Abstract
Background
Structural magnetic resonance imaging (MRI) is sensitive to neurodegeneration and can be used to estimate the risk of converting to Alzheimer’s disease (AD) in individuals with mild cognitive impairment (MCI). Brain changes in AD and prodromal AD involve a pattern of widespread atrophy. The use of multivariate analysis algorithms could enable the development of diagnostic tools based on structural MRI data. In this study we investigated the possibility of combining multiple MRI features in the form of a severity index.
Methods
We used baseline MRI scans from two large multicentre cohorts (AddNeuroMed and ADNI). Based on volumetric and cortical thickness measures at baseline with AD cases and healthy control (CTL) subjects as training sets, we generated an MRI-based severity index using the method of orthogonal projection to latent structures (OPLS). The severity index tends to be close to 1 for AD patients and 0 for CTL subjects. Values above 0.5 indicate a more AD-like pattern. The index was then estimated for subjects with MCI, and the accuracy of classification was investigated.
Results
Based on the data at follow-up, 173 subjects converted to AD, of whom 112 (64.7%) were classified as AD-like and 61 (35.3%) as CTL-like.
Conclusion
We found that joint evaluation of multiple brain regions provided accurate discrimination between progressive and stable MCI, with better performance than hippocampal volume alone, or a limited set of features. A major challenge is still to determine optimal cut-off points for such parameters and to compare their relative reliability.
Keywords: AD, MCI, MRI, multivariate analysis, progression to AD, sensitivity, specificity
Introduction
Alzheimer’s disease (AD) is a progressive age-related neurodegenerative disease and a growing health problem. Definite diagnosis can only be made post-mortem, and requires histopathological confirmation of amyloid plaques and neurofibrillary tangles. At the time of clinical manifestation of dementia, significant irreversible brain damage is already present. Therefore, an accurate diagnosis of AD at an early stage is a prerequisite for initiating disease-modifying treatments. Mild cognitive impairment (MCI) is a heterogeneous syndrome recently recognized as a diagnostic entity that includes the prodromal stage of AD [1]. Thus, subjects with MCI have a markedly increased risk of developing AD, with a conversion rate to AD of 15–20% per year in memory clinic settings (in the general population, the conversion rate is 1–2%) [2]. However, not all subjects with MCI go on to develop AD and some may even revert to normal cognition [3].
Neuroimaging biomarkers follow a dynamic model of change during different stages of the disease and could be valuable predictors of patient outcome. Structural magnetic resonance imaging (MRI) is sensitive to neurodegeneration and analysis of structural changes can be used to estimate the risk of converting to AD in individuals with MCI. The ability to identify an individual at risk of developing AD will be critical if disease-modifying treatments become available.
Brain changes in AD and prodromal AD lead to a pattern of widespread atrophy (measured as both volume and thickness), involving a number of different structures across the brain (e.g. hippocampus, entorhinal cortex, cingulate gyrus and frontal cortices) [4, 5].
Advances in statistical learning with the development of new multivariate and machine learning algorithms capable of dealing with high-dimensional data [e.g. support vector machines and orthogonal projection to latent structures (OPLS)] could enable the development of new diagnostic tools based on structural MRI data [6, 7]. These techniques, based on the principle of multivariate statistics, can be used to perform studies across multiple dimensions of data, while taking into account the effects of all variables on the responses of interest.
Published neuroimaging results are usually difficult to compare because of two main issues: sample size and unaligned MRI acquisition protocols. We have shown previously that the pattern of structural brain differences is similar when comparing two large cohorts with aligned MRI acquisition protocol, regardless of the demographic characteristics [8]. Results from combined large datasets with long follow-up periods would be easier to extrapolate to the general population, giving a more complete picture of dementia/AD. It is anticipated that this will lead to an earlier diagnosis for individual subjects and provide suitable markers for treatment response.
The main goal of this study was to derive an MRI-based severity index, based on multiple MRI features, with potential clinical value for estimating the future clinical progression of subjects with MCI. We used baseline MRI data from two large multicentre cohorts, the AddNeuroMed, a part of Innovative Medicines in Europe (InnoMed) project and the Alzheimer’s Disease Neuroimaging Initiative (ADNI) studies. Based on volumetric and cortical thickness measures, and using the AD cases and healthy control (CTL) subjects as training sets, we used the multivariate technique OPLS to generate a severity index. The index was then estimated for subjects with MCI and the accuracy of classification was evaluated using the available follow-up clinical diagnosis as a priori information. Analysis was performed in order to investigate the cognitive profile of the subjects and the influence of the apolipoprotein E4 (ApoE4) status in connection with the severity index. We also investigated whether additional characteristics of the study subjects (age, education, cognitive profile and ApoE4 status) validate the severity index in those with MCI who did not progress to AD during the study period.
Material and methods
Subjects
Data of subjects from two large multicentre studies, AddNeuroMed and ADNI, were used for the present study.
The AddNeuroMed project is part of the InnoMed European Union FP6 programme, designed to develop and validate novel surrogate markers in AD. It includes a human neuroimaging component [9, 10] which combines MRI data with other biomarker and clinical information. Data were collected from six different sites across Europe: University of Kuopio, Finland; University of Perugia, Italy; Aristotle University of Thessaloniki, Greece; King’s College London, UK; University of Łodz, Poland; and University of Toulouse, France. Written consent was obtained from research participants where possible; in those individuals in whom capacity was compromised by dementia, assent from the patient and written consent from a relative, according to local laws, was obtained. This study was approved by ethical review boards in each participating country. A total of 348 subjects from the AddNeuroMed project were included in the present study: 119 AD patients, 119 MCI patients and 110 healthy CTL subjects.
