Abstract
Age is a major risk factor for the development of cancer. Senescent fibroblasts, which accumulate with age, secrete pro-tumorigenic factors collectively referred to as the senescence-associated secretory phenotype (SASP). Here we examined the molecular mechanisms that control SASP activation, focusing on the known SASP factor osteopontin (OPN). We found that expression of the canonical SASP members IL6 and IL8, but not OPN, were dependent upon a persistent DNA damage response (DDR) as evidenced by ATM and NF-κB activation. Treatment with several histone deacetylase (HDAC) inhibitors robustly activated SASP in the absence of DNA breaks, suggesting that DDR-dependent SASP activation occurs in response to chromatin remodeling rather than physical breaks in DNA. In the setting of histone deacetylase inhibition, IL6 and IL8 expression remained dependent upon ATM and NF-κB, while OPN expression remained independent of these factors. Further analysis revealed that HDAC1 was sufficient to induce OPN expression, which is interesting given that loss of HDAC1 expression correlates with increased OPN expression within the stromal compartment of invasive breast cancers. Importantly, fibroblasts treated with HDAC inhibitors promoted tumor growth in vivo. Our findings therefore indicate that HDAC modulation plays an important role in stromal cell activation, with important implications for the use of HDAC inhibitors in the treatment of cancer.
Keywords: osteopontin, senescence, stroma, HDAC inhibitor, SASP
Introduction
Senescence within incipient tumor cells is a potent tumor suppressive mechanism. Cells undergoing senescence are characterized by a permanent cell cycle arrest, flattened cellular morphology, increased senescence-associated β-galactosidase (SA-βgal) activity, and in many instances the appearance of facultative heterochromatin domains known as senescence-associated heterochromatic foci (SAHFs) (1). Numerous oncogenic stimuli including activation of cellular oncogenes, persistent DNA damage, and continued cellular proliferation activate senescence (1). Yet the impact of senescence is context-dependent. Thus, when activated in an incipient tumor cell, senescence is a potent tumor suppressor that must be bypassed if cells are to complete the transformation process (2). However, when senescence occurs within surrounding stromal cells, those cells function as potent tumor-promoters (3, 4).
Extensive microarray analyses have revealed that senescent cells activate a conserved program that is characterized by increased expression of numerous factors including inflammatory cytokines, growth factors, and extracellular matrix (ECM) remodeling enzymes that are highly reminiscent of factors expressed in cancer-associated fibroblasts (CAFs). This expression profile is collectively referred to as the senescence-associated secretory phenotype (SASP) (5). Many SASP factors not only enforce the cell cycle arrest that characterizes senescence within the originating cell but they also impact the surrounding milieu. For example, secreted mitogens stimulate premalignant epithelial cell growth in vitro and in xenograft models (3, 4), ECM remodeling enzymes such as matrix metalloproteinases affect branching and migration (6), and other factors including cytokines promote invasion (7, 8). The ability of senescent fibroblasts to influence tumorigenesis has been documented in multiple systems; however, until recently, the underlying molecular mechanisms regulating SASP activation were unknown.
In human cells both the p53 and Rb pathways function redundantly to activate cellular senescence (9, 10). Abrogation of either pathway is insufficient to bypass senescence following a senescence-inducing stimulus. However, when both the p53 and Rb pathways are inactivated, cells bypass both telomere-driven replicative senescence and stress-induced premature senescence (SIPS), which can be induced by a wide range of cellular stresses. Given the importance of the senescence effector proteins in the activation of senescence it was hypothesized that their inhibition would result in loss of SASP activation. Surprisingly, when a senescence-inducing dose of DNA damage is delivered to p53/Rb deficient human cells, these cells continue to divide, yet still activate SASP factors IL6 and IL8 (7). Furthermore, when p53 and Rb are abrogated in already senescent, SASP-expressing cells, SASP expression remains (5), indicating that p53/Rb are not required to maintain SASP expression in senescent cells. Together these data indicate that senescent cells robustly express SASP but that the induction of senescence per se is not required to activate or maintain SASP expression.
Investigation into the cellular signaling pathways that activate the SASP indicate that a persistent DNA damage response (DDR) is sufficient to activate some SASP factors. Indeed, signaling downstream of ATM (including NBS1 and Chk2) controls a subset of SASP factors, including IL6 and IL8 (7). The mechanisms linking DDR to SASP activation remain unclear but DDR induces chromatin alterations that can impact numerous transcription pathways. Therefore, transcriptional changes that occur in senescent cells may result from specific chromatin modulations. Mounting evidence implicates chromatin remodeling in the establishment of the senescent state. In senescent cells, heterochromatic regions referred to as SAHFs appear at E2F promoters and functionally repress cellular proliferation (11). In replicative senescence, histone deacetylase (HDAC) activity diminishes (12) corresponding with an increase in histone acetylation. Additionally, a decline in global DNA methylation has been reported in senescent cells ((13) and references therein). Interestingly, treatment with HDAC inhibitors including sodium butyrate (NaB) or trichostatin A (TSA) induces senescence in some cell types, further supporting the hypothesis that chromatin relaxation plays a causative role in senescence (12, 14).
A role for transcriptional control in the regulation of SASP factors has also been suggested by recent work, particularly for a number of inflammatory factors including IL6, IL8 and CXCR2. Transcriptional regulation of such cytokines in other biological settings by NF-κB and CEBPβ applies to senescence as well. In fact, these transcription factors occupy the promoters of several cytokines in senescent cells (15, 16). However, it is unknown how these factors are activated in response to senescence-inducing stimuli and subsequently direct transcriptional changes in senescence.
Osteopontin (OPN), also known as secreted phosphoprotein 1 (SPP1), is a multifunctional signaling molecule (17). Originally identified in cancer cells (18), the physiological function of OPN is linked to matrix integrity and bone maintenance (19). Since its initial identification, OPN has been implicated in every stage of tumorigenesis and is a prognostic factor for several malignancies (20). We previously reported that OPN levels increase in senescent cells and showed that it is a critical mediator of stromal-epithelial interactions in tumorigenesis (4). In addition, OPN expression in the stromal compartment of human skin coincides with senescent markers, raising the possibility that it plays an important role in the early stages of tumorigenesis (21). Despite the importance of senescence-derived OPN, its regulation in senescent cells remains unknown. Here we report that following senescence-inducing stimuli, OPN upregulation occurs independent of the senescence effector pathways p53 and Rb, similar to what has been shown for other SASP members including IL6 and IL8 (5). This finding demonstrates that senescence induction is not a prerequisite for SASP activation following exposure to chronic cellular stress, including treatment with DNA damaging agents. Further investigation revealed that contrary to the signature SASP members, IL6 and IL8, OPN expression is insensitive to NF-κB and ATM signaling. Thus our results indicate that the SASP is controlled by at least two independent transcription programs. Importantly, agents capable of directly perturbing chromatin structure without inducing DNA breaks were potent inducers of SASP, including OPN expression. Specifically, ectopic expression of a dominant negative form of HDAC1 leads to OPN mRNA upregulation. Furthermore, treatment of fibroblasts with HDAC inhibitors led to a significant paracrine stimulation of tumor growth, which suggests that these inhibitors may adversely impact the stromal compartment in the therapeutic setting.
Materials and methods
Cell lines and treatments
Human foreskin BJ fibroblasts and 293T cells were grown as previously described (4). These cell lines were originally obtained from Dr. Robert Weinberg’s laboratory and were not recently reauthenticated. Human AT fibroblasts (GM09607) were purchased from Coriell Institute (Camden, NJ), used within six months of receipt and grown in MEM media supplemented with 15% non-heat inactivated FBS (Sigma, St. Louis, MO). WI38 fetal lung fibroblasts were grown in DME media supplemented with 10% heat-inactivated FBS (Sigma, St. Louis, MO) and transduced with telomerase (pBabe vector with hygromycin selection) and human papilloma virus 16 proteins E6 and E7 (expressed from the LXSN vector (22)). Fibroblasts were mock- or bleomycin sulfate-treated (100 µg/ml, Sigma, St. Louis, MO) for 24 hrs. After 72 hr serum-starvation, RNA was collected using TRI Reagent (Ambion/Applied Biosystems, Foster City, CA). Cells were incubated with two fresh changes of 4 mM sodium butyrate (Sigma) for 72 or 144 hr, 6 µM MS-275 (Santa Cruz Biotechnologies, Santa Cruz, CA) for 6 days, 3 µM of suberoylanilide hydroxamic acid (SAHA) or vorinostat (Selleck, Houston, TX) for 6 days, or 1 mM TSA (Sigma, St. Louis, MO) for 3 days.
