Abstract
PI2PE (http://pipe.sc.fsu.edu) is a suite of four web servers for predicting a variety of folding- and binding-related properties of proteins. These include the solvent accessibility of amino acids upon protein folding, the amino acids forming the interfaces of protein–protein and protein–nucleic acid complexes, and the binding rate constants of these complexes. Three of the servers debuted in 2007, and have garnered ∼2,500 unique users and finished over 30,000 jobs. The functionalities of these servers are now enhanced, and a new sever, for predicting the binding rate constants, has been added. Together, these web servers form a pipeline from protein sequence to tertiary structure, then to quaternary structure, and finally to binding kinetics.
Keywords: Solvent accessibility, Protein–protein interaction site, Protein–DNA interaction site, Association rate constant
Introduction
Recent years have seen dizzying advances in high-throughput technologies ranging from DNA sequencing to structural genomics to identification of protein–protein interactions. The resulting mountains of data have created ample opportunities for computational methods to mine the data for knowledge on the structure and function of proteins and to close the significant gaps left by the high-throughput approaches. In particular, as the number of completely sequenced genomes rapidly increases (Batley and Edwards 2009), and the number of protein sequences in public databases continues to grow exponentially (Magrane and Consortium 2011), the expansion rate of the Protein Data Bank (PDB) still pales in comparison, despite exciting progress of the worldwide Structural Genomics initiatives (Terwilliger 2011). Homology modeling has been able to generate structural models for a large number of protein domains (Pieper et al. 2009); for cases where close homology is lacking, fragment-based methods seem to be the most promising in structure prediction (Raman et al. 2009; Kinch et al. 2011; Xu et al. 2011).
Most cellular functions are carried out by large macromolecular complexes and regulated through an intricate network of short-lived protein–protein interactions. High-throughout techniques such as yeast two-hybrid system (Rual et al. 2005) have identified many of these interactions. Molecular characterization of cellular functions is premised on the structures of the protein complexes, which have been particularly challenging for structural biologists. Here again, homology modeling promises to close some of the gap (Aloy et al. 2004; Mosca et al. 2009; Tuncbag et al. 2011), but for the vast majority of protein complexes, docking based on unbound structures of protein domains seems to be the only viable option.
Quantitative modeling of protein interaction networks requires kinetic information on the binding and unbinding events, which at present is largely missing. The missing kinetic information has forced the use of the same values for all association and dissociation rate constants in a program called Pronet in simulating the dynamics of protein interaction networks (Bernaschi et al. 2007). Similarly, as a result of the unknown association and dissociation rate constants, Albert and Wang (2009) resorted to discrete modeling, specifically Boolean dynamics, in constructing signal transduction pathways.
The PI2PE (http://pipe.sc.fsu.edu) suite of web servers were designed to target the weak or missing links just identified. Its 2007 debut (Tjong et al. 2007) consisted of three servers: WESA, cons-PPISP, and DISPLAR. WESA (http://pipe.sc.fsu.edu/wesa/) predicts the solvent accessibility of amino acids from the protein sequence, based on a weighted ensemble of five separate methods (Chen and Zhou 2005b). The predictor was recognized as state of art, and the results are used in the fragment-based structure prediction method I-TASSER (Xu et al. 2011), the top performer in recent rounds of CASP.
cons-PPISP (http://pipe.sc.fsu.edu/ppisp/) predicts amino acids that form protein–protein interfaces, with the unbound structure of a protein as input (Chen and Zhou 2005a). The first version, PPISP, opened the area of protein–protein interaction site prediction (Zhou and Shan 2001). cons-PPISP has been cited as a benchmark for measuring the performance of new methods (Liang et al. 2006; Zhang et al. 2011; La and Kihara 2012). DISPLAR (http://pipe.sc.fsu.edu/displar/) predicts amino acids that form a DNA- or RNA-binding site, with the unbound structure of a nucleic acid-binding protein as input (Tjong and Zhou 2007). New methods for predicting nucleic acid-binding sites are still being developed (Ozbek et al. 2010; Xiong et al. 2011; Zhao et al. 2011), indicating continued interest in the value provided by such predictions for informing protein–nucleic acid interactions. cons-PPISP and DISPLAR can complement experimental techniques such as NMR chemical shift perturbation in characterizing protein–protein and protein–nucleic acid interfaces (Igarashi et al. 2008; Silva et al. 2011). Predicted interface residues can also help building structural models of protein complexes by the docking approach, either by guiding the docking process or by selecting models generated by a docking program (van Dijk et al. 2005; Qin and Zhou 2007, 2010, 2011; De Vries et al. 2007; Zhou and Qin 2007; Schneider and Zacharias 2012).
