SUMMARY
Approximately 15–30% of women diagnosed with ductal carcinoma in situ (DCIS) develop a subsequent tumor event within 10 years after surgical lumpectomy. To date, little is known about the molecular pathways that confer this differential risk for developing subsequent disease. In this study, we demonstrate that expression of biomarkers indicative of an abrogated response to cellular stress predicts DCIS with worse outcome and is a defining characteristic of basal-like invasive tumors. Mechanistic studies identify the Rb pathway as a key regulator of this response. Conversely, biomarkers indicative of an intact response to cellular stress are strongly associated with a disease-free prognosis. Assessment of these biomarkers in DCIS begins to allow prediction of tumor formation years before it actually occurs.
SIGNIFICANCE
Our ability to determine future tumor formation in women diagnosed with ductal carcinoma in situ (DCIS) is currently limited. Here we describe distinct subsets of molecular markers that identify women that have an increased risk or decreased risk of developing subsequent tumor events after diagnosis of DCIS. The markers for increased risk also characterize a subset of invasive tumors known as the “basal-like” subtype and provide a biological rationale for the aggressive malignant phenotypes associated with this subclassification of tumors. This information could be used in the clinic to determine which women should receive more or less aggressive therapy.
INTRODUCTION
Of the 62,000 women diagnosed with pure DCIS in 2006, only ~15–30% will develop a subsequent tumor event within the first decade after undergoing lumpectomy (Kerlikowske et al., 2003). In the absence of robust predictors for subsequent invasive disease, currently, women diagnosed with DCIS tend to be offered similar treatment options. A dwindling number of women elect to have complete mastectomies (~25%), while the majority of women opt for a lumpectomy with or without adjuvant treatments such as radiation, hormones or chemotherapy (~70%). A few women choose “watchful waiting” (~ 3–4%). Since the majority of DCIS lesions are not associated with subsequent invasive tumors, it is likely that many women diagnosed with DCIS who opt for surgery, with or without adjuvant treatment, are being over treated. Conversely, since even with this therapy some initially observed DCIS lesions are followed by subsequent invasive carcinomas, some women may be under treated. Identification of characteristics of DCIS and biomarkers that predict subsequent tumor development would allow us to stratify a women’s individual risk for subsequent invasive tumors and avoid over- and under-treatment of women with DCIS.
Historically, DCIS lesions are characterized by clinicopathological variables which are combined in several ways to provide classification systems (Van Nuys classification, Nottingham classification, etc.). An extensive effort over the last four decades has tried to identify additional clinicopathological variables (and more recently molecular markers) that may be important in predicting women who will develop subsequent tumors. Of these variables, several are routinely assessed in the clinic (ie size, nuclear grade, surgical margins, etc) and have been found to have predictive value. However, none have proven strong enough to fully support choices for intervention (hazards ratios ~2–5) (Bijker et al., 2006; Eusebi et al., 1994; Fisher et al., 1999; Kerlikowske et al., 2003).
Recent studies have focused on the differential expression of molecular markers to address this important clinical problem. Expression profiling and immunohistochemical studies confirm the presence of molecular subtypes in DCIS (Adeyinka et al., 2002; Bryan et al., 2006; Hannemann et al., 2006; Livasy et al., 2007) which may parallel the distinct molecular subtypes observed in invasive tumors. Features of molecular subtypes that can distinguish those pre-malignant lesions that will be followed by a subsequent tumor event from those that will not are currently lacking.
Our goal has been to evaluate molecular characteristics and their association with outcome in a cohort of women diagnosed with DCIS. In this translational study we tested the hypothesis that cells activated for stress-induced senescence would not form subsequent tumors while cells that bypass senescent signals could progress to tumor formation.
RESULTS
Determination of molecular markers associated with risk for subsequent tumor events in women diagnosed with DCIS
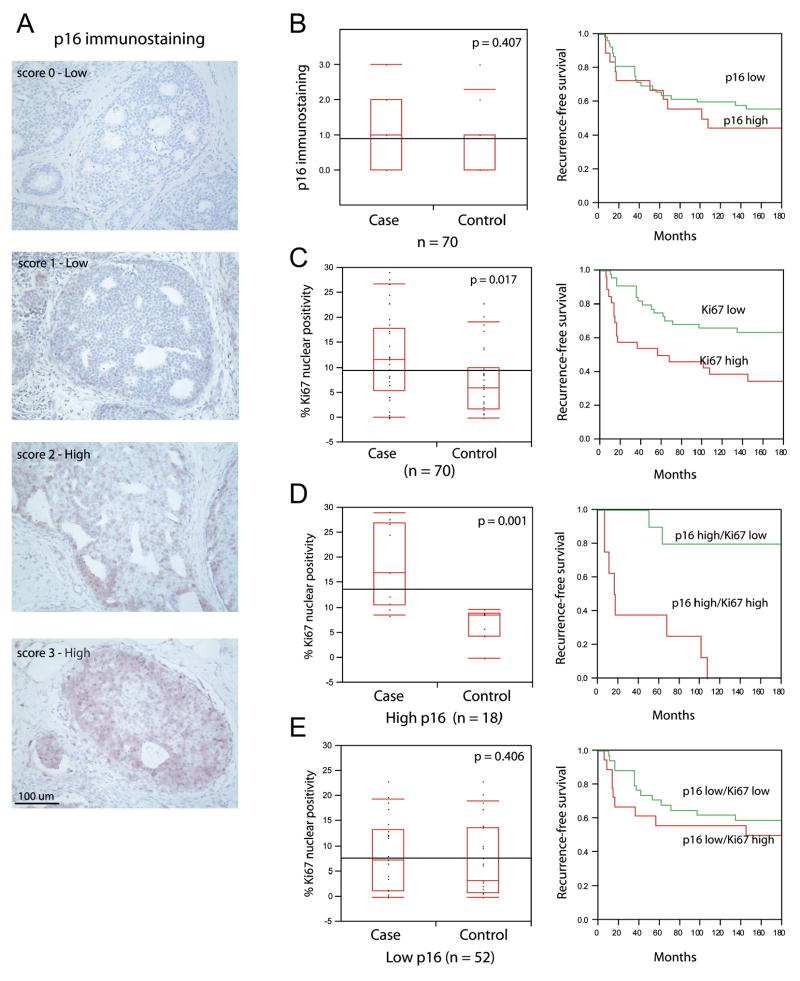
A well-recognized barrier to carcinogenesis is the induction of a senescent cellular response. To determine if this tumor suppressor pathway in DCIS lesions provides mechanistic insight about subsequent tumor events, we examined p16 expression in samples of women without subsequent disease (38 controls) matched to a sample of women with subsequent disease (32 cases), in a DCIS case-control study sufficiently powered to explore pathways that may provide risk stratification. A subsequent tumor event (recurrence) was defined as a subsequent DCIS lesion or invasive cancer lesion diagnosed in the ipsilateral breast or at a distant site at least 6 months following the initial diagnosis of DCIS. Representative p16 staining is illustrated in Figure 1A. We find 26% (18/70) of DCIS lesions show high p16 staining (Table 1). This p16 immunopositivity is not associated with any clinicopathological variables such as nuclear grade or hormone receptor status (Supplemental Table 1, 2). We would predict that high p16 expression induces a cellular growth arrest, and thus DCIS lesions overexpressing p16 would be less likely to precede subsequent disease. This prediction was not substantiated. We find that high p16 expression, as a univariate marker, does not significantly stratify a woman’s risk for developing a subsequent tumor event (DCIS and invasive cancer combined; HR=1.1, 95% CI, 0.5 to 2.5; Figure 1B, Table 1).
Figure 1. p16 overexpression coupled with proliferation increases the risk of subsequent tumor events among women with DCIS.
A) Representative p16 immunohistochemistry.
B) High p16 staining fails to stratify women with DCIS that develop subsequent disease. Recurrence-free survival plots demonstrate that women with DCIS staining high or low for p16 develop subsequent disease at the same rate.
C) Ki67 index labelling stratifies recurrence-free survival in women with DCIS.
D) DCIS exhibiting high p16 immunostaining and elevated Ki67 identifies women that have a reduced recurrence-free survival.
E) Ki67 does not differentiate risk in DCIS lesions with low p16. Box plots and corresponding p-values were determined using Wilcoxon/Kruskal-Wallis rank of sums test. Survival plots were generated using Kaplan-Meyer analysis.
