78 | R79 | R79A, K |
| K79 | K80 | K80Aa, R |
| Y81 | Y82 | Y82A, F |
| K88 | K89 | K89A, R |
| Y90 | Y91 | Y91A, F |
| Q91 | Q92 | Q92A, E |
| E125 | E125 | E125A, D, Q |
| E272c | D276 | D276A, N |
| D274 | D278 | * |
Site-directed mutagenesis was not successful.
Mutant could not be purified to homogeneity.
Residue is not rigorously conserved, but is typically glutamic or aspartic acid.
Not analyzed.
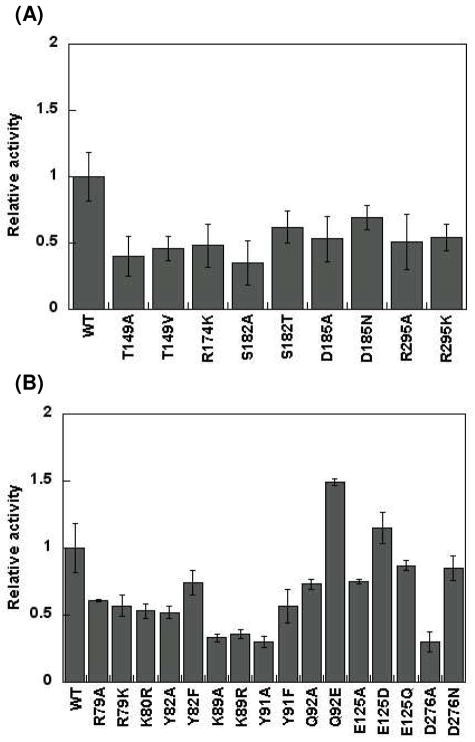
Each mutant enzyme was purified to homogeneity, with the exception of R174A, which could not be purified sufficiently for further analysis due to poor expression levels (see Figure S1 in Supporting Information). Initial rates of phosphorylation of Glu-tRNAGln were determined for each mutant enzyme using the coupled assay described above. Relative activities with respect to wild-type AdT were calculated and are shown in Figure 4A and 4B. No single mutation was sufficient to completely abolish phosphorylation activity, including mutations at K80 and Y82 (H. pylori numbering), which are located near the ATP binding site. These results show that the ATP binding site is remarkably tolerant of mutations at conserved residues. However, several point mutations did cause dramatic reductions in activity (Figure 4), with some leading to as much as a 75% drop in initial rates. Not surprisingly, in general, conservative mutations were less disruptive than the alanine screen. For example, the activity of Q92A is diminished by approximately 25% with respect to wild-type AdT; in contrast, the Q92E mutant is actually 50% more active than wild-type AdT, at least with respect to phosphorylation (Figure 4B). This increase might be due to simultaneous ATP hydrolysis caused by the mutation in parallel to the phosphorylation of Glu-tRNAGln, because Q92 is located near the ATP binding site.
Figure 4.
Steady-state initial rates of Glu-tRNAGln phosphorylation using wild-type H. pylori AdT and variants containing single point mutations along the ammonia tunnel. Initial rates, relative to wild-type AdT, are shown. See Table S2 in the Supporting Information for the raw initial rate data. The error bars represent standard deviation from triplicate assays. (A) Analysis of mutations in GatA; (B) Analysis of mutations in GatB.
Discussions in the literature have suggested that E125 may43 or may not44 act as a gate, blocking AdT’s ammonia tunnel to prohibit inadvertent ammonia release (e. g. in the absence of bound Glu-tRNAGln). Mutations at position E125 in GatB had little to no effect on AdT’s phosphorylation activity (Figure 4B). These results demonstrate that the identity of E125 does not impact the steady-state rate of phosphorylation of Glu-tRNAGln; further experiments are necessary to determine if E125 plays a different kinetic role. When D276 (which lies near the midpoint of the putative ammonia tunnel) was mutated to alanine, the phosphorylation activity of the resulting enzyme was only 30% of wild-type AdT. In contrast, the D276N mutation maintained 85% of the wild-type enzyme’s activity. S182A, in GatA, also showed a large reduction in phosphorylation activity (35% of wild-type) but this activity was partially recovered in the S182T mutant (to 62% of wild-type). Further analyses of these mutations are ongoing.
Mutagenesis at a few positions revealed a strong, deleterious response even upon the introduction of conservative mutations (e. g. T149 (GatA), R295 (GatA), K89 (GatB) and Y91 (GatB)). Three of these residues (R295, K89, and Y91) are localized at the interface between GatA and GatB (Figure 1). Remarkably, T149 is adjacent to the glutaminase active site, at the top of the tunnel in GatA, substantially removed from the GatB active site. Respectively, T149A AdT and T149V AdT demonstrated initial rates of phosphorylation of 40% and 46% that of wild-type AdT, while K89A and K89R AdT both caused a ~65% drop in the initial rates of phosphorylation. The kinase activity decrease caused by mutations at the distal T149 residue indicates the possibility of interdomain communication. The results with K89 mutations are also interesting because this residue is adjacent to E125, the putative gate. (See below for further analyses of these mutations).
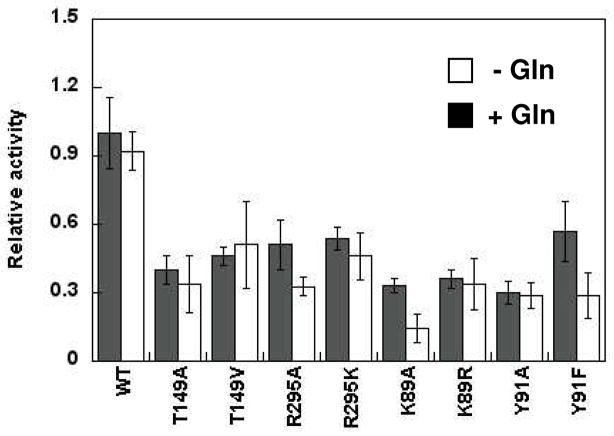
Effects of Glutamine on H. pylori AdT’s Kinase Activity
All AdT mutations were originally screened in the absence of glutamine (Figure 4). The phosphorylation activity of representative mutations and wild-type AdT were reevaluated in the presence of 5 mM glutamine to determine if this GatA active site substrate positively or negatively impacts the activity of any of these mutant enzymes (Figure 5). Wild-type AdT and all mutations showed a slight, reproducible reduction in activity in the presence of glutamine. However, this reduction was only significant for the R295A, K89A, and Y91F mutations, all of which lie at the interface of the two subunits.