Plasmids
The IκBα-mut in a pBabe construct (23) was purchased from Addgene (Boston, MA plasmid 15291). The NF-κB promoter luciferase construct, pκB5-Fluc (Stratagene, La Jolla, CA) and pRL-CMV (Promega, Madison, WI) were used in transient transfections. Two shRNA sequences targeting the human ATM gene (shATM-1:CCTTTCATTCAGCCTTTAGAA; shATM-2: TGATGGTCTTAAGGAACATCT) were provided in the pLKO.1 plasmid by the Washington University in St. Louis Children’s Discovery Institute and the RNAi Consortium. The p53-DD and DK constructs (22) were kindly provided by Robert Weinberg (Whitehead Institute, Boston, MA). The HDAC1-H140A cDNA in pBabe-puro (24) was kindly provided by Tso-Pang Yao (Duke University, Durham, NC). The HDAC3mut (HDAC3 1–401) cDNA (25) was kindly provided by Edward Seto (Moffit Cancer Center, Tampa, FL) and was subcloned into the EcoRI/PacI sites of pBabe-hygro-3xFLAG vector.
Quantitative PCR (qPCR)
Standard protocol was followed for cDNA and quantitative PCR (as previously reported in (4)) using the following primers (IDT, Coralville, IA): OPN, F 5′-TTGCAGCCTTCTCAGCCAA-3′, R 5′-CAAAAGCAAATCACTGCAATTCTC-3′; GAPDH, F 5′-GCATGGCCTTCCGTGTCC-3′, R 5′-AATGCCAGCCCCAGCGTCAAA-3′; I L 6, F 5′-ACATCCTCGACGGCATCTCA-3′, R 5′-TCACCAGGCAAGTCTCCTCA-3′; IL8, F 5′-GCTCTGTGTGAAGGTGCAGT-3′, R 5′-TGCACCCAGTTTTCCTTGGG-3′; ATM (Taqman, Applied Biosystems, Carlsbad, CA cat.# Hs01112317-g1). All qPCR results were analyzed using the method reported in Livak et al., (26).
Western Blot Analysis
Fibroblast cell pellets were lysed in buffer containing 50 mM HEPES pH 7.4, 150 mM NaCl, 1% Triton X-100, 1 M EDTA, 10% (v/v) glycerol) and protein was quantified by the Bradford Protein Assay (Bio-Rad, Hercules, CA). The following primary antibodies were used: p421, a p53 hybridoma supernatant kindly provided by Robert Weinberg (Whitehead Institute, Cambridge, MA) at 1:100; Cdk4 (Santa Cruz Biotechnologies, Santa Cruz, CA) at 1:200; β-actin (Sigma, St. Louis, MO) at 1:10,000; γ-actin (Novus, St. Louis, MO) at 1:5000; γH2AX (Millipore/Upstate, Billerica, MA) at 1:1000; FLAG (Sigma, St. Louis, MO) at 1:1000. All primary antibodies were detected by the appropriate HRP-conjugated secondary antibodies (Sigma, St. Louis, MO).
Virus production and retroviral infections
Virus was produced and target cells transduced as previously described (4).
Senescence-associated β-galactosidase (SA-βgal) staining
SA-βgal staining on cells was performed as previously described (4).
Xenograft model
All animal procedures were approved by the Washington University School of Medicine Animal Studies Committee. 5×105 BJ fibroblasts treated as described above and 5×105 HaCaTCBR were injected subcutaneously into the flanks of female NOD/SCID mice (NCI-Fredrick). In vivo imaging was performed on days 10 and 12 on an IVIS50 (Caliper; Living Image 3.2, 60 s exposure, binning 8, FOV 12cm, f/stop 1, open filter). For analysis, total photon flux (photons/sec) was measured from a fixed region of interest over the xenografts using Living Image 2.6.
HDAC1 IP and deacetylase assay
Vehicle and bleomycin-treated fibroblasts were lysed as described (27) and 2 mg of protein was immunoprecipitated with an antibody against HDAC1 (ab7028, Abcam, Cambridge, MA) or a negative control antibody (β-galactosidase, ab616, Abcam, Cambridge, MA) as described (27). HDAC activity was measured with the Fluor-de-Lys HDAC Activity Assay Kit (Enzo Life Sciences, Farmingdale, NY) per manufacturer’s recommendations.
Expression Analysis
Comparison of HDAC1 mRNA levels in normal vs. invasive breast cancer-associated stroma was exported from Oncomine (28) as extracted from the Finak et. al. study (29). Expression values are log-transformed and median-centered per array. Differential expression is identified by a permutation test and p-values are calculated by t-test and corrected for multiple comparisons by the method of false discovery rates (28).
Statistical analysis
Data are presented as the mean ± STDEV or SEM. Statistical significance was calculated using Student’s t-test. Values of p < 0.05 were considered significant.
Results
The senescence effector pathways p53 and Rb are not required to activate OPN
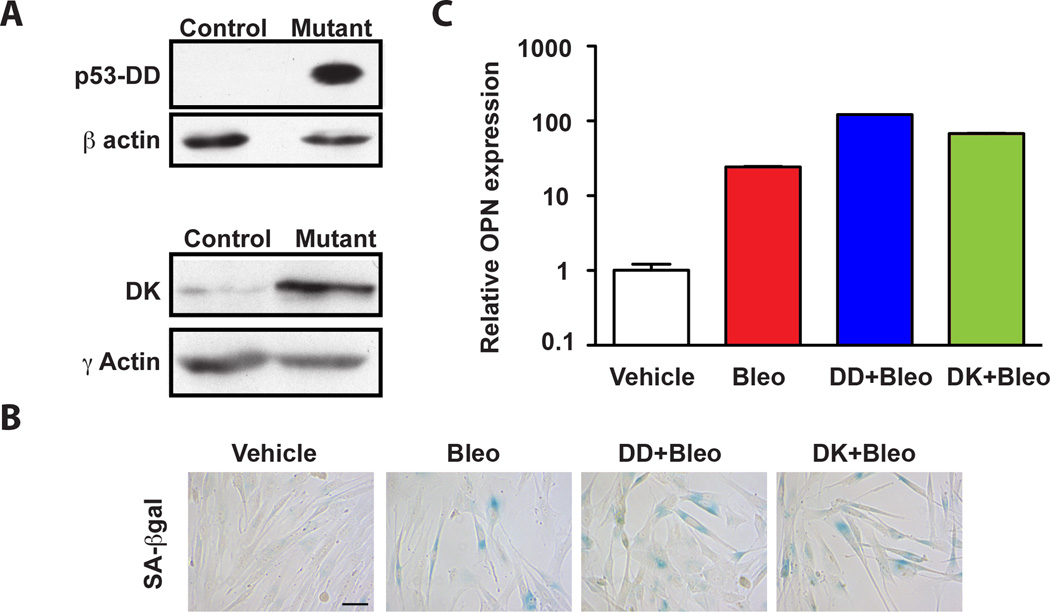
Activation of senescence is accompanied by upregulation of pro-tumorigenic factors collectively referred to as the SASP (5). Our previous work revealed that one member of this group, OPN, stimulates preneoplastic keratinocyte growth and is a critical stromal-derived pro-tumorigenic factor in a xenograft model (4). Given the significance of OPN in this model and the importance of the other senescence-secreted proteins, we investigated the regulation of OPN in senescence. We initially turned our attention to the senescence effector and tumor-suppressor proteins p53 and Rb. Importantly, in human cells, inhibition of either the p53 or Rb pathways alone is insufficient to bypass senescence while concomitant abrogation of these pathways allows cells to bypass senescence (30). Despite the importance of p53 and Rb in the activation of senescence, previous work has shown that they actively suppress the activation of some SASP factors including IL6 and IL8 (5). However, p53 regulates OPN expression in mouse embryonic fibroblasts, raising the possibility that it plays a role in the regulation of OPN in senescence (31). To investigate the requirement for p53 and Rb activity in senescence-associated OPN regulation, we ectopically expressed a well-characterized dominant negative p53 cDNA (p53-DD) (22) or a cDNA expressing a fusion protein consisting of a mutant form of cyclin dependent kinase 4R24C (Cdk4) and cyclin D1 (DK) that inhibits Rb (22) (Fig. 1A). Abrogation of either the p53 or Rb pathway alone had no impact on the induction of senescence. Indeed, exposure of control cells or cells expressing p53-DD or DK to bleomycin induced robust senescence as evidenced by growth inhibition, the appearance of large flattened cells and the induction of senescence-associated β-galactosidase (SA-βgal) (data not shown and Fig. 1B). We found that neither abrogation of p53 nor Rb reduced OPN transcripts upon activation of senescence (Fig. 1C). On the contrary, in the absence of p53 or Rb, OPN basal levels in vehicle-treated cells increased (Fig. S1B and C) and they were further augmented upon bleomycin treatment and the induction of senescence, arguing that p53 and Rb are not only dispensable for the senescence-associated stimulation of OPN expression, but that they suppress it. As previously reported, IL6 and IL8 levels were also increased in senescent cells when p53 and Rb function was compromised (Fig. S1A and (5)), indicating that the p53 and Rb pathways actively suppress a wide range of SASP members.