Table 1.
p16 and COX-2 expression coupled with proliferation increases the risk of developing subsequent disease among women with DCIS.
| Marker | n | Control | Case | HR (95% CI) |
|---|---|---|---|---|
| p16 | 70 | |||
| High | 24% (9/38) | 28% (9/32) | 0.17 (0.02,1.4) | |
| Low | 76% (29/38) | 72% (23/32) | ||
| COX-2 | 70 | |||
| High | 58% (22/38) | 53% (17/32) | 0.79 (0.4, 1.7) | |
| Low | 42% (16/38) | 47% (15/32) | ||
| Ki67 | 70 | |||
| High | 24% (9/38) | 53% (17/34) | 2.7 (1.2, 5.9) | |
| Low | 76% (29/38) | 47% (15/32) | ||
| p16 High | 18 | |||
| Ki67 High | 0 % (0/9) | 89% (8/9) | 15.1 (1.4, 161.4) | |
| K67 Low | 100% (9/9) | 11% (1/9) | ||
| p16 Low | 52 | |||
| Ki67 High | 31% (9/29) | 39% (9/23) | 1.1 (0.4, 3.4) | |
| Ki67 Low | 69% (20/29) | 61% (14/23) | ||
| COX-2 High | 39 | |||
| Ki67 High | 23% (5/22) | 76% (13/17) | 4.8 (0.8, 27.5) | |
| K67 Low | 77% (17/22) | 24% (4/17) | ||
| COX-2 Low | 31 | |||
| Ki67 High | 25% (4/16) | 27% (4/15) | 0.86 (0.2, 3.3) | |
| Ki67 Low | 75% (12/16) | 73% (11/15) | ||
| Marker | n | Control | Case | HR (95% CI) |
| p16 | 70 | |||
| High | 24% (9/38) | 28% (9/32) | 0.17 (0.02,1.4) | |
| Low | 76% (29/38) | 72% (23/32) | ||
| COX-2 | 70 | |||
| High | 58% (22/38) | 53% (17/32) | 0.79 (0.4, 1.7) | |
| Low | 42% (16/38) | 47% (15/32) | ||
| Ki67 | 70 | |||
| High | 24% (9/38) | 53% (17/34) | 2.7 (1.2, 5.9) | |
| Low | 76% (29/38) | 47% (15/32) | ||
| p16 High | 18 | |||
| Ki67 High | 0 % (0/9) | 89% (8/9) | 15.1 (1.4, 161.4) | |
| K67 Low | 100% (9/9) | 11% (1/9) | ||
| p16 Low | 52 | |||
| Ki67 High | 31% (9/29) | 39% (9/23) | 1.1 (0.4, 3.4) | |
| Ki67 Low | 69% (20/29) | 61% (14/23) | ||
| COX-2 High | 39 | |||
| Ki67 High | 23% (5/22) | 76% (13/17) | 4.8 (0.8, 27.5) | |
| K67 Low | 77% (17/22) | 24% (4/17) | ||
| COX-2 Low | 31 | |||
| Ki67 High | 25% (4/16) | 27% (4/15) | 0.86 (0.2, 3.3) | |
| Ki67 Low | 75% (12/16) | 73% (11/15) |
Paradoxically, overexpression of p16 can represent two different biological processes; a response to cellular stress or abrogation of functional Rb signaling (Bates et al., 1994; Parry et al., 1995; Serrano et al., 1993). A cell with functional p16/Rb signaling will initiate stress-induced overexpression of p16 resulting in a proliferative arrest characteristic of cellular senescence. On the other hand, a cell with a compromised Rb pathway will initiate a regulatory-induced overexpression of p16, due to unobstructed negative feedback regulation and disregard the many stress signals that induce senescence and cellular arrest. These later cells proliferate unimpeded and bypass senescence.
To distinguish between these two opposing phenotypes, we also stained serial sections for a proliferation marker, Ki67. Thirty-seven percent (26/70) and 63% (44/70) of the lesions within this case-control study express high and low Ki67, respectively (Table 1). High Ki67 alone provides a modest stratification for women that develop a subsequent tumor (DCIS and invasive combined; HR 2.7, 95% CI 1.2 to 5.9; Figure 1C and Table 1). Interestingly, almost half (8/18) of DCIS lesions exhibiting high Ki67 index labelling also show high p16 levels (Table 1). We determined if this phenotype, representing deregulated p16/Rb signaling, identifies DCIS associated with subsequent tumor events. We find that all women with DCIS expressing high p16 and high Ki67 develop a subsequent tumor (Table 1) and thus Ki67 stratifies high p16 expression into two groups: those who develop a subsequent breast cancer (case) versus those that do not (control; HR = 15.1, 95% CI, 1.4 to 161.4; Figure 1D). The tumors that develop following DCIS with high p16/high Ki67 are often invasive breast cancer (5 of 8; Supplemental Figure 1). The remaining DCIS lesions showing high p16 immunopositivity (9/18) exhibited low Ki67 index labelling, suggesting these cells have maintained p16/Rb checkpoint regulation. Indeed, most (9 of 10) lesions expressing a high p16/low Ki67 phenotype were not associated with a subsequent tumor event and thus may represent a protective signature (Supplemental Figure 1). Women with DCIS that exhibit low p16 irrespective of Ki67 are not likely to develop recurrent disease (Figure 1E; HR = 1.1, 95% CI, 0.4 to 3.4; Table 1).
High p16 mRNA levels defines the basal-like subtype of invasive tumors
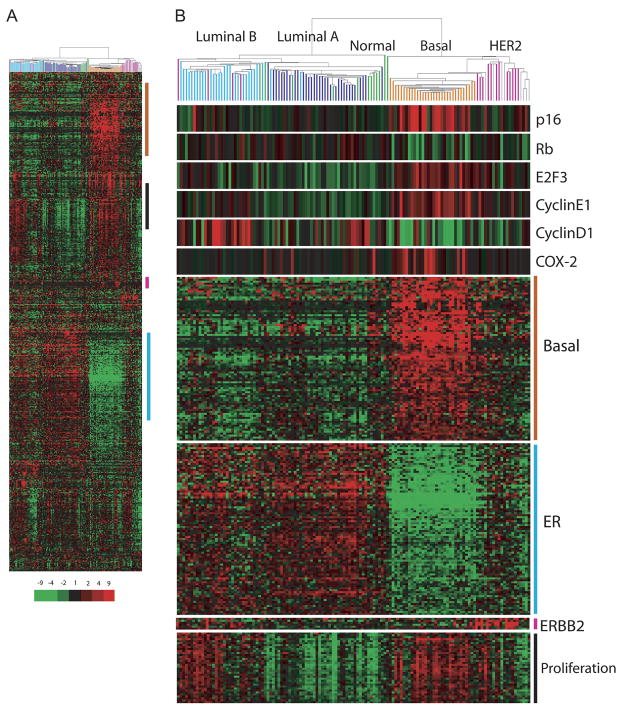
Previous reports have demonstrated that ipsilateral tumors that develop subsequent to DCIS share many histological and genetic alterations with the primary lesion suggesting a clonal relationship (Bijker et al., 2001; Lininger et al., 1998; Millis et al., 2004; Waldman et al., 2000). Therefore, one might anticipate that high p16/high Ki67 in the initial DCIS lesion would characterize the subsequent invasive carcinoma. To test this prediction, we explored the prevalence and subtype distribution of p16 and proliferation by evaluating a previously published dataset of gene expression profiles of 130 primary invasive breast tumors (Chin et al., 2006). Unsupervised clustering with a set of intrinsically variable genes identifies previously reported subtypes (Perou et al., 2000; Solie et al., 2003; Figure 2A, Supplemental Table 3). Molecular subtypes were additionally classified based upon a nearest centroid approach using molecular subtype training data as previously defined (see Supplemental File 1; (Hu et al., 2006).
Figure 2. High p16 mRNA defines the basal-like subtype of invasive tumors.
A) Unsupervised hierarchical clustering of 130 invasive tumors using 589 unique genes that are intrinsically variable (see Supplemental File 1) identifies luminal A, luminal B, normal-like, HER2 positive and basal-like clusters. Molecular subtypes, additionally classified by a nearest centroid approach are indicated by coloured bars (light blue: Luminal B; dark blue: Luminal B; green: Normal-like; orange: Basal-like; pink: HER2 positive tumors; grey: Unclassified).
B) Expression levels of p16, Rb, E2F3, cyclin E1, cyclin D1 and COX-2. The level of expression of each gene is relative to the median across all samples as represented by color saturation.
We find increased p16 mRNA expression preferentially characterizes the highly proliferative basal-like tumor subtype (Figure 2B). p16 expression falls within a gene cluster comprised of many well-established basal-like genes, such as keratin 5, 17, SFRP5, and MMP-7 (Supplemental Table 3). To determine if the increased expression of p16 in actively proliferating cells is a consequence of p16/Rb pathway deregulation, we examined the expression levels of Rb, E2F3, cyclin E and cyclin D1. We find the basal-like sample cluster tends to display relatively low levels of Rb and high levels of E2F3 (Figure 2B). Since E2F3 transcriptional upregulation is a consequence of Rb inactivation (Leone et al., 2000), we propose that deregulation of Rb signaling is a characteristic feature of basal-like tumors. Loss of Rb signaling can occur as a consequence of genetic alterations that has been observed to account for approximately 35% of breast tumors (Nielsen et al., 1997; Reis-Filho et al., 2006). Alternatively, Rb can be negatively regulated through phosphorylation by cyclin-dependent kinases. In examining the transcript levels of cyclin E and cyclin D1 among all tumor subtypes, we observe that cyclin E levels are among the highest and cyclin D1 among the lowest in basal-like tumors (Figure 2B). In concordance with previous observations (Loden et al., 2002; Reis-Filho et al., 2006), we find increased cyclin D1 levels to be most consistently elevated in the luminal B subtype. Although deregulation of p16/Rb signaling regulates genes involved in cell cycle progression, altered transcript levels of members of this pathway does not simply reflect proliferating tumor cells since highly proliferating Luminal B tumors do not exhibit the same transcriptional pattern.