We have now enhanced the functionalities of the three original servers, including more convenient input options and output displays. And we have added to PI2PE a new server, TransComp (http://pipe.sc.fsu.edu/transcomp/), for predicting the rate constants of protein–protein and protein–nucleic acid associations (Qin et al. 2011). The TransComp server implements our transient-complex theory (Alsallaq and Zhou 2008) and uses the structure of the protein native complex as input. The transient complex refers to a late intermediate along the association pathway, in which the two subunits have near-native separation and relative orientation but have yet to form the specific native contacts. It provides a practical solution to half of the protein association problem, i.e., for the diffusion-limited regime where the association rate constants fall in the high half of the rate-constant spectrum (above 104 M−1 s−1). With the addition of TransComp, PI2PE now becomes a pipeline that connects protein sequence (via tertiary and quaternary structures) to binding kinetics.
Using the PI2PE servers: enhanced and new functionalities
The three original PI2PE servers, WESA, cons-PPISP, and DISPLAR, have been widely used, both by scientists in the computational biology and bioinformatics communities and by experimentalists. Since 2008, the three servers have received jobs from ∼2,500 unique users (based on email addresses) and finished over 30,000 jobs. While ∼40 users were heavy users, each submitting over 100 jobs (possibly using our servers as benchmarks for new method developments), many others submitted a few jobs, probably targets in their specific projects. Users can now use these servers with enhanced functionalities and the new TransComp server.
WESA is accessed at http://pipe.sc.fsu.edu/wesa/. The user can either submit the protein sequence, in FASTA format, or enter the ID of a sequence in the UniProt Knowledgebase (http://www.uniprot.org/help/uniprotkb) (Magrane and Consortium 2011) to start the prediction of solvent accessibility. The output will be displayed in a web link.
cons-PPISP is accessed at http://pipe.sc.fsu.edu/ppisp/. The input is a protein structure in PDB format. The user can either upload a structure file or paste it directly on the submission page, and now has the third option of just entering the PDB ID; the server will retrieve the structure file from the PDB (http://www.rcsb.org/pdb/). Whatever the option, the user must specify the chain(s) in the structure file to be used for prediction of interface residues. The input for DISPLAR, accessed at http://pipe.sc.fsu.edu/displar/, is handled similarly. For both cons-PPISP and DISPLAR, we now use the Jmol plugin (http://jmol.sourceforge.net/) to interactively display the prediction results. The predicted interface residues are highlighted on the protein structure, and the user can choose spacefilling, cartoon, or wireframe for representation. cons-PPISP prediction raw scores can also be displayed by a coloring scheme (see Fig. 1 below for an example).
