Abstract
Cells that line major tissues in the body such as blood vessels, lungs and gastrointestinal tract experience deformation from mechanical strain with our heartbeat, breathing, and other daily activities. Tissues also remodel in both development and disease, changing their mechanical properties. Taken together, cells can experience vastly different mechanical cues resulting from the combination of these interdependent stimuli. To date, most studies of cellular mechanotransduction have been limited to assays in which variations in substrate stiffness and strain were not combined. Here, we address this technological gap by implementing a method that can simultaneously tune both substrate stiffness and mechanical strain. Substrate stiffness is controlled with different monomer and crosslinker ratios during polyacrylamide gel polymerization, and strain is transferred from the underlying silicone platform when stretched. We demonstrate this platform with polyacrylamide gels with elastic moduli at 6 kPa and 20 kPa in combination with two different silicone formulations. The gels remain attached with up to 50% applied strains. To validate strain transfer through the gels into cells, we employ particle-tracking methods and observe strain transmission via cell morphological changes.
Introduction
Human cells undergo a variety of deformations in normal and disease physiology. To study these processes in vitro, many macro- and microscale systems have been invented to apply both tensile and compressive strains to large cell populations.1–3 Using these systems, strain fields have been applied to a variety of biological models to affect cell behavior in wide-ranging ways, from differentiation4 to protein production5 as well as response to small molecules.6 Substrate stiffness has also been shown to dramatically affect cell behavior,7 spreading,8, 9 maturation,10 function,11, 12 and differentiation.13 This stiffness sensing mechanism is poorly understood among the complicated signaling cascades that control cell behavior.14–16 However, cellular responses to strain compared to stiffness cannot be readily distinguished since few existing systems are able to modulate stiffness and strain independently. While one group has managed to bond acrylamide to commercially available silicone culture plates, the authors report complicated and difficult bonding.17 Additionally, their reliance on commercial systems could be an expensive impediment to wide-ranging progress.
Here, we describe a simple and inexpensive way to incorporate interpenetrating networks (IPNs) of commonly used polyacrylamide hydrogels18, 19 onto silicone membranes.20 The method can be used on various silicone formulations with hydrogels with differing elastic moduli. We demonstrate up to 50% transfer of non-destructive uniaxial applied strain to gel substrates from two distinct silicone formulations to gels with elastic moduli of 6 kPa (“soft”) and 20 kPa (“stiff”).
To show the wide-ranging potential of this method for future mechanotransduction studies, we report on the strain transfer from gels to adherent vascular cells. We demonstrate both laminin and fibronectin to promote cell adhesion to the gel surface. These proteins facilitate cell adhesion through different binding domains.21–23 and surround vascular cells like endothelial cells (ECs) and smooth muscle cells (SMCs) in vivo.24, 25 The successful integration of these different proteins and cell types into our composite cell culture system suggests a wide variety of applications for this new method.
Materials and methods
Fabrication of composite acrylamide and silicone substrate
Two types of silicone were used to demonstrate the versatility of this method. Commercially available 250 µm thick silicone sheets (HT-6240, Stockwell Elastomerics, Philadelphia, PA) were cut into approximately 2.5 cm × 1.5 cm strips. Two-part polydimethylsiloxane (PDMS, Sylgard 182, Dow Corning, Midland, MI) was mixed at a 10:1 ratio per manufacturer’s directions and defoamed in a planetary centrifugal mixer (AR-100, ThinkyUSA, Laguna Hills, CA). We then spin coated 10 g of PDMS prepolymer on a silicon wafer at 250 RPM for 90 seconds and cured this membrane for 4 hours at 65 °C to achieve target thickness of 250 µm. The membrane was subsequently removed from the wafer and cut into approximately 2.5 cm × 1.5 cm strips.