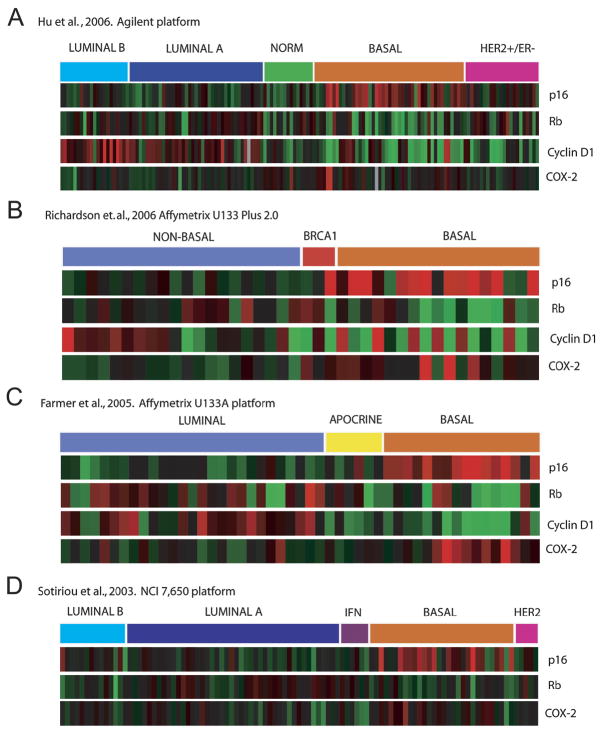
To confirm the reproducibility of the observed differential subtype specificity of p16/Rb/Cyclin D1, we analyzed gene expression levels in four publicly available datasets from three different platforms (Figure 3). In each case, tumors with overexpression of p16 and low transcript levels of both Rb and cyclinD1 were consistently found to be classified as basal-like tumors. Taken together, the observation that Rb transcript levels are among the lowest in basal-like tumors, and that E2F3 and cyclin E are among the highest suggests that loss of functional p16/Rb signaling may play a defining role in the biology of this tumor subtype.
Figure 3. High p16 and COX-2 mRNA levels are enriched in basal-like tumors across multiple microarray platforms.
mRNA expression for p16, Rb, cyclin D1 and COX-2 in the indicated four publicly available datasets utilizing different microarray platforms. Subtype classifications are as determined previously.
High COX-2 mRNA levels are enriched in basal-like tumors
To further analyze p16 and explore gene expression interactions, hierarchical clustering was performed on the top 6000 variable genes in 130 tumors (for details and data see Supplemental File 1, Supplemental Figure 2, and Supplemental Table 4). As expected, members of the p16/Rb/cyclinD1 pathway showed variable expression, along with many members of the E2F family. We find that COX-2 is a member of the basal-like gene cluster. It is well recognized that high levels of expression of many basal-like genes are also found in normal-like invasive tumors (Hu et al., 2006; Perou et al., 2000; Sorlie et al., 2003). Similarly, we find the 18% of tumors with elevated COX-2 mRNA are restricted to basal-like and normal-like subtypes. While 50% (16/32) of basal-like and 33% (4/12) of normal-like tumors overexpress COX-2, virtually all luminal and HER2 positive tumors express COX-2 mRNA levels below the median. The prevalence of COX-2 overexpression in basal and normal-like tumors is confirmed in independent data sets (Figure 3).
Concordance between mRNA and protein expression of p16 and COX-2 in tumors
The low levels of COX-2 mRNA expression in HER2 positive tumors is perplexing because previous studies had demonstrated that COX-2 protein levels are enriched in HER2 amplified tumors (Boland et al., 2004; Cho et al., 2006; Ristimaki et al., 2002). Notably, microarray analyses are typically average measurements of numerous cell types that often represent arbitrary units relative to a median value and fail to address post-transcriptional and post-translational regulation. It is therefore, critical to relate thresholds of detection of mRNA by microarray analysis to levels of protein expression as measured by immunohistochemistry (IHC). Further, it is important to determine the contributions of distinct cell types to overall levels of gene expression. We performed p16 and COX-2 IHC on paraffin-embedded tumor blocks representing 54 of the 130 tumors analyzed by microarray. These samples were chosen to represent all 5 molecular subtypes and span a continuum from the lowest to the highest levels of p16 and COX-2 microarray gene expression.
In samples of invasive tumors that showed elevated p16 via microarray analysis, immunopositivity is predominantly found in carcinoma cells. To a lesser extent, heterogeneous foci exhibiting p16 staining are detectable in the morphologically normal epithelial cells. Occasional p16 positivity is also observed in fibroblasts, predominantly those within desmoplastic appearing stroma. Cases with elevated COX-2 show abundant staining within the carcinoma cells as well as in the morphologically normal epithelia. In rare cases, we found intense COX-2 staining in macrophages infiltrating invasive tumors.
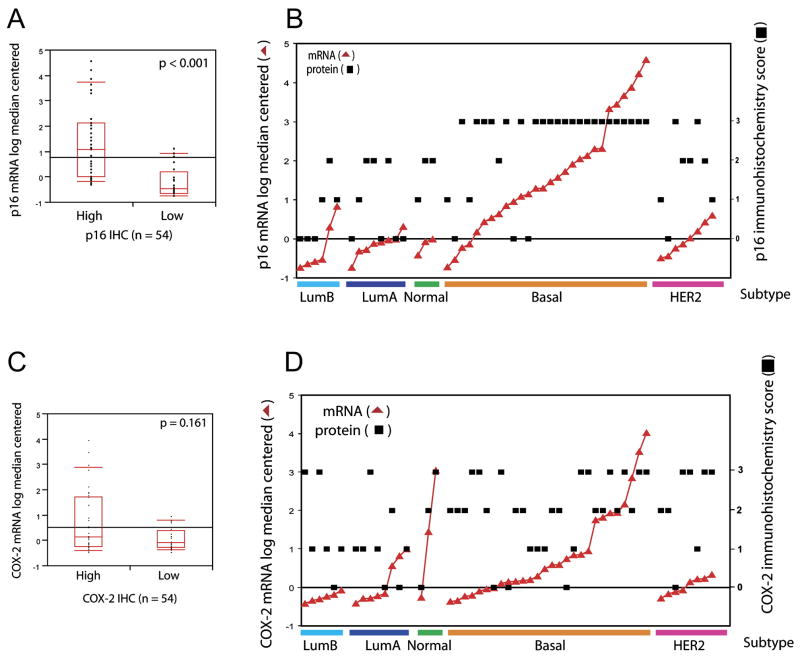
For p16, we observe a significant correlation (Wilcoxon rank of sums, p < 0.0001) between mRNA and protein expression (Figure 4A). Basal tumors that express the highest levels of p16 mRNA showed intense p16 protein staining by IHC (score 3+, Figure 4B). We did observe a fraction of HER2 positive tumors (2/8) that showed intense p16 staining despite low mRNA expression. This concordance between mRNA and protein suggests that p16 protein levels are primarily regulated at the transcriptional level and that p16 protein levels determined by IHC reflect the subtype specificity.
Figure 4. Concordance between p16 or COX-2 mRNA and protein expression in tumors.
A) p16 mRNA and protein levels show concordance using Wilcoxon rank of sums test.
B) p16 mRNA and protein concordance across molecular tumor subtypes. The log median centered mRNA level of p16 is plotted (red triangles) in comparison with the corresponding protein levels (black squares).
C) COX-2 mRNA and protein levels demonstrate poor concordance using Wilcoxon rank of sums test.
D) Discordance between COX-2 mRNA and protein levels is enriched in HER2 positive tumors. The mRNA level of COX-2 for each sample is plotted (red triangles) in comparison with corresponding protein levels (black squares).
In contrast, COX-2 demonstrated poor correlation between mRNA and protein levels (Wilcoxon rank of sums, p = 0.161; Figure 4C). Those cases with the highest levels of COX-2 mRNA, as defined by a greater than 2-fold increase over the median value, exhibited complete concordance and displayed high COX-2 immunoreactivity (Figure 4D). Ten of the 54 invasive tumors show COX-2 mRNA levels greater than 2-fold above the median, eight of these cases are basal and the remaining two are classified as normal-like. In remaining cases (44/54), 10 exhibit low mRNA and were discordant with elevated protein expression as measured by IHC (Figures 4D). We found that 80% of the discordant samples were in the HER2 subtype.