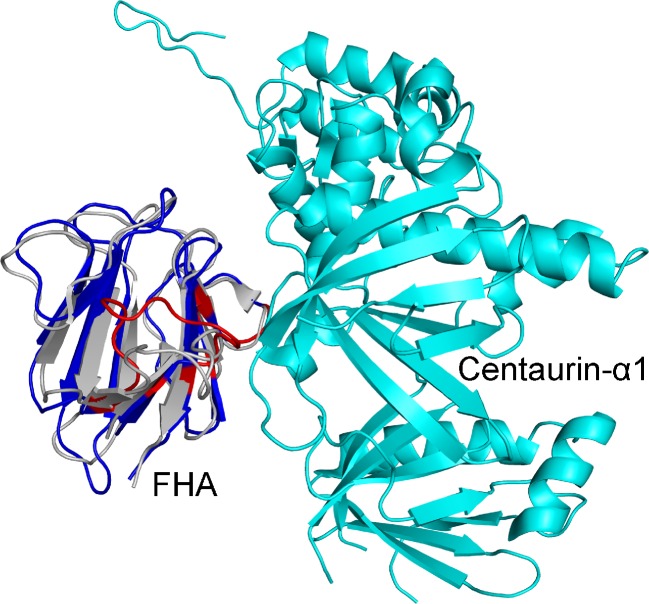
Fig. 1.
Residues of the FHA domain that are predicted by cons-PPISP to be in the interface with Centaurin-α1. The homology model and the bound structure of the FHA domain are shown in blue and gray, respectively. Predicted interface residues are shown in red
TransComp is accessed at http://pipe.sc.fsu.edu/transcomp/. The input is the structure of a protein complex in PQR format, which contains the partial charge and radius of each atom in addition to its coordinates. For users who are not familiar with the PQR format, a bypass (http://pipe.scfsu.edu/transcomp/frompdb.html) is provided to allow them to submit the input structure in PDB format (either by uploading a PDB file or by entering the PDB ID). The user must also specify the chain IDs of the two subunits for which the binding rate constant is to be predicted, and the ionic strength (default is 0.15 M) at which the electrostatic interaction energy is to be calculated. TransComp works for both protein–protein binding and for protein–nucleic acid binding; in the latter case one of the two subunits is an RNA or DNA molecule. The output includes the predicted association rate constant and its components (the basal rate constant for reaching the transient complex by random diffusion and the electrostatic interaction energy in the transient complex). Also displayed are the electrostatic surfaces of the subunits and the energy landscape generated for locating the transient complex (see Fig. 2 below for an example).
Fig. 2.
TransComp output for the association of CBFα and CBFβ. The input was the docked structure of the heterodimer. The two middle panels display the electrostatic surfaces (blue: positive; red: negative) of the two subunits. The light colors of the electrostatic surfaces within the interface of the two subunits and lack of blue-red complementarity across the interface are consistent with the moderate positive value of ΔG el*. The bottom panels illustrate how the transient complex is identified. The docked complex of CBFα and CBFβ has N c = 42. Upon sampling in the 6-dimensional space of relative translation and relative rotation, configurations with N c as large as 46 were obtained. The transient complex, with N c = 15, is located at the midpoint of the transition from the bound state (with large N c but a narrow range of the relative rotation angle, χ) to the unbound state (with small N c but a wide range of χ). Similar results were obtained using the crystal structure of the CBFα:CBFβ dimer as input, but in that case the crystal structure has the largest N c (= 50) during the sampling in the translational-rotational space
Each submitted job is put in a queue, and its status is displayed before the output or an error report is produced. To ensure private access, each submission is assigned a randomly generated ID. The user can optionally submit an e-mail address, where the output web link will be sent. At the submission sites of the four web servers, users can also browse input and output examples.
Illustrative applications
As noted above, WESA predictions can be used in methods for predicting protein tertiary structures in homology-model free cases. Nevertheless, homology models are increasingly used in many applications. In particular, they are now routinely used as substitutes for unbound structures in CAPRI (http://www.ebi.ac.uk/msd-srv/capri/) exercises, which aim to build structural models for protein complexes by docking the unbound structures of the subunits. Here, we illustrate the performance of cons-PPISP on a CAPRI target with a homology model for a subunit.
