Abstract
Purpose.
Lacritin is a prosecretory mitogen in tears and, although a tear protein, it promotes basal tearing and lacrimal gland secretion. Since scale up is relevant to its potential use in the treatment of dry eye, we explored various mutagenic strategies to alter the stability, solubility, and translational efficiency of nascent lacritin, and discovered 3′ clustering of rare human codons.
Methods.
Site-directed mutagenesis of lacritin coding cDNA “pLAC” generated 24 different nonsynonymous and 13 synonymous mutations. Nonsynonymous mutations altered amino acids with nonpolar, basic or acidic side chains to serine. Synonymous mutation progressively optimized human codons that are rare or uncommon in Escherichia coli without changing the amino acid specified. These changes were validated by sequencing and protein production, and analyzed via the “rare codon calculator” (RCC). Nonhuman primate and nonprimate lacritin coding sequences were extracted from Ensembl, and analyzed via RCC using codon usage appropriate for each species.
Results.
Superior yields were obtained by modification of individual hydrophobic residues or a predicted salt bridge, suggesting that production was limited by lacritin stability. Accordingly, elimination of rare codons increased yields less effectively. Importantly, RCC analysis of human, nonhuman primate (mouse lemur) and nonprimate (cat, tree shrew) lacritin coding sequences revealed remarkable 3′ clustering of rare codons, unlike human lipocalin-1 and 21 other widely expressed human tear genes.
Conclusions.
Lacritin protein yields were improved primarily by hydrophobic or salt bridge mutagenesis and less so by elimination of rare codons. The 3′ clustering of rare codons is conserved in all lacritin orthologs examined.
We used mutagenesis to increase recombinant lacritin yield, and discovered that almost all of lacritin's rare codons are clustered in its 3′ translated region. This remarkable and unusual feature is shared by all seven lacritin orthologs examined, but is absent from 21 tear proteins.
Introduction
Recombinant protein treatment of ocular surface disease is an unexplored area that could prove beneficial as functions of common and minor tear proteins are better appreciated,1 such as with lacritin, a small human tear protein. Recombinant lacritin generated in Escherichia coli displays several ocular surface-relevant activities.2 It promotes basal tearing in rabbits3 and constitutive, but not stimulated tear protein secretion by rat lacrimal acinar cells in primary cell culture.4 It also protects against dry eye–associated stress over a biphasic dose response with a 1 to 10 nM dose optimum (Wang N, et al. IOVS 2011;52:ARVO E-Abstract 3712). These observations suggest relevance to dry eye, in which lacritin appears to be the only prosecretory tear factor that is selectively deficient.2 Lacritin also is mitogenic for subconfluent human corneal epithelial cells5 and, therefore, might have benefit following refractive eye surgery. However, protein yields from E. coli production have been too low for clinical scale up.
Possible contributing factors include lacritin's C-terminal hydrophobicity, several predicted salt bridges, and/or suboptimal codon usage for translation of the human cDNA “pLAC” in E. coli. Hydrophobic residues can seed the aggregation and seclusion of nascent recombinant proteins into insoluble inclusion bodies. Salt bridges can be either stabilizing or nonstabilizing6 and, if destabilizing, can contribute to misfolding. Codons common in human protein translation are rare or uncommon in E. coli.7 With tRNA's limiting, translation is slowed. The latter is addressed routinely by synonymous mutation of rare or uncommon codons, or by overexpression of rare E. coli tRNA genes, although not addressing aggregation or misfolding for which a satisfactory solution rarely is achieved. Engineering selectively fewer hydrophobic residues by nonsynonymous mutagenesis could prove beneficial, at the risk of altering protein activity. Lacritin's C-terminal hydrophobic residues align on an α-helix as the binding face for the cell surface heparan sulfate proteoglycan syndecan-1.8 Syndecan-1 binding is a prerequisite for lacritin mitogenic and prosecretory activities.8 Distal from this site are other hydrophobic residues and a predicted salt bridge.
With transcription of lacritin in humans more eye-specific than α-crystallin,9 interesting regulatory mechanisms may be in play that may offer benefit in ocular tissue engineering. Rare codons can be arranged in clusters as a mechanism of translational regulation associated with restricted tissue expression in humans.10 Rare codon clustering also is thought by some to slow translation for more efficient Pfam domain protein folding11 or when 5′ for controlled ribosomal association.12,13 Here, we improved recombinant human lacritin production through nonsynonymous and synonymous site-directed mutagenesis. We also discovered 3′ clustering of rare codons in the LACRT coding sequence of humans and other vertebrates, to our knowledge the first apparent evidence of conserved rare codon clustering of mammalian non-Pfam domains.
Materials and Methods
Materials and Cells
The Wizard Plus SV DNA Purification System was purchased from Promega (Madison, WI). Restriction enzymes and chitin beads were obtained from New England BioLabs (Ipswich, MA). The QuikChange II XL Site Directed Mutagenesis kit was purchased from Stratagene (Santa Clara, CA). Competent BL21-CodonPlus E. coli cells containing rare E. coli tRNA genes on a chloramphenicol-resistant plasmid were purchased from Invitrogen (Grand Island, NY). Competent ER2566 cells were purchased from New England BioLabs. Diethylaminoethyl cellulose (DEAE)–Sepharose was purchased from GE Healthcare Bio-Science AB (Uppsala, Sweden). A BCA protein assay kit was purchased from Thermo Scientific (Waltham, MA).
