SUMMARY
Retrograde signalling from plastids to the nucleus is necessary to regulate the organelle’s proteome during the establishment of photoautotrophy and fluctuating environmental conditions. Studies that used inhibitors of chloroplast biogenesis have revealed that hundreds of nuclear genes are regulated by retrograde signals emitted from plastids. Plastid gene expression is the source of at least one of these signals, but the number of signals and their mechanisms used to regulate nuclear gene expression are unknown. To further examine the effects of plastid gene expression on nuclear gene expression, we analyzed Arabidopsis mutants that were defective in each of the six sigma factor (SIG) genes that encode proteins utilized by plastid-encoded RNA polymerase to transcribe specific sets of plastid genes. We showed that SIG2 and SIG6 have partially redundant roles in plastid transcription and retrograde signalling to control nuclear gene expression. The loss of GUN1 (a plastid-localized pentatricopeptide repeat protein) is able to restore nuclear (but not plastid) gene expression in both sig2 and sig6, whereas an increase in heme synthesis is able to restore nuclear gene expression in sig2 mutants only. These results demonstrate that sigma factor function is the source of at least two retrograde signals to the nucleus; one likely to involve the transcription of tRNAGlu. A microarray analysis showed that these two signals accounted for at least one subset of the nuclear genes that are regulated by the plastid biogenesis inhibitors norflurazon and lincomycin. Together these data suggest that such inhibitors can induce retrograde signalling by affecting transcription in the plastid.
Keywords: chloroplast, retrograde signalling, transcription, tetrapyrroles, sigma factors, photosynthesis
INTRODUCTION
Over 95% of the ~3000 proteins in the plastid are encoded in the nuclear genome, translated in the cytoplasm, and then imported into the organelle (Martin et al., 2002; Barbrook et al., 2006). Communication between the separate compartments is therefore necessary to regulate the stoichiometry of multi-subunit complexes encoded by both genomes, the proteome of the plastid, and to ensure that the organelle can meet the metabolic and energy demands of the cell (Woodson and Chory, 2008). Many plastid functions (e.g. DNA replication, division, and most gene regulation) are regulated by nuclear factors; this control is referred to as anterograde control or signalling. Conversely, the functional and developmental state of the plastid (e.g. undeveloped, stressed) allows the organelle to send retrograde signals to the nucleus to regulate gene expression. H2O2 (Laloi et al., 2006), singlet oxygen (Wagner et al., 2004), the plastid redox state (Pfannschmidt et al., 2003), and drought stress (Estavillo et al., 2011) act as ‘operational’ retrograde signals from mature photosynthesizing plastids that regulate nuclear genes to alter photosystem stoichiometry or to induce stress responses (Pogson et al., 2008).
At the same time, ‘developmental’ signals from plastids also regulate early chloroplast biogenesis as a seedling shifts from a heterotrophic to a photoautotrophic lifestyle. Such knowledge has been derived primarily from studies that used herbicide drugs that inhibit organelle function and development, e.g. norflurazon (NF), an inhibitor of phytoene desaturase and carotenoid biosynthesis that leads to photobleaching, or lincomycin (Linc), an inhibitor of 70S ribosomes and plastid translation (Nott et al., 2006). From these studies it was gleaned that hundreds of Photosynthetic Associated Nuclear Genes (PhANGs) are sensitive to the functional state of the plastid and, if plastid gene expression and/or plastid tetrapyrrole biosynthesis are attenuated, PhANGs are down-regulated as a response. Consequently, plastid RNA transcripts, proteins, and tetrapyrroles have all been posited to be potential signalling molecules (Fernandez and Strand, 2008; Pogson et al., 2008).
Specific and unbiased genetic screens in Arabidopsis thaliana have identified six genomes uncoupled (gun) mutants that still express PhANGs even when photobleached by NF (Susek et al., 1993). GUN2-6 encode for proteins clustered around the heme/chlorophyll branch point of the tetrapyrrole biosynthetic pathway (Mochizuki et al., 2001; Larkin et al., 2003). The one gain-of-function mutant, gun6, has an increase of ferrochelatase I (FC1) activity leading to the hypothesis that increasing FC1-produced heme in undeveloped chloroplasts was a positive signal to regulate PhANG expression (Woodson et al., 2011).
gun1 mutants were isolated from genetic screens that used both NF and Linc, and led to a model in which GUN1 is required for multiple-stress regulated pathways (Koussevitzky et al., 2007). Although the precise biochemical activity of GUN1 is not known, it is a chloroplast-localized pentatricopeptide repeat (PPR)-containing protein, which is believed to be one of the specific RNA binding factors that post-transcriptionally regulate gene expression by altering editing, splicing, stability, or translation (Delannoy et al., 2007). Therefore, a plausible model for GUN1 is that it regulates one or a small set of chloroplast RNAs (or protein product(s)) that is necessary for retrograde signalling.
Although clearly involved in retrograde signalling, the intertwined nature of transcription, translation, and tetrapyrrole synthesis within plastids has made it difficult to determine the exact sources of these signals. For instance, because much transcription in chloroplasts is performed by plastid-encoded RNA polymerase (PEP), inhibition of translation by Linc will also reduce most RNA transcripts within the plastid (Gray et al., 2003). Also, PEP-transcribed tRNAGlu is the starting precursor for all tetrapyrroles within the plastid (Schon et al., 1986). Therefore, tetrapyrrole metabolism is affected directly by inhibition of either transcription or translation. Because drug inhibitors used in these studies are likely to affect all of these processes, their usefulness in the study of retrograde signalling has been criticized (Pfannschmidt, 2010).
To analyze further the role that gene expression plays in plastid signalling, here we monitored retrograde signalling when chloroplast transcription had been specifically perturbed without inhibitors. To this end, we analyzed Arabidopsis mutants that were defective in each of the six sigma factor (SIG) genes that encode proteins utilized by PEP to transcribe specific sets of plastid genes. These studies allowed us to demonstrate that: (i) SIG2 and SIG6 are the source of two plastid retrograde signals—one is transcription of plastid tRNAGlu by SIG2 to increase tetrapyrrole synthesis and promote PhANG expression in the nucleus, (ii) these two signals affect many of the same genes as do drug inhibitor treatments, which suggested that a common signal is shared, and (iii) GUN1 and tetrapyrroles are important for genome co-ordination under physiologically relevant conditions.
RESULTS
Plastid transcription is necessary for full LHCB expression in Arabidopsis
Block of plastid transcription in barley, wheat, or mustard leads to repression of PhANGs (Lukens et al., 1987; Rapp and Mullet, 1991; Pfannschmidt and Link, 1997). To test if plastid transcription is important for regulation of PhANGs in Arabidopsis, we monitored the expression of PhANGs in seedlings that had been grown on media that contained the drug rifampicin, which selectively inhibits PEP activity. To this end, we used the genetic construct (6–3) that contained the canonical PhANG LHCB1.2 (Light Harvesting Chlorophyll Binding protein) minimal promoter driving the LUC gene (Koussevitzky et al., 2007). In agreement with previous studies, rifampicin treatment (50 μg ml−1) reduced LHCB expression in 2-day-old seedlings in a lightindependent manner (Figure 1a). Quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis confirmed that transcript levels of LHCB1.2, the PhANG Carbonic anhydrase 1 (CA1), and the PEP-dependent plastid genes psbA and psbD were reduced. Conversely, control genes [nuclear genes encoding plastid-localized proteins that are not regulated by plastid retrograde signals (Table S2) (Koussevitzky et al., 2007)] and the NEP-dependent plastid genes were unchanged or elevated (Figure S1). Together these results suggested that transcription of some plastid genes is necessary for the full expression of LHCB and CA1 in Arabidopsis.
Figure 1.
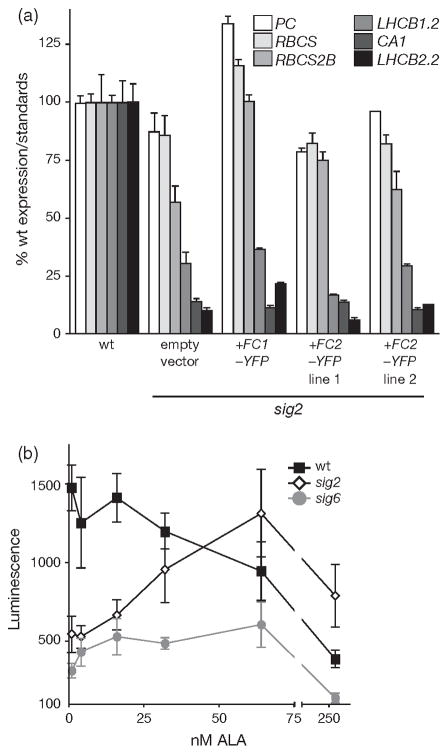
SIG2 and SIG6-mediated plastid transcription is required for PhANG expression. (a) The average luciferase activity of 2-day-old etiolated (− light) and de-etiolated (+ light) seedlings that harbour the Luc gene under the control of a minimal LHCB1.2 promoter (6–3) and that have been germinated in the presence or absence of 50 μg ml−1 rifampicin is shown. Data shown are the mean ± standard error of the mean (SEM) of 16 individual seedlings. (b) Image of 3-day-old sigma mutant seedlings. Quantitative polymerase chain reaction (qPCR) analysis of steady-state (c) plastid and (d) PhANG mRNA transcript levels of 2-day-old seedlings. Data shown are the mean ± SEM of triplicate reactions of a representative experiment.