Figure 1. The p53 and Rb pathways are dispensable for OPN upregulation in senescence.
A. Western Blot analysis of BJ fibroblasts stably transduced with a p53 mutant (DD) or CDK4/cyclinD1 (DK) fusion construct that inhibits wildtype p53 and Rb, respectively. β- and γ-actin serve as loading controls for the two cell lines. B. SA-βgal staining of BJ fibroblasts treated with vehicle alone (vehicle) or bleomycin (bleo) and BJ fibroblasts stably transduced with DD or DK treated with bleomycin (DD+Bleo and DK+Bleo, respectively). Scale bar is 100 microns. C. OPN mRNA levels were measured by quantitative PCR (qPCR) in cells described in A following treatment with vehicle or bleomycin. Vehicle-treated cells are defined as 1. The mean ± STDEV is shown (n=3).
Previous work had indicated that while SASP factors are robustly activated in senescent cells, the induction of senescence was dispensable for SASP activation (5, 7). We next wished to address whether OPN expression was dependent on senescence activation. To address this question, we treated immortalized WI38 fibroblasts expressing the papilloma virus E6 and E7 proteins, which abrogate p53 and Rb, respectively with bleomycin. As expected, bleomycin treatment of E6/E7-expressing cells did not induce senescence (Fig. S1D). We found that these cells failed to acquire the flattened cellular morphology of senescent cells and did not stain positive for SA-βgal (Fig. S1D). Despite the fact that E6/E7 cells failed to enter senescence, analysis of OPN, IL6, and IL8 revealed that these cells retained the ability to activate the SASP (Fig. S1E), indicating that SASP expression does not require the activation of the senescence effector proteins p53 and Rb nor entry into senescence.
SASP is controlled by distinct regulatory mechanisms
NF-κB is a master regulator of cytokine production in inflammatory responses (32) and was recently evoked in the transcriptional control of some SASP members, including IL6 and IL8 (16); (8). Like IL6, OPN can participate in cellular responses characterized by chronic inflammation (17) and NF-κB can directly activate OPN transcription under certain conditions (33). Therefore, we investigated whether NF-κB activates OPN transcription in senescent cells. NF-κB canonically resides in a complex with IκBα, which sequesters it in the cytoplasm. Upon stimulation, IκBα is phosphorylated and subsequently degraded, allowing NF-κB to translocate to the nucleus where it activates target gene transcription (32). We successfully blocked NF-κB signaling by stably expressing a mutant of IκBα that cannot be phosphorylated (IκBα-mut), thus trapping NF-κB in the cytoplasm (23). To confirm that the mutant was active, we examined its impact on an NF-κB reporter plasmid containing five tandem NF-κB binding elements. In BJ fibroblasts as expected, we found that expression of the reporter plasmid was inhibited in IκBα-mut cells compared to cells expressing a vector control (Fig. 2A).
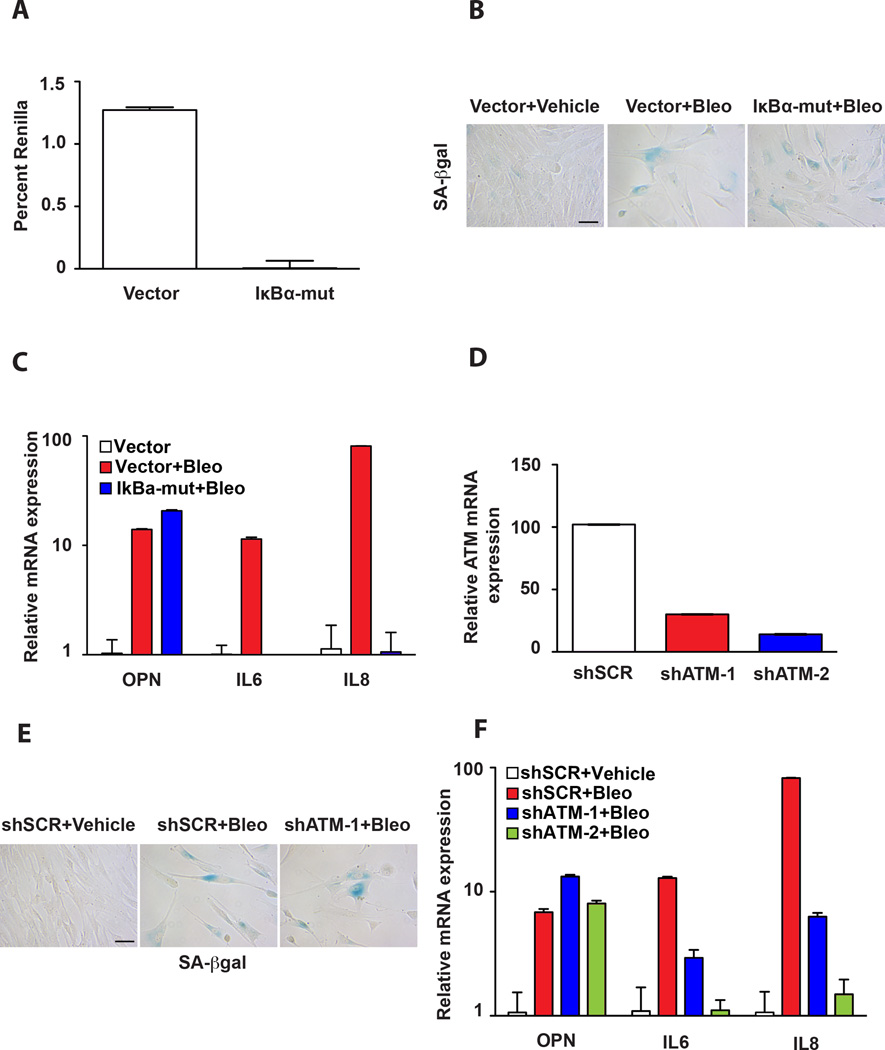
Figure 2. Transcriptional regulation of the SASP factors OPN versus IL6 and IL8 is governed by distinct mechanisms.