Lacritin Site-Directed Mutagenesis and Validation
PCR primers (Sigma-Aldrich, St. Louis, MO) as long as 48 base pairs (bp; see Supplementary Material and Supplementary Tables S1, S2, http://www.iovs.org/content/54/3/1979/suppl/DC1) were designed as recommended (QuikChange II XL Site-Directed Mutagenesis protocol; Stratagene). Primers were used to alter parent cDNA pLAC such that upon translation serine replaced 24 different C-terminal residues with nonpolar, acidic, or basic side chains (see Supplementary Material and Supplementary Table S1, http://www.iovs.org/content/54/3/1979/suppl/DC1). Other primers developed 13 different pLAC templates with progressively fewer codons used rarely or uncommonly in E. coli (see Supplementary Material and Supplementary Table S2, http://www.iovs.org/content/54/3/1979/suppl/DC1). QuikChange II XL Site-Directed Mutagenesis followed manufacturer's instructions. Briefly, we used 1.25 μL (125 ng) of forward and reverse primers, 2 μL template plasmid (initially parent plasmid pLAC5), 5 μL 10× reaction buffer, 1 μL dNTP mix, 36.5 μL distilled H20, 3 μL QuikSolution, and 1 μL Pfu high fidelity DNA polymerase in a 50 μL reaction volume. Thermocycler (BioRad, Hercules, CA) conditions were one cycle at 95°C for 30 seconds, followed by 18 cycles at 90°C (30 seconds), 55°C (1 minute), 68°C (8 minutes), and then one cycle at 68°C (7 minutes), ending with a 4°C hold. The PCR reactions were cleaned using the Promega Wizard clean up kit before transformation or additional mutagenic PCR. In the case of RC07, additional rounds of mutagenic PCR were performed before transformation (see Supplementary Material and Supplementary Table S3, http://www.iovs.org/content/54/3/1979/suppl/DC1). Amplicons were expanded by transformation and growth in ER2566 cells under ampicillin selection on LB-Amp plates, from which overnight cultures were established for plasmid mini-preps, and analysis of inserts following NdeI and HindIII digestion. cDNA from promising clones was sequenced by the University of Virginia DNA Sequencing Core on an Applied Biosystems 3730 DNA Analyzer (Applied Biosystems, Carlsbad, CA).
Lacritin Production and Purification
Lacritins from unaltered or mutagenized pLAC in transformed ER2566 E. coli cells, or pLAC in BL21-CodonPlus E. coli cells were generated and affinity purified on chitin beads using the intein fusion tag as described previously.5 Subsequent addition of β-mercaptoethanol separated lacritin from intein. Eluted lacritin was concentrated by Amicon ultraspin filtration (EMD Millipore Corp., Billerica, MA) on a 3 kDa molecular weight cutoff cartridge, dialyzed versus 5 L of PBS, and then purified further on DEAE Sepharose. DEAE binds a C-terminal lacritin cleavage fragment, contaminating E. coli chaperone DnaK and endotoxin. Purified lacritin was quantitated by the BCA protein assay, assessed by SDS-PAGE, and then aliquotted and lyophilized for −70° storage.
Lacritin Coding Sequence Analyses
Human lacritin coding sequence from pLAC and coding sequence from mutagenized pLAC-RC13 were entered into the “rare codon calculator” (RCC)14 (available in the public domain at http://www.codons.org/) set for E. coli codon usage. Complete coding sequences for human, chimp (Pan troglodytes), mouse lemur (Microcebus murinus), cat (Felis catus), tree shrew (Tupaia belangeri), shrew (Sorex araneus), and horse (Equus caballus) lacritin were analyzed similarly using species-appropriate nonmitochondrial codon usage tables obtained from the Codon Usage Database (available in the public domain at http://www.kazusa.or.jp/codon/). For comparison, coding sequences for LCN1 and a number of other human tear proteins were obtained from the human hORFeome Database (available in the public domain at http://horfdb.dfci.harvard.edu/), and also assessed by RCC using the human nonmitochondrial codon usage tables. Selected were 21 of the first 27 tear proteins of the human tear proteome (as presented in Table 1 of Ref. 1).
Results
Enhanced Lacritin Yield
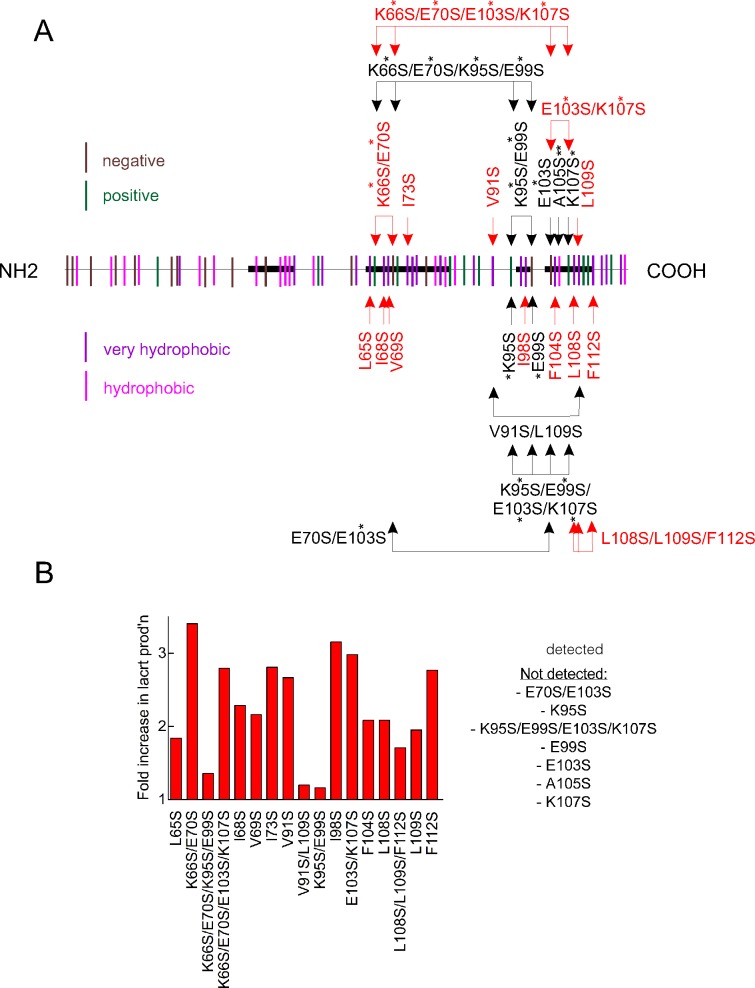