SIG2 and SIG6 are necessary for full PhANG expression in seedlings
In land plants, prokaryotic-derived nuclear-encoded sigma factors are involved in PEP promoter recognition and are important for the transcription of certain sets of PEP-transcribed plastid genes (Lysenko, 2007; Schweer et al., 2010). The Arabidopsis nuclear genome encodes six sigma factors with partially redundant functions in plastid transcription. Because the previous experiment showed that PEP activity is required for full LHCB expression, we surmised that a certain sigma factor(s) might also be required. To investigate this possibility, we obtained T-DNA lines and disrupted each of the six SIG genes (Table S1). Quantitative real-time PCR (qPCR) analysis showed that, except for sig1, each of the T-DNAs severely reduced the level of steady-state RNA transcript (Figure S2a). Except for the sig2 T-DNA, all the insertions were in exons, a finding that suggested that even the low levels of transcripts are not expected to encode functional proteins.
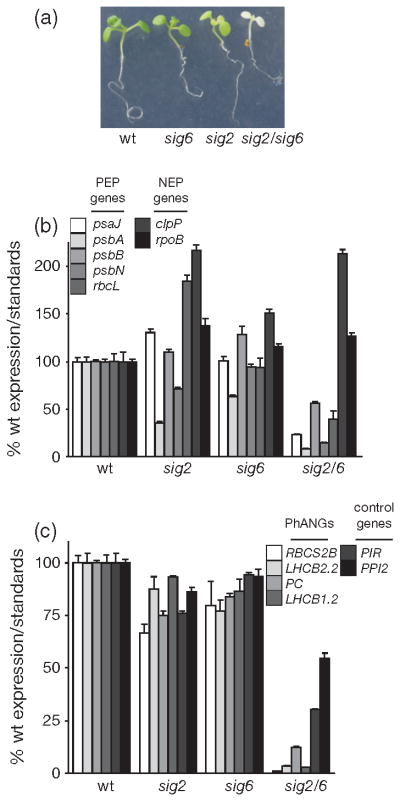
In agreement with previous reports, mutations that affected SIG2 (Shirano et al., 2000) or SIG6 (Ishizaki et al., 2005) resulted in pale seedlings that accumulated significantly less chlorophyll (Figures 1b and S2f). Mutants that lacked the other four sigma factors had a wild-type (wt)- like physical appearance as has been reported for sig1 (Lai et al., 2011), sig3 (Zghidi et al., 2007), sig4 (Favory et al., 2005), and sig5 (Nagashima et al., 2004b). Several plastid transcripts failed to accumulate to wt levels in these mutants. As has been reported previously, sig2 had lower levels of psaJ, psbA, and psbD (Hanaoka et al., 2003; Privat et al., 2003; Nagashima et al., 2004a), sig3 had reduced levels of psbN (Zghidi et al., 2007), sig4 had reduced levels of ndhF (Favory et al., 2005), and sig6 had lower transcript levels of all the PEP-dependent genes that we tested (Ishizaki et al., 2005; Loschelder et al., 2006) (Figure 1c). In addition, we observed that sig2, sig3, and sig4 mutants failed to accumulate normal levels of additional PEP-transcribed transcripts (psbB and psbN in sig2, psaJ in sig3, psaJ and rbcL in sig4); the sig2 mutant had the most severe defect. sig1 and sig5 seedlings had essentially wt levels of plastid transcripts. Only PEP-transcribed genes appeared to be affected as nuclear-encoded polymerase (NEP) transcribed transcripts (clpP, rpoB, rpoC) were either at wt levels or even elevated in sig2 and sig6 lines (Figure S2b).
Next, we tested if the reduced plastid transcripts levels led to reduced PhANG expression in any of the sig mutants, i.e. is there retrograde signalling? Although the PhANG RBCS2B (RuBisCO small subunit 2B) was slightly reduced in sig3 and sig4, the most drastic effects were observed in sig2 and sig6 (Figure 1d). In these two lines, the PhANGs LHCB1.2, LHCB2.2, CA1, RBCS2B and Plastocyanin (PC) levels were reduced compared with wt, whereas levels of control gene transcripts were unchanged or elevated (Figure S2c). This phenotype was not light dependent as etiolated sig2 and sig6 seedlings also had reduced levels of PhANG and plastid transcripts (Figures 2a and S3a). In the sig2 and sig6 mutants, the observed defects in chlorophyll synthesis (Figure S2f,g), plastid (Figure S2d) and PhANG transcript levels (Figure S2e) were complemented by reintroduction of a wt copy of either SIG2 or SIG6, and confirmed causality of the T-DNA insertions. As predicted, both the overexpressed SIG2-YFP-HA and SIG6-YFP-HA appear to be localized only to chloroplasts (Figure S2h).
Figure 2. Genetic interactions between sigma and gun mutants.
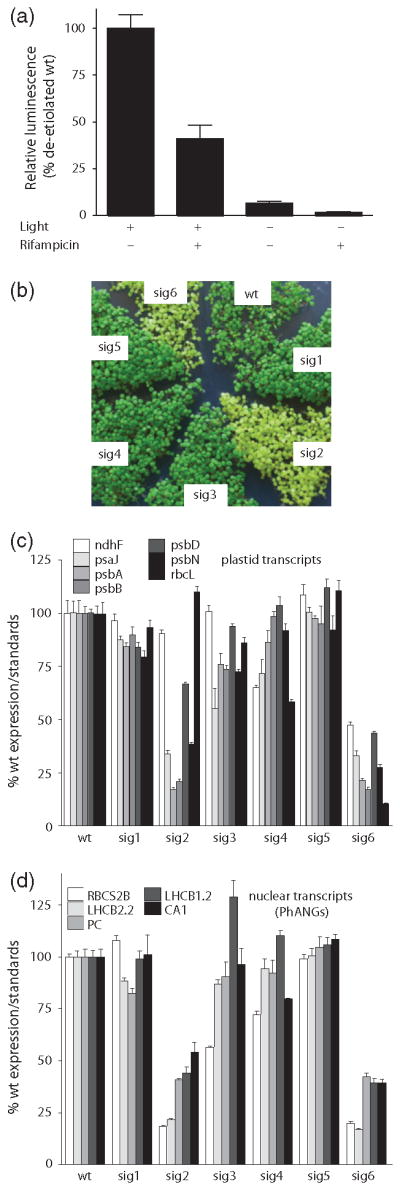
Quantitative polymerase chain reaction (qPCR) analysis of steady-state PhANG mRNA transcript levels of 2-day-old (a) etiolated and (b) de-etiolated seedlings. Data shown are the mean ± standard error of the mean (SEM) of triplicate reactions of a representative experiment. (c) Western blot analysis of plastid proteins. 20 μg of total protein was fractioned by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) and blotted. Filters were probed with antibodies that were specific for the proteins indicated to the right.
SIG2 and SIG6 are partially redundant for genome co-ordination
Plants that lack SIG2 or SIG6 are virescent (leaves emerge yellow or pale green and then eventually turn green (Hanaoka et al., 2003; Ishizaki et al., 2005)). We therefore performed a time course analysis of seedlings grown from 2 to 10 days. Although there were individual differences between both the genes and the genotypes, the general trend (with one exception, psaJ) was that both plastid and PhANG transcripts were lowest at day 2 and accumulated to or near to wt levels by day 10 in the sig2 and sig6 mutants (Figure S4a). In agreement with the transcript data, three plastid-encoded proteins accumulated more slowly in the sig2 or sig6 mutants compared with wt (Figure S4b).
To assess whether there was genetic redundancy between SIG2 and SIG6, we generated a sig2/sig6 double mutant. The sig2/sig6 mutant was severely pale (Figure 3a) even after 5 weeks of growth on medium supplemented with 1% sucrose (Figure S5b). Although the phenotype of the sig2 or sig6 single mutant was similar to wt after 1 week, plastid PEP (but not NEP) dependent transcripts and PhANG transcripts failed to accumulate in sig2/sig6 (Figure 3b,c). A small reduction in the level of control gene transcripts suggested that the double mutant had a broader effect on nuclear genes encoding plastid proteins.
Figure 3. SIG2 and SIG6 are partially redundant for genome coordination.
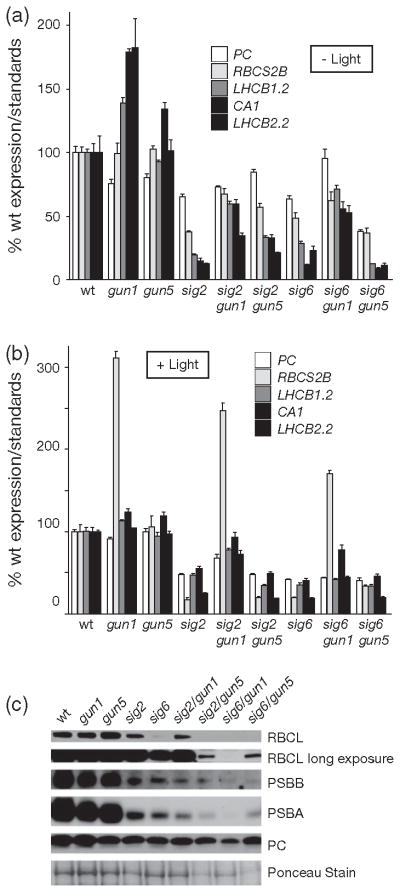
(a) Image of 7-day-old sigma mutant seedlings that were grown on medium supplemented with 1% sucrose. Quantitative polymerase chain reaction (qPCR) analysis of steady-state (b) plastid and (c) nuclear mRNA transcript levels of 7-day-old seedlings. Data shown are the mean ± standard error of the mean (SEM) of triplicate reactions of a representative experiment. The sig2/sig6 was propagated as heterozygous for the sig6 allele.