Figure 5.
Effect of glutamine on the kinase activity of wild-type AdT and selected mutants (T149A, T149V, R295A, and R295K in GatA; K89A, K89R, Y91A, and Y91F in GatB). Initial rates relative to wild-type AdT were determined in the absence and presence of glutamine (5 mM). Error bars represent standard deviation from triplicate assays. See Table S3 in the Supporting Information for the raw initial rate data.
Molecular Dynamics Simulations of Wild-Type, T149V and K89R AdT
T149 is located in the GatA active site, 37 Å from the site of ATP binding and Glu-tRNAGln phosphorylation in GatB. The fact that the kinase activity of AdT is sensitive to both valine and alanine substitutions of T149, despite the distance between this residue and the GatB active site, suggests that the hydroxyl group in the T149 side chain may play an essential role for proper domain-domain communication. (Attempts to produce a T149S mutation, to further evaluate this hypothesis, were unsuccessful; data not shown).
Mutagenesis at residue K89, located in GatB near the interface between GatA and GatB, also resulted in a deleterious effect on kinase activity even upon the introduction of conservative mutation (K89R). One possible explanation is that the bulkier side chain of arginine (compared to lysine) might block the AdT ammonia tunnel, resulting in the observed decrease in enzymatic activity.
To further understand the role of these mutations on AdT, wild-type, T149V and K89R AdT models were subjected to molecular dynamics (MD) simulations. MD simulations were performed on six systems, and they were examined using the AMBER software suite: 1) wild-type AdT without glutamine bound to GatA; 2) wild-type AdT with bound glutamine; 3) T149V AdT without glutamine; 4) T149V with bound glutamine; 5) K89R AdT without glutamine; and, 6) K89R with bound glutamine. Simulations were conducted as described in Materials and Methods.
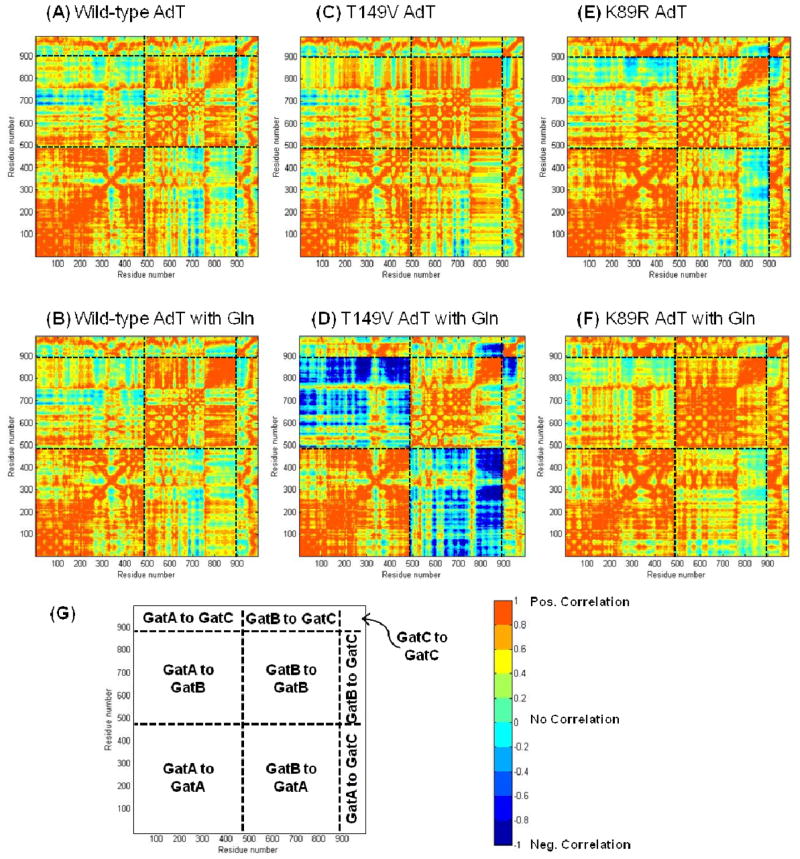
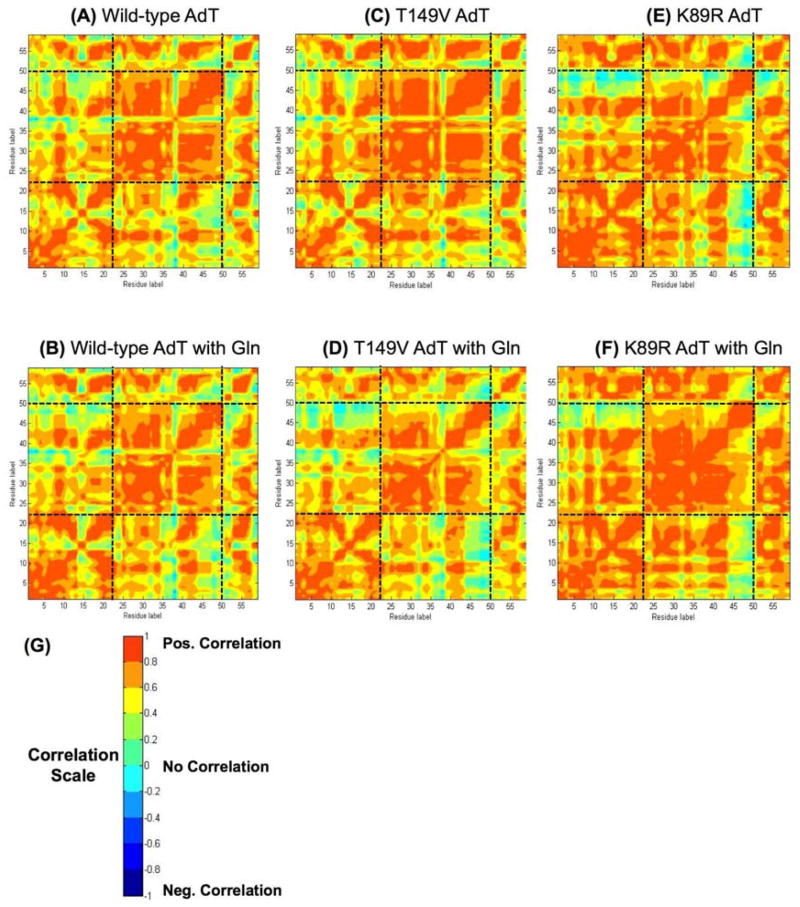
Residue-wise correlation studies were carried out for each of the six simulated systems (Figure 6). The figure is colored to separately to denote both positive (red) and negative (blue) correlations. A positive correlation result indicates that the movement of the two residues is concerted and they both move in the same direction (e. g. both side chains have rotated in a clockwise direction compared to the original crystal structure). A negative correlation result also indicates concerted movement; however, in this case, the two residues move in opposing directions (e. g. one side chain rotates clockwise and the other rotates counterclockwise). In other words, correlation scores of +0.7 and −0.7 indicate the same extent of correlation, but in synergistic versus opposing directions, respectively.