Lacritin is a pluripotent tear protein that promotes basal tearing,3 is cytoprotective against inflammatory cytokines, and promotes the proliferation of subconfluent human corneal epithelial cells.5 Although lacritin purified from monkey tears is prosecretory,15 all of lacritin's known activities were discovered with recombinant human lacritin generated by E. coli, historically the most efficient protein manufacturing approach. Current yields are sufficient for research, but not for scale up. Problems could derive from hydrophobic, acidic, or basic residues. For example, hydrophobic residues can promote misfolding. Misfolded proteins aggregate and become secluded in inclusion bodies.16 Secreted lacritin has 61 residues with nonpolar side chains (Fig. 1A; hydrophobic, magenta; very hydrophobic, purple), including 34 of 59 residues in the C-terminal half where all known lacritin activities have been attributed.2 Of these, 19 (of 22 for all of lacritin) are highly hydrophobic. We mutated 11 to serine. Lacritin A105S was insoluble. However, lacritins I68S, I68S/I78S, V69S, I73S, V91S, I98S, F104S, L108S, L109S, and F112S were completely soluble and were generated with yields two or three times that of unaltered lacritin (Fig. 1B). Lacritin protects human corneal epithelial cells that have been stressed with the inflammatory cytokines interferon-γ and tumor necrosis factor (Wang N, et al. IOVS 2011;52:ARVO E-Abstract 3712), and requires syndecan-1 for cell targeting.8 Lacritins I98S, F104S, and F112S were inactive in cytoprotection assays (Wang N, et al. IOVS 2011;52:ARVO E-Abstract 3712), and I73S, L108S, L109S, and F112S failed to bind syndecan-1 (Zhang Y, et al. IOVS 2010;51:ARVO E-Abstract 4179). However, lacritins V69S, I73S, L108S, and L109S each were fully cytoprotective (Wang N, et al. IOVS 2011;52:ARVO E-Abstract 3712), and lacritins I68S, V69S, V91S, I98S, and F104S fully bound syndecan-1 (Zhang Y, et al. IOVS 2010;51:ARVO E-Abstract 4179).
Figure 1.
Selective point mutagenesis of lacritin's hydrophobic residues, and a charged pair, substantially improves protein yield in E. coli. (A) Distribution of all hydrophobic (magenta) and very hydrophobic (purple) residues, as well as positively (green) and negatively (brown) charged residues along the length of lacritin. Shown is lacritin without the signal peptide. Point mutants under study are indicated. Double and single asterisks, respectively, indicate conservative and semiconservative substitutions. All other substitutions are radical. (B) Fold increase in purified mutant lacritin generated versus unaltered lacritin from 1 L productions runs of each that had been purified on chitin columns, released without tag with β-mercaptoethanol, concentrated by Amicon ultraspin filtration (EMD Millipore Corp.) on a 3 kDa molecular weight cutoff cartridge, dialyzed versus 5 L of PBS and then purified further on DEAE Sepharose. Purified lacritin was quantitated by the BCA protein assay.
Salt bridges between proximal amino acids with opposite charge can either stabilize or destabilize protein folding.6,17 Lacritin lacking the signal peptide has 15 negatively and 13 positively charged residues (Fig. 1A, respectively, brown, green), mostly glutamic acid and lysine. Of these, 5 negatively and 10 positively charged residues are located in the C-terminal half. To initiate this analysis, we generated single and double point mutations affecting 4 glutamic acids [E] and 4 lysines [K] (see Supplementary Material and Supplementary Table S1, http://www.iovs.org/content/54/3/1979/suppl/DC1); thus, 4 of 5 negatively charged residues. Lacritins K66S/E70S, K95S, E99S, E103S, E70S/E103S, and K107S were each insoluble, in keeping with the charge destabilizing effect of a single mutation on salt bridges.17 However, lacritins K66S/E70S/K95S/E99S, K66S/E70S/E103S/K107S, E99S/K95S, and E103S/K107S were soluble. Lacritins K66S/E70S/K95S/E99S and E99S/K95S were generated at a level equivalent to unaltered lacritin. Remarkably, the lacritin K66S/E70S was four times greater, and yields of lacritins K66S/E70S/E103S/K107S and E103S/K107S were almost three times greater (Fig. 1B). Although lacritin K66S/E70S has not yet been tested in either functional assay, lacritin lacking 71 amino acids from the N-terminus is fully cytoprotective (Wang N, et al. IOVS 2011;52:ARVO E-Abstract 3712) and fully binds syndecan-1 (Zhang Y, et al. IOVS 2010;51:ARVO E-Abstract 4179). Lacritin K66S/E70S/E103S/K107S also has not yet been tested; however, lacritin E103S/K107S does not bind syndecan-1 (Zhang Y, et al. IOVS 2010;51:ARVO E-Abstract 4179). Thus, engineering fewer paired charges and nonpolar side chains significantly improves recombinant lacritin yield.
Slightly Enhanced Lacritin Yield
Human codons that are rare or uncommon in E. coli can slow translation by promoting ribosomal stacking, particularly when mRNA is in excess.18,19 Of lacritin's 119 codons represented in pLAC, 42 are rare or uncommon in E. coli (Fig. 2A, blue). Since available E. coli tRNA's are limited accordingly, protein production should be less. Accordingly, generation of recombinant lacritin should be hindered.
Figure 2.