GUN1 is required in the plastid transcription signalling pathway
To test if GUN1, which is involved in multiple retrograde signalling pathways (Koussevitzky et al., 2007), was also involved in the retrograde signalling observed in the sig2 and sig6 mutants, we generated sig2/gun1 and sig6/gun1 double mutants. The gun1 mutation partially restored the expression of several PhANGs in both sig2 and sig6 mutants, and this effect was light independent (Figure 2a, b) and confirmed that GUN1 acts in a light-independent signalling pathway (Ruckle et al., 2007). The restoration of nuclear gene expression by gun1 could not be explained by a concurrent increase in plastid transcripts (Figure S3a, b) or protein (Figure 2c). The gun1 mutation had little effect on plastid transcripts and proteins in the sig2 mutant, but further reduced their accumulation in the sig6 mutant. At the visual level, the sig2/gun1 mutant was similar in appearance to sig2, but gun1/sig6 was noticeable paler and sicker than sig6 after 7 days (Figure S3c).
We considered the possibility that GUN1 affects the abundance of a specific transcript or small set of transcripts not included in our qPCR analysis. We, therefore, analyzed plastid transcripts in these mutants by strandspecific RNA sequencing. Because the sig2 mutation primarily affects tRNA transcription (Kanamaru et al., 2001) and such an analysis is not practical for monitoring small structured RNAs, we tested wt, gun1, sig6 and sig6/gun1 plants. Of the 72 plastid genes that we could reliably monitor, the sig6 mutation reduced steady-state transcript levels of most of the PEP-dependent genes (Figures 4 and S6a). This analysis showed that there was a strong correlation to the expression changes of plastid genes that we monitored by qRT-PCR (Figure S6c). The gun1 allele alone had no significant effect on any plastid transcript and the addition of gun1 did not restore expression of any single plastid transcript in sig6 mutants (Figure S6b). Instead, gun1 addition led to a global reduction of all 72 plastid transcripts, both PEP and NEP-dependent, in this background. Although our analysis could not account for the effect of small or structured RNA molecules (tRNAs, rRNAs, etc.) we concluded that the gun1 mutation does not restore PhANG expression in the sig6 mutant by globally restoring plastid transcript levels or recoupling genome expression.
Figure 4. The gun1 mutation further reduces plastid transcripts in sig6 mutants.
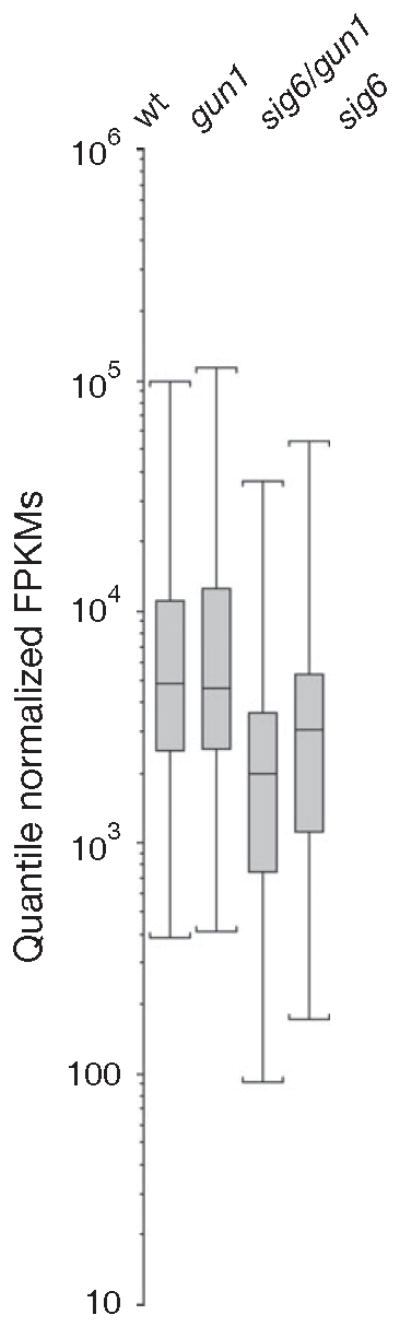
Analysis of steady-state plastid transcripts from 2-day-old seedlings as determined by strand-specific RNA-Seq analysis. Box-plot representation of plastid gene expression levels (Quantile Normalized Fragments Per Kilobase of exon model per Million mapped fragments (FPKMs) reveals that the plastid transcriptome is globally reduced in sig6 mutants and that this reduction is further enhanced in sig6/gun1 mutants.
Tetrapyrroles are involved in SIG2 dependent genome co-ordination
Our published genetic studies have previously linked plastid tetrapyrrole synthesis to retrograde signalling to the nucleus (Mochizuki et al., 2001; Larkin et al., 2003; Woodson et al., 2011). The starting substrate for all tetrapyrroles is glutamyl-tRNA that is enzymatically produced from glutamate and the PEP-transcribed tRNAGlu. We, therefore, investigated the possibility that perturbations in the tetrapyrrole pathway may be responsible for lowered PhANG expression in sig2 and sig6. Indeed, both tRNAGlu and glutamyl- tRNA levels were decreased in sig6 and especially in sig2 (Figures 5a and S7). This decrease of the tetrapyrrole starting substrate is likely to be responsible for the reduced levels of the other tetrapyrroles we measured. The rate-limiting early intermediate δ-aminolevulinic acid (ALA) (Figure 5c), the dark-accumulating late intermediate protochlorophyllide (Pchlide) (Figure 5b), and chlorophyll (Figure S2f) were reduced in both sig2 and sig6. Interestingly, total non-covalently attached heme levels (Figure 5d) were decreased in sig2, but not in sig6 mutants, suggesting that these sigma factors may contribute differently to the tetrapyrrole pathway(s).
Figure 5. Analysis of tRNAGlu and tetrapyrrole levels in sig2 and sig6 mutants.
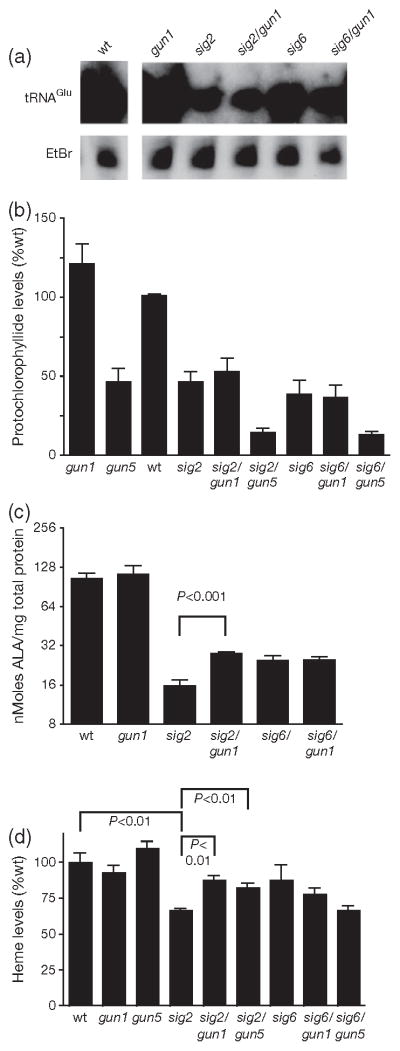
(a) Northern blot analysis of tRNAGlu; (b) steady-state levels of Pchlide; (c) ALA biosynthetic capacity; and (d) steady-state levels of non-covalently bound heme in 2-day-old seedlings. Error bars indicate the mean ± standard error of the mean (SEM) of two independent experiments with biological duplicates.
Next, we tested if the gun1 mutation affected tetrapyrrole metabolism in either sigma mutant. Indeed, gun1 increased ALA, heme, and Chl levels in the sig2 (but not the sig6) mutant (Figures 5c,d and S2f). Pchlide levels, however, were not elevated significantly (P-value = 0.18; Figure 5b). The ability of gun1 to increase tetrapyrrole synthesis in sig2, however, was not achieved by an increase in total tRNAGlu transcripts or glutamyl-tRNA levels (Figures 5a and S7).
If reduced PhANG expression in the sig2 mutant was due to decreased tetrapyrrole synthesis, as suggested by the previous experiments, then exogenous feeding of the precursor ALA should be able to restore PhANG expression. Indeed, when low levels of ALA (32–64 nM) were added to the medium, LHCB expression increased in etiolated sig2 seedlings (Figure 6b). A similar trend was not observed in sig6 mutants, in which LHCB was unresponsive to low levels of ALA. At higher levels of ALA (>250 nM) LHCB expression was repressed in all genotypes. This finding may be due to the increase of toxic tetrapyrrole intermediates, which led to photobleaching after light exposure (Figure S8d) and are known to affect chloroplast DNA levels even in the dark (Vinti et al., 2000).