A. NF-κB activity was examined in BJ fibroblasts stably expressing a vector control or IκBα-mut allele. Cells were transiently transfected with an NF-κB-responsive promoter driving Firefly luciferase. Data are presented as percent Renilla Luciferase, a control for transfection efficiency. The mean ± SEM is shown (n=3). B. SA-βgal staining of BJ fibroblasts stably transduced with a vector control or IκBα-mut allele and treated with vehicle (Vector+Vehicle) or bleomycin (Vector+Bleo and IκBα-mut+Bleo). Scale bar is 100 microns. C. The mRNA levels of OPN, IL6, and IL8 as indicated were measured by qPCR in vector control BJ fibroblasts treated with vehicle (Vector) (set to 1) or bleomycin (Vector+Bleo) or BJ fibroblasts expressing the IκBα-mut cDNA and treated with bleomycin (IκBα-mut+Bleo). The mean ± STDEV is shown (n=3). D. ATM mRNA levels were quantified by qPCR in BJ fibroblasts expressing one of two short-hairpin RNAi constructs targeting ATM (shATM-1 or shATM-2) or a control hairpin (shSCR), which was defined as 100 percent. The mean ± STDEV is shown (n=3). E. SA-βgal staining of control hairpin cells treated with vehicle alone (shSCR+Vehicle) or bleomycin (shSCR+Bleo) and shATM expressing BJ fibroblasts treated with bleomycin (shATM-1+Bleo). Scale bar is 100 microns. F. The mRNA levels of OPN, IL6, and IL8 were measured by qPCR in BJ fibroblasts expressing a control hairpin treated with vehicle or bleomycin (shSCR+Vehicle and shSCR+Bleo) or a short-hairpin targeting ATM and treated with bleomycin (shATM-1+Bleo and shATM-2+Bleo, respectively). Expression levels in shSCR+Vehicle cells were set to 1 for each factor measured. The mean ± STDEV is shown (n=3).
To determine whether NF-κB activity was required to activate OPN in senescent cells, BJ fibroblasts expressing a control vector or the IκBα mutant (IκBα-mut) were treated with bleomycin. We found that bleomycin treatment induced robust senescence in the control and IκBα mutant-expressing cells as evidenced by the induction of a flattened morphology and the induction of SA-βGal (Fig. 2B), indicating that this mode of NF-κB inhibition does not abrogate the induction of senescence in these cells. In agreement with previous reports (16), we found that NF-κB activation is essential for the upregulation of IL6 and IL8 in senescence (Fig. 2C). Indeed, we found that IκBα-mut cells treated with bleomycin failed to upregulate IL6 and IL8. In contrast, OPN levels remained unperturbed in IκBα-mut cells treated with bleomycin (Fig. 2C). Because the expression of the IκBα mutant precedes the exposure to bleomycin, our findings indicate that NF-κB signaling is neither required for the initiation nor maintenance of OPN levels in response to DNA damage. Together these findings also indicate that SASP is regulated by at least two distinct transcriptional pathways.
Having established that neither the p53 or Rb pathways nor activation of NF-κB plays a role in OPN regulation following bleomycin treatment and SASP activation, we next turned our attention to the putative role of the DNA damage response (DDR). Senescence is characterized by a robust and persistent DNA damage response (34) that includes the activation of the ATM kinase, which has been implicated in SASP activation (7). To address whether ATM activation was required for OPN upregulation in senescent cells, we utilized ATM-specific short hairpin (shRNA) constructs to deplete cells of ATM to greater than 80% (Fig. 2D). As expected (7), ATM depletion had no impact on the induction of senescence (Fig. 2E) but resulted in a significant reduction in IL6 and IL8 levels (Fig. 2F). In contrast, ATM depletion had no impact on OPN expression following bleomycin treatment (Fig. 2F). These results indicate that OPN expression, unlike IL6 and IL8, is not controlled by DDR or NF-κB signaling in senescence, but instead is regulated by a distinct mechanism.
Histone deacetylase (HDAC) inhibition induces SASP
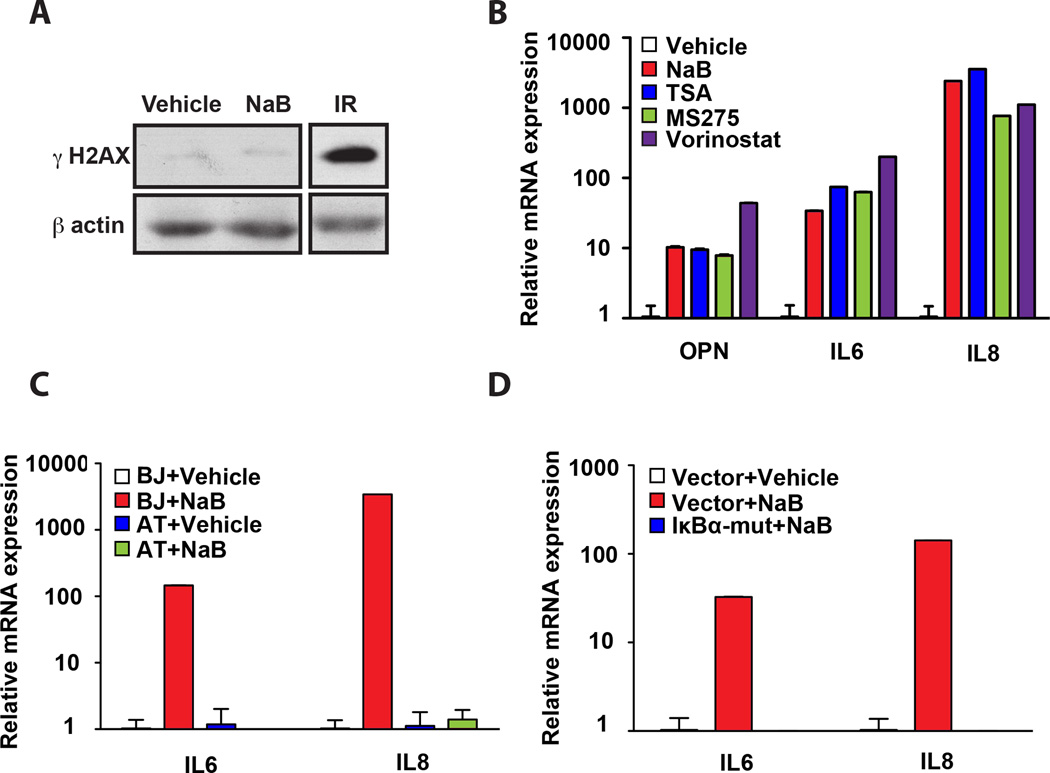
A wide variety of cellular stresses can induce senescence. Previous work demonstrated agents that impact histone deacetylase (HDAC) activity, including sodium butyrate (NaB) and trichostatin A (TSA), induce senescence or a senescence-like state in the absence of DNA damage (12, 14). These findings raised the possibility that chromatin modulation rather than bona fide DNA breaks were responsible for activation of the SASP. Therefore, we examined whether double strand DNA breaks were required for SASP expression. BJ fibroblasts were treated with NaB, which resulted in a robust cell cycle arrest and flattened cellular morphology but failed to activate SA-βgal activity (Fig. S2). To assess the presence of DNA double strand breaks, we evaluated levels of γH2AX, a phosphorylated form of the histone variant H2AX that is widely recognized as a marker of double strand breaks and active DDR (35). As a control for γH2AX induction we analyzed irradiated cells by Western blot analysis and noted a robust increase in γH2AX as expected. In contrast, NaB treatment did not increase γH2AX levels compared to vehicle-treated cells (Fig. 3A). The comet assay, a sensitive technique utilized to detect DNA breaks in individual cells (36), corroborated these results (data not shown) in agreement with previous findings (14). Next, we examined OPN mRNA levels and observed robust induction despite the lack of detectable DNA damage (Fig. 3B). In agreement with previous findings, both IL6 and IL8 also increased in NaB-treated cells (8). Together these findings indicate that DNA breaks are not required for SASP induction.
Figure 3. HDAC inhibitors activate the SASP.
A. Western Blot analysis of BJ fibroblasts treated with vehicle or sodium butyrate (NaB) for 72 hr were analyzed for γ-H2AX. Cells irradiated with 10 Gy (IR) serve as a positive control and β-actin was used as a loading control. B. The mRNA levels of OPN, IL6, and IL8 were measured by qPCR in representative vehicle-treated BJ fibroblasts or BJ fibroblasts treated independently with several HDAC inhibitors as indicated. Expression levels in Vehicle-treated cells were set to 1 for each factor measured. The mean ± STDEV is shown (n=3). C. BJ fibroblasts and AT fibroblasts were treated with vehicle or NaB, and IL6 and IL8 mRNA levels were measured by qPCR. The mean ± STDEV is shown (n=3). Expression levels in vehicle-treated cells were set to 1 for each cell line. D. BJ fibroblasts expressing a vector control (Vector) or an IκBα-mut cDNA (IκBα-mut) were treated with vehicle or NaB. IL6 and IL8 mRNA levels were quantified by qPCR and expression levels of IL6 and IL8 in the Vector+Vehicle were set to 1. The mean ± STDEV is shown (n=3).