Comparative coding sequences of human lacritin before and after synonymous mutation for enhanced transcription and translation in E. coli. (A) Human lacritin mRNA without signal peptide contains 42 codons that are uncommonly (blue, 11%–20% frequency) or rarely (blue underline, <10%) used in E. coli (E. coli codon table). (B) Mutagenized coding sequence (pLAC-RC13) in which 29 rare or uncommon codons (green underline) were modified to common E. coli codons.
Site-directed mutagenesis of pLAC's rare or uncommon codons (Fig. 2B) was initiated in a progressive manner with repeated validation by sequencing at each step. Particular focus was on grouped rare or uncommon codons appropriate for simultaneous mutation with 48 nucleotide primer pairs (see Supplementary Material and Supplementary Table S2, http://www.iovs.org/content/54/3/1979/suppl/DC1). Codons 74 to 76 were altered first (pLAC-RC01), followed successively by codons 62, 64 (pLAC-RC02); 34 to 36 (pLAC-RC03); 51 and 54 (pLAC-RC04); 114 and 115 (pLAC-RC05); 37, 39, and 40 (pLAC-RC06); 59, 84, 86, 108, and 110 (pLAC-RC07); and 68 and 70 (pLAC-RC08). Two unexpected mutations in RC07 (codons 81, 83) were corrected respectively in pLAC-RC09. Subsequent mutagenesis involved codons 43 and 44 (pLAC-RC10); 92 and 94 (pLAC-RC11); 17 and 20 (pLAC-RC12); and codon 101 (pLAC-RC13; see Supplementary Material and Supplementary Table S3, http://www.iovs.org/content/54/3/1979/suppl/DC1). A total of 29 codons was mutated synonymously.
Lacritin protein production was compared (Fig. 3A, left). Surprisingly, synonymous mutagenesis of rare or uncommon codons only slightly benefitted yield, unlike mutagenesis of hydrophobic or charged residues. Best results were obtained for lacrt-RC05 and -RC06 with yields decreasing with further mutagenesis. Transformation of pLAC into BL21-CodonPlus cells that carry tRNA genes on a T7 promoter-driven plasmid was not beneficial, although production of C-25 (lacritin lacking 25 amino acids from the C-terminus) did benefit almost 2-fold (Fig. 3A, right). Yield similarly was improved by synonymous mutation of N-65 (lacritin lacking 65 amino acids from the N-terminus; Fig. 3A, middle). Together, these data were in keeping with the possibility that aberrant folding, not translation, was the primary factor limiting lacritin production. Combining nonsynonymous mutation, that is of K66S/E70S, with synonymous mutagenesis may be the ultimate solution.
Figure 3.
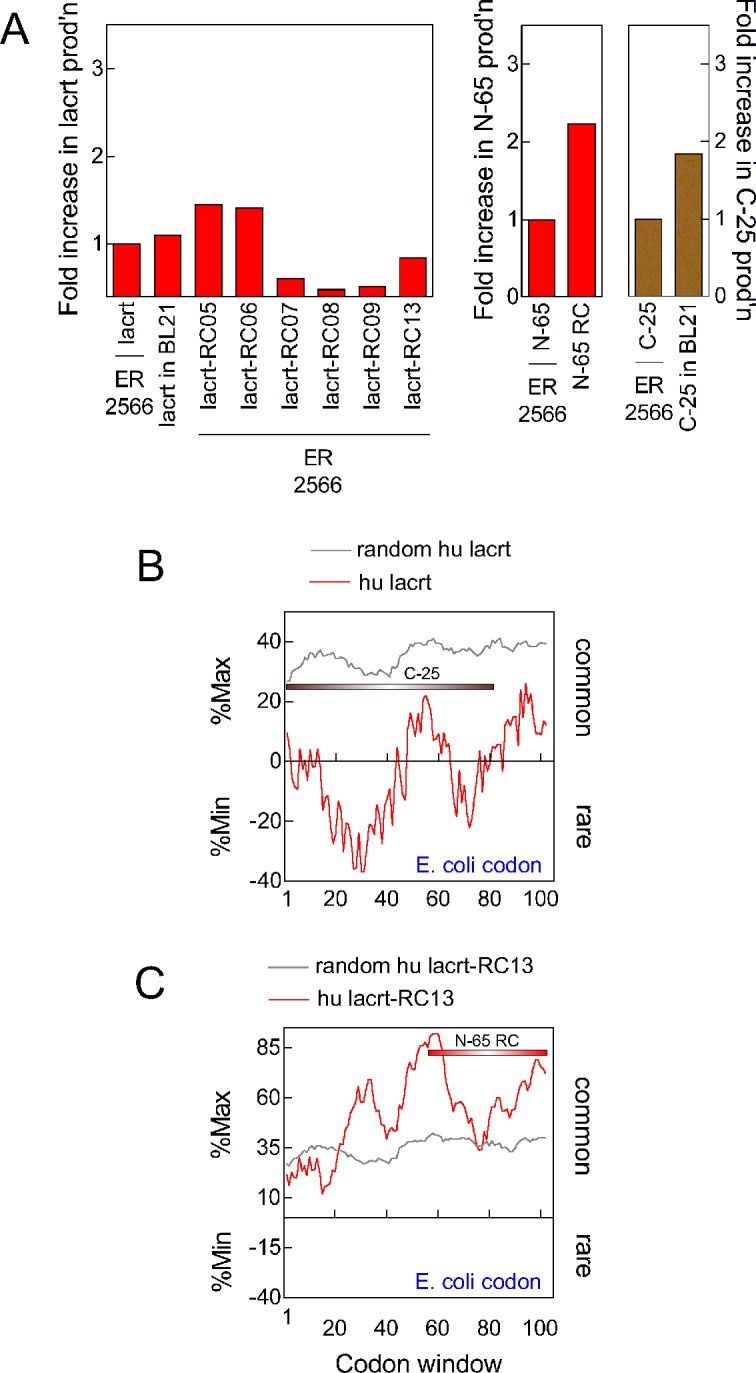
Protein yield after mutagenesis of rare/uncommon (“rare”) and analysis of rare or common codon window values. (A) Left: 13 successive generations of mutagenized lacritin were prepared. Compared is lacritin protein production (as per Fig. 1B) by pLAC versus lacrt-RC05, -RC06, -RC07, -RC08, -RC09, -RC13, or lacrt-RC13 transfected E. coli ER2566 cells. Also compared is pLAC in E. coli BL21 cells. Middle: lacritin N-65 production versus synonymously mutated N-65 in ER 2566 cells. Right: lacritin C-25 production in ER2566 cells versus in BL21 cells. (B) RCC analysis of human pLAC using E. coli codon usage. The algorithm calculates average rare or common codon prevalence within consecutive windows of 18 nucleotides, with expression (red tracing) as the fraction of each window in which amino acids are coded by the most common (%Max) or most rare (%Min) codons. Windows with rare codons predominate. Grey tracing indicates control values from 200 “random reverse translations” in which codon table-biased substitutions were performed randomly for each amino acid. (C) After mutagenesis of 29 codons, windows with common codons are now more prevalent.