Figure 6.
Residue by residue correlation analysis for all simulated structures. (A) Wild-type AdT; (B) Wild-type AdT with glutamine bound to GatA; (C) T149V AdT; (D) T149V AdT with glutamine; (E) K89R AdT; and (F) K89R AdT with glutamine. (G) Cartoon showing different regions of correlation; residues proceed from left to right (N-terminal to C-terminal) with GatA residues numbered 1–485, GatB residues numbered 486–897, and GatC residues numbered 898–991. All panels apply the same color-coded scale representing the correlation coefficient that stands for how one residue moves with respect to another. Correlation coefficients of +1 and −1 both indicate the highest level of correlation observed between any two residues; negative correlations indicate residue movements correlated in opposing directions.
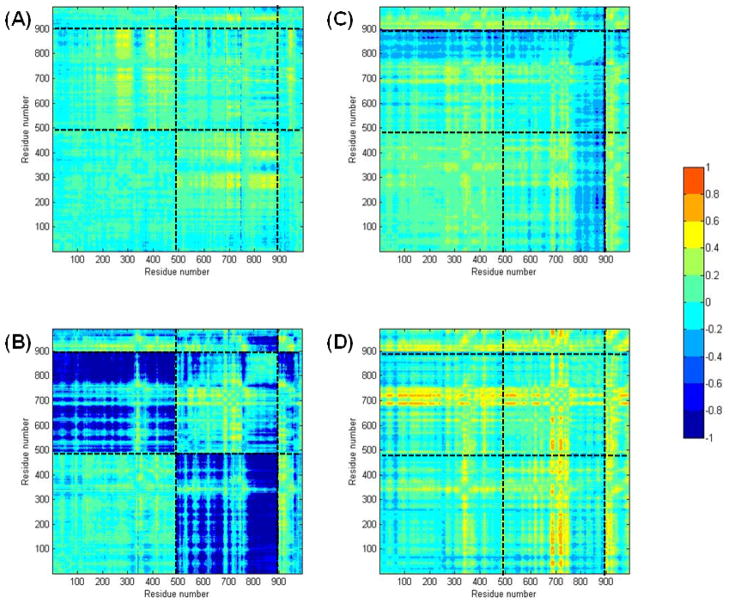
To more readily visualize the specific correlation changes throughout each mutant enzyme, correlation differences were calculated by subtracting the wild-type correlation values from those of each mutant (Figure 7). These correlation difference maps show that there are changes in correlation between the wild-type and the mutant structures in all cases. These changes are most significant for the T149V mutant with glutamine bound to GatA, where strong anti-correlations between GatA and GatB are observed. The intensity of these correlation changes demonstrates that the identity of the T149 residue in GatA (in this case, the T149V mutation) is propagated throughout much of GatB, clearly demonstrating a physical connection between the two active sites. Although changes in correlation are observed in the T149V without glutamine in the GatA active site, these changes are less dramatic. This observation is in contrast to the experimental results for the phosphorylation rates with Glu-tRNAGln where there were no changes observed in the presence or absence of glutamine. It is important to note that the correlations shown in Figures 6 and 7 are relevant on a nanosecond time scale whereas the steady-state kinetic experiments reported in Figure 5 were acquired on a time scale of seconds to minutes. Consequently, the biochemical and computational results are not necessarily in contradiction with each other. In fact, together these results offer a two-pronged explanation for why T149 and K89 are rigorously conserved in AdT.
Figure 7.
Differences in correlation for each mutant compared to the corresponding wild-type simulation results (wild-type values were subtracted from those of each mutant). (A) Differences between wild-type and T149V AdT; (B) Differences between wild-type and T149V AdT, both in the presence of glutamine; (C) Differences between wild-type and K89R AdT; (D) Differences between wild-type and K89R, both in the presence of glutamine. All panels apply the same color-coded scale (right panel) representing the calculated correlation differences. See Figure 6 for a key to the different panel sub-sections.
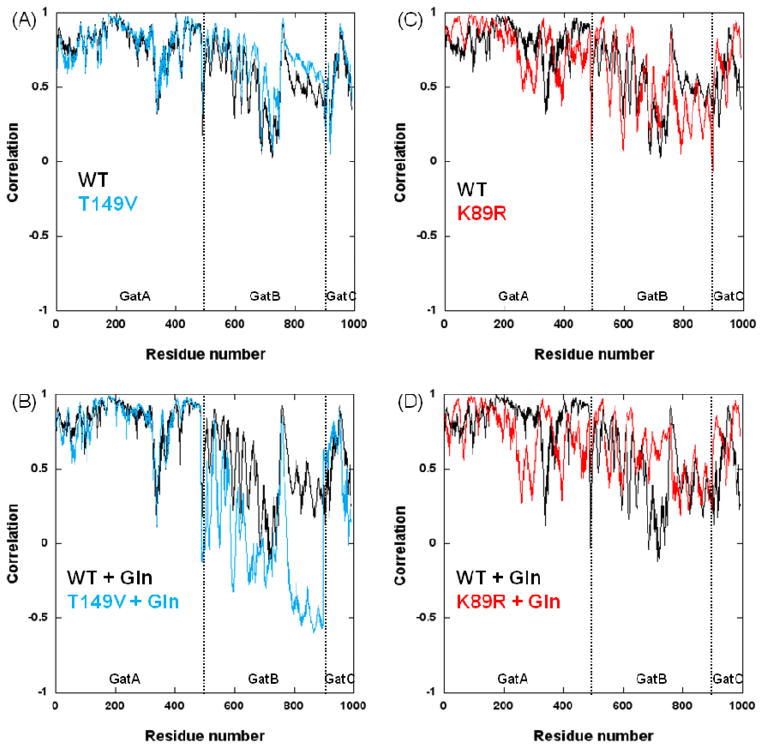
Correlations between the residues at the two mutation sites and every other residue for all AdT models give further insight into the physical connection between GatA and GatB (Figure 8). The T149V mutation shows a significant change in correlation between residues in GatB and position 149 with respect to wild-type AdT; differences between the T149V and wild-type enzymes are much less significant in GatA and GatC (Figure 8A and 8B). These differences in correlation are enhanced when glutamine is bound to GatA during the simulations. In contrast, a noticeable change in correlation is observed in all three subunits for the K89R mutant compared to wild-type (Figure 8C and 8D). This change is also enhanced in the presence of glutamine in GatA. These global variations throughout K89R AdT are consistent with its structural location at the interface between GatA and GatB. Biochemical assays of the K89R mutation show that it has the lowest rate of phosphorylation of all mutations examined in this work (2.8 fold decrease compared to wild-type AdT). Combined, these results suggest that K89 plays a vital role in the apparent connectivity between the two active sites.