CAPRI Target 38 is the complex between centaurin-α1 and the forkhead-associated (FHA) domain of KIF13B. The structure of the unbound FHA domain given to predictors was a homology model using PDB entry 2G1L as template (with 38 % sequence identity). The structure of the Target 38 complex is now available (PDB entry 3FM8). The homology model and the bound structure of the FHA domain aligned [by Dalilite (http://www.ebi.ac.uk/Tools/dalilite/)] to an RMSD of 1.7 Å. Using the homology model of the FHA domain as input, cons-PPISP predicted 20 interface residues, covering 12 of the 16 residues found in the interface of the actual complex. The prediction results are displayed in Fig. 1, along with the structure of the complex. In comparison, using the bound structure of the FHA domain as input, cons-PPISP predicted 26 interface residues, covering 13 of the actual 16 interface residues. The two sets of predictions have 18 residues in common, of which 11 are correct. However, cons-PPISP failed to predict any of the residues of centaurin-α1 that interact with the FHA domain. No structural models submitted by any of the CAPRI groups for the the Target 38 complex were correct.
In our 2007 report (Tjong et al. 2007), we used interface predictions of cons-PPISP and DISPLAR to assist the docking of a transcription factor heterodimer and of its DNA-bound ternary complex. The transcription factor is a core binding factor (CBF) with the ALM1/RUNX1 Runt domain as the DNA-contacting CBFα subunit. Here, we use the previously built structural models for the heterodimer and the ternary complex to illustrate the prediction of association rate constants by TransComp.
The structural model for the CBFα:CBFβ heterodimer (docked from the unbound structures in PDB entries 1EAQ and 1ILF, respectively) has an RMSD of 2.2 Å from the crystal structure (PDB entry 1E50). Using this structural model (after sidechain refinement by energy minimization) as input, TransComp predicted an association rate constant (k a) of 1.3 × 105 M–1 s–1 for forming the heterodimer. The basal rate constant (k a0) is 5.5 × 105 M–1 s–1 and the electrostatic interaction energy (ΔG el*) is 0.88 kcal/mol (at ionic strength = 0.15 M). The server output is displayed in Fig. 2, which, in addition to the details of the predicted rate constant just listed, contains the electrostatic surfaces of the two subunits and the interaction energy surface generated for locating the transient complex. In comparison, using the crystal structure of the heterodimer as input, the predicted k a is 2.4 × 105 M–1 s–1, along with values of 7.0 × 105 M–1 s–1 for k a0 and 0.63 kcal/mol for ΔG el*. So the predicted k a results are very similar using the docked structure and using the crystal structure. The electrostatic surfaces of the two subunits and the interaction energy surface generated for locating the transient complex are also similar (not shown). There is a subtle difference at the bottom of the interaction energy surface: whereas the crystal structure is located at the very bottom (with a contact number, N c, of 50), the docked structure has a smaller N c of 42, and upon sampling in the 6-dimensional space of relative translation and relative rotation, configurations with larger N c values were obtained (maximum N c = 46). No experimental value is available to test the TransComp k a predictions, but the predicted basal rate constant falls in the middle of the 104 to 106 M–1 s–1 range found for many protein–protein complexes (Qin et al. 2011), and a moderate positive ΔG el* is consistent with the mixed electrostatic surfaces of the subunits (Pang et al. 2011).