Figure 6. SIG2-mediated signals involve tetrapyrrole synthesis.
(a) Quantitative polymerase chain reaction (qPCR) analysis of steady-state PhANG mRNA transcript levels of 2-day-old seedlings. Data shown are the mean ± standard error of the mean (SEM) of triplicate reactions of a representative experiment.
(b) LHCB1.2 promoter-driven luciferase activity in 2-day-old etiolated seedlings grown in the presence of the indicated the amount of δ-aminolevulinic acid (ALA). Mean ± standard error of the mean (SEM) of eight individual seedlings is shown.
As there appeared to be a correlation between tetrapyrrole levels in PhANG expression in the sig2 mutant, we next tested the genetic interaction of the sig mutants with gun5 (a weak allele of chlH encoding the H subunit of the Mg-chelatase, the first enzyme of the Mg branch of the tetrapyrrole pathway). Analysis of the double mutants demonstrated that the gun5 mutation partially restored PhANG expression in etiolated sig2, but not sig6 mutants (Figure 2a). Plastid transcript levels, on the other hand, did not change significantly (Figure S3a). To observe the effect of gun5 on tetrapyrrole synthesis we measured the end products of the branched tetrapyrrole pathway. As expected, combining gun5 with either sig mutant reduced Chl (Figure S2f) and Pchlide (Figure 5b) levels further due to blocking the Mg branch of the tetrapyrrole pathway. Heme levels, however, were increased significantly (P-value < 0.01) in the sig2 (but not the sig6) mutant by the addition of gun5 (Figure 5d). Together these data suggested that by diverting flow of the tetrapyrrole pathway towards heme synthesis, the gun5 mutation was able to partially restore PhANG expression in the sig2 mutant.
FC1-produced heme has previously been hypothesized to be a signalling molecule from developing chloroplasts that have been photobleached with NF (Woodson et al., 2011). To test if a similar situation exists in sig2 mutants, we constructed transgenic lines that overexpressed either FC1 or FC2. As during a norflurazon treatment, FC1 overexpression increased PhANG expression in a sig2 mutant without affecting plastid gene expression (Figure 6a, S8a). FC2 overexpression, however, was unable to affect PhANG expression even though comparable levels of transcript and protein were synthesized (Figure S8b,c).
Plastid transcription plays a role in GUN signalling in herbicide-treated plants
The above results suggested that the effects of the sig2 and sig6 mutations on retrograde signalling are similar to those of herbicides and plastid biogenesis inhibitors such as NF and Linc, which also reduce plastid transcript levels (Gray et al., 2003). This finding led us to test if the sigma factors were playing a role in regulation of PhANGs in NF-treated seedlings. Indeed, photobleaching wt seedlings with NF reduced transcript levels of all six SIG genes (Figure 7a), which may have contributed to a reduction of plastid transcript levels (Figure S9). In addition, overexpression of either SIG2 or SIG6 was able to partially restore PhANG expression in these photobleached seedlings (Figure 7b).
Figure 7. Relationship between SIG-mediated and herbicide-inhibitor signals.
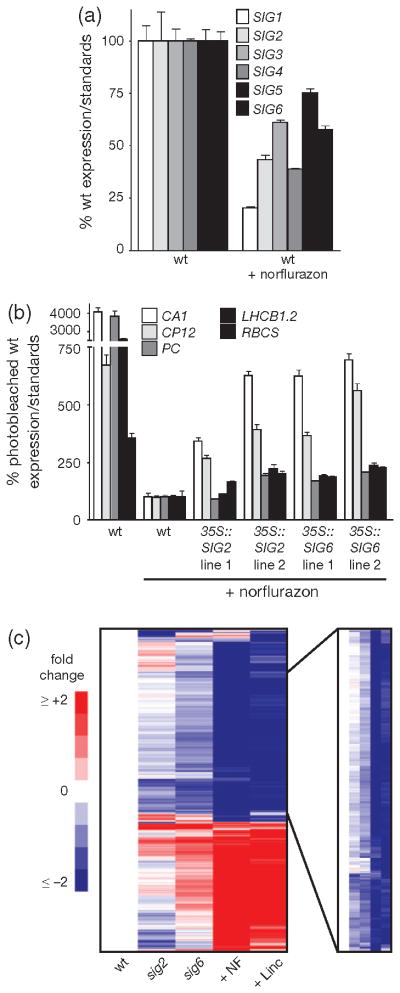
Quantitative polymerase chain reaction (qPCR) analysis of steady-state (a) Sigma and (b) PhANG mRNA transcript levels of 2-day-old seedlings. Data shown are the mean ± standard error of the mean (SEM) of triplicate reactions of a representative experiment. (c) A heat-map based on a microarray analysis depicts 1638 genes differentially (≥twofold) expressed in the sigma mutants (sig2 or sig6) or by herbicide-inhibitor treatment (NF or Linc). Average values were calculated using untreated wild type (wt) as a reference. To the right is a zoomed image of one cluster of 714 similarly expressed genes that was subjected to gene ontology analysis, 353 of which encode plastid proteins.
To determine the extent to which SIG-mediated signals overlap with each other and with drug inhibitor-induced signals, we performed a microarray analysis of wt, sig2, sig6, wt treated with NF, and wt treated with Linc. Compared with untreated wt seedlings, 1638 nuclear genes were differentially expressed at least twofold in at least one group. A cluster analysis of these genes showed that while the drug treatments had a broader affect on nuclear gene expression, there is significant correlation between the groups (Figure 7c). One major cluster of 714 down-regulated genes was enriched significantly for plastid (353 genes), and photosynthesis-related genes [photosynthesis (81 genes), photosystem (42 genes), thylakoid (68 genes), and tetrapyrrole metabolic process (20 genes)]. A comparison of genes that were differentially expressed in each group (cutoff = twofold change in expression) revealed that there were significant overlaps between the sigma mutants and drug treatments (Table S5 and Figure S10e,f). For instance, 76% of the Linc group genes were shared with the NF group and 49% of the sig2 group genes were shared with the sig6 group. Furthermore, 43% and 60% of genes up- and down-regulated, respectively, in sig2 were similarly affected by at least one of the drug treatments and 55% and 96% of genes up- and down-regulated, respectively, in sig6 were similarly affected by at least one of the drug treatments. Together these results suggest that at least part of the NF and Linc responses are due to a decrease in SIG function and/or plastid transcription.
DISCUSSION
SIG2 and SIG6 coordinate plastid and nuclear gene expression in developing seedlings
In this study we aimed to determine the role of plastid transcription in retrograde signalling. To this end, we analyzed mutants that were defective in all six SIG genes and hypothesized that each mutant, defective in transcription of a certain set of plastid genes, may exhibit retrograde signalling without additional stresses. Although most of the sig mutants were defective in the expression of at least one plastid gene that we monitored, sig2 and sig6 mutants were clearly the most affected and the only mutants to have pale phenotypes (Figure 1b–d). Two other mutants, sig1 and sig5 appeared to be essentially wt in both appearance and plastid transcript accumulation even though SIG1 and SIG5 are normally present in the conditions that we tested (Demarsy et al., 2006). Previous reports, however, have shown that these two proteins may have more specialized functions: SIG1 being involved in WRKY-mediated pathogen response (Lai et al., 2011) and redox control of psaA expression (Shimizu et al., 2010) and SIG5 being a stress-induced sigma factor that directs transcription of psbD at a blue light-responsive promoter (Nagashima et al., 2004b).
Analysis of nuclear gene expression in the sig mutants revealed that both sig2 and sig6 had a significant reduction of PhANG transcripts, a finding that suggested that the reduced transcription of a certain plastid transcript or set of transcripts was causing a retrograde response. The double mutant, sig2/sig6, had a much more severe reduction of both plastid and PhANG transcripts even after 7 days, at which time the single mutants would have recovered (Figure 3a–c). Such a phenotype supports the hypothesis that the sigma factors are partially redundant and utilize the same or overlapping promoters for some genes (Lysenko, 2007). Also, the increased accumulation of SIG6 has been proposed to partially complement the SIG2 deficiency (Privat et al., 2003; Nagashima et al., 2004a; Ishizaki et al., 2005). Alternatively, the severe phenotype of the double mutant may be due to the additive affect of a larger number of transcripts that do not accumulate properly. Given that SIG2 and SIG6 contributed to retrograde signalling differently (see below), such an explanation seems to be responsible at least partly. In any case, these data suggest that the roles of SIG2 and SIG6 in plastid biogenesis and retrograde signalling cannot be replaced by the other endogenously expressed sigma factors. SIG3 and SIG4 may also contribute to retrograde signalling to some extent as their respective mutations caused a mild reduction in some PhANG transcripts (RBS2B, CA1). These two sigma factors are perhaps the least well characterized of the six in Arabidopsis (Lysenko, 2007), and a more detailed analysis (e.g. combination mutants) would be required to determine their role in retrograde signalling.