To appreciate better how rare codon mutagenesis alters the codon usage landscape, we used the RCC,14 which calculates average rare (%Min) or common (%Max) codon prevalence within consecutive windows of 18 nucleotides (Figs. 3B, 3C). Analysis was done using the E. coli codon usage table on pLAC that lacks coding for the signal peptide. Rare codons occupy much of the 5′ half of pLAC as a large cluster. A smaller cluster is 3′. Neither was observed in random reverse translated pLAC in which codon table-biased substitutions were performed randomly for each amino acid (Fig. 3B, grey tracing). The mutagenized pLAC-RC13 profile differed substantially, with common codons dominating all codon windows.
Rare Codon Clustering in Human LACRT and Orthologs
Rare codons can be arranged in clusters as a mechanism of translational regulation associated with restricted tissue expression in humans.10 The lacritin coding LACRT gene is one of the most eye-specific genes,9 and could be a candidate for this form of translational regulation. Rare codon clustering also is thought to slow translation for more efficient Pfam domain protein folding11 or when 5′ for controlled ribosomal association.12,13 We analyzed the complete LACRT coding sequence (including coding for the signal peptide) using the RCC set instead for human codon usage (Fig. 4A), rather than E. coli codon usage (above). As a negative control, similar analysis was performed on LACRT coding sequence that had been random reverse translated (grey tracing). Interestingly, the 3′ region (red tracing) is represented largely by rare or uncommon codons, with very few common. These code for three α-helices (Fig. 4A, red rectangles), including the C-terminal amphipathic α-helix necessary for targeting lacritin's cell surface co-receptor syndecan-1,8 and in turn all of lacritin's known activities. Between α-helices, codons generally are more common (Fig. 4A).
Figure 4.
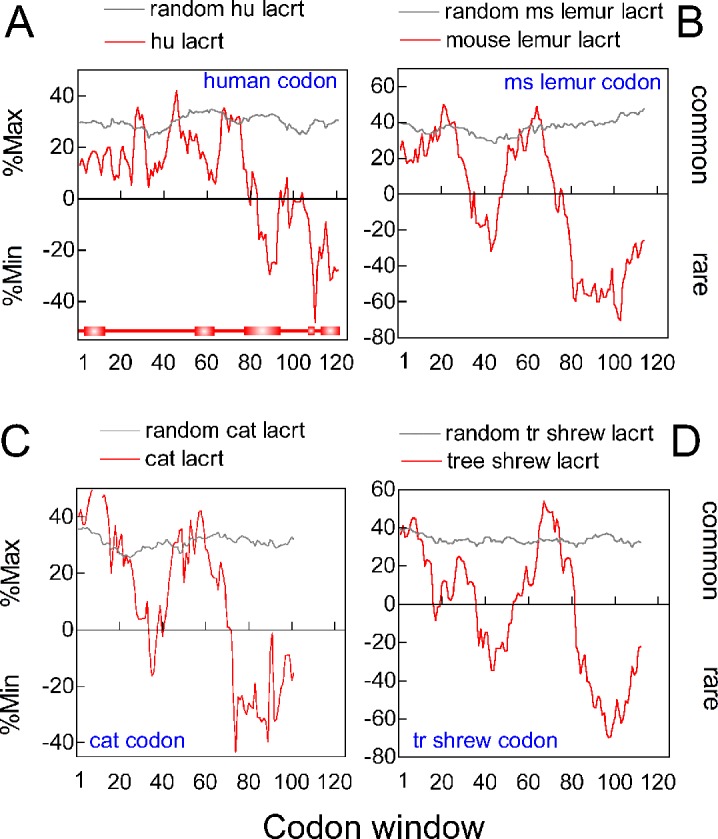
Rare codon clustering in the 3′ region of primate and nonprimate LACRT coding sequences. (A) RCC analysis of human LACRT coding sequence (red tracing) versus randomized control (grey tracing). Analysis was with the human codon frequency table. Bottom: linear diagram of lacritin with signal peptide and PSIPRED predicted α-helices aligned by codon number, but lacking eight C-terminal amino acids as per the last codon window (codons 112–130, centered on 121), in keeping with an 18 nucleotide window. (B) RCC analysis of mouse lemur (Microcebus murinus) LACRT coding sequence, using the M. murinus codon frequency table. (C) RCC analysis of cat (Felis catus) LACRT coding sequence with the F. catus codon frequency table. (D) RCC analysis of tree shrew (Tupaia belangeri) using T. belangeri codon frequencies. (A) through (D) were calculated with coding sequence for the signal peptide included.