Figure 8.
Correlations between each enzyme residue with the T149 and K89 positions (wild-type and mutant AdTs). (A) Comparison between T149 (black) and T149V (cyan) correlations; (B) Comparison between T149 (black) and T149V (cyan) correlations when simulated in the presence of glutamine bound to GatA; (C) Comparison between K89 (black) and K89R (red) correlations; (D) Comparison between K89 (black) and K89R (red) correlations in the presence of glutamine bound to GatA.
Root mean square deviations (RMSDs) for each system were calculated with respect to the crystal structure using the last 10 ns in each simulation. RMSD calculations were carried out on three different levels: a) the entire heterotrimeric protein; 2) the individual subunits (GatA, GatB and GatC); and, 3) on individual active site residues in GatA (residues 80, 131, 154, 155, 156, 174, 176, 177, 178, 179, 207, 310, 311, 359, 426; termed AS1) and GatB (residues 6, 10, 12, 79, 91, 124, 150; termed AS2) (see Figure S5). (Note: Residues are numbered according to the wild-type AdT sequence from S. aureus, not as recorded in the crystal structure PDB file.) The RMSD of GatB showed considerable fluctuation over time, mainly due to movement of the tRNA-binding subdomain (GatB residues 275–412). This subdomain has been proposed to serve as a clamp for Glu-tRNAGln/Asp-tRNAAsn binding.26 The high mobility is probably due to the absence of tRNA in the initial crystal structure as deletion of the contributions of this domain in the RMSD calculation results in significant reduction of the calculated RMSD of GatB (Figure S5).
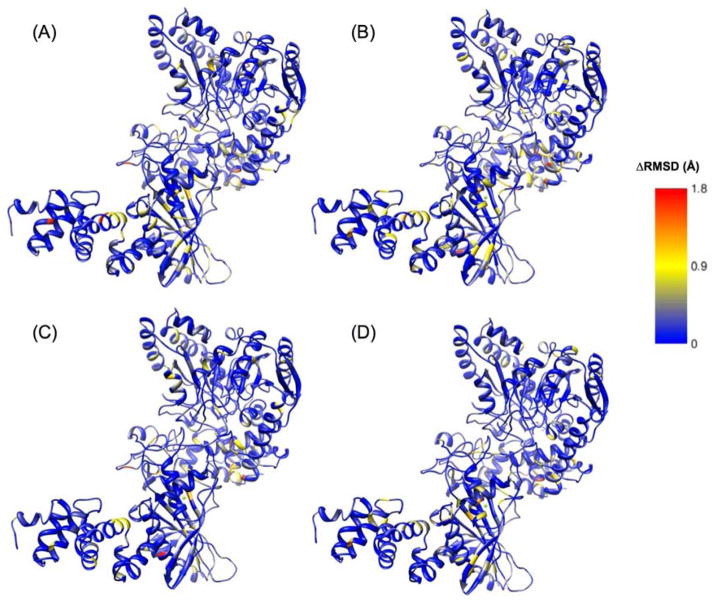
Average RMSDs (RMSDave) were calculated for each residue in each system over the last 10 ns of each simulation. The difference in RMSDave between each residue in the mutant versus the wild-type simulations were determined to identify residues that exhibited greater movement in either of the mutant enzymes compared to wild-type AdT. In other words, ΔRMSDave = Mutant RMSDave − WT RMSDave. These RMSD differences were mapped onto the corresponding simulated structures of AdT (Figure 9).
Figure 9.
Differences in RMSD (ΔRMSD) for each mutant versus wild-type AdT (wild-type values were subtracted from mutant values; see text for details). (A) ΔRMSD between T149V AdT simulation, with glutamine bound to GatA, compared to AdT structure; (B) ΔRMSD between T149V AdT simulation in the absence of bound glutamine, compared to AdT structure; (C) ΔRMSD between K89R AdT simulation, with glutamine bound to GatA, compared to AdT structure; (D) ΔRMSD between K89R AdT simulation and AdT structure, in the absence of bound glutamine. Right panel shows the color scheme for panels A–D in Angstroms. Larger representations of these figures are provided in the Supporting Information, Figure S6.
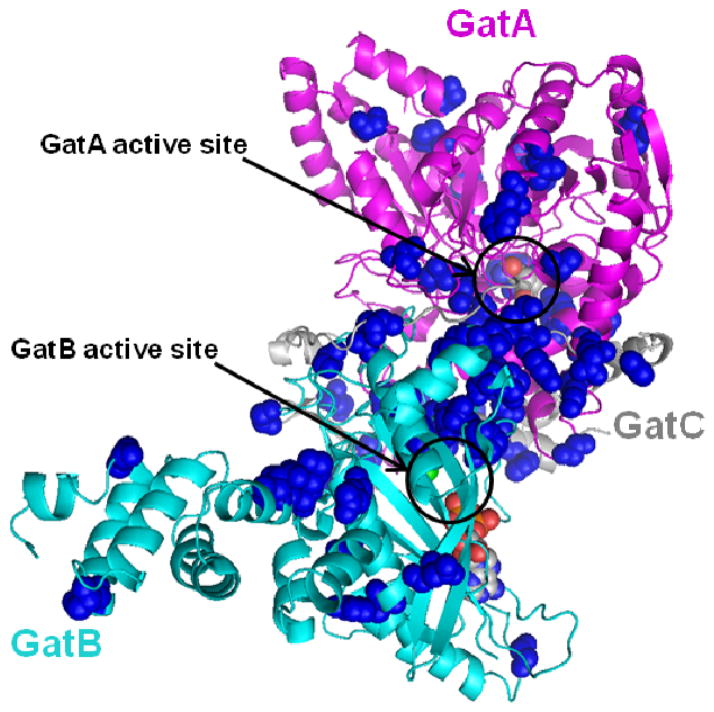
Percentage RMSD changes were calculated for every residue in both mutant simulations with respect to the corresponding wild-type residue by dividing ΔRMSDave by WT RMSDave. This analysis showed that around 300 residues (< 30% of the entire structure) present an RMSDave change of 20% or more. The residues that showed the highest percentage change from each mutant structure were selected using a 20 percent change in RMSDave as an approximate cutoff value for significant change. Out of these 300 residues, a subset of 59 residues was identified that is common to both mutant structures. These residues were mapped onto the structure of AdT (Figure 10). These amino acids are distributed throughout all three subunits of AdT and across the full enzyme structure.