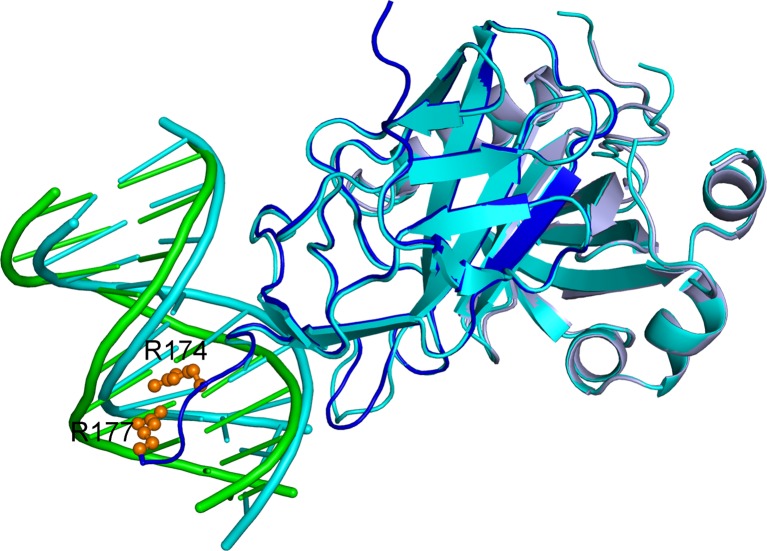
The docked ternary complex (from the structure of the heterodimer in PDB entry 1E50 and a DNA molecule with standard B-DNA conformation) has an RMSD of 1.2 Å from the crystal structure (PDB entry 1H9D). Using this docked structure (after sidechain refinement by energy minimization) as input, TransComp predicted a k a of 6.9 × 107 M–1 s–1 for DNA binding by the heterodimer (with k a0 = 2.4 × 106 M–1 s–1 and ΔG el* = −2.0 kcal/mol at ionic strength = 0.15 M). In comparison, using the crystal structure of the ternary complex as input, TransComp predicted k a = 5.2 × 108 M–1 s–1, with k a0 = 5.2 × 104 M–1 s–1, and ΔG el* = −7.9 kcal/mol). An important difference between the docked structure and the crystal structure is that the docked structure lacks four residues, R174–R177, in the C-terminus of CBFα. In the crystal structure, the two C-terminal Arg residues form close interactions with the DNA (Fig. 3). Mutations of R177 abolished CBF binding to DNA and resulted in loss of activity (Osato et al. 1999), and mutations of both R174 and R177 are found in patients with AML1-related leukemogenesis (Roumier et al. 2003). The tight fit of the CBFα C-terminal four residues into the major groove of the DNA explains both the relatively low k a0 and the extremely strong ΔG el* predicted by TransComp. Crute et al. (1996) used surface plasmon resonance (SPR) measurements to obtain a k a value of 2.5 × 106 M–1 s–1, but cautioned that this value “is considerably underestimated.” Indeed, with a dissociation rate constant (k d) of 0.1 s–1, the resulting dissociation constant (K d) of 4 × 10–8 M, is four orders of magnitude higher than the result obtained by the same group using electrophoretic mobility shift assay (EMSA) (Tang et al. 2000). The latter method is more reliable for low K d. Using EMSA, the same group had previously measured a k d ∼0.01 s–1 (Wang et al. 1993). Combined with the K d by EMSA method, we can deduce a k a ∼ 109 M–1 s–1, which is comparable to the TransComp prediction using the crystal structure of the ternary complex.
Fig. 3.
Comparison of docked and crystal structures of the DNA:CBFα:CBFβ ternary complex. The docked structure is in cyan; in the crystal structure, DNA, CBFα, and CBFβ are shown in green, dark blue, and light blue, respectively, and the sidechains of R174 and R177 are shown as spheres
The last application shows that reliable prediction of k a requires high-quality structure at the binding interface. For the CBFα:CBFβ heterodimer, the observation that sampling in the 6-dimensional space of relative translation and relative rotation can generate configurations that are more native-like (i.e., with higher contact numbers; Fig. 2) suggests that such sampling may be used to refine the docked structure. In the case of the DNA:CBFα:CBFβ ternary complex, adding the CBFα C-terminal tail four residues to the docked structure and further refinement may lead to a better structure.
In addition to protein–protein and protein–DNA complexes, TransComp works equally well for predicting the association rate constants of protein–RNA complexes. The methodology implemented in TransComp was successful in quantitatively rationalizing the association rate constants of several protein–RNA complexes (Qin and Zhou 2008, 2009). Here, we present application to another two protein–RNA complexes, formed by the C-terminal domain of RIG-I, a cytosolic sensor of viral RNA, and double-stranded RNAs with and without 5′ triphosphate. This protein–RNA interaction plays essential role in mediating innate immune responses toward vial infection. Using the crystal structures of these complexes (PDB entries 3LRN and 3OG8), TransComp predicted very high association rate constants, 4.3 × 109 and 4.0 × 109 M–1 s–1 (with k a0 = 4.3 × 105 and 4.6 × 105 M–1 s–1 and ΔG el* = −10.9 and −6.5 kcal/mol at ionic strength = 0.16 M), respectively. Lu et al. (2010) used SPR to measure these rate constants and the results, ∼107 M–1 s–1, are at the detection upper limit of this technique, suggesting that the actual rate constants could be much higher. Many RNA molecules gain tertiary structures only after binding to proteins. In some cases, Mg2+ are involved in mediating protein–RNA interactions. These factors can complicate the application of TransComp to protein–RNA association.