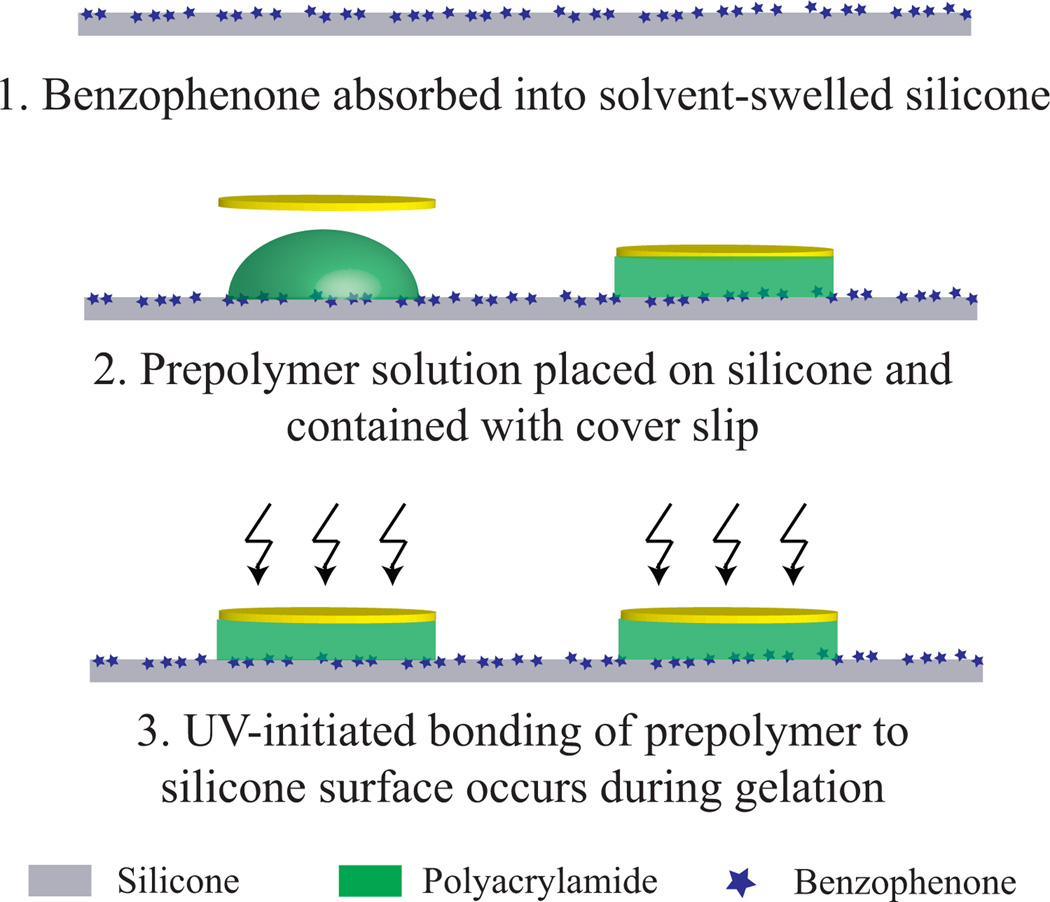
Both silicone formulations were then impregnated with the UV-reactive molecule benzophenone according to previously reported methods.26 Briefly, a solution of 10 wt%/vol benzophenone (99% pure, Acros Organics, NJ) dissolved in a water/acetone mixture (35:65 w/w) was placed on the silicone strip for 60 s (Fig. 1). The silicone strips were immediately rinsed with methanol and dried with nitrogen.
Figure 1.
Straightforward fabrication process for siliconeacrylamide substrate. Surfaces of the silicone and/or cover slip can be coated with fiducial markers for tracking and calibration.
To form a composite network of acrylamide and silicone, we mixed a prepolymer solution of 10% w/v acrylamide (99% pure, Bio-Rad, Hercules, CA), 0.05 or 0.5% w/v bisacrylamide (Molecular grade, Promega Corporation, Madison WI), ammonium persulfate (500 µg/ml, Bio-Rad), tetramethylethylenediamine (1:2000 vol/vol, Bio-Rad) and distilled water. Ten µl of this solution was pipetted onto the center of the silicone and covered with a 12 mm diameter round coverslip. The assembly was then treated with ultraviolet light (10.8 J/cm2, λUV = 383 nm). Coverslips were removed, and gels were allowed to equilibrate in phosphate buffered saline (PBS, pH 7.4) overnight at room temperature.