Although on the surface contradictory, adoption and conservation of rare codons is beneficial evolutionarily.20 We were curious as to whether rare codon clustering is conserved accordingly in lacritin orthologs from primates and nonprimates. Mouse lemur (M. murinus), cat (F. catus), and tree shrew (T. belangeri) lacritins are, respectively, 48%, 44%, and 38% identical with human. All displayed substantial 3′ rare codon clustering (Figs. 4B–D) when examined with species appropriate codon tables (Codon Usage Database, available in the public domain at http://www.kazusa.or.jp/codon/). We extracted codon frequency data for comparison (Fig. 5). Codon usage is well-conserved across human, chimp (P. troglodytes), mouse lemur, horse (E. caballus), cat, tree shrew, and common shrew with the exception of the more common use of CUC, GUC, and ACC by common shrew (Fig. 5), possibly a reflection of currently limited codon data for this species.
Figure 5.
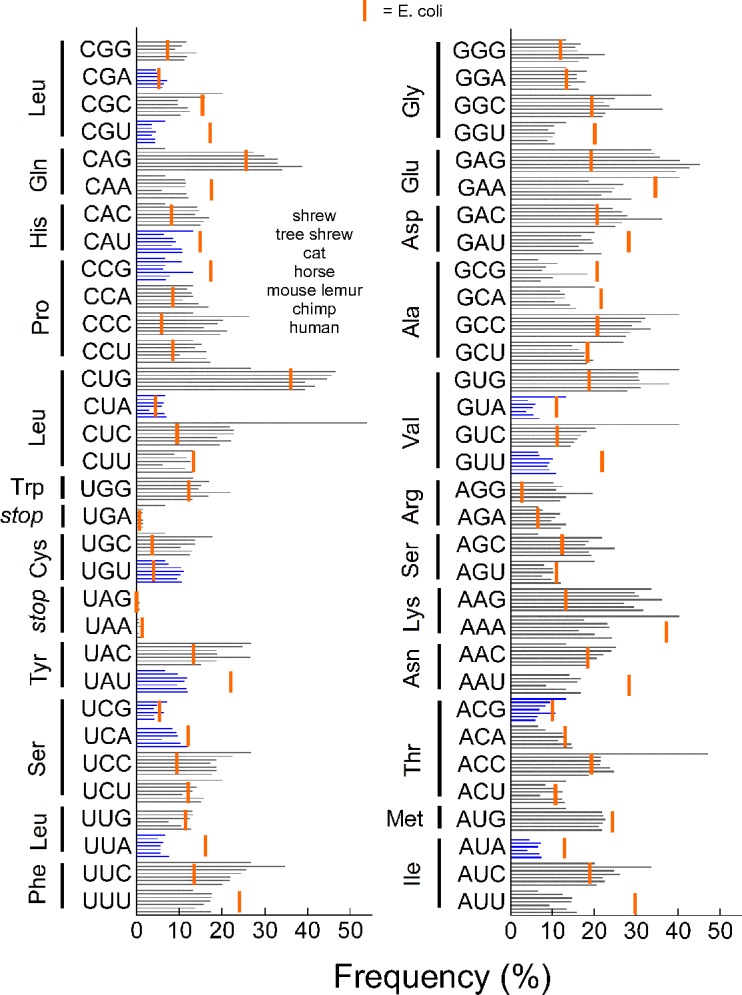
Frequency of codon usage is similar among common shrew, tree shrew, cat, horse, mouse lemur, chimp, and human (order from top down for each codon). Rare codon usage for each is indicated in blue. These data contrast with E. coli (vertical orange bars). Data were assembled from the Codon Usage Database.
Several other lacritin orthologs were examined. Substantial 3′ rare codon clustering also was apparent in chimp and common shrew (not shown), 99% and 41%, respectively, identical with human. Recently, we discovered lacritin in horse tears (45% identical)21 and from extracted sequence could determine that the C-terminal predicted α-helical structure was conserved, including a likely amphipathic α-helix. Horse lacritin coding sequence also displays strong 3′ rare codon clustering (#7 in Fig. 6B). Thus, in all lacritin orthologs examined, 3′ rare codon clustering is conserved remarkably.
Figure 6.
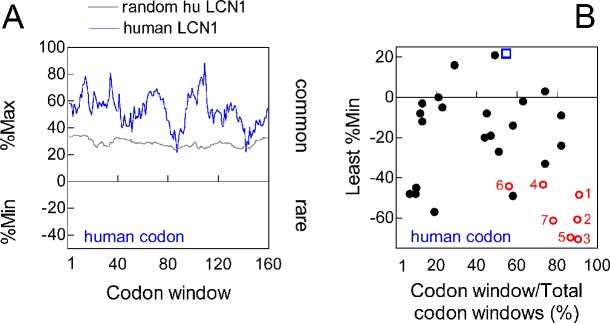
3′ rare codon clustering is not a general property of tear proteins. (A) RCC analysis of LCN1 coding sequence with the human codon frequency table. (B) The least %Min for each the first 21 tear proteins of the human tear proteome (see Table 1 in Ref. 1) was plotted as a fraction of coding sequence length (closed circles). Included was the least %Min for LCN1 (open square), and each of seven LACRT orthologs (#1, human; #2, chimp; #3, mouse lemur; #4, cat; #5, tree shrew; #6, shrew; #7, horse). Each was analyzed by the RCC, respectively, using human, chimp, mouse lemur, cat, tree shrew, shrew, and horse codon frequency tables. (A) and (B) were calculated with coding sequence for the signal peptide included.