In summary, the four web servers described here form a pipeline from protein sequence to tertiary structure, then to quaternary structure, and finally to binding kinetics. This opens the door to quantitative modeling of the dynamics of protein interaction networks.
Acknowledgment
We acknowledge the staff at the Florida State University High-Performance Computing Facility for assistance. This work was supported in part by Grant GM58187 from the National Institutes of Health.
Conflict of interest
None
Contributor Information
Sanbo Qin, Email: sqin@fsu.edu.
Huan-Xiang Zhou, Email: hzhou4@fsu.edu.
References
- Albert R, Wang RS. Discrete dynamic modeling of cellular signaling networks. Methods Enzymol. 2009;467:281–306. doi: 10.1016/S0076-6879(09)67011-7. [DOI] [PubMed] [Google Scholar]
- Aloy P, Bottcher B, Ceulemans H, Leutwein C, Mellwig C, Fischer S, Gavin AC, Bork P, Superti-Furga G, Serrano L, Russell RB. Structure-based assembly of protein complexes in yeast. Science. 2004;303:2026–2029. doi: 10.1126/science.1092645. [DOI] [PubMed] [Google Scholar]
- Alsallaq R, Zhou HX. Electrostatic rate enhancement and transient complex of protein-protein association. Proteins. 2008;71:320–335. doi: 10.1002/prot.21679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batley J, Edwards D. Genome sequence data: management, storage, and visualization. Biotechniques. 2009;46:333–336. doi: 10.2144/000113134. [DOI] [PubMed] [Google Scholar]
- Bernaschi M, Castiglione F, Ferranti A, Gavrila C, Tinti M, Cesareni G. ProtNet: a tool for stochastic simulations of protein interaction networks dynamics. BMC Bioinforma. 2007;8:S4. doi: 10.1186/1471-2105-8-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zhou HX. Prediction of interface residues in protein-protein complexes by a consensus neural network method: test against NMR data. Proteins. 2005;61:21–35. doi: 10.1002/prot.20514. [DOI] [PubMed] [Google Scholar]
- Chen H, Zhou HX. Prediction of solvent accessibility and sites of deleterious mutations from protein sequence. Nucleic Acids Res. 2005;33:3193–3199. doi: 10.1093/nar/gki633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crute BE, Lewis AF, Wu Z, Bushweller JH, Speck NA. Biochemical and biophysical properties of the core-binding factor alpha2 (AML1) DNA-binding domain. J Biol Chem. 1996;271:26251–26260. doi: 10.1074/jbc.271.42.26251. [DOI] [PubMed] [Google Scholar]
- De Vries SJ, van Dijk ADJ, Krzeminski M, van Dijk M, Thureau A, Hsu V, Wassenaar T, Bonvin AMJJ. HADDOCK versus HADDOCK: new features and performance of HADDOCK2.0 on the CAPRI targets. Proteins. 2007;69:726–733. doi: 10.1002/prot.21723. [DOI] [PubMed] [Google Scholar]
- Igarashi S, Osawa M, Takeuchi K, Ozawa S, Shimada I. Amino acid selective cross-saturation method for identification of proximal residue pairs in a protein-protein complex. J Am Chem Soc. 2008;130:12168–12176. doi: 10.1021/ja804062t. [DOI] [PubMed] [Google Scholar]
- Kinch L, Yong Shi S, Cong Q, Cheng H, Liao Y, Grishin NV. CASP9 assessment of free modeling target predictions. Proteins. 2011;79:59–73. doi: 10.1002/prot.23181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La D, Kihara D. A novel method for protein-protein interaction site prediction using phylogenetic substitution models. Proteins. 2012;80:126–141. doi: 10.1002/prot.23169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S, Zhang C, Liu S, Zhou Y. Protein binding site prediction using an empirical scoring function. Nucleic Acids Res. 2006;34:3698–3707. doi: 10.1093/nar/gkl454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Xu H, Ranjith-Kumar CT, Brooks MT, Hou TY, Hu F, Herr AB, Strong RK, Kao CC, Li P. The structural basis of 5' triphosphate double-stranded RNA recognition by RIG-I C-terminal domain. Structure. 2010;18:1032–1043. doi: 10.1016/j.str.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magrane M and Consortium U (2011) UniProt Knowledgebase: a hub of integrated protein data. Database 2011:bar009 [DOI] [PMC free article] [PubMed]