HDAC inhibition in tumor cells results in reduced cell growth and tumor cell death. Thus, recent work in xenografts and human clinical trials has focused on the therapeutic potential of HDAC inhibition (37). These findings have led to the approval of one such compound, suberoylanilide hydroxamic acid (SAHA) or vorinostat, for the treatment of cutaneous T-cell lymphoma, with several other classes of HDAC inhibitors currently in clinical trials (38). Given the putative clinical importance of these inhibitors and our findings with NaB, we tested whether other HDAC inhibitors activated the SASP. Indeed, we found that other HDAC inhibitors including TSA, MS275 and vorinostat similarly increased OPN, IL6, and IL8 levels (Fig. 3B). We obtained identical results when we treated primary breast fibroblasts with NaB and vorinostat (Fig. S3), indicating that HDAC inhibition also elicits a SASP response in primary stromal cells. Together, these findings raise concern that HDAC inhibitors may impact the tumor microenvironment.
Above we show that the SASP is induced in the absence of DNA double strand breaks; however, ATM is activated in cells treated with chromatin relaxers (39), raising the possibility that the DDR required to activate IL6 and IL8 is initiated by a more general mechanism such as chromatin modulation independently of physical damage. Given that in bleomycin-induced senescence, ATM and NF-κB are required for IL6 and IL8 upregulation (Fig. 2), we next investigated whether both ATM and NF-κB were also required to regulate IL6 and IL8 in NaB-treated cells. We treated cells that express the IκBα-mut or AT cells (genetically deficient in ATM activity) with NaB as above. Although NaB does not induce DNA double strand breaks ((14) and data not shown), we found that IL6 and IL8 upregulation retained their requirement for both NF-κB and ATM (Fig. 3C and D). In contrast, we observed a robust upregulation of OPN in IκBα-mut expressing fibroblasts and AT cells following NaB treatment (data not shown). Together these findings indicate that the DNA damage signaling that is required for the upregulation of IL6 and IL8 does not emanate from physical DNA breaks (8) and suggest instead that chromatin modulation is central to the regulation of SASP factors.
HDAC inhibition creates a pro-tumorigenic microenvironment
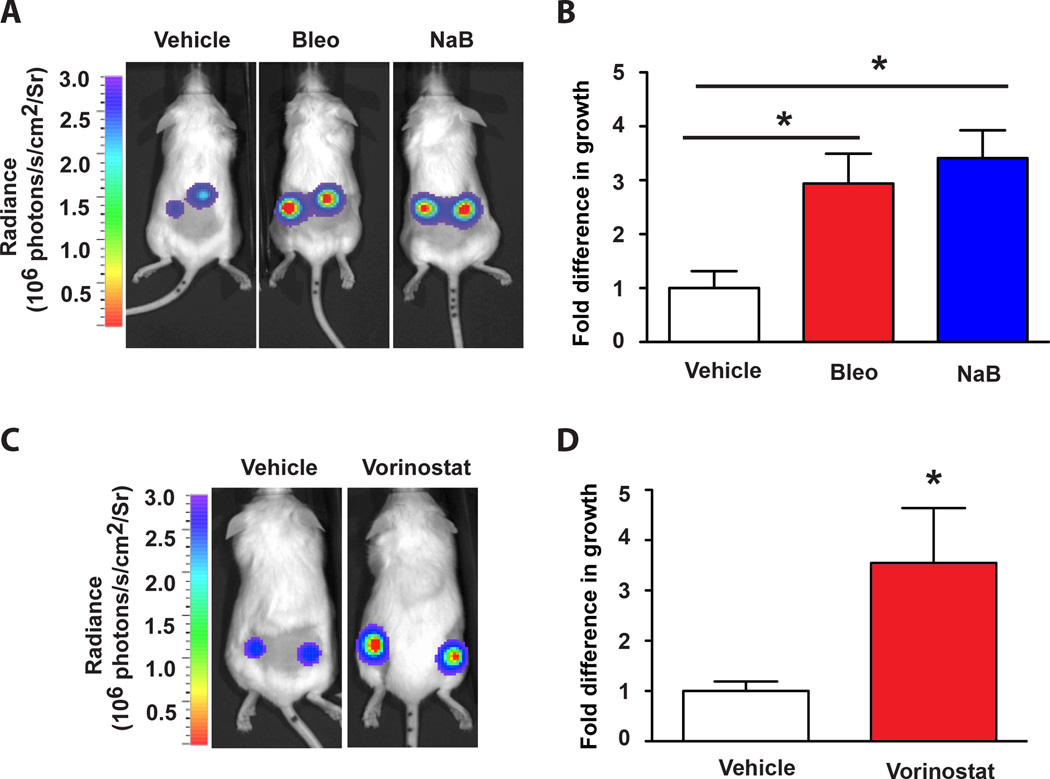
Our finding that HDAC inhibition stimulates the tumor-promoting SASP (Fig. 3B) led us to investigate their impact in vivo. To investigate this, we treated BJ fibroblasts with NaB, which led to the robust upregulation of OPN, IL6, and IL8 (Fig. S4A) and examined whether these fibroblasts promoted tumor growth in vivo. Similar to our previous report, we found that bleomycin-treated fibroblasts significantly promote preneoplastic HaCaTCBR cell growth when co-injected in xenografts (Fig. 4A and B). When cells were injected in combination with vehicle-treated fibroblasts, there was minimal growth. Significantly, the presence of fibroblasts treated with NaB also led to a substantial enhancement of HaCATCBR cell growth compared to cells injected with vehicle-treated fibroblasts (Fig. 4A and B), arguing that HDAC inhibition is a potent SASP inducer in vivo. To further corroborate our findings, we tested the only HDAC inhibitor currently used in the clinic – vorinostat. Upon treatment with vorinostat, BJ fibroblasts activated the SASP (Fig. S4B) and promoted HaCATCBR cell growth compared to their vehicle counterparts (Fig. 4C and D). Together, our results indicate that HDAC inhibition in the stroma activates a protumorigenic profile and leads to increased tumor growth in vivo.
Figure 4. Fibroblasts treated with HDAC inhibitors promote preneoplastic cell growth.
A. Bioluminescent images (day 10) of representative mice injected in the flanks with HaCaTCBR cells in combination with vehicle-treated (Vehicle), bleomycin-treated (Bleo), or NaB–treated (NaB) fibroblasts. B. Quantification of the luminescence in panel A (measured as photons/second/cm2/steradian (photons/s/cm2/Sr), vehicle-treated cells are defined as 1. The mean ± SEM is shown, * p < 0.05 (n=8). C. Bioluminescent images (day 12) of representative mice injected in the flanks with a mixture of HaCaTCBR cells with either vehicle-treated BJ fibroblasts or vorinostat-treated BJ fibroblasts. D. Quantification of the luminescence in the groups described in panel C, vehicle-treated cells are defined as 1. The mean ± SEM is shown, * p < 0.05 (n=8).