SIG2 retrograde signalling involves tetrapyrrole synthesis
SIG2 has a specialized role in transcription of tRNAs in the plastid, including tRNAGlu, the starting substrate for all tetrapyrroles in the cell (Kanamaru et al., 2001). As a result, ALA, Chl, Pchlide, and heme all accumulate to lower levels in sig2 mutants (Figures 5b–d and S2f). Such a defect in transcription is relevant considering that tetrapyrroles are candidate retrograde signalling molecules in yeast (Zhang and Hach, 1999; heme), green algae (Vasileuskaya et al., 2005; heme, Mg-Protoporphyrin IX), red algae (Kobayashi et al., 2009; Mg-Protoporphyrin IX), and humans (Yin et al., 2007; heme). In Arabidopsis, analysis of the gun mutants revealed that PhANGs are regulated by increased heme synthesis by FC1 in NF-photobleached seedlings (Woodson et al., 2011). This situation led us to speculate if the decrease of tRNAGlu transcripts and tetrapyrrole synthesis in sig2 mutants was responsible for the decreased PhANG expression.
ALA feeding (Figure 6b), addition of the gun5 allele (Figure 2a), or overexpression of FC1 (Figure 6a) increased PhANG expression in the sig2 mutant, a finding that suggested that increasing heme synthesis can overcome the sig2 retrograde phenotype. Although ALA feeding or FC1 overexpression would increase heme levels directly, the gun5 mutation may be able to increase heme levels by blocking the Mg branch of the tetrapyrrole pathway and by redirecting flow towards heme synthesis. Indeed, gun5 did increase steady-state heme levels in the sig2 mutant (Figure 5d). Somewhat curiously, however, the gun5 mutation only increased PhANG transcripts in etiolated sig2 seedlings. This situation may be due to HY5-mediated light signalling masking the effect of gun5 and increased heme synthesis (Ruckle et al., 2007). Alternatively, the increased cellular demand for metabolic heme in de-etiolated seedlings may reduce the pool that can act as a signal. Interestingly, overexpression of FC2 was unable to affect PhANG expression in sig2 mutants, a finding that was consistent with the hypothesis that only FC1-produced heme is able to induce PhANG expression in undeveloped seedlings (Woodson et al., 2011).
sig6 mutants also have decreased levels of tRNAGlu, ALA, Pchlide and Chl. Heme levels, however, appear to be unaffected (Figures 5a–d and S2f). This factor may be due to the less severe reduction in both tRNAGlu and ALA compared with the sig2 mutant. Alternatively, the tRNAGlu supplied by SIG2 and SIG6 may contribute to different pools of ALA used for heme and Chl synthesis (Czarnecki et al., 2011). Various attempts to further increase heme levels (ALA feeding, gun5 mutation) did not increase PhANG expression in sig6 mutants as they did in sig2 mutants (Figures 2a and 6b). Together these results indicated that the retrograde signals mediated by SIG2 and SIG6 are distinct and that SIG2 signals to the nucleus by increasing tetrapyrrole synthesis through tRNAGlu transcription.
SIG2- and SIG6-mediated retrograde signalling requires GUN1
The plastid-localized PPR protein GUN1 is necessary for the repression of PhANGs during a variety of stresses and conditions, leading to a model in which GUN1 integrates various chloroplast signals (Koussevitzky et al., 2007). Consistent with such a role, GUN1 is necessary for both the SIG2 and SIG6 retrograde signals to regulate PhANG expression (Figure 2a). Although PhANG expression was partially restored by the gun1 mutation in sig2 and sig6, plastid transcript levels (Figure S3a), plastid protein levels (Figure 2c), and the pale appearances (Figure S3c) of these mutants were not restored. This situation is particularly apparent in the sig6 mutant, in which the addition of gun1 resulted in a decrease in both plastid transcript and proteins levels. Of the 72 plastid genes that we could reliably monitor by strand-specific RNA-sequencing, steady-state levels of all 72 were decreased to some extent (Figures 4 and S6b). Presumably this situation led to the sick and pale visual phenotype of sig6/ gun1 mutants. Together, these data indicate that the gun1 mutation does not restore SIG2 and SIG6 function and that GUN1 either (i) affects a smaller or structured RNA(s) not included in our analysis, (ii) affects translation of a specific RNA(s) like some PPR proteins (Schmitz-Linneweber and Small, 2008), or (iii) affects a plastid process other than RNA metabolism.
These data are consistent with the model that GUN1 acts downstream of plastid stresses and developmental defects (Koussevitzky et al., 2007). However, as gun1 is able to increase steady-state heme levels in sig2 mutants (Figure 5d), it is tempting to speculate that GUN1 may also be affecting tRNAGlu transcripts (Figure 5a). Northern blot analysis demonstrated that gun1 does not increase the steady-state levels of this transcript. GUN1, however, may be involved in regulating the fate of the transcript (i.e. translation versus tetrapyrrole synthesis) as there is only one tRNAGlu gene in the plastid genome. Alternatively, as a recent report has shown that separate pools of ALA exist for heme and Chl biosynthesis (Czarnecki et al., 2011), GUN1 may help dictate the pool of ALA to which tRNAGlu transcripts contribute. This approach is especially interesting, considering that gun1 is able to increase heme but not Pchlide levels in sig2. Our laboratory is currently investigating these possibilities.
Recent research from other laboratories has suggested that the regulation of PEP and sigma factors is quite complex. The PPR protein Delayed Greening 1 (DG1) interacts with SIG6 and is required for the expression of many PEP-dependent genes in seedlings (Chi et al., 2010). The possibility that DG1 regulates retrograde signals such as GUN1 has not been investigated. Phosphorylation studies were carried out with SIG1 and SIG6 that demonstrated that such post-translational modifications can change their affinity to different plastid promoters. In the case of SIG1, phosphorylation in regulated by the redox state of plastoquinone (Schweer et al., 2010; Shimizu et al., 2010; Turkeri et al., 2012). How this additional level of sigma regulation will also affect retrograde signalling should be a topic of subsequent studies. Furthermore, the redox state of photosynthetic electron transport chain has a more general affect on PEP activity in photosynthesizing chloroplasts (Arsova et al., 2010; Kindgren et al., 2012). Under high light intensities, PEP activity is the source of a signal to the nucleus to regulate LHCB, but this signalling does not appear to involve GUN1 or tetrapyrroles. Lastly, a role for SIG6 in retrograde signalling during singlet oxygen stress has been reported (Coll et al., 2009). A genetic screen uncovered a sig6 mutant (soldat8) that was able to survive the high levels of singlet oxygen in the Arabidopsis flu mutant that has uncontrolled tetrapyrrole synthesis. Together these studies demonstrate that the effect of plastid transcription on plastid-nuclear communication is quite complex, involves multiple signals, and needs further examination.
sigma mutants validate drug inhibitor studies
Our studies strongly suggested that the retrograde signals in the sig2 and sig6 mutants are similar to those induced by drug inhibitors. Similar to NF-photobleached seedlings, the gun5 mutation (Figure 2a), FC1 overexpression (Figure 6a), and ALA feeding (Figure 6b) restored PhANG expression in sig2 mutants and the gun1 mutation restored PhANG expression in both sig2 and sig6 mutants (Figure 2a,b; Koussevitzky et al., 2007; Mochizuki et al., 2001; Woodson et al., 2011). SIG2 and SIG6 overexpression increased PhANG expression in NF-treated seedlings (Figure 7b). Lastly, a microarray analysis demonstrated that within the broad effect that NF and Linc have on nuclear genes expression, there is a core response shared with the sigma mutants. This relative weakness of the SIG2 and SIG6 signals may be due to their partial redundancy and that the sig2 and sig6 mutants still have photosynthesizing chloroplasts. Taken together, however, these data suggest that at least part of the NF and Linc responses are due to a decrease in SIG function and/or plastid transcription.
Although pharmacology has played an important role in pathway analysis in metazoans (Liu and Butow, 2006; Su, 2006; Conradt, 2009) and genetic screens that used chloroplast inhibitors have identified specific processes involved in retrograde signalling (Nott et al., 2006), these studies have recently been under scrutiny (Pfannschmidt, 2010). Therefore, the ability of the sig mutants to phenocopy the ‘GUN signal’ is significant. It validates previous studies by demonstrating NF (and likely Linc) can elicit specific plastid signals by decreasing plastid transcription. We provide additional evidence that these signals are not dependent on light (Ruckle et al., 2007) or that these signals are due to reactive oxygen species (ROS) or severe plastid/cell damage that has been speculated by some groups (Mochizuki et al., 2008; Saini et al., 2011). At the same time, the use of sig mutants offers an alternative system to monitor retrograde signalling when only some plastid genes are affected. The ability to study these signals independent of light is also an advantage as light signalling networks are known to complicate the analysis of retrograde signalling mechanisms (Ruckle et al., 2007). New genetic screens and in-depth analyses that used these mutants could provide clues as to the mechanisms by which signals are transmitted to the nucleus.
CONCLUSIONS
As seeds germinate, the nucleus must provide sigma factors to interact with the PEP core enzyme to start transcription in the developing chloroplast. The work presented here demonstrates that the successful PEP-mediated expression of tRNAGlu by SIG2 and other photosynthesis genes by SIG6 in the plastid are required to establish a communication pathway with the nucleus to regulate chloroplast development. This signalling network is independent of light and requires GUN1. The ability to elicit specific retrograde signals with sigma mutations offers a novel way to elucidate the mechanisms by which plastids communicate with the nucleus.