COX-2 overexpression coupled with proliferation is associated with subsequent tumor events among women with DCIS
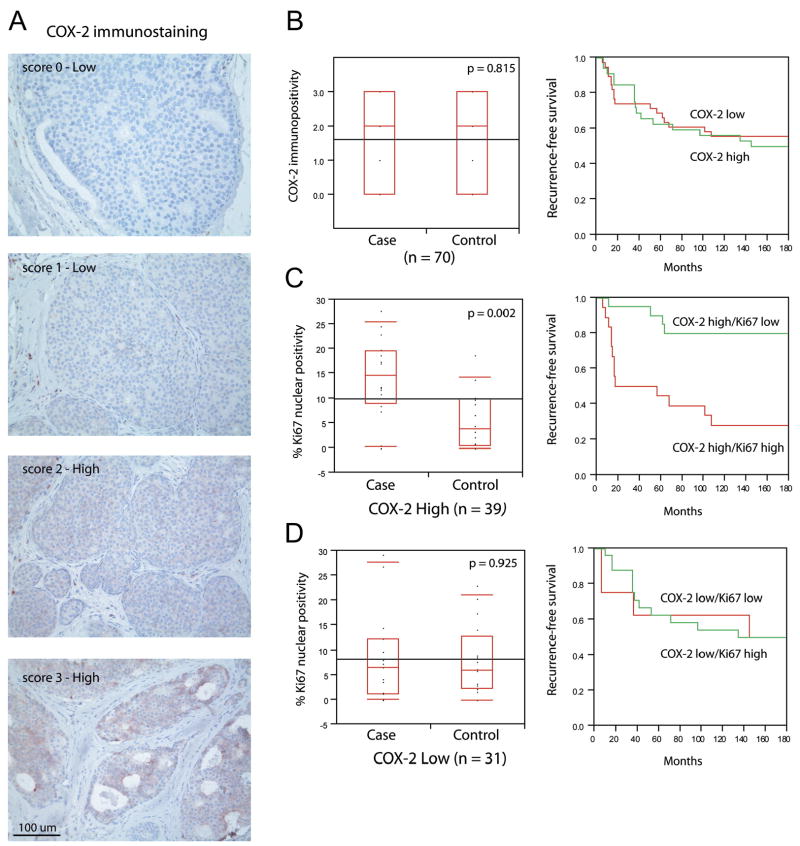
To determine if elevated COX-2 protein expression in DCIS is also associated with subsequent tumor events, COX-2 IHC was performed in serial sections of 70 cases previously analyzed for p16 and Ki67 (representative staining in Figure 5A). We find that 56% (39/70) of DCIS lesions show high COX-2 protein levels (Table 1) which by itself does not stratify risk for subsequent tumor formation and, similar to p16 overexpression by itself, is equally distributed among women that develop subsequent DCIS or invasive cancer (case) and those that do not (control; HR=0.79, 95% CI, 0.4 to 1.7; Table 1; Figure 5B).
Figure 5. COX-2 overexpression coupled with proliferation increases the risk of subsequent tumor events among women with DCIS.
A) Representative COX-2 immunohistochemistry.
B) COX-2 measured as a single variable fails to stratify risk of recurrent disease or recurrence-free survival.
C) DCIS lesions high for COX-2 and Ki67 identify women that develop subsequent breast cancer and have reduced recurrence-free survival.
D) Ki67 does not differentiate risk in DCIS lesions with low COX-2. Box plots and corresponding p-values were determined using Wilcoxon/Kruskal-Wallis rank of sums test. Survival plots were generated using Kaplan-Meyer analysis.
As with p16, stratifying high and low COX-2 DCIS lesions by proliferation identifies those more and less likely to have a subsequent tumor event. A significantly higher fraction of women (13 of 17) with high COX-2/high Ki67 develop a subsequent tumor (Figure 5C; Wilcoxon rank test; p = 0.002) as compared to lesions that show high COX-2/low Ki67 (HR = 4.8, 95% CI, 0.8 to 27.5, Table 1). Correspondingly, we did not observe an increase in subsequent tumor events in women that exhibit low COX-2 expressing DCIS irrespective of Ki67 (Figure 5D; Wilcoxon rank test p = 0.925; HR = 0.86, 95% CI, 0.2 to 3.3; Table 1). In examining the lesions that develop subsequent to high COX-2/high Ki67 DCIS, 7 of 13 cases are invasive breast cancer (Supplemental Figure 1).
Similar to our observation that high p16 in the absence of proliferation identifies a protective signature (Table 1), 81% (17/21) of women with high COX-2/low Ki67 DCIS do not develop a subsequent tumor event (Table 1). Most DCIS lesions (6/7) expressing both high p16 and high COX-2 in the absence of proliferation are not associated with subsequent disease (Supplemental Figure 1). These observations suggest high COX-2 and/or high p16 mark two clinically different populations of cells in DCIS that can be stratified by proliferation.
COX-2 overexpression causes cell cycle arrest in cells that maintain functional p16/Rb signaling
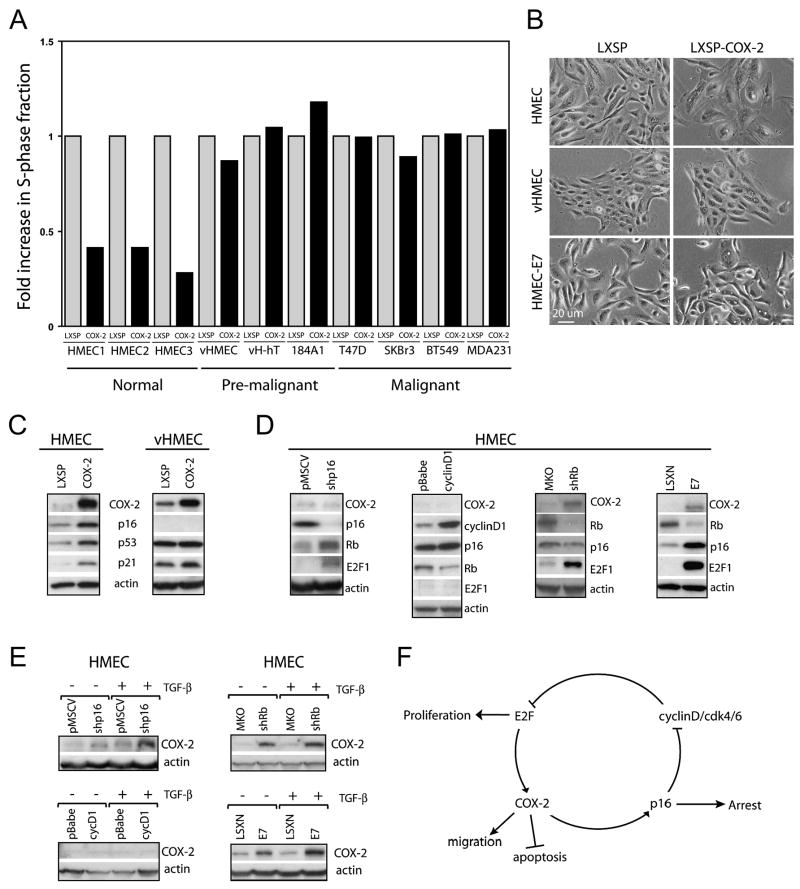
To determine the cellular context that governs if COX-2 is associated with quiescence or proliferation, we investigated the S-phase fraction in normal mammary epithelial cells and in a series of mammary cell lines (both premalignant and malignant), engineered to constitutively overexpress COX-2. Normal human mammary epithelial cells (HMEC) were propagated from disease-free reduction mammoplasty tissue from three different individuals. Premalignant cells studied include a subpopulation of HMEC (variant HMEC or vHMEC) with an extended but finite lifespan (Crawford et al., 2004; Gauthier et al., 2005; McDermott et al., 2006; Romanov et al., 2001), vHMEC-hTert, vHMEC immortalized by stably expressing human telomerase, and 184A1, non-tumorigenic immortalized mammary epithelial cells. In addition, we examined 4 malignant cell lines (T47D, MDA-MB-231, BT549, SkBr3) that have compromised p16/Rb signaling through diverse mechanisms including p16 hypermethylation, p16 deletion, and/or Rb deletion.
In HMEC, sustained COX-2 overexpression significantly reduces the number of cycling cells (Figure 6A) and produces enlarged flattened morphology (Figure 6B). The proliferative arrest phenocopies that observed with overexpression of p16 (data not shown). In contrast, COX-2 overexpression neither induces nor diminishes proliferation in any of premalignant and malignant cells examined (Figure 6A). Functional p16/Rb signaling is one of the distinguishing features of normal cells compared to all other cells we examined. Therefore, we hypothesized that COX-2-induced growth arrest is dependent on functional p16/Rb signaling. Indeed, we find HMEC overexpressing COX-2 exhibited elevated protein levels of p16, p53 and p21 (Figure 6C). This is in contrast to vHMEC, which did not express p16, where overexpression of COX-2 did not alter the level of p53 or p21. Additionally, targeted degradation of Rb, p107 and p130 (all three Rb family members) by HPV16-E7 (human papilloma virus16-E7) in HMEC resulted in ongoing proliferation in the presence of COX-2 overexpression (Figure 6B). Thus, COX-2 overexpression in cells with functional p16/Rb signaling induces a p16-dependent growth arrest, while cells with disrupted p16/Rb signaling continue to proliferate in the presence of COX-2 overexpression.
Figure 6. p16/Rb pathway regulates COX-2 expression and COX-2-dependent cell cycle arrest.
A) Cells of varying malignant potential were retrovirally infected with a constitutive expressing COX-2 construct or an empty vector control (LXSP). Cells were pulsed for 4 h with BrdU and analyzed by flow cytometry following propidium iodide staining. The S-phase fraction is expressed as fold increase of COX-2 expressing cells compared to vector control cells.
B) Phase contrast micrographs of HMEC, vHMEC and HMEC expressing HPV16-E7. Cells either retrovirally infected with a constitutive COX-2 expressing construct or an empty vector control (LXSP).
C) Western blot analysis of COX-2, p16, p53 and p21 in cell lysates from COX-2 overexpressing and vector control cells.