Figure 10.
The K89R and T149V AdT mutations induce substantial RMSD variations in a common set of 59 residues (RMSD > 20% in both mutant enzymes). These residues were mapped onto the S. aureus Mu50 AdT crystal structure and are highlighted in blue spheres. GatA, GatB, and GatC are colored as in Figure 1 (magenta, cyan, and grey, respectively). The 59 residues are listed in Supporting Information, Table S4.
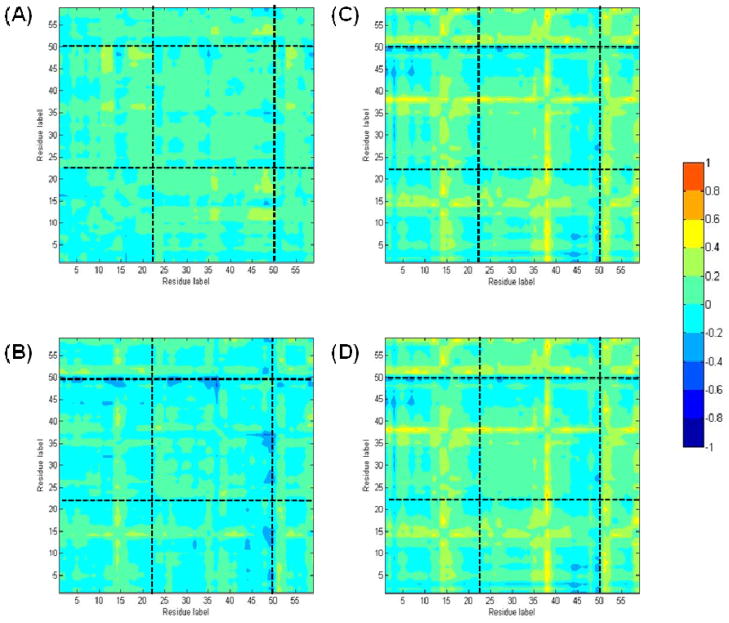
The identification of this small subset of residues common to both mutants suggests that they comprise all or part of a pathway for communication between GatA and GatB. For this hypothesis to be correct, these 59 residues should show common patterns of correlation not only between the two mutant enzymes but also in simulations of wild-type AdT. Consequently, correlation analyses were performed for these 59 residues, with and without glutamine in the active site (Figure 11). As expected, these residues are also highly correlated between GatA and GatB in wild-type AdT (Figure 11A and B). Correlation difference plots were also constructed comparing each these residues from each mutant enzyme to wild-type AdT (Figure 12). These data show that the correlation results demonstrated in Figure 11 do not vary widely between the two mutant enzymes and wild-type AdT. This observation directly supports the hypothesis that these residues are involved in domain-domain communication in AdT, connecting GatA to GatB.
Figure 11.
A residue-by-residue correlation of wild-type, T149V and K89R AdT for the 59 common residues shown in Figure 10. The sub-section divisions are the same as in Figure 6. Note: these residues are dispersed throughout the primary sequence of AdT; they are not consecutive. See Supporting Information, Table S4, for the identity of each residue. (A) wild-type AdT; (B) wild-type AdT with glutamine bound to GatA; (C) T149V AdT; (D) T149V with glutamine bound; (E) K89R AdT; (F) K89R AdT with bound glutamine. (G) Correlation color scale. As in Figure 6, correlation coefficients of +1 and −1 both indicate the highest level of correlation observed between any two residues; negative correlations indicate residue movements that are correlated in opposing directions.
Figure 12.
The correlation difference of the 59 common residues for wild-type and K89R AdT. The sub-section divisions are the same as in Figure 6. The difference color scale is shown on the right. Note: these residues are dispersed throughout the primary sequence of AdT; they are not consecutive. See Supporting Information, Table S4, for the identity of each residue. A) wild-type AdT; (B) wild-type AdT with glutamine bound to GatA; (C) T149V AdT; (D) T149V with glutamine bound; (E) K89R AdT; (F) K89R AdT with bound glutamine.
DISCUSSION
Previous enzymological studies of H. pylori AdT have predominately focused on its glutaminase and transamidase activities.25, 46 Results from these studies suggest the enhancement of glutaminase activity resulting from ATP and Glu-tRNAGln binding to the opposing active site. In other words, substrate-binding events in the GatB active site are communicated to and impact the efficiency of the GatA active site. This observation suggests the existence of essential connectivities between the GatA and GatB active sites that are sensitive to steps in AdT’s reaction pathway. The fact that AdT uses a molecular tunnel to connect its two active sites suggests that this tunnel would be a likely component of AdT’s communication network. In this study, mutagenic analyses were used to evaluate conserved residues within the AdT ammonia tunnel, and molecular dynamics simulations were used to further evaluate interdomain communication in this system.
Glu-tRNAGln was used as the sole aminoacyl-tRNA substrate for AdT (instead of both Glu-tRNAGln and Asp-tRNAAsn). As reported previously, the identity elements for enzyme recognition of the two aa-tRNAs are similar; in particular, they share a common U1:A72 base pair as a key identity element in their acceptor stems.26, 47 To simplify analysis, we chose to focus our efforts on Glu-tRNAGln as a representative example of the two aa-tRNA substrates for AdT. The adapted enzyme-coupled assay afforded a streamlined way to examine wild-type AdT and its mutants. This approach provided informative data that enabled us to select the T149V and K89R mutations for molecular dynamics simulations.