Characterization of composite substrates
We used atomic force microscopy (alpha300A, WITec, Knoxville, TN) to investigate the elastic moduli of assembled gels, and details can be found in electronic supplementary information (SI). Indentation results (10 µm/s indentation rate) are fit to the Oliver-Pharr model27, 28 to yield elastic modulus.
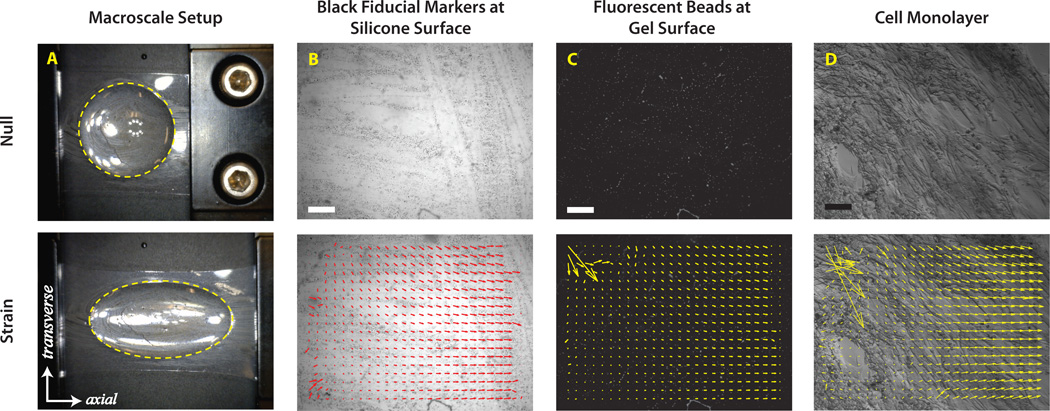
Silicone-acrylamide strips were loaded into a linear actuator (ST- Japan, Fig. 2A). To characterize the macroscale behavior of the system, the silicone strip was stretched to approximately 150% its original length for 50% engineering strain. “Macroscale” strain of the attached gel was assessed by taking pictures of initial and stretched samples (DinoLite Pro, AnMo Electronics, Torrance, CA) and using ImageJ to calculate the engineering strain (additional details in SI).
Figure 2.
(A) Images and schematic of macroscale calibration process; gels are 12 mm across in null condition. Ellipses fit to edge of gels (yellow dashed lines) are used to calculate engineering strain of the gel. (B–D) Images and representative analysis for microscale calibration pictures depicts displacement vectors calculated for image pair characterizing the (B) HT-6240 silicone surface, (C) 20 kPa gel surface, and (D) SMCs attached to fibronectin-coated 20 kPa gel. Inconsistent vectors along the outer perimeter of the image were disregarded during strain calculations by using vectors in a consistent 0.25 mm2 region. Scale bar = 100 µm.
Microscale strain transfer characterization
To confirm microscale strain transfer to the gels was similar to observed macroscale strain, conventional image correlation algorithms (implemented in PIVlab29) were used to track fiducial markers at the surface of the silicone and at the surface of the beads. The sample was strained approximately 10%, and we evaluated the Lagrangian strain tensor for the displacement results of the image correlation algorithms (Fig. 2B–C, additional information in ESI). To assess strain transfer from the gel to the cell monolayer, random cell morphologies were adequate fiducial markers to use PIVlab again on the paired-paired images (Fig. 2C–D), and the engineering strain was calculated from these data. Detailed methods are described in SI.