- Mosca R, Pons C, Fernandez-Recio J, Aloy P. Pushing structural information into the yeast interactome by high-throughput protein docking experiments. PLoS Comput Biol. 2009;5:e1000490. doi: 10.1371/journal.pcbi.1000490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osato M, Asou N, Abdalla E, Hoshino K, Yamasaki H, Okubo T, Suzushima H, Takatsuki K, Kanno T, Shigesada K, Ito Y. Biallelic and heterozygous point mutations in the runt domain of the AML1/PEBP2aB gene associated with myeloblastic leukemias. Blood. 1999;93:1817–1824. [PubMed] [Google Scholar]
- Ozbek P, Soner S, Erman B, Haliloglu T. DNABINDPROT: fluctuation-based predictor of DNA-binding residues within a network of interacting residues. Nucleic Acids Res. 2010;38:W417–W423. doi: 10.1093/nar/gkq396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang X, Qin S, Zhou HX. Rationalizing 5,000-fold differences in receptor-binding rate constants of four cytokines. Biophys J. 2011;101:1175–1183. doi: 10.1016/j.bpj.2011.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieper U, Eswar N, Webb BM, Eramian D, Kelly L, Barkan DT, Carter H, Mankoo P, Karchin R, Marti-Renom MA, et al. MODBASE, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2009;37:D347–D354. doi: 10.1093/nar/gkn791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S, Zhou H-X. A holistic approach to protein docking. Proteins. 2007;69:743–749. doi: 10.1002/prot.21752. [DOI] [PubMed] [Google Scholar]
- Qin S, Zhou HX. Prediction of salt and mutational effects on the association rate of U1A protein and U1 small nuclear RNA stem/loop II. J Phys Chem B. 2008;112:5955–5960. doi: 10.1021/jp075919k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S, Zhou HX. Dissection of the high rate constant for the binding of a ribotoxin to the ribosome. Proc Natl Acad Sci USA. 2009;106:6974–6979. doi: 10.1073/pnas.0900291106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S, Zhou HX. Selection of near-native poses in CAPRI rounds 13-19. Proteins. 2010;78:3166–3173. doi: 10.1002/prot.22772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S, Zhou HX. Structural models of protein-DNA complexes based on interface prediction and docking. Curr Protein Pept Sci. 2011;12:531–539. doi: 10.2174/138920311796957694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S, Pang X, Zhou HX. Automated prediction of protein association rate constants. Structure. 2011;19:1744–1751. doi: 10.1016/j.str.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman S, Vernon R, Thompson J, Tyka M, Sadreyev R, Pei J, Kim D, Kellogg E, DiMaio F, Lange O, et al. Structure prediction for CASP8 with all-atom refinement using Rosetta. Proteins. 2009;77:89–99. doi: 10.1002/prot.22540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roumier C, Fenaux P, Lafage M, Imbert M, Eclache V, Preudhomme C. New mechanisms of AML1 gene alteration in hematological malignancies. Leukemia. 2003;17:9–16. doi: 10.1038/sj.leu.2402766. [DOI] [PubMed] [Google Scholar]