Our assay results show that AdT’s kinase activity is sensitive to mutations in the GatA glutaminase active site (T149V and T149A) and at the interface between GatA and GatB (R295A, K89A, and Y91F). Mutations near the ATP binding site had little to no effect on activity. Remarkably, the kinase activity of GatB was slightly more susceptible to mutations in GatA, on average, than those in GatB. These observations suggest two things. First, it is unlikely that the observed reductions in kinase activity are due to structural alterations in either active site. In this case, one would expect that GatB mutations would have greater impact than GatA mutations. Second, these mutations appear to have unmasked communication between GatA and GatB, even though the phosphorylation of misacylated Glu-tRNAGln is not particularly sensitive to the presence or absence of glutamine in the GatA active site. Previous work on H. pylori AdT’s glutaminase activity showed that the binding of ATP and Glu-tRNAGln to GatB enhances the distal glutaminase activity in GatA by about 30-fold.25 When combined, these analyses have important ramifications for cellular pH homeostasis. They suggest that AdT has evolved to keep its glutaminase activity in check until an aminoacyl-tRNA substrate is available; at this point, glutamine is hydrolyzed to produce ammonia, which is efficiently transferred to the enzyme-bound substrate, rather than released to the intracellular environment.
According to our molecular dynamic simulations, the T149V and K89R AdT mutants showed changes in correlation between GatA and GatB residues compared to wild-type (Figure 6). The presence of glutamine bound to GatA increases these changes. The highest level of correlations occurs between GatA and GatB residues in the T149V mutant enzyme with glutamine bound to GatA (Figure 7). Similar patterns of correlations are present for T149V and K89R AdT in the absence of glutamine, albeit at significantly lower levels. For wild-type AdT and K89R, this observation is consistent with our kinase assay results that show only slight changes in activity upon the addition of 5 mM glutamine. Conversely, the T149V mutant shows a much higher level of correlation between these motifs in the presence of glutamine, in contrast to the minor reduction observed in kinase activity upon the addition of glutamine. It appears that the T149V mutation has reached its maximum impact on kinase activity in the absence of glutamine. However, the simulation results in the presence of glutamine emphasize the extended significance of this mutation on AdT structure and mobility. T149 is immediately adjacent to the glutamine-binding pocket of GatA. Our results suggest that the isosteric conversion of the T149 hydroxyl group to the valine methyl group in the T149V mutation is not only enough to perturb distal kinase activity but also to have a dramatic effect on glutamine binding and communication between the two active sites. Efforts to characterize this enzyme’s glutaminase activity are underway.
The correlation patterns of K89 (wild-type) and K89R AdT with all other residues in the enzyme reveal a significant correlation change in all three subunits (Figure 8). Combining the experimental results that the K89A and K89R mutations both significantly reduce the initial rate of phosphorylation of Glu-tRNAGln suggests that K89 is also involved in interdomain communication.
In addition, significant changes in correlation were observed for both T149V and K89R in areas that connect GatA and GatB (Figure 7), which further supports the hypothesis that a communication pathway exists between these two subunits and that K89 and T149 are influential members in this pathway. The fact that a mutation in GatB (K89R) and a mutation in GatA (T149V) separately and independently change the correlation pattern between GatA and GatB suggests bidirectionality in communication between GatA and GatB.
The RMSD comparison analysis identified 59 common residues that showed appreciable deviations in both mutant enzymes compared to wild-type. The fact that these 59 residues are common to both mutant enzymes strongly suggests their involvement in the communication pathway between GatA and GatB. The locations of these 59 residues (see Figure 10) show that they are located in and around the GatA and GatB active sites as well as throughout the region connecting the two sites. A few of the residues are in GatC as well, hinting at a possible role for GatC in mediating communication between GatA and GatB.
Importantly, further analyses of these 59 residues revealed a common correlation pattern connecting GatA to GatB not only in both mutant enzymes but also in wild-type AdT (Figure 11). This commonality is highlighted by the fact that only moderate differences in correlation were observed when comparing each mutant directly to wild-type (Figure 12). Consequently, these results demonstrate that our MD simulations have unmasked these residues as all or part of an interactive, dynamic, connective path for domain-domain communication in wild-type AdT. Given that K89 and T149 are members of this network and are essential for wild-type AdT activity, it seems probable that these 59 residues offer the first clear evidence of an extensive network of essential interactions throughout AdT.
In conclusion, structural studies work well for observing substantial motion in enzyme domains. For example, allosteric rearrangements, loop movements and domain rotations are often readily visualized by comparing co-crystals structures obtained in the presence of different substrates or ligands. Subtle structural changes can also be examined by crystallography but are sometimes more difficult to assess because of the challenges faced in tying observed residue movement to function. Molecular dynamics simulations are particularly useful for looking at domain-domain communication on a short time scale (nanoseconds). In this work, computational methods allowed us to readily visualize and compare movements in multiple simulations and to assess the impact of functionally interesting mutations. In this way, we were able to identify mobile elements in AdT that might have been difficult to isolate in a crystal structure. The importance of this work goes beyond simply advancing our understanding of how AdT functionally delivers ammonia from GatA to GatB; these results also have implications for other GATs and even other enzymes with apparent domain-domain communication mechanisms.
Supplementary Material
Acknowledgments
The authors thank Anastayisa Bordyukova for help in the construction of the pKM049 and pKM050 plamids, Drs. Stéphane Skouloubris and Pitak Chuawong for plasmids pSS003 and pPTC032, and Wayne State C&IT for computing time.
This work was financially supported by N.I.H. grant GM071480 to T. L. H. and by Wayne State University funds to G. A. C. and T. L. H.