Is 3′ rare codon clustering a property of other tear proteins? No clustering is detected in the coding sequence of the lipocalin-1 gene (LCN1, Fig. 6A). Lipocalin-1 is a common tear protein.22 We then extracted coding sequences from 21 of the first 27 tear proteins in the tear proteome1 (see Table 1 of Ref. 1), including ANG, SERPINF1, ATP5B, B4GALT1, PDIA6, AHSG, ANXA2, ANXA5, CALR, CALU, CANT1, NUCB1, AGL, CHI3L2, ENO1, GAPDH, LGALS3, LGALS3BP, PKM, ALB, and AZGP1. Although some contain rare codon clusters, data expressed as the fractional position of the least “%Min” for each coding sequence revealed no overall 5′ or 3′ preference (black closed circles, Fig. 6B). This contrasts with the least %Min of human LACRT and six lacritin orthologs (red open circles, Fig. 6B), and of LCN1 (blue open square). Thus 3′ rare codon clustering is not a common feature of other tear proteins.
Discussion
We set out to increase recombinant lacritin production by E. coli using a newly generated library of hydrophobic, charge, and rare codon mutants. Animal studies to date suggest that topical lacritin may be beneficial for dry eye, thus efficiency of production for scale up is an important pursuit. We improved yields through mutagenesis of individual hydrophobic residues and two putative salt bridges. Less effective was mutagenesis of rare codons, whose potential benefits may become apparent in hydrophobic or salt bridge mutants. In the course of this analysis, we unexpectedly discovered an evolutionarily conserved 3′ clustering of rare codons in human lacritin and its orthologs, a pattern associated with restricted tissue expression in humans,10 and slowed translation for efficient folding in some proteins11 or when 5′ for controlled ribosomal association.12,13
Although E. coli historically is the most efficient vehicle for generating recombinant protein, aggregation of misfolded proteins in insoluble inclusion bodies often is limiting.16 Lowering the temperature of pLAC induction to suppress hydrophobic interactions was successful and has been used for several years, but cooler induction suffers from decreased transcription and translation.16 Coexpression with molecular chaperones might help folding, although enhanced proteolysis can be problematic,23 and could complicate purification, since endogenous E. coli chaperone DnaK appears to be bound to lacritin through 0.5 M NaCl washes, since they co-elute, but subsequently is removed by DEAE ion exchange chromatography. Less often attempted is selective mutagenesis of hydrophobic and charged residues. This was successful and compatible in some cases with function (I68S, V69S, V91S), or not yet tested but likely compatible with function (L65S, K66S/E70S), as per mutations distal to the lacritin mitogenic5 and syndecan-1 binding8 domain centered on amino acids 100 to 109. Parallel studies have explored lacritin production in baculovirus-transduced insect cells, in P. pastoris (Coffman G, McKown R, Laurie G, unpublished data, 2005), in 293E cell spin culture transduced with pTT22S4 lacritin constructs, as a lacritin-His construct in CHO cells, or alternatively as a fusion construct with the N-terminus of co-receptor SDC1 (Delafosse, Durocher, Laurie, unpublished). None has yet been as productive as expression in E. coli, now improved by selective primary structural mutatagenesis. A contributing factor could be lacritin's latent bactericidal activity that is embodied by N-65, but lacking from C-25 (McKown RL, et al. IOVS 2009;ARVO E-Abstract 4264). However, if so, C-25 yields should be superior, suggesting that nascent lacritin largely is retained in cells under the production conditions used.
Synonymous mutagenesis of human codons rare or uncommon in E. coli, or expression in BL21-CodonPlus cells was less beneficial. Through 13 rounds of site-directed mutagenesis, addressing 29 of 42 rare codons, lacritin E. coli production did not increase substantially. Rather, synonymous mutagenesis beyond lacrt-RC05 and -RC06 displayed decreased yields. This result underlined the need to improve stability first (as likely was achieved with the successful hydrophobic and salt bridge mutants), and then optimize codons for E. coli.
Further coupling of human LACRT coding sequence with the human codon usage table, and of LACRT orthologs with species appropriate codon usage tables revealed a remarkable level of conserved rare codon clustering in the 3′ region. Evolutionary conservation of rare codon clusters in functional non-Pfam domains of higher vertebrates has not been noted previously to our knowledge. Slowed translation for acquisition of local protein structure, widely studied in bacteria, is available in eukaryotes, including human for which tRNA availability is important.24 Rare codons can slow translation of the leader sequence to enhance thereby subsequent translation efficiency in rat neuronal cells,25 and similarly rare tRNA's widely suppress translation efficiency of the first 50 codons.12 Recent natural selection for rare codon use has been noted in several eukaryotes, particularly affecting regulatory and signaling genes.20
Others recently have documented how the approximately 450 to 500 human tRNA genes26,27 are not expressed uniformly across different tissues, indicating an additional form of translational control28 that extends beyond translation.27,29 Lacritin is expressed most highly and selectively in the lacrimal gland.4,9 Unfortunately, to our knowledge no tRNA information is available comparing lacrimal gland with other tissues, although Ortwerth and Chu-Der have documented tRNA differences between bovine lens and brain.30 Exploration of all forms of lacritin transcriptional and translational control could prove rewarding as efforts expand to bioengineer solutions to dry eye.
Conclusions
Lacritin protein production in E. coli was improved successively via nonsynonymous site-directed mutagenesis. Previously unappreciated rare codon clustering in the 3′ region is conserved in all examined orthologs.
Supplementary Material
Footnotes
Supported by NIH Grants RO1EY013143, RO1EY018222 (GWL), and a grant from the Virginia Commonwealth Health Research Board (RLM).