- Rual J-F, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Schneider S, Zacharias M. Scoring optimisation of unbound protein-protein docking including protein binding site predictions. J Mol Recogn. 2012;25:15–23. doi: 10.1002/jmr.1165. [DOI] [PubMed] [Google Scholar]
- Silva JL, Vieira T, Gomes MPB, Rangel LP, Scapin SMN, Cordeiro Y. Experimental approaches to the interaction of the prion protein with nucleic acids and glycosaminoglycans: Modulators of the pathogenic conversion. Methods. 2011;53:306–317. doi: 10.1016/j.ymeth.2010.12.002. [DOI] [PubMed] [Google Scholar]
- Tang YY, Crute BE, Kelley JJ, Huang X, Yan J, Shi J, Hartman KL, Laue TM, Speck NA, Bushweller JH. Biophysical characterization of interactions between the core binding factor alpha and beta subunits and DNA. FEBS Lett. 2000;470:167–172. doi: 10.1016/S0014-5793(00)01312-0. [DOI] [PubMed] [Google Scholar]
- Terwilliger T. The success of structural genomics. J Struct Funct Genom. 2011;12:43–44. doi: 10.1007/s10969-011-9114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjong H, Zhou HX. DISPLAR: an accurate method for predicting DNA-binding sites on protein surfaces. Nucleic Acids Res. 2007;35:1465–1477. doi: 10.1093/nar/gkm008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjong H, Qin S, Zhou HX. PI2PE: protein interface/interior prediction engine. Nucleic Acids Res. 2007;35:W357–W362. doi: 10.1093/nar/gkm231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuncbag N, Gursoy A, Nussinov R, Keskin O. Predicting protein-protein interactions on a proteome scale by matching evolutionary and structural similarities at interfaces using PRISM. Nat Protoc. 2011;6:1341–1354. doi: 10.1038/nprot.2011.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk AD, de Vries SJ, Dominguez C, Chen H, Zhou HX, Bonvin AM. Data-driven docking: HADDOCK’s adventures in CAPRI. Proteins. 2005;60:232–238. doi: 10.1002/prot.20563. [DOI] [PubMed] [Google Scholar]
- Wang S, Wang Q, Crute BE, Melnikova IN, Keller SR, Speck NA. Cloning and characterization of subunits of the T-cell receptor and murine leukemia virus enhancer core-binding factor. Mol Cell Biol. 1993;13:3324–3339. doi: 10.1128/mcb.13.6.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Liu JA, Wei DQ. An accurate feature-based method for identifying DNA-binding residues on protein surfaces. Proteins. 2011;79:509–517. doi: 10.1002/prot.22898. [DOI] [PubMed] [Google Scholar]
- Xu D, Zhang J, Roy A, Zhang Y. Automated protein structure modeling in CASP9 by I-TASSER pipeline combined with QUARK-based ab initio folding and FG-MD-based structure refinement. Proteins. 2011;79:147–160. doi: 10.1002/prot.23111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang QC, Deng L, Fisher M, Guan J, Honig B, Petrey D. PredUs: a web server for predicting protein interfaces using structural neighbors. Nucleic Acids Res. 2011;39:W283–W287. doi: 10.1093/nar/gkr311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Yang Y, Zhou Y. Structure-based prediction of RNA-binding domains and RNA-binding sites and application to structural genomics targets. Nucleic Acids Res. 2011;39:3017–3025. doi: 10.1093/nar/gkq1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HX, Qin S. Interaction-site prediction for protein complexes: a critical assessment. Bioinformatics. 2007;23:2203–2209. doi: 10.1093/bioinformatics/btm323. [DOI] [PubMed] [Google Scholar]
- Zhou HX, Shan Y. Prediction of protein interaction sites from sequence profile and residue neighbor list. Proteins. 2001;44:336–343. doi: 10.1002/prot.1099. [DOI] [PubMed] [Google Scholar]