Cell culture methods
To bond proteins to polyacrylamide gels, we pooled 1 µM N-sulfosuccinimidyl- 6-(4’-azido-2’-nitrophenylamino) hexanoate (sulfoSANPAH, G Biosciences, St. Louis, MO) suspended in PBS onto the gels and treated with ultraviolet light (10.8 J/cm2, λUV = 383 nm). We then rinsed the gels with PBS and repeated the sulfoSANPAH treatment. PBS solution (100 µl) containing either 172 µg/ml fibronectin (Human plasma, BD Biosciences, San Jose, CA) or 172 µg/ml laminin (Ultrapure mouse, BD Biosciences) was pooled on the gels for 2 hours. Between two and four composite substrates were arranged in a 100 mm petri dish. Human dermal microvascular ECs30 (courtesy of the Cooke Lab, Stanford) or human aortic SMCs (Cell Systems, Kirkland, WA) at a density of ~3 million cells/ml were then plated on fibronectin- or laminin-coated gels and allowed to attach for 30 mins. Media (10 ml) was added to remove unattached cells, and the substrates were incubated at 37 °C overnight. Only regions of >80% confluence were included for analysis.
Results and discussion
Composite structures can be formed from a variety of silicone and acrylamide combinations
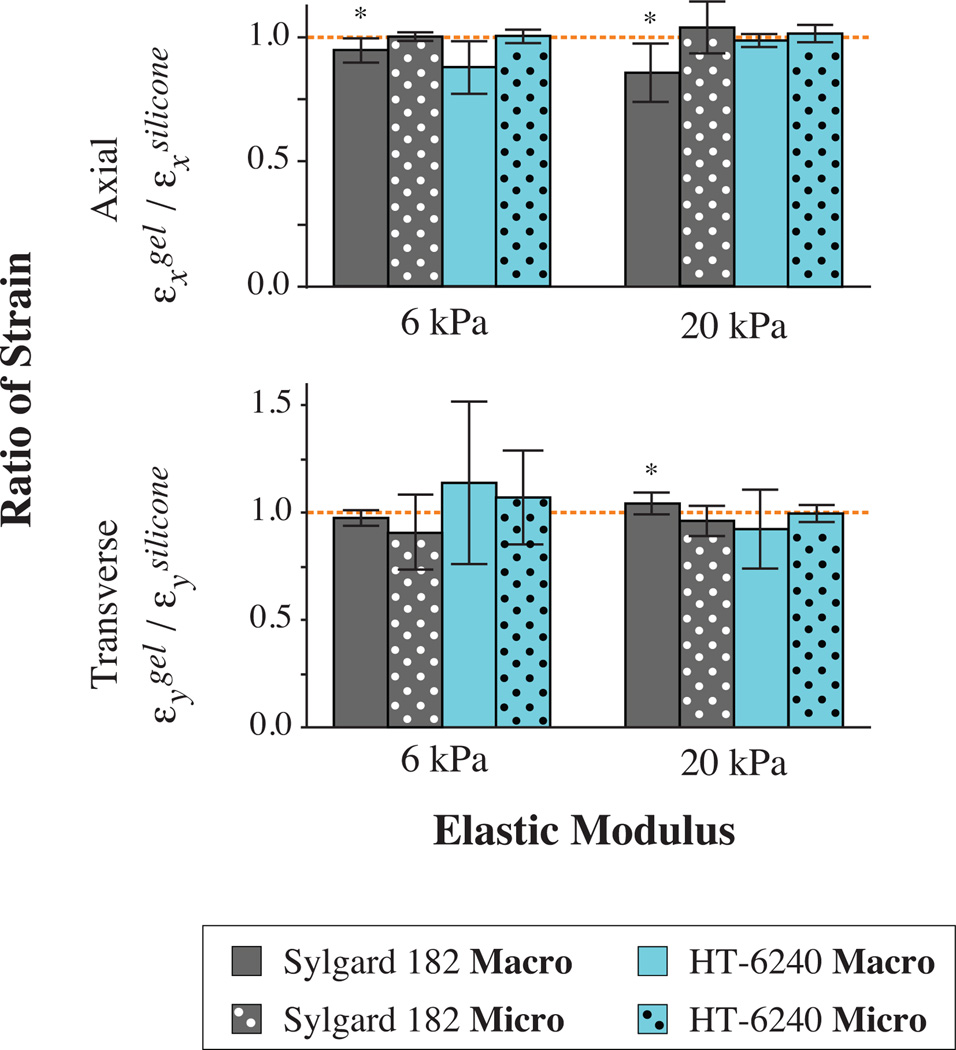
Commercially available silicone sheets and fabricated PDMS membranes were both used successfully to generate stretchable devices. For both types of silicone, strong bonds to the hydrogels resisted up to 50% strain during macroscale calibration (Fig. 3). We were able to fabricate composite structures with gels of different stiffnesses exhibiting elastic moduli E0.05% = 5.9 ± 0.2 kPa and E0.05% = 20.1 ± 0.8 (mean ± s.d., n = 24–25). The level of gel crosslinking, which affects stiffness, does not seem to impact the device functionality in the range of tested strains (Fig. 3).