D and E) HMEC were infected with retrovirus expressing a short hairpin RNA targeting p16 (shp16), cyclin D1 (cycD1), a short hairpin RNA targeting Rb (shRb), HPV16-E7, or their corresponding empty vector controls. (D) COX-2, p16, Rb, cyclin D1, E2F1 and actin were detected by western blot. (E) COX-2 protein expression was determined by immunoblotting following 24 h exposure to 1ng/ml TGF-β.
F) Diagram representing the connection between p16/Rb pathway and COX-2.
Deregulation of Rb signaling causes COX-2 overexpression
Our finding that the majority of high p16/high Ki67 DCIS lesions overexpress COX-2 suggests that deregulation of p16/Rb may drive COX-2 expression. To test this hypothesis, we modulated the p16/Rb pathway in HMEC generated from three different reduction mammoplasties using different approaches then determined the effect on basal and induced COX-2 protein levels. Downregulation of p16 protein level in HMEC by infecting them with a virus expressing p16-targeted short hairpin RNA (shp16) lead to upregulation of both Rb and E2F1 as expected. (Figure 6D and (Zhang et al., 2006)). Reducing p16 protein level did not change the basal level of COX-2 expression appreciable but increased TGF-β treatment-induced COX-2 expression (Figure 6E). Overexpression of cyclin D1 alone did not cause hyperphosphorylation of Rb or alter E2F1 protein levels (Figure 6D), consistent with previous findings (Lundberg and Weinberg, 1998). Overexpression of cyclin D1 did not increase either basal or TGF-β treatment-induced COX-2 expression (Figure 6E). Silencing of Rb expression by infecting cells with a virus expressing an established short hairpin RNA against Rb (shRb) (Boehm et al., 2005) resulted in the upregulation of not only E2F1 (Figure 6D) but also both basal and induced levels of COX-2 protein (Figure 6E). Similarly, expression of HPV16-E7 elevated both basal and induced levels of COX-2 protein (Figure 6D,E). These data demonstrate that abrogation of p16/Rb signaling through genetic silencing of p16, Rb and Rb family members sensitizes cells to COX-2 upregulation. Thus, the propensity for COX-2 overexpression in DCIS lesions that exhibit high p16/high Ki67 is most likely a consequence of deregulation of the p16/Rb pathway.
DISCUSSION
In this report, we demonstrate that expression of biomarkers indicative of an abrogated response to cellular stress occurs in DCIS associated with subsequent tumor events and is a defining characteristic of basal-like invasive tumors. Conversely, expression of biomarkers indicative of an intact response to cellular stress and the induction of senescence are strongly associated with DCIS lesions that are not associated with subsequent tumor events. These phenotypes may predict tumor events that would or would not happen up to 10 years in advance. The clinical significance of these biological phenotypes is currently being validated in a large independent cohort of women previously diagnosed with DCIS.
Biological Rationale for Initial Selection of Stress-Associated Biomarkers
Normal cellular responses to stress are important barriers to carcinogenesis and therefore provide molecular candidates to identify lesions that will not progress to malignancy (Bartkova et al., 2005; Campisi, 2005; Gorgoulis et al., 2005; Mooi and Peeper, 2006; Schmitt, 2003). Genotoxic, oxidative, metabolic stress as well as oncogene-driven mitogenic signals all engage cellular stress response programs in normal cells. If the level of damage is substantial, an apoptotic or senescent program limits the propagation of damaged cells. Activated p16 signaling drives premature and replicative senescence. (Lowe and Sherr, 2003; Serrano et al., 1995; Serrano et al., 1996). Through inhibition of cyclin-dependent kinase (cdk4/6), p16 blocks phosphorylation of Rb and thus inhibits E2F targets essential for cell cycle progression (Dyson, 1998; Harbour and Dean, 2000; Lukas et al., 1995; Medema et al., 1995; Narita et al., 2003; Nevins, 1998; Sellers et al., 1995).
Stress Associated Biomarkers are Conditional
Cells that have abrogated apoptotic or senescent pathways may exhibit unimpeded growth despite signals that indicate activation of cellular stress. For example, in cervical cancer, high p16 expression represents inactivation of the Rb pathway through the HPV E6/E7 proteins (Kalof and Cooper, 2006). Therefore, biological markers of stress activation are conditional and can reflect two biologically different processes that can be distinguished by the absence or presence of proliferation. We demonstrate that two stress-activated proteins, p16 and COX-2, identify different groups among women with DCIS.
The first group, high p16 and/or COX-2 in the absence of proliferation, reflects stress activation and a protective anti-proliferative response. These lesions have maintained intact p16/Rb checkpoint regulation and may reflect an initiated senescent program. This DCIS phenotype begins to identify women less likely to have a subsequent tumor event adding support to the hypothesis that the senescent program is a barrier to tumorigenesis (Braig et al., 2005; Chen et al., 2005; Collado and Serrano, 2005; Michaloglou et al., 2005).
The second group, high p16 and/or COX-2 in the presence of ongoing proliferation, reflects an abrogated response to cellular stress. These lesions have lost functional p16/Rb signaling allowing cells to bypass senescence, extend proliferation and drive chromosome instability (Hernando et al., 2004; McDermott et al., 2006; Romanov et al., 2001) ultimately leading to increased risk of malignant conversion. This phenotype identifies women that are more likely to develop a subsequent tumor.
Previously, others have sought to determine if aberrations in G1/S checkpoint regulation could stratify women with DCIS that have an increased risk of developing subsequent disease (Jirstrom et al., 2003). In examining the levels of cyclin D1, cyclin E, p16 and p27 by IHC, only cyclin D1 showed an association with local recurrence. Interestingly, the authors found that cyclin D1 immunopositivity was strongly and inversely associated with developing subsequent disease. Our results are consistent with this observation. The data presented here also support the previous finding that p16, measured as a single variable, fails to identify women at high risk for subsequent tumor development.
COX-2 immunostaining has also been examined as a biomarker for DCIS lesions that progress to malignancy. Recently COX-2 immunopositivity in DCIS has been demonstrated to correlate with disease recurrence when measured as a single variable (Barnes et al., 2006). This apparent discrepancy with our reported results is likely due to the relative over-representation of actively proliferating high grade DCIS lesions in the Barnes et. al. cohort and therefore a relative under-representation of quiescent COX-2-positive lesions. The proportion of high-grade DCIS lesions in Barnes et. al. (60%) is significantly higher than the proportion of high grade DCIS lesions in this study (37%)
Functional Parallels Between p16/Rb and COX-2 activities and their Regulatory Connection
Our studies reveal functional parallels between p16/Rb and COX-2 activities. Similar to p16 induction, cells induce COX-2 expression following DNA damage, oncogenic activity or in response to inflammatory cytokines. Although COX-2 has tumor-promoting effects in cell lines, we show that COX-2 overexpression in finite lifespan primary mammary epithelial cells induces a p16-dependent cell cycle arrest and morphologic changes, analogous to cellular senescence (Figure 6). Cells that have lost p16/Rb checkpoint regulation are refractive to COX-2-induced cell cycle arrest. These cells continue to proliferate in the presence of high COX-2 and manifest COX-2-dependent tumorigenic phenotypes. These data have important consequences because COX-2 has both anti-tumorigenic and pro-tumorigenic properties that are dependent on the genetic composition of the cell.
Underlying the functional parallels between p16 and COX-2 lies a regulatory relationship between abrogation of the p16/Rb pathway and regulation of COX-2. To unmask this link we examined normal non-tumorigenic cells that are not confounded by the genomic alterations in cell lines. In finite lifespan normal epithelial cells, loss of p16 or Rb activity through genetic manipulation leads to COX-2 upregulation (Figure 6). These observations are consistent with previous reports demonstrating increased COX-2 expression and activity in murine prostate epithelial cells with engineered deletion of Rb (Davis et al., 2005) and HPV E6/E7 infected cervical cancer cell lines (Subbaramaiah and Dannenberg, 2007). Although primary mammary epithelial cells with engineered loss of p16/Rb signaling proliferate in the presence of high COX-2, they maintain a finite lifespan and are non-tumorigenic. Finally, this relationship also provides a molecular rationale for why basal-like DCIS lesions are associated with worse outcome. The overexpression of COX-2 in these samples and the consequential acquisition of the malignant phenotypes that it controls, would predict poor prognosis.
Additional Mechanisms to Overexpress COX-2 are Found in the HER-2 Overexpressing Tumor Subtype
In a representative subset of our invasive tumor series, we have directly correlated mRNA expression by microarray to protein expression by IHC within the same sample. In a number of samples, we observe high COX-2 protein expression in the absence of high COX-2 mRNA (Figure 4D). This apparent discordance has a number of possible explanations that are technical (i.e. inconsistent sampling of tumor tissue in microarray samples) and biological (i.e. post-transcriptional or post-translational regulation). Inconsistent sampling of tumor tissue in microarray samples appears an unlikely explanation as all of the samples exhibiting low COX-2 mRNA still showed robust basal-like tumor microarray signatures (Figure 4B,C and data not shown). Furthermore, a number of the samples discordant for COX-2 mRNA and protein levels showed concordant elevations in p16 mRNA and protein levels (data not shown). We favor a biological explanation for the discrepant protein/IHC and mRNA/microarray levels of COX-2 because the discordance is enriched in the HER2 positive tumors. Our results suggest that HER2 positive invasive tumors target protein stabilization or increased translation of COX-2 as a distinct and important mechanism of achieving COX-2 protein overexpression.