HDAC1 inhibition activates OPN
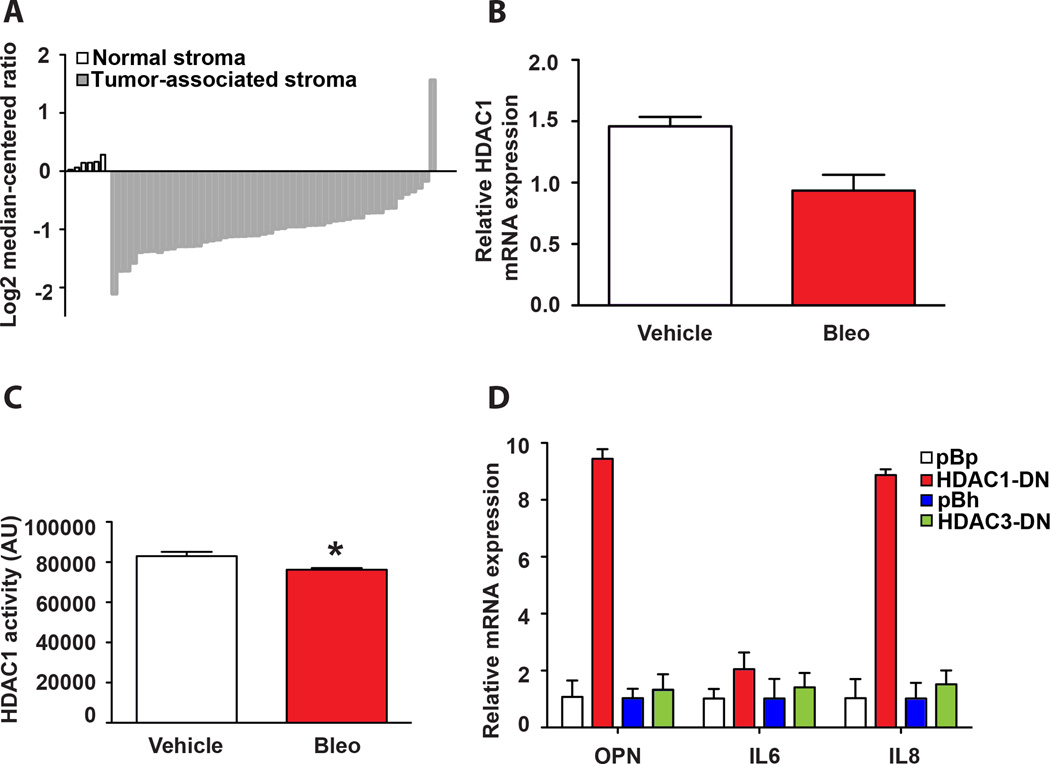
There are three major groups of HDACs (excluding sirtuins) and 18 HDACs are present in the human genome. Most HDAC inhibitors target multiple HDACs; we next examined whether a specific HDAC was critical for OPN activation. A recent study analyzing stromal changes in breast cancer identified OPN as a key component of a stroma-derived prognostic predictor (SDPP) that successfully clustered tumors by clinical outcome (29). Oncomine-based interrogation of the same dataset revealed that HDAC1 levels were significantly (p < 0.05) lower in breast cancer-associated stroma compared to normal stroma (Fig. 5A). Given that the SASP is reminiscent of the expression profile observed in cancer-associated fibroblasts and that the HDAC inhibitors used in our study target HDAC1, we examined whether HDAC1 played an important role in OPN activation.
Figure 5. Inhibition of HDAC1 leads to OPN upregulation.
A. Oncomine expression analysis of HDAC1 levels between normal stroma and invasive breast cancer-associated stroma extracted from the Finak et. al. study (29). Expression values are log transformed and median-centered per array (see Materials and Methods). B. HDAC1 mRNA levels were measured by qPCR in vehicle and bleomycin-treated BJ fibroblasts. The mean ± STDEV is shown (n=3). C. HDAC1 activity was measured in lysates from vehicle or bleomycin-treated cells after immunoprecipitation of HDAC1 with an HDAC1 antibody. The mean ± SEM is shown, * p < 0.05, (n=3). D. OPN, IL6 and IL8 mRNA levels were quantified by qPCR in BJ fibroblasts transduced with a vector control (pBp or pBh) or a dominant negative cDNA of HDAC1 (HDAC-DN) or HDAC3 (HDAC3-DN). The mean ± STDEV is shown (n=3).
To determine whether HDAC1 levels changed in vehicle versus bleomycin-treated cells, we examined HDAC1 mRNA levels by quantitative PCR (qPCR). In agreement with our previous microarray data (4), we observed a decrease in HDAC1 mRNA levels in bleomycin-treated fibroblasts compared to vehicle-treated cells (Fig. 5B). When we immunoprecipitated HDAC1 from vehicle and bleomycin-treated fibroblasts and assayed for deacetylase activity, we observed a consistent decrease in cells treated with bleomycin (Fig. 5C), raising the possibility that loss of HDAC1 activity specifically contributes to OPN activation following bleomycin treatment.
To directly test whether HDAC1 activity was important for the increase in OPN expression, we ectopically expressed a dominant-negative mutant of HDAC1 (HDAC1-DN) (24) in fibroblasts (Fig. S5A). While overexpression of HDAC1-DN significantly reduced cell proliferation it did not result in growth arrest or SA-βgal expression ((40) and Fig. S5B). However, we observed that ectopic expression of HDAC1-DN was sufficient to induce significant upregulation in OPN mRNA levels (Fig. 5D), mirroring the response to treatments with HDAC inhibitors (Fig. 3B). This finding further demonstrates that the induction of the SASP can be decoupled from the induction of senescence. Importantly, this is an HDAC1-specific effect, because ectopic expression of an HDAC3 mutant (Fig. S4C) did not alter OPN levels (Fig. 5D). Analysis of IL6 and IL8 expression in HDAC1-DN expressing cells revealed that loss of HDAC1 activity was also sufficient to increase IL8 and to a lesser extent IL6 while HDAC3-DN had no impact on either IL6 or IL8. Together, these findings suggest that chromatin alterations via manipulation of HDAC1 activity impact the regulation of the SASP independent of the activation of senescence.
Discussion
Senescent fibroblasts promote tumorigenesis in multiple models (3, 4). In efforts to understand how this is accomplished, several groups have examined the expression profile of senescent cells and uncovered a signature secretory program enriched in growth factors, cytokines, and proteases termed the senescence-associated secretory phenotype (SASP) (4, 5, 15, 16). Specific SASP components have been directly implicated in senescent stromal-promoted tumorigenesis (4, 6, 8, 15). However, it is still unclear how the SASP is activated. Initial findings demonstrated that the transcription factors NF-κB and C/EBPβ directly bind the promoters of some of the inflammatory cytokines expressed in senescent cells (8, 15, 16). Additionally, the DNA damage response (DDR) was identified as an upstream SASP inducer (7). Indeed, the above-mentioned transcription factors can be activated in response to DNA damage (41, 42), although the mechanism of activation in senescence is not known. Utilizing osteopontin (OPN) as a surrogate for SASP regulation, we found that the SASP is not characterized by a single transcriptional axis, but is instead controlled by at least two independent signaling cascades both of which may be activated in response to chromatin changes.
OPN impacts a vast array of signaling pathways, hence its transcription is governed by multiple mechanisms in a tissue- and context-dependent manner (17). We demonstrate that following a senescence-inducing dose of bleomycin, OPN expression is not controlled by p53, Rb, or NF-κB (31, 33). The lack of p53 and Rb involvement is in agreement with previous work showing that these essential senescence effector pathways are dispensable for activation of the SASP factors IL6 and IL8 (5). Furthermore, our work demonstrates that OPN, like other SASP factors including IL6 and IL8, is expressed in cells that are unable to senesce due to abrogation of the p53 and Rb pathways. This finding is significant because it demonstrates that while senescent cells express SASP factors, the induction of senescence is not required to activate the SASP. Thus, SASP activation may occur in a wide variety of stromal cells undergoing cellular stress. It is interesting to note that the SASP is highly reminiscent of the expression profile observed in cancer associated fibroblasts (CAFs), which like senescent cells stimulate tumor formation in xenograft models (43).
This work and that of others demonstrates p53 and Rb are uniformly dispensable for SASP expression, yet it is clear that SASP regulation is complex and several distinct mechanisms drive expression of specific SASP factors. Indeed, we show that while NF-κB directly activates IL6 and IL8 (Fig. 2 and data not shown), it is not required for OPN upregulation in senescence (Fig. 2 and data not shown). Similarly, ATM knockdown has a profound effect on IL6 and IL8 expression, yet OPN increases to the same levels as in control cells (Fig. 2). Together, these results indicate that IL6 and IL8 are activated by the DDR and NF-κB as has been demonstrated (7, 8), and OPN is not. Therefore, it is reasonable to speculate that subsets of SASP are regulated by discrete signaling pathways. In fact, the SASP is defined by distinct classes of proteins such as growth factors, mitogens, and extracellular remodeling enzymes (4) whose activation may be governed by several mechanisms. Indeed, our work demonstrates that OPN regulation is distinct from the inflammatory cytokines IL6 and IL8 (15, 16). Discovery of putative activators of OPN and whether they control the expression of other proteins belonging to the extracellular matrix core of the SASP is the subject of ongoing work (4).