EXPERIMENTAL PROCEDURES
Biological material, growth conditions, and treatments
The Arabidopsis transgenic line 6-3 (Col-0/6-3) (Koussevitzky et al., 2007) in the Columbia background was used as the wt and parent line for transgenic constructs. This line contains the LUC (Luciferase) marker under the control of a minimal LHCB1.2 promoter. gun1-9, gun5-1, sig1-1, sig3-2, and sig6-1 were described previously (Ishizaki et al., 2005; Koussevitzky et al., 2007; Lai et al., 2011; Susek et al., 1993; Zghidi et al., 2007; Methods S1). The T-DNA insertion lines (Alonso et al., 2003) sig1-1 (SALK_147985), sig2-2 (SALK_045706), sig3-2 (SALK_009166), sig4- 2 (SALK_078760), sig5-3 (SALK_141383) and sig6-1 (SAIL_893_C09) were obtained from the Arabidopsis Biological Resource Center (http://abrc.osu.edu/). Double mutants were obtained by crossing mutant lines and all genotypes were confirmed by PCR-based markers. Primer sequences for PCR are listed in Methods S1.
Seeds were surface sterilized using chlorine gas for 16 h and plated on Linsmaier and Skoog medium pH 5.7 (Caisson Laboratories, North Logan, UT, USA) with 0.6% micropropagation type-1 agar powder (Caisson Laboratories). After a 4–6 day stratification in the dark at 4°C, plates were moved to constant light conditions of 25 μmol m−2 s−1. To grow etiolated seedlings, plates were wrapped in aluminum foil after a 2-h exposure to light. These dark-grown seedlings were harvested under dim green light. For plastid transcription inhibition studies, rifampicin powder (Sigma) [(final) 50 μg ml−1] was added directly to cool medium before solidification. For photobleaching experiments, medium was supplemented with 5 μM NF (Supelco, Bellafonte, PA, USA) or 220 μg ml−1 Linc (Sigma). For experiments with the seedling lethal sig2/sig6 mutant, medium was supplemented with 1% sucrose (w/v). To minimize differences in seed quality for a given experiment, all seeds from all genotypes were harvested from plants grown concurrently in the same growth chambers.
For cloning purposes, E. coli and Agrobacterium tumefaciens strains were grown in liquid Miller nutrient broth or solid medium that contained 1.5% agar (w/v). Cells were grown at 37°C (E. coli) or 28°C (A. tumefaciens) with the appropriate antibiotics and liquid medium was shaken at 225 rpm.
RNA extraction, real-time quantitative PCR, and northern blotting
Total RNA was extracted from whole seedlings using the Spectrum Plant Total RNA kit (Sigma-Aldrich, http://www.sigmaaldrich.com) and cDNA was synthesized using the Maxima first strand cDNA synthesis kit (Fermentas, http://www.fermentas.com) following the manufacturer’s instructions. Real-time PCR was performed using a C1000 Thermal Cycler and CFX384 R-T System detection instrument (Biorad, http://www.bio-rad.com). The following standard thermal profile was used for all PCRs: 95°C for 3 min, 40 cycles of 95°C for 10 s and 60°C for 30 s. Expression levels for all genes were normalized using Actin2 and 18S rRNA as standards. The primers used for qPCR are listed in Methods S1.
Northern blot analysis of tRNAGlu and glutamyl-tRNA were performed as follows: for tRNAGlu (deacetylated form), total RNA was isolated from whole seedlings using the mirVanaTM miRNA Isolation Kit (Ambion, http://www.invitrogen.com/site/us/en/home/brands/ambion.html). Then, 0.5 μg of total RNA was separated on denaturing 6% urea polyacrylamide gel electrophoresis (PAGE) and transferred to a nylon membrane. Hybridization was carried out following standard procedures previously described (Chory et al., 1991). tRNAGlu DNA probes were generated from a plasmid that contained the trnE genomic DNA. The primers used for probe amplification are listed in Methods S1. For the aminoacylated form of tRNAGlu (glutamyl-tRNA), total RNA isolation was carried out under acidic conditions using the Trizol reagent (Invitrogen http://www.invitrogen.com). Then, 0.5 μg of total RNA was separated by 6.5% acid urea PAGE and transferred as previously described (Kohrer and Rajbhandary, 2008). Membrane was subjected to northern blot hybridization as described above.
RNA-Seq library construction and analysis
Total RNA was isolated from 2-day-old seedlings as described above. For each sample, 10 μg of RNA was depleted of rRNAs following the manufacturer’s protocol included in the Ribominus kit from Life Technologies, http://www.lifetechnologies.com/uk/en/home.html. 50 ng of rRNA-depleted samples were used for strand-specific library construction according to the ‘Directional mRNA-Seq Library Prep Pre-Release’ protocol (Illumina, CA, USA). Libraries were sequenced on an Illumina GAIIx instrument. Sequencing data were aligned to the Arabidopsis TAIR10 reference genome using TOPHAT (version 1.3.3)/BOWTIE (version 0.12.5) using default commands with the exception of the –segment-length 20 and the –library-type fr-second strand flags (Langmead et al., 2009; Trapnell et al., 2009). Cufflinks software was used for quantification of transcript abundance (Trapnell et al., 2010).
Microarray expression and cluster analysis
Total RNA was extracted from 2-day-old seedlings from three separate biological experiments. The Affymetrix GeneChip Arabidopsis ATH1 Genome Array (Affymetrix, Santa Clara, CA, USA) was used to analyze the expression of ~24 000 genes according to the manufacturer’s instructions. Laboratory procedures and data analysis are described in detail in Methods S1.
Luciferase activity measurements
Single 2-day-old seedlings were placed in a microplate well that contained 100 μl Linmaier and Skoog medium pH 5.7. Next, 100 μl of water that contained 2 mM D–luciferin and 0.1% Tween 20 were added followed by a 1 h incubation at room temperature. Luminescence was measured with a Promega (http://www.promega.com/) Glomax 96 microplate luminometer.
Chlorophyll measurements
Two-day-old seedlings were frozen in liquid N2 and homogenized. Chlorophyll was extracted twice in ice-cold 80% acetone and cell debris was removed by centrifugation at 10 000 g for 20 min at 4°C. Protein levels were determined by Bradford assay after solubilizing the dried pellet in 0.1 N NaOH. Chlorophyll was measured spectrophotometrically and levels were calculated according to Hendry and Price (1993).
Heme measurements
Two-day-old seedlings were frozen in liquid N2 and homogenized. Total non-covalently bound heme was extracted twice using acetone with 2% HCL (v:v) and measured by the chemiluminescence-based method described by Masuda and Takahashi (2006) using a Promega Glomax 96 microplate luminometer that measures reconstituted activity from apo-horseradish peroxidase (Biozyme, Blaenovan, Wales). Protein levels were determined by Bradford assay after solubilizing the dried pellet in 0.1 N NaOH.
Pchlide measurements
Pchlide was extracted and quantified as previously described (Shin et al., 2009). Pchlide levels were determined by measuring fluorescence in a Tecan Safire2 fluorometer.
ALA measurements
ALA accumulation was measured as described previously (Beator and Kloppstech, 1993) with some modifications. Two-day-old Arabidopsis seedlings were transferred to liquid Linsmaier and Skoog (LS) medium that contained 1.3 mM levulinic acid (Sigma) and incubated for 16 h. Tissue was frozen in liquid nitrogen, homogenized in 1 N trichloroacetic acid (TCA), 1% sodium dodecyl sulphate (SDS) and centrifuged at 65 000 g for 10 min at 4°C. The resulting protein pellet was washed twice with acetone, resuspended in 0.1 N NaOH, centrifuged at 65 000 g for 10 min at 4°C, and the protein concentration of the supernatant was determined using a bicinchoninic acid (BCA) assay (Pierce, Rockford, IL, USA). The cleared extract was neutralized with an equal volume of 0.5 M NaH2PO4, pH 7.5 and condensed with 50 μl of ethylacetoacetate (Sigma) by boiling for 10 min. The cooled extract was treated with an equal volume of modified Ehrlich’s reagent (0.2 g of p-dimethylaminobenzaldehyde (Sigma), 8.4 ml of acetic acid, 1.6 ml of 70% perchloric acid), centrifuged for 5 min and assayed spectrophotometrically at 553 nm using a Tecan Saphire plate reader. The amount of porphobilinogen formed from ALA was determined using a coefficient of extinction of 7.45 × 104 M−1 cm−1 (Harel and Klein, 1972) and normalized to protein content.
Confocal imaging
Confocal microscopy was performed with a Leica SP/2 inverted microscope. Image analysis was performed with the Leica SP/2 software package and the ImageJ bundle provided by the Wright Cell Imaging facility.
Protein extraction and western blot analysis
Protein extraction and western blot analysis were performed using standard laboratory procedures and described in detail in Methods S1.
Construction of overexpression plasmids
Plasmids and plant lines used in this study are listed in Methods S1.