Overexpression of p16 is a Characteristic of the Basal-like Subtype
This report highlights that elevated p16 mRNA is specific to the basal-like subtype as measured and defined by microarray analysis. Increased p16 mRNA as a defining feature of basal-like tumors has been underappreciated largely due to technical limitations of the microarray platforms used to define intrinsic sets and molecular subtypes in previous studies. p16 gene probesets were either absent or unreliable such that resulting datapoints were empty spots or flagged as erroneous. Our findings that mRNA levels of p16, cyclin E and E2F3 are among the highest and levels of Rb and cyclin D1 are among the lowest in the basal-like tumors strongly suggest that inactivation of Rb is mechanistically linked to the basal-like subtype. In support of this link, p16 overexpression at the immunohistochemical level has been previously identified in poorly differentiated tumors through associations with high nuclear grade, high Ki67, increased p53, and low ER/PR expression (Emig et al., 1998; Han et al., 2001; Hui et al., 2000; Milde-Langosch et al., 2001; Singh et al., 2004). In addition, increased immunodetection of p16 correlates with decreased Rb in breast carcinomas (Dublin et al., 1998; Gorgoulis et al., 1998; Nielsen et al., 1997).
Relationship between the Basal-like Subtype in DCIS and Nuclear Grade
Molecular phenotypes are of particular clinical significance if they outperform traditional histopathological parameters. Consistent with our previous findings, and that of others (Bijker et al., 2001; Kerlikowske et al., 2003; Millis et al., 2004; Silverstein et al., 1995) high nuclear grade predicts risk for a subsequent tumor event in a significant manner (HR 5.6, 95% CI 1.2 to 25.5). Although nuclear grade statistically stratifies a subpopulation of women with increased risk for a subsequent tumor event in this pilot set, roughly one third of women with high nuclear grade DCIS do not develop subsequent disease; while roughly one quarter of women with low grade DCIS develop subsequent disease. In this pilot study, we do not have the statistical power to determine to what extent an abrogated response to cellular stress is independent from nuclear grade. However, it is worth noting that previous studies have demonstrated that less than one-half of high grade DCIS lesions exhibit basal-like characteristics (Bryan et al., 2006; Livasy et al., 2007). Consistent with these previous reports, we find that a minority of high-grade lesions demonstrate an abrogated response to cellular stress (6/26 high grade lesions show high p16/Ki67). In this report, all high nuclear grade/high p16/high Ki67 lesions develop subsequent disease (100% or 6/6). Therefore, this phenotype appears to identify a subset of high nuclear grade DCIS lesions, all of which develop subsequent disease and thus, evaluation in a large population based study may considerably refine the predictive value of nuclear grade.
Abrogated Response to Cellular Stress (ARCS) is a property of ER negative and ER positive DCIS: Implications for the Origin of the Basal-Like Subtype
In extension of these data, we have examined a larger series of DCIS lesions and find that over one-half of those cases that exhibit ARCS (either high p16/high Ki67 and/or high COX-2/high Ki67) were clinically classified as ER positive (data not shown). This is in contrast to invasive tumors where we find ARCS limited to the ER negative, basal-like subtype. This discrepancy could be explained by differential risk for subsequent tumor formation: ER negative ARCS DCIS lesions would be followed by ER negative basal-like invasive carcinomas whereas ER positive ARCS DCIS would be “dead-end” lesions not linked to subsequent disease. Alternatively all DCIS lesions, both ER negative and positive, with ARCS may give rise to basal-like tumors. Our data argue for this later possibility since nearly all triple positive (high p16/high COX-2/high Ki67) DCIS lesions, both ER positive and negative, are linked to subsequent invasive tumors.
Since ARCS appears to be a defining feature of basal-like tumors and ER expression defines luminal tumors, our findings suggest that a subset of DCIS lesions exist of mixed basal and luminal character. If true, efforts to identify basal-like DCIS lesions based upon an ER-negative restricted definition would underestimate the prevalence of the basal-like subtype in DCIS. This may account for recent reports from the Carolina Breast Cancer Study that find 16% of invasive tumors are of the basal molecular subtype compared to only 8% of DCIS lesions (Livasy et al., 2007; Millikan et al., 2007).
Finally, the finding of premalignant lesions with mixed basal and luminal character may hold important clues to the origins of the basal-like subtype. In particular, basal-like invasive carcinomas may be linked to precursor lesions that display variable degrees of ER positivity and luminal differentiation. In so much as an abrogated response to cellular stress may characterize such precursor lesions, the combination of stress-activation and deregulation of p16/Rb signalling may represent a defining signature of basal-like carcinogenesis that can be assayed far in advance to the development of invasive disease and present clinical opportunities.
Experimental Procedures
Cells and cell culture
Human mammary epithelial cells (HMEC) and variant HMEC (vHMEC) were isolated from reduction mammoplasties (RM) of multiple individuals RM13, RM 15, RM16, RM18, RM21. All RM tissue was acquired with patient consent and Institutional Review Board approval. Cells were propagated in modified MCDB 170 media (MEGM, Cambrex) as previously described (Hammond et al., 1984; Romanov et al., 2001). Non-tumorigenic immortalized 184A1 breast cells were a kind gift from M. Stampfer (Lawrence Berkeley National Laboratories). Breast cancer cell lines T47D, SKBr3, BT549 and MDA-MB-231 were obtained from the ATCC.
DNA constructs
DNA constructs used in this study are as follows: pMSCV, pMSCV-shp16 (G. Hannon and S. Lowe, Cold Spring Harbor Laboratories); pLXSN, pLXSN-HPV16 E7 (D. Galloway, Fred Hutchinson Cancer Center); pMKO, pMKO-shRb (W. Hahn, Harvard Medical School and Dana-Farber Cancer Institute), pBabe; pBabe-cyclinD1 (O. Tetsu, UCSF Cancer Center), pBabe, pBabe-hTert (K. Collins, UC-Berkeley); LXSP and LXSP-COX-2 (D. Dixon, Vanderbilt University Medical Center).
Western blot
Cell lysates containing 15 – 20 μg total protein were electrophoretically separated according to standard procedures. Antisera against COX-2 (160107; Cayman Chemical, MI), Rb (554136, BD Pharmingen), p16 (16P07 Neomarkers), E2F1 (sc-251 Santa Cruz), cyclinD1 (2926, Cell Signaling), p53 (sc-126, Santa Cruz), p21 (SC-6246 Santa Cruz) were used according to manufacturers protocols.
Tumor samples
Primary gene expression analyses were performed on 130 primary invasive breast cancers from UCSF and California Pacific Medical Center (CPMC). Details of this cohort have been previously described (Chin et al., 2006) Raw microarray data and additional sample information is available (http://cancer.lbl.gov/breastcancer/data.php). Paraffin-embedded tumor samples corresponding to 61 of the 130 cases were obtained with patient consent and Institutional Review Board approval.
Gene expression profiling analyses and identification of molecular subtypes
For details of microarray methods and derivation of molecular subtypes please see Supplemental File 1.
Pre-malignant samples
DCIS samples comprise a subset of women treated by lumpectomy or lumpectomy/radiation from UCSF and CPMC (n= 28) and a large population-based cohort study of women treated by lumpectomy alone between 1984 and 1996 (n= 42). To determine if response to adjuvant therapy affected the predictive value of the phenotypes studied, we removed the samples from patients who had been treated with radiation and reanalyzed the remaining subset of samples. We conclude, in this study, there is no correlation with treatment type. All tissue was acquired with patient consent and Institutional Review Board approval. Patients were identified through anonymous reference numbers.
Tissue preparation and immunohistochemistry
Five μM sections from formalin-fixed paraffin embedded tissue were stained with antisera against COX-2 (Dako M3617, 1/200), p16 (Neomarkers MS218, 1/150), or Ki67 (Dako M7250, 1/80) overnight at 4°C. Antigen-antibody complexes were labelled using the Vectastain Elite ABC (Vector Laboratories, CA), visualised using 2.5% 3-amino-9-ethyl-carbazole in 50mM acetate buffer pH5, with 0.05% hydrogen peroxide and counterstained in Mayers hematoxylin.
Evaluation of immunohistochemistry staining
Using a condensed Allred score (Allred et al., 1998), COX-2 staining was evaluated on a scale of 0, 1, 2, 3 with each value corresponding to a combination of 2 Allred classes (ie, 0=0,1; 1=2,3; 2=5,6; 3=7,8). COX-2 showed predominantly a cytoplasmic pattern of immunopositivity with occasional membranous staining. p16 staining was scored on a 0, 1, 2, 3 scale based on the extent of immunopositive cells (0: no staining; 1: < 25%; 2: 25 – 75%; 3: > 75%). p16 showed predominantly a cytoplasmic pattern of immunopositivity with occasional nuclear staining. In the manuscript where indicated for both COX-2 and p16, “high” immunostaining refers to a score of ≥ 2. Ki67 index was determined by manually counting a minimum of 1000 nuclei within at least three 40x fields. High Ki67 is defined as greater than 10%.