Despite the fact that IL6 and IL8 are regulated independently of OPN, following the induction of senescence, all three factors increase at the mRNA level, arguing that there must be a common inducer of SASP. Our work shows that ATM and NF-κB are not the sole SASP regulators, raising the possibility that an upstream stimulus initiates multiple signaling cascades. Our experiments with histone deacetylase inhibitors demonstrate that in the absence of DNA breaks (as measured by H2AX phosphorylation and comet assays (see Fig. 3 and (14)), the SASP is still activated. Interestingly, ATM and NF-κB are still required for the upregulation of IL6 and IL8 (Fig. 3) in HDAC inhibitor-induced SASP, thus lending additional support to the argument that DNA damage signaling, but not breaks per se, activates the SASP. HDAC inhibition modifies the chromatin by inducing hyperacetylation of histone and non-histone proteins (44), resulting in sustained transcriptional changes (44). The data presented here fit a model wherein ATM activation upon bleomycin and NaB treatment occurs in response to chromatin alteration. It is known that DNA breaks induce changes in the surrounding chromatin, which facilitates signaling and repair; conversely, chromatin modifications trigger the DNA damage checkpoint (45). Indeed, because treatment with an HDAC inhibitor activates ATM (39), it is plausible that chromatin relaxation in senescence induces ATM activation and subsequent IL6 and IL8 expression.
While it is unclear how chromatin changes are instituted and maintained in senescence, there is ample evidence for their presence (11, 46). It is conceivable that widespread chromatin modifications impact promoter activity globally. However, only two percent of the genes are differentially regulated in senescence (4) making this unlikely. Furthermore, specific transcription factors (e.g., NF-κB and C/EBPβ) are required for expression of some SASP factors, pointing to a regulated mechanism of expression. Finally, our results demonstrate that only specific chromatin modifiers impact OPN expression. Specifically, inhibition of HDAC1 but not HDAC3, upregulates OPN, IL6, and IL8 mRNA levels. Furthermore, we observe a reduction in HDAC1 levels and activity in senescent cells compared to their vehicle-treated counterparts. Together these data argue that HDAC1 containing complexes drive chromatin alterations that then drive OPN expression. HDACs role in transcriptional regulation is not limited to SASP. Indeed, HDAC1 inhibition results in an altered transcriptional profile in HGPS cells (27). Additionally, HDAC1 is decreased in breast cancer stroma (Fig. 5A and (29)), which also undergoes profound transcriptional changes that alter the microenvironment and contribute to tumorigenesis. What initiates the changes in chromatin in these different biological settings is currently unknown and the triggers are likely to vary depending on the biological context. It is known that HDAC1 binds Rb (47) and interestingly, individual inhibition of either Rb or HDAC1 leads to marked upregulation of OPN mRNA levels (Figs. 1 and 5), raising the possibility that in non-senescent cells HDAC1 complexes occupy SASP promoters and actively repress transcription, but upon senescence induction, they translocate preferentially to E2F targets (47). It remains to be determined if the Rb-HDAC1 complex is targeted to the promoters of OPN and other SASP members or whether it controls upstream regulators.
Our results add an additional layer of complexity to HDACs and their putative involvement in the tumor microenvironment. Recently, significant effort has been invested in designing HDAC inhibitors as anticancer agents leading to the approval of one compound, vorinostat (also known as SAHA), for the treatment of cutaneous T-cell lymphoma (48) and several others currently in clinical trials (49). Most studies focus on the antiproliferative or apoptotic effects of these reagents on tumor cells and report a minimal impact on normal cells (48, 50, 51). Our studies suggest that the impact of HDAC inhibition on normal cells is quite different. When normal cells, including fibroblasts are treated with HDAC inhibitors they are relatively resistant to apoptosis (52) but enter a senescence-like state (14). Moreover, regardless of the effect on cell cycle progression, all the HDAC inhibitors we tested (Fig. 3B) robustly upregulate SASP expression and promote tumor growth in vivo in a paracrine fashion (Fig. 4). The half-life of the HDAC inhibitors used in the clinic is relatively short (52), limiting the therapeutic window for solid malignancies (53). However, the molecular responses as measured by overall hyperacetylation are sustained following treatment with HDAC inhibitors. Our results indicate that the transcriptional changes associated with HDAC inhibitor treatment are maintained in normal fibroblasts, and previous studies have demonstrated that the SASP is a chronic response (7). Therefore, it will be important to assess sustained molecular changes in the stroma that may lead to increased tumor formation following HDAC therapy. Our work underscores the importance of examining the tumor microenvironment when analyzing the profile of therapeutic agents.
Supplementary Material
Acknowledgements
We thank Dr. Cindy Malone and the Stewart laboratory for helpful discussions and Daniel Teasley, Megan Ruhland, and Hayley Moore for critical reviews of the manuscript. We also thank Dr. Joseph Roti Roti and Robert Vanderwaal for carrying out the COMET assay. We are grateful to Adnan Elhammali for invaluable help with bioinformatic analysis.
Financial Support: This work was supported by the NIH Molecular Oncology Training Grant T32 CA113275 (EP), NIH Cellular Biochemical and Molecular Sciences Predoctoral Training Grant T32 GM007067-35 (EA), the HHMI Medical Research Fellows Program (AM), Molecular Imaging Center P50 CA94056 (DPW), and NIH 5 R01 CA130919 (SAS).
Footnotes
Conflicts of interest: None
References
- 1.Pazolli E, Stewart SA. Senescence: the good the bad and the dysfunctional. Curr Opin Genet Dev. 2008;18:42–47. doi: 10.1016/j.gde.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 2.Braig M, Lee S, Loddenkemper C, Rudolph C, Peters AH, Schlegelberger B, et al. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005;436:660–665. doi: 10.1038/nature03841. [DOI] [PubMed] [Google Scholar]
- 3.Krtolica A, Parrinello S, Lockett S, Desprez PY, Campisi J. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc Natl Acad Sci U S A. 2001;98:12072–12077. doi: 10.1073/pnas.211053698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pazolli E, Luo X, Brehm S, Carbery K, Chung JJ, Prior JL, et al. Senescent Stromal-Derived Osteopontin Promotes Preneoplastic Cell Growth. Cancer Res. 2009;69:1230–1239. doi: 10.1158/0008-5472.CAN-08-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coppe JP, Patil CK, Rodier F, Sun Y, Munoz DP, Goldstein J, et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parrinello S, Coppe JP, Krtolica A, Campisi J. Stromal-epithelial interactions in aging and cancer: senescent fibroblasts alter epithelial cell differentiation. J Cell Sci. 2005;118:485–496. doi: 10.1242/jcs.01635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodier F, Coppe JP, Patil CK, Hoeijmakers WA, Munoz DP, Raza SR, et al. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat Cell Biol. 2009;11:973–979. doi: 10.1038/ncb1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orjalo AV, Bhaumik D, Gengler BK, Scott GK, Campisi J. Cell surface-bound IL-1alpha is an upstream regulator of the senescence-associated IL-6/IL-8 cytokine network. Proc Natl Acad Sci U S A. 2009;106:17031–17036. doi: 10.1073/pnas.0905299106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campisi J. Aging and cancer cell biology. 2007. Aging Cell. 2007;6:261–263. doi: 10.1111/j.1474-9726.2007.00292.x. [DOI] [PubMed] [Google Scholar]
- 10.Campisi J. The biology of replicative senescence. Eur J Cancer. 1997;33:703–709. doi: 10.1016/S0959-8049(96)00058-5. [DOI] [PubMed] [Google Scholar]