Disclosure: R.L. McKown, EyeRx Research, Inc. (E); R.W. Raab, None; P. Kachelries, None; S. Caldwell, None; G.W. Laurie, EyeRx Research, Inc. (C), P
References
- 1. Laurie GW, Olsakovsky LA, Conway BP, et al. Dry eye and designer ophthalmics. Optom Vis Sci. 2008; 85: 643–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McKown RL, Wang N, Raab RW, et al. Lacritin and other new proteins of the lacrimal functional unit. Exp Eye Res. 2009; 88: 848–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Samudre SS, Lattanzio FA, Lossen V, et al. Lacritin, a novel human tear glycoprotein, promotes sustained basal tearing and is well tolerated. Invest Ophthalmol Vis Sci. 2011; 52: 6265–6270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sanghi S, Kumar R, Lumsden A, et al. cDNA and genomic cloning of lacritin, a novel secretion enhancing factor from the human lacrimal gland. J Mol Biol. 2001; 310: 127–139 [DOI] [PubMed] [Google Scholar]
- 5. Wang J, Wang N, Xie J, et al. Restricted epithelial proliferation by lacritin via PKCα-dependent NFAT and mTOR pathways. J Cell Biol. 2006; 174: 689–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Missimer JH, Steinmetz MO, van Gunsteren WF, Dolenc J. Influence of 63Ser phosphorylation and dephosphorylation on the structure of the stathmin helical nucleation sequence: a molecular dynamics study. Biochem. 2012; 51: 8455–8463 [DOI] [PubMed] [Google Scholar]
- 7. Dana A, Tuller T. Efficient manipulations of synonymous mutations for controlling translation rate: an analytical approach. J Comput Biol. 2012; 92: 200–231 [DOI] [PubMed] [Google Scholar]
- 8. Ma P, Beck SL, Raab RW, et al. Heparanase deglycanation of syndecan-1 is required for binding of the epithelial-restricted prosecretory mitogen lacritin. J Cell Biol. 2006; 174: 1097–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ozyildirim AM, Wistow GJ, Gao J, et al. The lacrimal gland transcriptome is an unusually rich source of rare and poorly characterized gene transcripts. Invest Ophthalmol Vis Sci. 2005; 46: 1572–1580 [DOI] [PubMed] [Google Scholar]
- 10. Parmley J, Huynen M. Clustering of codons with rare cognate tRNAs in human genes suggests an extra level of expression regulation. PLoS Genet. 2009; 5: e1000548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chartier M, Gaudreault F, Najmanovich R. Large-scale analysis of conserved rare codon clusters suggests an involvement in co-translational molecular recognition events. Bioinformatics. 2012; 28: 1438–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tuller T, Carmi A, Vestigian K, et al. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010; 141: 344–354 [DOI] [PubMed] [Google Scholar]
- 13. Man O, Pilpel Y. Differential translation efficiency of orthologous genes is involved in phenotypic divergence of yeast species. Nat Genet. 2007; 39: 415–421 [DOI] [PubMed] [Google Scholar]
- 14. Clarke T, Clark P. Rare codons cluster. PLoS ONE. 2008; 3: e341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fujii A, Morimoto-Tochigi A, Walkup RD, Shearer TR, Azuma M. Lacritin-induced secretion of tear proteins from cultured monkey lacrimal acinar cells. Invest Ophthalmol Vis Sci. In Press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Baneyx F, Mujacic M. Recombinant protein folding and misfolding in Escherichia coli. Nat Biotechnol. 2004; 22: 1399–1408 [DOI] [PubMed] [Google Scholar]
- 17. Bosshard HR, Marti DN, Jelesarov I. Protein stabilization by salt bridges: concepts, experimental approaches and clarification of some misunderstandings. J Mol Recognit. 2004; 17: 1–16 [DOI] [PubMed] [Google Scholar]
- 18. Varenne S, Buc J, Lloubes R, Lazdunski C. Translation is a non-uniform process. Effect of tRNA availability on the rate of elongation of nascent polypeptide chains. J Mol Biol. 1984; 180: 549–576 [DOI] [PubMed] [Google Scholar]
- 19. Pedersen S. Escherichia coli ribosomes translate in vivo with variable rate. EMBO J. 1984; 3: 2895–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Neafsey DE, Galagan JE. Positive selection for unpreferred codon usage in eukaryotic genomes. BMC Evol Biol. 2007; 7: 119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Laurie DE, Splan RK, Green K, et al. Detection of prosecretory mitogen lacritin in nonprimate tears primarily as a C-terminal-like fragment. Invest Ophthalmol Vis Sci. 2012; 53: 6130–6136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Glasgow BJ, Gasymov OK. Focus on molecules: tear lipocalin. Exp Eye Res. 2011; 92: 242–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martínez-Alonso M, García-Fruitós E, Ferrer-Miralles N, et al. Side effects of chaperone gene co-expression in recombinant protein production. Microb Cell Fact. 2010; 9: 64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Deane CM, Saunders R. The imprint of codons on protein structure. Biotechnol J. 2011; 6: 641–649 [DOI] [PubMed] [Google Scholar]
- 25. Fernandez J, Yaman I, Huang C, et al. Ribosome stalling regulates IRES-mediated translation in eukaryotes, a parallel to prokaryotic attenuation. Mol Cell. 2005; 17: 405–416 [DOI] [PubMed] [Google Scholar]
- 26. Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009; D93–D97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Geslain R, Pan T. tRNA: vast reservoir of RNA molecules with unexpected regulatory function. Proc Natl Acad Sci U S A. 2011; 108: 16489–16890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dittmar KA, Goodenbour JM, Pan T. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2006; 2: e221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rudinger-Thirion J, Lescure A, Paulus C, Frugier M. Misfolded human tRNA isodecoder binds and neutralizes a 3′ UTR-embedded Alu element. Proc Natl Acad Sci U S A. 2011; 108: E794–E802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ortwerth BJ, Chu-Der OM. Studies on the specialized transfer RNA population of the lens. Exp Eye Res. 1974; 19: 521–532 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.