Figure 3.
Characterization of strain transfer from silicone to gel surface demonstrates minimal loss. Consistent strain transfer was observed for commercially available silicone membranes (HT-6240) in both macroscale and microscale tests. While small but statistically significant losses were observed for macroscale strain of custom-made membranes (Sylgard 182), microscale calibration shows complete strain transfer in both the axial and transverse directions. (*p < 0.05 for one-sided t- test against hypothesized 100% strain transfer depicted as dashed line)
Applied strain is transmitted to the gel with minimal loss
While modulating hydrogel stiffness is important, system functionality depends on the transfer of strain from the silicone handle to the top surface of the gel upon which the cells are cultured. Macroscale evaluation of strain propagated from the silicone handle layer to the gel reveals minimal loss (Fig. 3). For assemblies made with HT-6240 silicone sheets, no significant losses were detected (n = 5). Variation in measurements seems to originate from edge effects at the outer boundary of the gel since microscale strain analysis in the interior of the gel indicates undetectable losses at both stiffnesses (Fig. 3, n = 7).
Macroscale analysis of the strain transfer from Sylgard 182-based fabricated membranes to gels revealed small but statistically significant strain transfer losses in the lateral direction of 5.5 ± 4.9% and 13.9 ± 10.9% for soft and stiff gels, respectively (n = 5). Thinner gels may mitigate this strain loss; however, we found the volume of prepolymer solution used necessary to ensure uniform gel polymerization. Other initiator ratios and coverslip setups may be tested in the future if thinner gels are desired. Furthermore, microscale calibration of fabricated membrane assemblies reveals consistent strain transfer for both stiffnesses (Fig. 3, n = 12–14). This suggests that there are more prominent edge effects for gels bonded to Sylgard 182 membranes but that these edge effects minimally affect strain transfer in the interior of the assembly.
We confirmed this finding with a simple two-dimensional finite element model in Abaqus (Dassault Systèmes, Waltham, MA). When applying 10% strain to the silicone handle (solid homogenous isotropic linear elastic, E = 1 MPa, ν = 0.45), the strain transfer to the gel (solid homogenous isotropic linear elastic, E = 6 kPa or 20 kPa, ν = 0.45) was around 95%. Thus, strain at the surface of the gel interior can be estimated as the strain applied to the silicone handle for the two silicone formulations and two tested gel stiffnesses.