ABBREVIATIONS
- AdT
Asp-tRNAAsn/Glu-tRNAGln amidotransferase
- AsnRS
asparaginyl-tRNA synthetase
- GAT
glutamine-dependent amidotransferase
- GlnRS
glutaminyl-tRNA synthetase
- His6
six-histidine tag
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- LDH
L-lactate dehydrogenase
- MD
molecular dynamics
- NAD+
nicotinamide adenine dinucleotide (oxidized)
- NADH
nicotinamide adenine dinucleotide (reduced)
- ND-AspRS
non-discriminating AspRS
- PEP
phosphoenolpyruvate
- PK
pyruvate kinase
- RMSD
root mean square deviation
Footnotes
SUPPORTING INFORMATION AVAILABLE
Figures S1-S6, showing SDS-PAGE analyses of wild-type AdT and all mutations, the effects of NADH concentration on S/N in the kinase enzyme-coupled assay, assay calibration curve, alignment of S. aureus Mu50 and H. pylori AdT primary sequences, RMSD variation with time, and close-ups of Figure 9; and Tables S1-S4, listing the primers used for site-directed mutagenesis, observed initial rates for wild-type AdT and all mutations in the absence and presence of bound glutamine, and the identities of the 59 common residues analyzed in Figures 11 and 12. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Hyde CC, Ahmed SA, Padlan EA, Miles EW, Davies DR. Three-dimensional structure of the tryptophan synthase α2β2 multienzyme complex from Salmonella typhimurium. J Biol Chem. 1988;263:17857–17871. [PubMed] [Google Scholar]
- 2.Weeks A, Lund L, Raushel FM. Tunneling of intermediates in enzyme-catalyzed reactions. Curr Opin Chem Biol. 2006;10:465–472. doi: 10.1016/j.cbpa.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 3.Raushel FM, Thoden JB, Holden HM. Enzymes with molecular tunnels. Acc Chem Res. 2003;36:539–548. doi: 10.1021/ar020047k. [DOI] [PubMed] [Google Scholar]
- 4.Huang X, Holden HM, Raushel FM. Channeling of substrates and intermediates in enzyme-catalyzed reactions. Annu Rev Biochem. 2001;70:149–180. doi: 10.1146/annurev.biochem.70.1.149. [DOI] [PubMed] [Google Scholar]
- 5.Miles EW, Rhee S, Davies DR. The molecular basis of substrate channeling. J Biol Chem. 1999;274:12193–12196. doi: 10.1074/jbc.274.18.12193. [DOI] [PubMed] [Google Scholar]
- 6.Fitzpatrick TB, Amrhein N, Kappes B, Macheroux P, Tews I, Raschle T. Two independent routes of de novo vitamin B6 biosynthesis: not that different after all. Biochem J. 2007;407:1–13. doi: 10.1042/BJ20070765. [DOI] [PubMed] [Google Scholar]
- 7.Williams L, Fresquet V, Santander PJ, Raushel FM. The multiple amidation reactions catalyzed by Cobyric acid synthetase from Salmonella typhimurium are sequential and dissociative. J Am Chem Soc. 2007;129:294–295. doi: 10.1021/ja067962b. [DOI] [PubMed] [Google Scholar]
- 8.Wojcik M, Seidle HF, Bieganowski P, Brenner C. Glutamine-dependent NAD+ synthetase. How a two-domain, three-substrate enzyme avoids waste. J Biol Chem. 2006;281:33395–33402. doi: 10.1074/jbc.M607111200. [DOI] [PubMed] [Google Scholar]
- 9.Ibba M, Becker HD, Stathopoulos C, Tumbula DL, Soll D. The adaptor hypothesis revisited. Trends Biochem Sci. 2000;25:311–316. doi: 10.1016/s0968-0004(00)01600-5. [DOI] [PubMed] [Google Scholar]
- 10.Klem TJ, Davisson VJ. Imidazole glycerol phosphate synthase: the glutamine amidotransferase in histidine biosynthesis. Biochemistry. 1993;32:5177–5186. doi: 10.1021/bi00070a029. [DOI] [PubMed] [Google Scholar]
- 11.Richards NG, Kilberg MS. Asparagine synthetase chemotherapy. Annu Rev Biochem. 2006;75:629–654. doi: 10.1146/annurev.biochem.75.103004.142520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teplyakov A, Leriche C, Obmolova G, Badet B, Badet-Denisot MA. From Lobry de Bruyn to enzyme-catalyzed ammonia channelling: molecular studies of D-glucosamine-6P synthase. Nat Prod Rep. 2002;19:60–69. doi: 10.1039/b103713g. [DOI] [PubMed] [Google Scholar]
- 13.Kappock TJ, Ealick SE, Stubbe J. Modular evolution of the purine biosynthetic pathway. Curr Opin Chem Biol. 2000;4:567–572. doi: 10.1016/s1367-5931(00)00133-2. [DOI] [PubMed] [Google Scholar]
- 14.Evans DR, Guy HI. Mammalian pyrimidine biosynthesis: fresh insights into an ancient pathway. J Biol Chem. 2004;279:33035–33038. doi: 10.1074/jbc.R400007200. [DOI] [PubMed] [Google Scholar]
- 15.Binda C, Bossi RT, Wakatsuki S, Arzt S, Coda A, Curti B, Vanoni MA, Mattevi A. Cross-talk and ammonia channeling between active centers in the unexpected domain arrangement of glutamate synthase. Structure. 2000;8:1299–1308. doi: 10.1016/s0969-2126(00)00540-2. [DOI] [PubMed] [Google Scholar]
- 16.Chaudhuri BN, Lange SC, Myers RS, Chittur SV, Davisson VJ, Smith JL. Crystal structure of imidazole glycerol phosphate synthase: a tunnel through a (beta/alpha)8 barrel joins two active sites. Structure. 2001;9:987–997. [PubMed] [Google Scholar]
- 17.Larsen TM, Boehlein SK, Schuster SM, Richards NG, Thoden JB, Holden HM, Rayment I. Three-dimensional structure of Escherichia coli asparagine synthetase B: a short journey from substrate to product. Biochemistry. 1999;38:16146–16157. doi: 10.1021/bi9915768. [DOI] [PubMed] [Google Scholar]
- 18.Muchmore CR, Krahn JM, Kim JH, Zalkin H, Smith JL. Crystal structure of glutamine phosphoribosylpyrophosphate amidotransferase from Escherichia coli. Protein Sci. 1998;7:39–51. doi: 10.1002/pro.5560070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teplyakov A, Obmolova G, Badet B, Badet-Denisot MA. Channeling of ammonia in glucosamine-6-phosphate synthase. J Mol Biol. 2001;313:1093–1102. doi: 10.1006/jmbi.2001.5094. [DOI] [PubMed] [Google Scholar]
- 20.Thoden JB, Raushel FM, Benning MM, Rayment I, Holden HM. The structure of carbamoyl phosphate synthetase determined to 2.1 A resolution. Acta Crystallogr D Biol Crystallogr. 1999;55:8–24. doi: 10.1107/S0907444998006234. [DOI] [PubMed] [Google Scholar]