Supplementary Material
Acknowledgments
We thank Dr. Samantha S. Orchard for useful discussions about our findings and for the seedling images presented in this manuscript and Dr. Chris Benner for assistance with the microarray analysis. J.D.W. and R.J.S. were supported by K fellowships from the NIH, J.M.P.-R. was funded by the Spanish Ministry of Education and HHMI. The authors acknowledge the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the U.S. Department of Energy through grant DE-FG02- 04ER15540 and support from the Howard Hughes Medical Institute and the Gordon and Betty Moore foundation. J.C. and J.R.E. are investigators of the Howard Hughes Medical Institute. All of the sequencing data has been deposited to the National Center for Biotechnology Information (SRA accession number SRA052959).
Footnotes
Additional Supporting Information may be found in the online version of this article.
Figure S1. Plastid transcription is required for PhANG expression in Arabidopsis.
Figure S2. SIG2 and SIG6-mediated plastid transcription is required for PhANG expression.
Figure S3. Genetic interaction between sigma and gun mutants.
Figure S4. Correlation of PhANG and plastid gene expression in sig2 and sig6.
Figure S5. SIG2 and SIG6 are partially redundant for genome coordination.
Figure S6. The gun1 mutation further reduces plastid transcripts in sig6 mutants.
Figure S7. Analysis of glutamyl-tRNA levels in sig2 and sig6 mutants.
Figure S8. SIG2-mediated signals involve tetrapyrrole synthesis.
Figure S9. NF reduces plastid gene expression.
Figure S10. Microarray analysis of sigma mutants and drug inhibitor treatments.
Table S1. Arabidopsis mutants used in study.
Table S2. List of control genes used in this study.
Table S3. Primers used in study.
Table S4. Plasmids used in study.
Table S5. Overlap of retrograde-responsive nuclear genes identified by microarray analysis
Table S6. List of genes up-regulated by retrograde signals as identified by microarray analysis.
Table S7. List of genes down-regulated by retrograde signals as identified by microarray analysis.
Methods S1. Supplemental experimental procedures: construction of overexpression plasmids; protein extraction and Western Blot analysis; microarray expression and cluster analysis.
Please note: As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer-reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
References
- Alonso JM, Stepanova AN, Leisse TJ, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Arsova B, Hoja U, Wimmelbacher M, Greiner E, Ustun S, Melzer M, Petersen K, Lein W, Bornke F. Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol-dependent manner: evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana. Plant Cell. 2010;22:1498–1515. doi: 10.1105/tpc.109.071001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbrook AC, Howe CJ, Purton S. Why are plastid genomes retained in non-photosynthetic organisms? Trends Plant Sci. 2006;11:101–108. doi: 10.1016/j.tplants.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Beator J, Kloppstech K. The circadian oscillator coordinates the synthesis of apoproteins and their pigments during chloroplast development. Plant Physiol. 1993;103:191–196. doi: 10.1104/pp.103.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi W, Mao J, Li Q, Ji D, Zou M, Lu C, Zhang L. Interaction of the pentatricopeptide-repeat protein DELAYED GREENING 1 with sigma factor SIG6 in the regulation of chloroplast gene expression in Arabidopsis cotyledons. Plant J. 2010;64:14–25. doi: 10.1111/j.1365-313X.2010.04304.x. [DOI] [PubMed] [Google Scholar]
- Chory J, Nagpal P, Peto CA. Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in Arabidopsis. Plant Cell. 1991;3:445–459. doi: 10.1105/tpc.3.5.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll NS, Danon A, Meurer J, Cho WK, Apel K. Characterization of soldat8, a suppressor of singlet oxygen-induced cell death in Arabidopsis seedlings. Plant Cell Physiol. 2009;50:707–718. doi: 10.1093/pcp/pcp036. [DOI] [PubMed] [Google Scholar]
- Conradt B. Genetic control of programmed cell death during animal development. Annu Rev Genet. 2009;43:493–523. doi: 10.1146/annurev.genet.42.110807.091533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarnecki O, Hedtke B, Melzer M, Rothbart M, Richter A, Schroter Y, Pfannschmidt T, Grimm B. An Arabidopsis GluTR binding protein mediates spatial separation of 5-aminolevulinic acid synthesis in chloroplasts. Plant Cell. 2011;23:4476–4491. doi: 10.1105/tpc.111.086421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delannoy E, Stanley WA, Bond CS, Small ID. Pentatricopeptide repeat (PPR) proteins as sequence-specificity factors in post-transcriptional processes in organelles. Biochem Soc Trans. 2007;35:1643–1647. doi: 10.1042/BST0351643. [DOI] [PubMed] [Google Scholar]
- Demarsy E, Courtois F, Azevedo J, Buhot L, Lerbs-Mache S. Building up of the plastid transcriptional machinery during germination and early plant development. Plant Physiol. 2006;142:993–1003. doi: 10.1104/pp.106.085043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estavillo GM, Crisp PA, Pornsiriwong W, et al. Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell. 2011;23:3992–4012. doi: 10.1105/tpc.111.091033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favory JJ, Kobayshi M, Tanaka K, Peltier G, Kreis M, Valay JG, Lerbs-Mache S. Specific function of a plastid sigma factor for ndhF gene transcription. Nucleic Acids Res. 2005;33:5991–5999. doi: 10.1093/nar/gki908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez AP, Strand A. Retrograde signaling and plant stress: plastid signals initiate cellular stress responses. Curr Opin Plant Biol. 2008;11:509–513. doi: 10.1016/j.pbi.2008.06.002. [DOI] [PubMed] [Google Scholar]
- Gray JC, Sullivan JA, Wang JH, Jerome CA, MacLean D. Coordination of plastid and nuclear gene expression. Philos Trans R Soc Lond B Biol Sci. 2003;358:135–145. doi: 10.1098/rstb.2002.1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka M, Kanamaru K, Takahashi H, Tanaka K. Molecular genetic analysis of chloroplast gene promoters dependent on SIG2, a nucleus-encoded sigma factor for the plastid-encoded RNA polymerase, in Arabidopsis thaliana. Nucleic Acids Res. 2003;31:7090–7098. doi: 10.1093/nar/gkg935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel E, Klein S. Light dependent formation of -aminolevulinic acid in etiolated leaves of higher plants. Biochem Biophys Res Commun. 1972;49:364–370. doi: 10.1016/0006-291x(72)90419-6. [DOI] [PubMed] [Google Scholar]
- Hendry G, Price AH. Stress indicators: chlorophylls and carotenoids. In: Hendry G, Grime JP, editors. Methods in Comparative Plant Ecology – A Laboratory Manual. London: Chapman and Hall; 1993. pp. 148–152. [Google Scholar]
- Ishizaki Y, Tsunoyama Y, Hatano K, Ando K, Kato K, Shinmyo A, Kobori M, Takeba G, Nakahira Y, Shiina T. A nuclearencoded sigma factor, Arabidopsis SIG6, recognizes sigma-70 type chloroplast promoters and regulates early chloroplast development in cotyledons. Plant J. 2005;42:133–144. doi: 10.1111/j.1365-313X.2005.02362.x. [DOI] [PubMed] [Google Scholar]
- Kanamaru K, Nagashima A, Fujiwara M, Shimada H, Shirano Y, Nakabayashi K, Shibata D, Tanaka K, Takahashi H. An Arabidopsis sigma factor (SIG2)-dependent expression of plastid-encoded tRNAs in chloroplasts. Plant Cell Physiol. 2001;42:1034–1043. doi: 10.1093/pcp/pce155. [DOI] [PubMed] [Google Scholar]
- Kindgren P, Kremnev D, Blanco NE, de Dios Barajas Lopez J, Fernandez AP, Tellgren-Roth C, Small I, Strand A. The plastid redox insensitive 2 mutant of Arabidopsis is impaired in PEP activity and high light-dependent plastid redox signalling to the nucleus. Plant J. 2012;70:279–291. doi: 10.1111/j.1365-313X.2011.04865.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Kanesaki Y, Tanaka A, Kuroiwa H, Kuroiwa T, Tanaka K. Tetrapyrrole signal as a cell-cycle coordinator from organelle to nuclear DNA replication in plant cells. Proc Natl Acad Sci USA. 2009;106:803–807. doi: 10.1073/pnas.0804270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohrer C, Rajbhandary UL. The many applications of acid urea polyacrylamide gel electrophoresis to studies of tRNAs and aminoacyltRNA synthetases. Methods. 2008;44:129–138. doi: 10.1016/j.ymeth.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J. Signals from chloroplasts converge to regulate nuclear gene expression. Science. 2007;316:715–719. [PubMed] [Google Scholar]
- Lai Z, Li Y, Wang F, Cheng Y, Fan B, Yu JQ, Chen Z. Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. Plant Cell. 2011;23:3824–3841. doi: 10.1105/tpc.111.090571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloi C, Przybyla D, Apel K. A genetic approach towards elucidating the biological activity of different reactive oxygen species in Arabidopsis thaliana. J Exp Bot. 2006;57:1719–1724. doi: 10.1093/jxb/erj183. [DOI] [PubMed] [Google Scholar]