Statistical analysis
Chi-square tests were used to determine associations between p16, COX-2, Ki67, nuclear grade and combinations therein with subsequent tumor development among women with DCIS. JMP statistical package (SAS Institute) was used for all analyses. We used a Cox Proportional Hazards Model stratified by year of diagnosis to study the ability of four markers (grade and expression of COX-2, p16 and Ki67) to predict recurrence during follow-up. Controls were matched to cases by year of diagnosis. There were too few cases (recur) and controls (non-recur) for several of the years of diagnosis, so years were grouped as shown for the stratified analyses (Supplemental Table 5). We analyzed biomarkers separately and in combination. Results are expressed as Hazard Ratios representing time to subsequent tumor event.
Supplementary Material
Acknowledgments
This work was supported by the NCI-funded Breast Cancer Specialized Program of Research Excellence P50 CA58207 (K.K. and T.D.T), RO1 CA097214 (T.D.T.) and The Cancer League. We thank Kim Stewart and Annette Molinaro for their helpful suggestions, Joe Gray, Fred Waldman, and Paul Spellman for providing access to primary microarray data and patient advocates and our SPORE colleagues for their support and encouragement.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adeyinka A, Emberley E, Niu Y, Snell L, Murphy LC, Sowter H, Wykoff CC, Harris AL, Watson PH. Analysis of gene expression in ductal carcinoma in situ of the breast. Clin Cancer Res. 2002;8:3788–3795. [PubMed] [Google Scholar]
- Allred DC, Harvey JM, Berardo M, Clark GM. Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod Pathol. 1998;11:155–168. [PubMed] [Google Scholar]
- Barnes N, Haywood P, Flint P, Knox WF, Bundred NJ. Survivin expression in in situ and invasive breast cancer relates to COX-2 expression and DCIS recurrence. Br J Cancer. 2006;94:253–258. doi: 10.1038/sj.bjc.6602932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- Bates S, Parry D, Bonetta L, Vousden K, Dickson C, Peters G. Absence of cyclin D/cdk complexes in cells lacking functional retinoblastoma protein. Oncogene. 1994;9:1633–1640. [PubMed] [Google Scholar]
- Bijker N, Meijnen P, Peterse JL, Bogaerts J, Van Hoorebeeck I, Julien JP, Gennaro M, Rouanet P, Avril A, Fentiman IS, et al. Breast-conserving treatment with or without radiotherapy in ductal carcinoma-in-situ: ten-year results of European Organisation for Research and Treatment of Cancer randomized phase III trial 10853--a study by the EORTC Breast Cancer Cooperative Group and EORTC Radiotherapy Group. J Clin Oncol. 2006;24:3381–3387. doi: 10.1200/JCO.2006.06.1366. [DOI] [PubMed] [Google Scholar]
- Bijker N, Peterse JL, Duchateau L, Robanus-Maandag EC, Bosch CA, Duval C, Pilotti S, van de Vijver MJ. Histological type and marker expression of the primary tumour compared with its local recurrence after breast-conserving therapy for ductal carcinoma in situ. Br J Cancer. 2001;84:539–544. doi: 10.1054/bjoc.2000.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm JS, Hession MT, Bulmer SE, Hahn WC. Transformation of human and murine fibroblasts without viral oncoproteins. Mol Cell Biol. 2005;25:6464–6474. doi: 10.1128/MCB.25.15.6464-6474.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland GP, Butt IS, Prasad R, Knox WF, Bundred NJ. COX-2 expression is associated with an aggressive phenotype in ductal carcinoma in situ. Br J Cancer. 2004;90:423–429. doi: 10.1038/sj.bjc.6601534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braig M, Lee S, Loddenkemper C, Rudolph C, Peters AH, Schlegelberger B, Stein H, Dorken B, Jenuwein T, Schmitt CA. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005;436:660–665. doi: 10.1038/nature03841. [DOI] [PubMed] [Google Scholar]
- Bryan BB, Schnitt SJ, Collins LC. Ductal carcinoma in situ with basal-like phenotype: a possible precursor to invasive basal-like breast cancer. Mod Pathol. 2006;19:617–621. doi: 10.1038/modpathol.3800570. [DOI] [PubMed] [Google Scholar]
- Campisi J. Suppressing cancer: the importance of being senescent. Science. 2005;309:886–887. doi: 10.1126/science.1116801. [DOI] [PubMed] [Google Scholar]
- Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, Lapuk A, Neve RM, Qian Z, Ryder T, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Cho MH, Yoon JH, Jaegal YJ, Choi YD, Lee JS, Lee JH, Nam JH, Choi C, Lee MC, Park CS, et al. Expression of cyclooxygenase-2 in breast carcinogenesis and its relation to HER-2/neu and p53 protein expression in invasive ductal carcinoma. Breast. 2006;15:390–398. doi: 10.1016/j.breast.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Collado M, Serrano M. The senescent side of tumor suppression. Cell Cycle. 2005;4:1722– 1724. doi: 10.4161/cc.4.12.2260. [DOI] [PubMed] [Google Scholar]
- Crawford YG, Gauthier ML, Joubel A, Mantei K, Kozakiewicz K, Afshari CA, Tlsty TD. Histologically normal human mammary epithelia with silenced p16(INK4a) overexpress COX-2, promoting a premalignant program. Cancer Cell. 2004;5:263–273. doi: 10.1016/s1535-6108(04)00023-6. [DOI] [PubMed] [Google Scholar]
- Davis JN, McCabe MT, Hayward SW, Park JM, Day ML. Disruption of Rb/E2F pathway results in increased cyclooxygenase-2 expression and activity in prostate epithelial cells. Cancer Res. 2005;65:3633–3642. doi: 10.1158/0008-5472.CAN-04-3129. [DOI] [PubMed] [Google Scholar]
- Dublin EA, Patel NK, Gillett CE, Smith P, Peters G, Barnes DM. Retinoblastoma and p16 proteins in mammary carcinoma: their relationship to cyclin D1 and histopathological parameters. Int J Cancer. 1998;79:71–75. doi: 10.1002/(sici)1097-0215(19980220)79:1<71::aid-ijc14>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- Emig R, Magener A, Ehemann V, Meyer A, Stilgenbauer F, Volkmann M, Wallwiener D, Sinn HP. Aberrant cytoplasmic expression of the p16 protein in breast cancer is associated with accelerated tumour proliferation. Br J Cancer. 1998;78:1661–1668. doi: 10.1038/bjc.1998.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eusebi V, Feudale E, Foschini MP, Micheli A, Conti A, Riva C, Di Palma S, Rilke F. Long-term follow-up of in situ carcinoma of the breast. Semin Diagn Pathol. 1994;11:223–235. [PubMed] [Google Scholar]
- Fisher ER, Dignam J, Tan-Chiu E, Costantino J, Fisher B, Paik S, Wolmark N. Pathologic findings from the National Surgical Adjuvant Breast Project (NSABP) eight-year update of Protocol B-17: intraductal carcinoma. Cancer. 1999;86:429–438. doi: 10.1002/(sici)1097-0142(19990801)86:3<429::aid-cncr11>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Gauthier ML, Pickering CR, Miller CJ, Fordyce CA, Chew KL, Berman HK, Tlsty TD. p38 regulates cyclooxygenase-2 in human mammary epithelial cells and is activated in premalignant tissue. Cancer Res. 2005;65:1792–1799. doi: 10.1158/0008-5472.CAN-04-3507. [DOI] [PubMed] [Google Scholar]
- Gorgoulis VG, Koutroumbi EN, Kotsinas A, Zacharatos P, Markopoulos C, Giannikos L, Kyriakou V, Voulgaris Z, Gogas I, Kittas C. Alterations of p16-pRb pathway and chromosome locus 9p21–22 in sporadic invasive breast carcinomas. Mol Med. 1998;4:807–822. [PMC free article] [PubMed] [Google Scholar]
- Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA, Jr, Kastrinakis NG, Levy B, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- Hammond SL, Ham RG, Stampfer MR. Serum-free growth of human mammary epithelial cells: rapid clonal growth in defined medium and extended serial passage with pituitary extract. Proc Natl Acad Sci U S A. 1984;81:5435–5439. doi: 10.1073/pnas.81.17.5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Ahn SH, Park K, Bae BN, Kim KH, Kim HJ, Kim YD, Kim HY. P16INK4a protein expression is associated with poor survival of the breast cancer patients after CMF chemotherapy. Breast Cancer Res Treat. 2001;70:205–212. doi: 10.1023/a:1013047413895. [DOI] [PubMed] [Google Scholar]
- Hannemann J, Velds A, Halfwerk JB, Kreike B, Peterse JL, van de Vijver MJ. Classification of ductal carcinoma in situ by gene expression profiling. Breast Cancer Res. 2006;8:R61. doi: 10.1186/bcr1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour JW, Dean DC. Rb function in cell-cycle regulation and apoptosis. Nat Cell Biol. 2000;2:E65–67. doi: 10.1038/35008695. [DOI] [PubMed] [Google Scholar]
- Hernando E, Nahle Z, Juan G, Diaz-Rodriguez E, Alaminos M, Hemann M, Michel L, Mittal V, Gerald W, Benezra R, et al. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature. 2004;430:797–802. doi: 10.1038/nature02820. [DOI] [PubMed] [Google Scholar]
- Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, Livasy C, Carey LA, Reynolds E, Dressler L, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui R, Macmillan RD, Kenny FS, Musgrove EA, Blamey RW, Nicholson RI, Robertson JF, Sutherland RL. INK4a gene expression and methylation in primary breast cancer: overexpression of p16INK4a messenger RNA is a marker of poor prognosis. Clin Cancer Res. 2000;6:2777–2787. [PubMed] [Google Scholar]
- Jirstrom K, Ringberg A, Ferno M, Anagnostaki L, Landberg G. Tissue microarray analyses of G1/S-regulatory proteins in ductal carcinoma in situ of the breast indicate that low cyclin D1 is associated with local recurrence. Br J Cancer. 2003;89:1920–1926. doi: 10.1038/sj.bjc.6601398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalof AN, Cooper K. p16INK4a immunoexpression: surrogate marker of high-risk HPV and high-grade cervical intraepithelial neoplasia. Adv Anat Pathol. 2006;13:190–194. doi: 10.1097/00125480-200607000-00006. [DOI] [PubMed] [Google Scholar]
- Kerlikowske K, Molinaro A, Cha I, Ljung BM, Ernster VL, Stewart K, Chew K, Moore DH, 2nd, Waldman F. Characteristics associated with recurrence among women with ductal carcinoma in situ treated by lumpectomy. J Natl Cancer Inst. 2003;95:1692–1702. doi: 10.1093/jnci/djg097. [DOI] [PubMed] [Google Scholar]
- Leone G, Nuckolls F, Ishida S, Adams M, Sears R, Jakoi L, Miron A, Nevins JR. Identification of a novel E2F3 product suggests a mechanism for determining specificity of repression by Rb proteins. Mol Cell Biol. 2000;20:3626–3632. doi: 10.1128/mcb.20.10.3626-3632.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lininger RA, Fujii H, Man YG, Gabrielson E, Tavassoli FA. Comparison of loss heterozygosity in primary and recurrent ductal carcinoma in situ of the breast. Mod Pathol. 1998;11:1151–1159. [PubMed] [Google Scholar]
- Livasy CA, Perou CM, Karaca G, Cowan DW, Maia D, Jackson S, Tse CK, Nyante S, Millikan RC. Identification of a basal-like subtype of breast ductal carcinoma in situ. Hum Pathol. 2007;38:197–204. doi: 10.1016/j.humpath.2006.08.017. [DOI] [PubMed] [Google Scholar]
- Loden M, Stighall M, Nielsen NH, Roos G, Emdin SO, Ostlund H, Landberg G. The cyclin D1 high and cyclin E high subgroups of breast cancer: separate pathways in tumorogenesis based on pattern of genetic aberrations and inactivation of the pRb node. Oncogene. 2002;21:4680–4690. doi: 10.1038/sj.onc.1205578. [DOI] [PubMed] [Google Scholar]
- Lowe SW, Sherr CJ. Tumor suppression by Ink4a-Arf: progress and puzzles. Curr Opin Genet Dev. 2003;13:77–83. doi: 10.1016/s0959-437x(02)00013-8. [DOI] [PubMed] [Google Scholar]
- Lukas J, Parry D, Aagaard L, Mann DJ, Bartkova J, Strauss M, Peters G, Bartek J. Retinoblastoma-protein-dependent cell-cycle inhibition by the tumour suppressor p16. Nature. 1995;375:503–506. doi: 10.1038/375503a0. [DOI] [PubMed] [Google Scholar]
- Lundberg AS, Weinberg RA. Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol Cell Biol. 1998;18:753–761. doi: 10.1128/mcb.18.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott KM, Zhang J, Holst CR, Kozakiewicz BK, Singla V, Tlsty TD. p16(INK4a) prevents centrosome dysfunction and genomic instability in primary cells. PLoS Biol. 2006;4:e51. doi: 10.1371/journal.pbio.0040051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema RH, Herrera RE, Lam F, Weinberg RA. Growth suppression by p16ink4 requires functional retinoblastoma protein. Proc Natl Acad Sci U S A. 1995;92:6289–6293. doi: 10.1073/pnas.92.14.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, Majoor DM, Shay JW, Mooi WJ, Peeper DS. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- Milde-Langosch K, Bamberger AM, Rieck G, Kelp B, Loning T. Overexpression of the p16 cell cycle inhibitor in breast cancer is associated with a more malignant phenotype. Breast Cancer Res Treat. 2001;67:61–70. doi: 10.1023/a:1010623308275. [DOI] [PubMed] [Google Scholar]
- Millikan RC, Newman B, Tse CK, Moorman PG, Conway K, Smith LV, Labbok MH, Geradts J, Bensen JT, Jackson S, et al. Epidemiology of basal-like breast cancer. Breast Cancer Res Treat. 2007 doi: 10.1007/s10549-007-9632-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millis RR, Pinder SE, Ryder K, Howitt R, Lakhani SR. Grade of recurrent in situ and invasive carcinoma following treatment of pure ductal carcinoma in situ of the breast. Br J Cancer. 2004;90:1538–1542. doi: 10.1038/sj.bjc.6601704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooi WJ, Peeper DS. Oncogene-induced cell senescence--halting on the road to cancer. N Engl J Med. 2006;355:1037–1046. doi: 10.1056/NEJMra062285. [DOI] [PubMed] [Google Scholar]
- Narita M, Nunez S, Heard E, Narita M, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- Nevins JR. Toward an understanding of the functional complexity of the E2F and retinoblastoma families. Cell Growth Differ. 1998;9:585–593. [PubMed] [Google Scholar]
- Nielsen NH, Emdin SO, Cajander J, Landberg G. Deregulation of cyclin E and D1 in breast cancer is associated with inactivation of the retinoblastoma protein. Oncogene. 1997;14:295–304. doi: 10.1038/sj.onc.1200833. [DOI] [PubMed] [Google Scholar]
- Parry D, Bates S, Mann DJ, Peters G. Lack of cyclin D-Cdk complexes in Rb-negative cells correlates with high levels of p16INK4/MTS1 tumour suppressor gene product. Embo J. 1995;14:503–511. doi: 10.1002/j.1460-2075.1995.tb07026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Reis-Filho JS, Savage K, Lambros MB, James M, Steele D, Jones RL, Dowsett M. Cyclin D1 protein overexpression and CCND1 amplification in breast carcinomas: an immunohistochemical and chromogenic in situ hybridisation analysis. Mod Pathol. 2006;19:999–1009. doi: 10.1038/modpathol.3800621. [DOI] [PubMed] [Google Scholar]
- Ristimaki A, Sivula A, Lundin J, Lundin M, Salminen T, Haglund C, Joensuu H, Isola J. Prognostic significance of elevated cyclooxygenase-2 expression in breast cancer. Cancer Res. 2002;62:632–635. [PubMed] [Google Scholar]
- Romanov SR, Kozakiewicz BK, Holst CR, Stampfer MR, Haupt LM, Tlsty TD. Normal human mammary epithelial cells spontaneously escape senescence and acquire genomic changes. Nature. 2001;409:633–637. doi: 10.1038/35054579. [DOI] [PubMed] [Google Scholar]
- Schmitt CA. Senescence, apoptosis and therapy--cutting the lifelines of cancer. Nat Rev Cancer. 2003;3:286–295. doi: 10.1038/nrc1044. [DOI] [PubMed] [Google Scholar]
- Sellers WR, Rodgers JW, Kaelin WG., Jr A potent transrepression domain in the retinoblastoma protein induces a cell cycle arrest when bound to E2F sites. Proc Natl Acad Sci U S A. 1995;92:11544–11548. doi: 10.1073/pnas.92.25.11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M, Gomez-Lahoz E, DePinho RA, Beach D, Bar-Sagi D. Inhibition of ras- induced proliferation and cellular transformation by p16INK4. Science. 1995;267:249–252. doi: 10.1126/science.7809631. [DOI] [PubMed] [Google Scholar]
- Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho RA. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- Silverstein MJ, Poller DN, Waisman JR, Colburn WJ, Barth A, Gierson ED, Lewinsky B, Gamagami P, Slamon DJ. Prognostic classification of breast ductal carcinoma-in-situ. Lancet. 1995;345:1154–1157. doi: 10.1016/s0140-6736(95)90982-6. [DOI] [PubMed] [Google Scholar]
- Singh M, Parnes MB, Spoelstra N, Bleile MJ, Robinson WA. p16 expression in sentinel nodes with metastatic breast carcinoma: evaluation of its role in developing triaging strategies for axillary node dissection and a marker of poor prognosis. Hum Pathol. 2004;35:1524–1530. doi: 10.1016/j.humpath.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaramaiah K, Dannenberg AJ. Cyclooxygenase-2 transcription is regulated by human papillomavirus 16 E6 and E7 oncoproteins: evidence of a corepressor/coactivator exchange. Cancer Res. 2007;67:3976–3985. doi: 10.1158/0008-5472.CAN-06-4273. [DOI] [PubMed] [Google Scholar]
- Waldman FM, DeVries S, Chew KL, Moore DH, 2nd, Kerlikowske K, Ljung BM. Chromosomal alterations in ductal carcinomas in situ and their in situ recurrences. J Natl Cancer Inst. 2000;92:313–320. doi: 10.1093/jnci/92.4.313. [DOI] [PubMed] [Google Scholar]
- Zhang J, Pickering CR, Holst CR, Gauthier ML, Tlsty TD. p16INK4a modulates p53 in primary human mammary epithelial cells. Cancer Res. 2006;66:10325–10331. doi: 10.1158/0008-5472.CAN-06-1594. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.