- 21.Curnow AW, Hong K, Yuan R, Kim S, Martins O, Winkler W, Henkin TM, Soll D. Glu-tRNAGln amidotransferase: a novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc Natl Acad Sci USA. 1997;94:11819–11826. doi: 10.1073/pnas.94.22.11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ibba M, Soll D. Aminoacyl-tRNA synthesis. Ann Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 23.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, Nelson K, Quackenbush J, Zhou L, Kirkness EF, Peterson S, Loftus B, Richardson D, Dodson R, Khalak HG, Glodek A, McKenney K, Fitzegerald LM, Lee N, Adams MD, Hickey EK, Berg DE, Gocayne JD, Utterback TR, Peterson JD, Kelley JM, Cotton MD, Weidman JM, Fujii C, Bowman C, Watthey L, Wallin E, Hayes WS, Borodovsky M, Karp PD, Smith HO, Fraser CM, Venter JC. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 24.Cathopoulis T, Chuawong P, Hendrickson TL. Novel tRNA aminoacylation mechanisms. Mol Biosyst. 2007;3:408–418. doi: 10.1039/b618899k. [DOI] [PubMed] [Google Scholar]
- 25.Sheppard K, Akochy PM, Salazar JC, Soll D. The Helicobacter pylori amidotransferase GatCAB is equally efficient in glutamine-dependent transamidation of Asp-tRNAAsn and Glu-tRNAGln. J Biol Chem. 2007;282:11866–11873. doi: 10.1074/jbc.M700398200. [DOI] [PubMed] [Google Scholar]
- 26.Nakamura A, Yao M, Chimnaronk S, Sakai N, Tanaka I. Ammonia channel couples glutaminase with transamidase reactions in GatCAB. Science. 2006;312:1954–1958. doi: 10.1126/science.1127156. [DOI] [PubMed] [Google Scholar]
- 27.Bailly M, Blaise M, Lorber B, Becker HD, Kern D. The transamidosome: a dynamic ribonucleoprotein particle dedicated to prokaryotic tRNA-dependent asparagine biosynthesis. Mol Cell. 2007;28:228–239. doi: 10.1016/j.molcel.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 28.Huot JL, Balg C, Jahn D, Moser J, Emond A, Blais SP, Chenevert R, Lapointe J. Mechanism of a GatCAB amidotransferase: aspartyl-tRNA synthetase increases its affinity for Asp-tRNA(Asn) and novel aminoacyl-tRNA analogues are competitive inhibitors. Biochemistry. 2007;46:13190–13198. doi: 10.1021/bi700602n. [DOI] [PubMed] [Google Scholar]
- 29.Horiuchi KY, Harpel MR, Shen L, Luo Y, Rogers KC, Copeland RA. Mechanistic studies of reaction coupling in Glu-tRNAGln amidotransferase. Biochemistry. 2001;40:6450–6457. doi: 10.1021/bi002599l. [DOI] [PubMed] [Google Scholar]
- 30.Kiianitsa K, Solinger JA, Heyer WD. Rad54 protein exerts diverse modes of ATPase activity on duplex DNA partially and fully covered with Rad51 protein. J Biol Chem. 2002;277:46205–46215. doi: 10.1074/jbc.M207967200. [DOI] [PubMed] [Google Scholar]
- 31.Kowalczykowski SC, Krupp RA. Effects of Escherichia coli SSB protein on the single-stranded DNA-dependent ATPase activity of Escherichia coli RecA protein.. Evidence that SSB protein facilitates the binding of RecA protein to regions of secondary structure within single-stranded DNA. J Mol Biol. 1987;193:97–113. doi: 10.1016/0022-2836(87)90630-9. [DOI] [PubMed] [Google Scholar]
- 32.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein Identification and Analysis Tools on the ExPASy Server. In: Walker JM, editor. The Proteomics Protocols Handbook. Human Press; Totowa, NJ: 2005. pp. 571–607. [Google Scholar]
- 33.Skouloubris S, Ribas de Pouplana L, De Reuse H, Hendrickson TL. A noncognate aminoacyl-tRNA synthetase that may resolve a missing link in protein evolution. Proc Natl Acad Sci USA. 2003;100:11297–11302. doi: 10.1073/pnas.1932482100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang KM, Hendrickson TL. Recognition of tRNAGln by Helicobacter pylori GluRS2--a tRNAGln-specific glutamyl-tRNA synthetase. Nucleic Acids Res. 2009;37:6942–6949. doi: 10.1093/nar/gkp754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng L, Sheppard K, Tumbula-Hansen D, Soll D. Gln-tRNAGln formation from Glu-tRNAGln requires cooperation of an asparaginase and a Glu-tRNAGln kinase. J Biol Chem. 2005;280:8150–8155. doi: 10.1074/jbc.M411098200. [DOI] [PubMed] [Google Scholar]
- 36.Case DA, Cheatham TE, 3rd, Darden T, Gohlke H, Luo R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical-Integration of Cartesian Equations of Motion of a System with Constraints - Molecular-Dynamics of N-Alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 38.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A Smooth Particle Mesh Ewald Method. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 39.York DM, Darden TA, Pedersen LG. The Effect of Long-Range Electrostatic Interactions in Simulations of Macromolecular Crystals - a Comparison of the Ewald and Truncated List Methods. J Chem Phys. 1993;99:8345–8348. [Google Scholar]
- 40.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 42.Berendsen HJC, Postma JPM, Vangunsteren WF, Dinola A, Haak JR. Molecular-Dynamics with Coupling to an External Bath. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 43.Sheppard K, Akochy PM, Salazar JC, Soll D. The Helicobacter pylori amidotransferase GatCAB is equally efficient in glutamine-dependent transamidation of Asp-tRNAAsn and Glu-tRNAGln. J Biol Chem. 2007;282:11866–11873. doi: 10.1074/jbc.M700398200. [DOI] [PubMed] [Google Scholar]
- 44.Wu J, Bu W, Sheppard K, Kitabatake M, Kwon ST, Soll D, Smith JL. Insights into tRNA-dependent amidotransferase evolution and catalysis from the structure of the Aquifex aeolicus enzyme. J Mol Biol. 2009;391:703–716. doi: 10.1016/j.jmb.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaledin M, Kaledin AL, Brown A, Bowman JM. Driven Molecular Dynamics for Normal Modes of Biomolecules without the Hessian, and Beyond. In: Cui Q, Bahar I, editors. Normal Mode Analysis: Theory and Applications to Biological and Chemical Systems. Chapman and Hall/CRC; Boca Raton: 2006. pp. 281–300. [Google Scholar]
- 46.Cathopoulis TJ, Chuawong P, Hendrickson TL. A thin-layer electrophoretic assay for Asp-tRNAAsn/Glu-tRNAGln amidotransferase. Anal Biochem. 2007;360:151–153. doi: 10.1016/j.ab.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bailly M, Giannouli S, Blaise M, Stathopoulos C, Kern D, Becker HD. A single tRNA base pair mediates bacterial tRNA-dependent biosynthesis of asparagine. Nucleic Acids Res. 2006;34:6083–6094. doi: 10.1093/nar/gkl622. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.