Strain is transmitted to cells with minimal loss for various cell type and matrix protein combinations
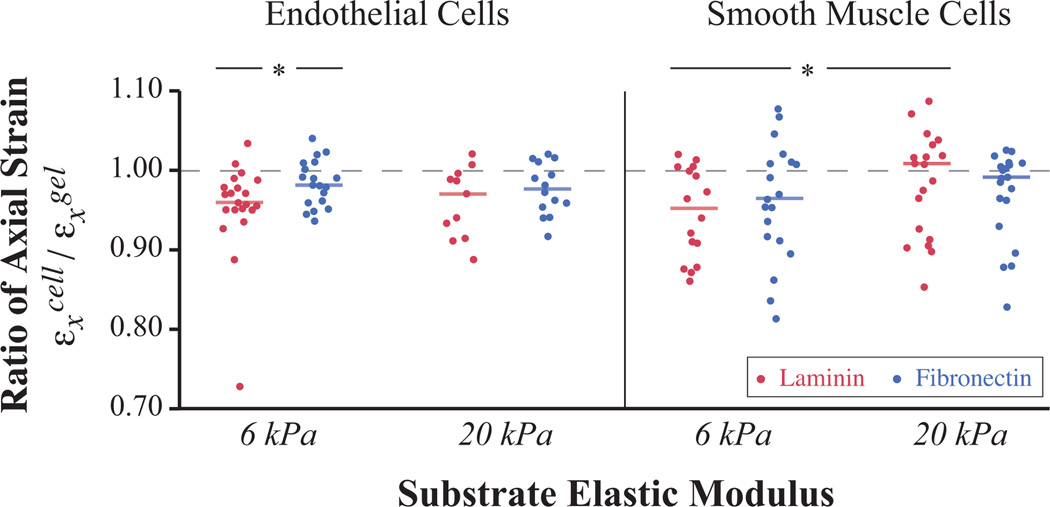
While past studies have used applied strain to test the effect of a dynamic environment on cells, it is often unclear how strain is transferred to cells in the specific culture system utilized. To assess the strain transfer for our specific system, we quantified the amount of strain experienced within a monolayer of vascular cells, both ECs and SMCs, cultured on the device’s hydrogel surfaces. To characterize the effect of matrix protein coatings, we tested two different proteins, laminin and fibronectin, covalently crosslinked to the gel. Strain transfer from the surface of the gels to cultured vascular cells averaged 96% for all combinations tested. Slight differences could be detected for certain protein, stiffness, and cell type combinations (Fig. 4). Seven of the eight combinations tested, with SMCs on 20 kPa laminin-coated gels being the exception, showed significant but small (< 5%) losses in strain transfer (p <= 0.01 for one-sided t-test hypothesizing less than 100% strain transfer).
Figure 4.
Strain transfer from gels to adherent cells shows statistically significant but minimal losses. Graph depicts strain transfer in axial direction with each horizontal bar representing the mean and the dashed line indicating 100% strain transfer (n > 10 each condition). Attached cells exhibited significant strain losses up to 5% when tested against a hypothesized 100% strain transfer (p < 0.01 for one-sided t-test on all populations except SMCs on 20 kPa laminin-coated gels where p = 0.3). SMCs, on average, exhibited less strain transfer when attached through laminin to soft gels than stiff gels, and ECs exhibited less strain transfer when attached to soft gels with laminin coating than with fibronectin (*p < 0.1 for two-sided t-test).
Among the eight groups, two significant differences in distribution were detected. SMCs showed less strain transfer when attached through laminin to 6 kPa gels than to 20 kPa gels (p = 0.090 for two-sided t-test), and ECs exhibited less strain transfer when attached to 6 kPa gels with laminin coating than with fibronectin (p = 0.073 for two-sided t-test).
We suspect that values greater than one arise from error in the cross-correlation algorithm. As the cells are stretched, their volume is conserved and the cells thin in the vertical z-direction. This changes the optical properties of the monolayer and thus the light microscopy images captured.
With the presented substrate and cell culture system, transfer of strain to cells is accurate for gels of different tested stiffnesses. Our system can be used in new research moving forward to better understand how applied strain and stiffness affect biological results.
Conclusions
This work demonstrates a cell culture system that transmits applied strain from a silicone handle layer through variable gel stiffnesses to adherent cells. The chemical functionalization process described strongly binds gels to the surface of two different silicone formulations. At the microscale level of the cell, strain is transmitted robustly from the silicone surface to the gel surface with undetectable loss.
Furthermore, we have shown that the applied strain is transferred from the gel surface through matrix proteins to both endothelial and smooth muscle cells with high efficiency. This is critical to successful future investigations into cell mechanics and mechanotransduction, as we describe below.