- Langmead B, Schatz MC, Lin J, Pop M, Salzberg SL. Searching for SNPs with cloud computing. Genome Biol. 2009;10:R134. doi: 10.1186/gb-2009-10-11-r134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin RM, Alonso JM, Ecker JR, Chory J. GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science. 2003;299:902–906. doi: 10.1126/science.1079978. [DOI] [PubMed] [Google Scholar]
- Liu Z, Butow RA. Mitochondrial retrograde signaling. Annu Rev Genet. 2006;40:159–185. doi: 10.1146/annurev.genet.40.110405.090613. [DOI] [PubMed] [Google Scholar]
- Loschelder H, Schweer J, Link B, Link G. Dual temporal role of plastid sigma factor 6 in Arabidopsis development. Plant Physiol. 2006;142:642–650. doi: 10.1104/pp.106.085878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukens JH, Mathews DE, Durbin RD. Effect of tagetitoxin on the levels of ribulose 1,5-bisphosphate carboxylase, ribosomes, and RNA in plastids of wheat leaves. Plant Physiol. 1987;84:808–813. doi: 10.1104/pp.84.3.808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysenko EA. Plant sigma factors and their role in plastid transcription. Plant Cell Rep. 2007;26:845–859. doi: 10.1007/s00299-007-0318-7. [DOI] [PubMed] [Google Scholar]
- Martin W, Rujan T, Richly E, Hansen A, Cornelsen S, Lins T, Leister D, Stoebe B, Hasegawa M, Penny D. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc Natl Acad Sci USA. 2002;99:12246–12251. doi: 10.1073/pnas.182432999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda T, Takahashi S. Chemiluminescent-based method for heme determination by reconstitution with horseradish peroxidase apoenzyme. Anal Biochem. 2006;355:307–309. doi: 10.1016/j.ab.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J. Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc Natl Acad Sci USA. 2001;98:2053–2058. doi: 10.1073/pnas.98.4.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N, Tanaka R, Tanaka A, Masuda T, Nagatani A. The steady-state level of Mg-protoporphyrin IX is not a determinant of plastid-to-nucleus signaling in Arabidopsis. Proc Natl Acad Sci USA. 2008;105:15184–15189. doi: 10.1073/pnas.0803245105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima A, Hanaoka M, Motohashi R, Seki M, Shinozaki K, Kanamaru K, Takahashi H, Tanaka K. DNA microarray analysis of plastid gene expression in an Arabidopsis mutant deficient in a plastid transcription factor sigma, SIG2. Biosci Biotechnol Biochem. 2004a;68:694–704. doi: 10.1271/bbb.68.694. [DOI] [PubMed] [Google Scholar]
- Nagashima A, Hanaoka M, Shikanai T, Fujiwara M, Kanamaru K, Takahashi H, Tanaka K. The multiple-stress responsive plastid sigma factor, SIG5, directs activation of the psbD blue light-responsive promoter (BLRP) in Arabidopsis thaliana. Plant Cell Physiol. 2004b;45:357–368. doi: 10.1093/pcp/pch050. [DOI] [PubMed] [Google Scholar]
- Nott A, Jung HS, Koussevitzky S, Chory J. Plastid-to-nucleus retrograde signaling. Annu Rev Plant Biol. 2006;57:739–759. doi: 10.1146/annurev.arplant.57.032905.105310. [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T. Plastidial retrograde signalling–a true “plastid factor” or just metabolite signatures? Trends Plant Sci. 2010;15:427–435. doi: 10.1016/j.tplants.2010.05.009. [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T, Link G. The A and B forms of plastid DNA-dependent RNA polymerase from mustard (Sinapis alba L.) transcribe the same genes in a different developmental context. Mol Gen Genet. 1997;257:35–44. doi: 10.1007/s004380050621. [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T, Schutze K, Fey V, Sherameti I, Oelmuller R. Chloroplast redox control of nuclear gene expression – a new class of plastid signals in interorganellar communication. Antioxid Redox Signal. 2003;5:95–101. doi: 10.1089/152308603321223586. [DOI] [PubMed] [Google Scholar]
- Pogson BJ, Woo NS, Forster B, Small ID. Plastid signalling to the nucleus and beyond. Trends Plant Sci. 2008;13:602–609. doi: 10.1016/j.tplants.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Privat I, Hakimi MA, Buhot L, Favory JJ, Mache-Lerbs S. Characterization of Arabidopsis plastid sigma-like transcription factors SIG1, SIG2 and SIG3. Plant Mol Biol. 2003;51:385–399. doi: 10.1023/a:1022095017355. [DOI] [PubMed] [Google Scholar]
- Rapp JC, Mullet JE. Chloroplast transcription is required to express the nuclear genes rbcS and cab. Plastid DNA copy number is regulated independently. Plant Mol Biol. 1991;17:813–823. doi: 10.1007/BF00037063. [DOI] [PubMed] [Google Scholar]
- Ruckle ME, DeMarco SM, Larkin RM. Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis. Plant Cell. 2007;19:3944–3960. doi: 10.1105/tpc.107.054312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini G, Meskauskiene R, Pijacka W, Roszak P, Sjogren LL, Clarke AK, Straus M, Apel K. ‘Happy on norflurazon’ (hon) mutations implicate perturbance of plastid homeostasis with activating stress acclimatization and changing nuclear gene expression in norflurazon-treated seedlings. Plant J. 2011;65:690–702. doi: 10.1111/j.1365-313X.2010.04454.x. [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Small I. Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 2008;13:663–670. doi: 10.1016/j.tplants.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Schon A, Krupp G, Gough S, Berry-Lowe S, Kannangara CG, Soll D. The RNA required in the first step of chlorophyll biosynthesis is a chloroplast glutamate tRNA. Nature. 1986;322:281–284. doi: 10.1038/322281a0. [DOI] [PubMed] [Google Scholar]
- Schweer J, Turkeri H, Kolpack A, Link G. Role and regulation of plastid sigma factors and their functional interactors during chloroplast transcription – recent lessons from Arabidopsis thaliana. Eur J Cell Biol. 2010;89:940–946. doi: 10.1016/j.ejcb.2010.06.016. [DOI] [PubMed] [Google Scholar]
- Shimizu M, Kato H, Ogawa T, Kurachi A, Nakagawa Y, Kobayashi H. Sigma factor phosphorylation in the photosynthetic control of photosystem stoichiometry. Proc Natl Acad Sci USA. 2010;107:10760–10764. doi: 10.1073/pnas.0911692107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Kim K, Kang H, Zulfugarov IS, Bae G, Lee CH, Lee D, Choi G. Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc Natl Acad Sci USA. 2009;106:7660–7665. doi: 10.1073/pnas.0812219106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirano Y, Shimada H, Kanamaru K, et al. Chloroplast development in Arabidopsis thaliana requires the nuclear-encoded transcription factor sigma B. FEBS Lett. 2000;485:178–182. doi: 10.1016/s0014-5793(00)02216-x. [DOI] [PubMed] [Google Scholar]
- Su TT. Cellular responses to DNA damage: one signal, multiple choices. Annu Rev Genet. 2006;40:187–208. doi: 10.1146/annurev.genet.40.110405.090428. [DOI] [PubMed] [Google Scholar]
- Susek RE, Ausubel FM, Chory J. Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell. 1993;74:787–799. doi: 10.1016/0092-8674(93)90459-4. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkeri H, Schweer J, Link G. Phylogenetic and functional features of the plastid transcription kinase cpCK2 from Arabidopsis signify a role of cysteinyl SH-groups in regulatory phosphorylation of plastid sigma factors. FEBS J. 2012;279:395–409. doi: 10.1111/j.1742-4658.2011.08433.x. [DOI] [PubMed] [Google Scholar]
- Vasileuskaya Z, Oster U, Beck CF. Mg-protoporphyrin IX and heme control HEMA, the gene encoding the first specific step of tetrapyrrole biosynthesis, in Chlamydomonas reinhardtii. Eukaryot Cell. 2005;4:1620–1628. doi: 10.1128/EC.4.10.1620-1628.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinti G, Hills A, Campbell S, Bowyer JR, Mochizuki N, Chory J, Lopez-Juez E. Interactions between hy1 and gun mutants of Arabidopsis, and their implications for plastid/nuclear signalling. Plant J. 2000;24:883–894. doi: 10.1046/j.1365-313x.2000.00936.x. [DOI] [PubMed] [Google Scholar]
- Wagner D, Przybyla D, Op den Camp R, et al. The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science. 2004;306:1183–1185. doi: 10.1126/science.1103178. [DOI] [PubMed] [Google Scholar]
- Woodson JD, Chory J. Coordination of gene expression between organellar and nuclear genomes. Nat Rev Genet. 2008;9:383–395. doi: 10.1038/nrg2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Perez-Ruiz JM, Chory J. Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Curr Biol. 2011;21:897–903. doi: 10.1016/j.cub.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Wu N, Curtin JC, et al. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- Zghidi W, Merendino L, Cottet A, Mache R, Lerbs-Mache S. Nucleus-encoded plastid sigma factor SIG3 transcribes specifically the psbN gene in plastids. Nucleic Acids Res. 2007;35:455–464. doi: 10.1093/nar/gkl1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Hach A. Molecular mechanism of heme signaling in yeast: the transcriptional activator Hap1 serves as the key mediator. Cell Mol Life Sci. 1999;56:415–426. doi: 10.1007/s000180050442. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.