- 11.Narita M, Nunez S, Heard E, Lin AW, Hearn SA, Spector DL, et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 12.Place RF, Noonan EJ, Giardina C. HDACs and the senescent phenotype of WI-38 cells. BMC cell biology. 2005;6:37. doi: 10.1186/1471-2121-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calvanese V, Lara E, Kahn A, Fraga MF. The role of epigenetics in aging and age-related diseases. Ageing research reviews. 2009;8:268–276. doi: 10.1016/j.arr.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 14.Pospelova TV, Demidenko ZN, Bukreeva EI, Pospelov VA, Gudkov AV, Blagosklonny MV. Pseudo-DNA damage response in senescent cells. Cell Cycle. 2009;8:4112–4118. doi: 10.4161/cc.8.24.10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuilman T, Michaloglou C, Vredeveld LC, Douma S, van Doorn R, Desmet CJ, et al. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133:1019–1031. doi: 10.1016/j.cell.2008.03.039. [DOI] [PubMed] [Google Scholar]
- 16.Acosta JC, O'Loghlen A, Banito A, Guijarro MV, Augert A, Raguz S, et al. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 2008;133:1006–1018. doi: 10.1016/j.cell.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 17.Rangaswami H, Bulbule A, Kundu GC. Osteopontin: role in cell signaling and cancer progression. Trends Cell Biol. 2006;16:79–87. doi: 10.1016/j.tcb.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 18.Senger DR, Wirth DF, Hynes RO. Transformed mammalian cells secrete specific proteins and phosphoproteins. Cell. 1979;16:885–893. doi: 10.1016/0092-8674(79)90103-x. [DOI] [PubMed] [Google Scholar]
- 19.Rittling SR, Denhardt DT. Osteopontin function in pathology: lessons from osteopontin-deficient mice. Exp Nephrol. 1999;7:103–113. doi: 10.1159/000020591. [DOI] [PubMed] [Google Scholar]
- 20.Fedarko NS, Jain A, Karadag A, Van Eman MR, Fisher LW. Elevated serum bone sialoprotein and osteopontin in colon, breast, prostate, and lung cancer. Clin Cancer Res. 2001;7:4060–4066. [PubMed] [Google Scholar]
- 21.Luo P, Tresini M, Cristofalo V, Chen X, Saulewicz A, Gray MD, et al. Immortalization in a normal foreskin fibroblast culture following transduction of cyclin A2 or cdk1 genes in retroviral vectors. Exp Cell Res. 2004;294:406–419. doi: 10.1016/j.yexcr.2003.11.030. [DOI] [PubMed] [Google Scholar]
- 22.Hahn WC, Dessain SK, Brooks MW, King JE, Elenbaas B, Sabatini DM, et al. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol Cell Biol. 2002;22:2111–2123. doi: 10.1128/MCB.22.7.2111-2123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boehm JS, Zhao JJ, Yao J, Kim SY, Firestein R, Dunn IF, et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 2007;129:1065–1079. doi: 10.1016/j.cell.2007.03.052. [DOI] [PubMed] [Google Scholar]
- 24.Ito K, Adcock IM. Histone acetylation and histone deacetylation. Mol Biotechnol. 2002;20:99–106. doi: 10.1385/MB:20:1:099. [DOI] [PubMed] [Google Scholar]
- 25.Yang WM, Tsai SC, Wen YD, Fejer G, Seto E. Functional domains of histone deacetylase-3. J Biol Chem. 2002;277:9447–9454. doi: 10.1074/jbc.M105993200. [DOI] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Pegoraro G, Kubben N, Wickert U, Gohler H, Hoffmann K, Misteli T. Ageing-related chromatin defects through loss of the NURD complex. Nat Cell Biol. 2009;11:1261–1267. doi: 10.1038/ncb1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia (New York, NY. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 30.Shay JW, Pereira-Smith OM, Wright WE. A role for both RB and p53 in the regulation of human cellular senescence. Exp Cell Res. 1991;196:33–39. doi: 10.1016/0014-4827(91)90453-2. [DOI] [PubMed] [Google Scholar]
- 31.Morimoto I, Sasaki Y, Ishida S, Imai K, Tokino T. Identification of the osteopontin gene as a direct target of TP53. Genes Chromosomes Cancer. 2002;33:270–278. doi: 10.1002/gcc.10020. [DOI] [PubMed] [Google Scholar]
- 32.Karin M. NF-kappaB as a critical link between inflammation and cancer. Cold Spring Harbor perspectives in biology. 2009;1 doi: 10.1101/cshperspect.a000141. a000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Renault MA, Jalvy S, Potier M, Belloc I, Genot E, Dekker LV, et al. UTP induces osteopontin expression through a coordinate action of NFkappaB, activator protein-1, and upstream stimulatory factor in arterial smooth muscle cells. J Biol Chem. 2005;280:2708–2713. doi: 10.1074/jbc.M411786200. [DOI] [PubMed] [Google Scholar]
- 34.Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 35.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 36.Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res. 1988;175:184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- 37.Wagner JM, Hackanson B, Lubbert M, Jung M. Histone deacetylase (HDAC) inhibitors in recent clinical trials for cancer therapy. Clinical epigenetics. 2010;1:117–136. doi: 10.1007/s13148-010-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grant S, Dent P. Simultaneous interruption of signal transduction and cell cycle regulatory pathways: implications for new approaches to the treatment of childhood leukemias. Curr Drug Targets. 2007;8:751–759. doi: 10.2174/138945007780830764. [DOI] [PubMed] [Google Scholar]
- 39.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 40.Lagger G, O'Carroll D, Rembold M, Khier H, Tischler J, Weitzer G, et al. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. Embo J. 2002;21:2672–2681. doi: 10.1093/emboj/21.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang H, Ye QH, Ren N, Zhao L, Wang YF, Wu X, et al. The prognostic significance of preoperative plasma levels of osteopontin in patients with hepatocellular carcinoma. J Cancer Res Clin Oncol. 2006;132:709–717. doi: 10.1007/s00432-006-0119-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elkon R, Rashi-Elkeles S, Lerenthal Y, Linhart C, Tenne T, Amariglio N, et al. Dissection of a DNA-damage-induced transcriptional network using a combination of microarrays, RNA interference and computational promoter analysis. Genome Biol. 2005;6:R43. doi: 10.1186/gb-2005-6-5-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olumi AF, Grossfeld GD, Hayward SW, Carroll PR, Tlsty TD, Cunha GR. Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Res. 1999;59:5002–5011. doi: 10.1186/bcr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buchwald M, Kramer OH, Heinzel T. HDACi--targets beyond chromatin. Cancer Lett. 2009;280:160–167. doi: 10.1016/j.canlet.2009.02.028. [DOI] [PubMed] [Google Scholar]
- 45.van Attikum H, Gasser S. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 2009;19:207–217. doi: 10.1016/j.tcb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 46.Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain JP, et al. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 48.Dokmanovic M, Clarke C, Marks PA. Histone deacetylase inhibitors: overview and perspectives. Mol Cancer Res. 2007;5:981–989. doi: 10.1158/1541-7786.MCR-07-0324. [DOI] [PubMed] [Google Scholar]
- 49.Tan J, Cang S, Ma Y, Petrillo RL, Liu D. Novel histone deacetylase inhibitors in clinical trials as anti-cancer agents. Journal of hematology & oncology. 2010;3:5. doi: 10.1186/1756-8722-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qiu L, Burgess A, Fairlie DP, Leonard H, Parsons PG, Gabrielli BG. Histone deacetylase inhibitors trigger a G2 checkpoint in normal cells that is defective in tumor cells. Mol Biol Cell. 2000;11:2069–2083. doi: 10.1091/mbc.11.6.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ungerstedt JS, Sowa Y, Xu WS, Shao Y, Dokmanovic M, Perez G, et al. Role of thioredoxin in the response of normal and transformed cells to histone deacetylase inhibitors. Proc Natl Acad Sci U S A. 2005;102:673–678. doi: 10.1073/pnas.0408732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kelly WK, O'Connor OA, Krug LM, Chiao JH, Heaney M, Curley T, et al. Phase I study of an oral histone deacetylase inhibitor, suberoylanilide hydroxamic acid, in patients with advanced cancer. J Clin Oncol. 2005;23:3923–3931. doi: 10.1200/JCO.2005.14.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Munster PN, Thurn KT, Thomas S, Raha P, Lacevic M, Miller A, et al. A phase II study of the histone deacetylase inhibitor vorinostat combined with tamoxifen for the treatment of patients with hormone therapy-resistant breast cancer. Br J Cancer. 2011;104:1828–1835. doi: 10.1038/bjc.2011.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.