Future outlook
The enabling innovation crucial to the developed system is the strong attachment of soft hydrogels to a robust silicone handle layer. The silicone layer allows researchers to easily apply external strain using a custom or commercial micromanipulator while culturing cells on soft substrates of interest. Additionally, the versatile silicone layer can be reversibly or permanently bonded into other systems made of silicone,31 rigid plastics32, 33 or glass,34 increasing the variety and complexity of mechanotransduction experiments possible.
Furthermore, the strong adhesion of polyacrylamide-based gels to the silicone surface suggests this technique may enable additional hydrogel studies. Evidence that ligand density35 and surface charge36, 37 may also affect cell behavior can be concurrently investigated alongside stiffness and strain using this broadly enabling technology.
Supplementary Material
Acknowledgements
The authors thank Gadryn Higgs for his help with AFM measurements and Ngan Huang for her gift of ECs. We also wish to acknowledge the funding support of NSF EFRI MIKS-1136790, NIH R33 HL089027, and NIH R01 EB006745-01A1. C.S. recognizes individual support from NSF GRFP, Stanford VPGE DARE Fellowship, and Stanford CVI Smittcamp Fellowship.
Footnotes
† Electronic Supplementary Information (ESI) available describing detailed characterization methods.
Notes and references
- 1.Brown TD. Journal of Biomechanics. 2000;33:3–14. doi: 10.1016/s0021-9290(99)00177-3. [DOI] [PubMed] [Google Scholar]
- 2.Lim C, Zhou E, Quek S. Journal of Biomechanics. 2006;39:195–216. doi: 10.1016/j.jbiomech.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 3.Simmons CS, Petzold BC, Pruitt BL. Lab Chip. 2012;12:3235–3248. doi: 10.1039/c2lc40308k. [DOI] [PubMed] [Google Scholar]
- 4.Estes B, Gimble J, Guilak F. Current Topics in Developmental Biology. 2004;60:91–126. doi: 10.1016/S0070-2153(04)60004-4. [DOI] [PubMed] [Google Scholar]
- 5.Gupta V, Grande-Allen KJ. Cardiovascular research. 2006;72:375–383. doi: 10.1016/j.cardiores.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 6.Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE. Science. 2010;328:1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Discher D, Janmey P, Wang Y. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 8.Pelham R, Jr, Wang Y. Proceedings of the National Academy of Sciences. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Califano JP, Reinhart-King CA. Journal of Biomechanics. 2010;43:79–86. doi: 10.1016/j.jbiomech.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 10.Jacot JG, McCulloch AD, Omens JH. Biophysical Journal. 2008;95:3479–3487. doi: 10.1529/biophysj.107.124545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engler AJ, Carag-Krieger C, Johnson CP, Raab M, Tang H-Y, Speicher DW, Sanger JW, Sanger JM, Discher DE. Journal of cell science. 2008;121:3794–3802. doi: 10.1242/jcs.029678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacot GJ, Kita-Matsuo H, Wei KA, Chen HSV, Omens JH, Mercola M, McCulloch AD. Ann N Y Acad Sci, 2010. 1188:121–127. doi: 10.1111/j.1749-6632.2009.05091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engler A, Sen S, Sweeney H, Discher D. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 14.Pan CQ, Sudol M, Sheetz M, Low BC. Cellular signalling. 2012;24:2143–2165. doi: 10.1016/j.cellsig.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 15.Ehrlich PJ, Lanyon LE. Osteoporos Int. 2002;13:688–700. doi: 10.1007/s001980200095. [DOI] [PubMed] [Google Scholar]
- 16.Rottner K, Hall A, Small JV. Curr Biol. 1999;9:640–648. doi: 10.1016/s0960-9822(99)80286-3. [DOI] [PubMed] [Google Scholar]
- 17.Throm Quinlan AM, Sierad LN, Capulli AK, Firstenberg LE, Billiar KL. PLoS ONE. 2011;6:e23272. doi: 10.1371/journal.pone.0023272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kandow CE, Georges PC, Janmey PA, Beningo KA. Methods Cell Biol. 2007;83:29–46. doi: 10.1016/S0091-679X(07)83002-0. [DOI] [PubMed] [Google Scholar]
- 19.Nemir S, West J. Ann Biomed Eng. 2010;38:2–20. doi: 10.1007/s10439-009-9811-1. [DOI] [PubMed] [Google Scholar]
- 20.Makamba H, Kim JH, Lim K, Park N, Hahn JH. Electrophoresis. 2003;24:3607–3619. doi: 10.1002/elps.200305627. [DOI] [PubMed] [Google Scholar]
- 21.Hohenester E, Engel J. Matrix Biol. 2002;21:115–128. doi: 10.1016/s0945-053x(01)00191-3. [DOI] [PubMed] [Google Scholar]
- 22.Clyman RI, McDonald KA, Kramer RH. Circ Res. 1990;67:175–186. doi: 10.1161/01.res.67.1.175. [DOI] [PubMed] [Google Scholar]
- 23.Schnaper HWH, Kleinman HKH, Grant DSD. Kidney Int. 1993;43:20–25. doi: 10.1038/ki.1993.5. [DOI] [PubMed] [Google Scholar]
- 24.Jaffe EAE, Mosher DFD. J Exp Med. 1978;147:1779– 1791. doi: 10.1084/jem.147.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grant DSD, Kleinman HKH. Conserv Genet. 1997;79:317–333. [Google Scholar]
- 26.Wang Y, Lai H, Bachman M, Sims C, Li G, Allbritton N. Anal Chem. 2005;77:7539–7546. doi: 10.1021/ac0509915. [DOI] [PubMed] [Google Scholar]
- 27.Oliver W, Pharr G. Journal of materials research. 1992;7:1564–1583. [Google Scholar]
- 28.Carrillo F, Gupta S, Balooch M, Marshall S, Marshall G, Pruitt L, Puttlitz C. Journal of materials research. 2005;20:2820–2830. [Google Scholar]
- 29.Thielicke W, Stamhuis E. doi: 10.1088/1748-3190/aad5a3. www.pivlab.blogspot.com. [DOI] [PubMed]
- 30.Ades EWE, Candal FJF, Swerlick RAR, George VGV, Summers SS, Bosse DCD, Lawley TJT. J Invest Dermatol. 1992;99:683–690. doi: 10.1111/1523-1747.ep12613748. [DOI] [PubMed] [Google Scholar]
- 31.Eddings MA, Johnson MA, Gale BK. Journal of Micromechanics and Microengineering. 2008;18:067001. [Google Scholar]
- 32.Mehta G, Lee J, Cha W, Tung Y-C, Linderman JJ, Takayama S. Analytical Chemistry. 2009;81:3714–3722. doi: 10.1021/ac802178u. [DOI] [PubMed] [Google Scholar]
- 33.Lee KS, Ram RJ. Lab Chip. 2009;9:1618–1624. doi: 10.1039/b820924c. [DOI] [PubMed] [Google Scholar]
- 34.Beh CW, Zhou W, Wang T-H. Lab Chip. 2012;12:4120– 4127. doi: 10.1039/c2lc40315c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trappmann B, Gautrot JE, Connelly JT, Strange DGT, Li Y, Oyen ML, Stuart MAC, Boehm H, Li B, Vogel V, Spatz JP, Watt FM, Huck WTS. Nature Materials. 2012;11:1–8. doi: 10.1038/nmat3339. [DOI] [PubMed] [Google Scholar]
- 36.Lu HH, Guo LL, Kawazoe NN, Tateishi TT, Chen GG. J Biomater Sci Polym Ed. 2009;20:577–589. doi: 10.1163/156856209X426402. [DOI] [PubMed] [Google Scholar]
- 37.Akbar N, Mohamed T, Whitehead D, Azzawi M. Biotechnol Appl Biochem. 2011;58:353–362. doi: 10.1002/bab.46. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.