Data from the ADNI cohort were obtained from the ADNI database (www.loni.ucla.edu/ADNI). ADNI was launched in 2003 by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, the Food and Drug Administration, private pharmaceutical companies and non-profit organizations as a 5-year public–private partnership. The primary goal of ADNI has been to test whether serial MRI, of MCI and early AD could establish a set of sensitive and specific markers of very early AD progression in order to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the duration and cost of clinical trials. Subjects aged 55 to 90 years from more than 50 sites across the USA and Canada participated in the ADNI study; more detailed information is available at www.adni-info.org. For the present study, 716 subjects were included from the ADNI cohort: 176 AD patients, 315 MCI patients and 225 healthy CTL subjects.
Inclusion and exclusion criteria
For the AddNeuroMed cohort, inclusion criteria for the AD group were the National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer's Disease and Related Disorders Association and Diagnostic and Statistical Manual of Mental Disorders (DSM-IV) [11] criteria for probable AD, Mini Mental State Examination (MMSE) score between 12 and 28, age 65 years or above. Exclusion criteria were significant neurological or psychiatric illness other than AD, significant unstable systematic illness or organ failure. All AD subjects had a Clinical Dementia Rating (CDR) scale score of ≥0.5.
Criteria for inclusion in the CTL and MCI groups were MMSE score between 24 and 30, Geriatric Depression Scale score of <5, age 65 years or above, stable medication and good general health, whereas exclusion criteria were DSM-IV criteria for dementia, significant neurological or psychiatric illness other than AD, significant unstable systematic illness or organ failure. Discrimination between patients with MCI and CTL subjects was based on two criteria: (i) CDR score of 0 for CTL subjects and CDR of 0.5 for those with MCI; and (ii) reported occurrence of memory problems (by subject or informant) for MCI patients.
CDR, MMSE and the Consortium to Establish a Registry for Alzheimer’s disease (CERAD) cognitive battery scores were assessed for each subject. The CERAD cognitive battery was replaced with the Alzheimer's disease Assessment Scale (ADAS-Cog) for patients with AD. This cognitive test is specially designed for AD trials [12]. Both the ADAS-Cog and the CERAD battery use the same 10-word recall task, although the scoring in the two tests is in the opposite direction. The mean number of words that were not recalled in the word list of the CERAD immediate recall task was calculated. The variable obtained was termed ADAS1, corresponding to the first subtest of ADAS-Cog. This was performed to provide comparable measures for the ADNI and AddNeuroMed cohorts.
For the ADNI cohort, a detailed description of the inclusion criteria can be found at http://www.adni-info.org/Scientists/AboutADNI.aspx#. Subjects were between 55 and 90 years of age, had a study partner who was able to provide independent evaluation of functioning, and spoke either English or Spanish. All subjects were willing and able to undergo all test procedures including neuroimaging and agreed to longitudinal follow-up. Use of specific psychoactive medications was excluded.
Inclusion criteria for the AD group were MMSE score between 20 and 26, CDR scale score of 0.5 or 1 and NINCDS/ADRDA criteria for probable AD. For inclusion in the MCI group, criteria were MMSE score between 24 and 30, memory problems with objective memory loss measured with the Wechsler Memory Scale Logical Memory II (education-adjusted scores), CDR score of 0.5, absence of significant levels of impairment in other cognitive domains, preservation of activities of daily living and absence of dementia. Inclusion criteria for the CTL group were MMSE score between 24 and 30, CDR score of 0, and absence of depression, MCI and dementia.
A total of 1064 subjects were included in the current study. Ten MCI subjects were classified as CTLs at follow-up and were excluded from the analysis. The characteristics of the study subjects are presented in Table 1, with the demographics of the individual ADNI and AddNeuroMed cohorts shown in Supplementary Table 1. Although both studies have a longitudinal design, only baseline MRI data were analysed in the present study.
Table 1.
Baseline characteristics
| CTL | MCI-s | MCI-c | AD | |
|---|---|---|---|---|
| Number | 335 | 261 | 173 | 295 |
| Age (years) | 74.95±5.74 | 74.79±6.59 | 74.33±6.90 | 75.25±6.81 |
| Gender (male/female) | 167/168 | 152/109 | 102/71 | 129/166 |
| Education (years) | 14.31±4.37 | 13.20±4.91* | 14.83±3.68* | 11.92±4.76 |
| MMSE score | 29.12±1.05 | 27.31±1.69* | 26.61±1.71* | 22.35±3.57 |
| CDR-SOB score | 0.04±0.14 | 1.34±0.76* | 1.80±0.96* | 5.19±2.62 |
| ADAS1 score | 3.1±1.3 | 4.6±1.4 | 5.4±1.3 | 6.3±1.5 |
| ApoE 4+ | 26.3% | 36.0% | 64.0% | 60.3% |
Data are mean±standard deviation. AD, Alzheimer's disease; MCI, mild cognitive impairment (c, converter; s, stable); CTL, healthy control; MMSE, Mini Mental State Examination; CDR-SOB, Clinical Dementia Rating – Sum of Boxes; ADAS1, word list non-learning. Subjects with MCI are divided into MCI-s and MCI-c subgroups based on their follow-up diagnosis at 12, 18, 24 and 36 months.
Statistically significant difference between MCI-s and MCI-c, ANOVA.
Based on the follow-up diagnosis (12 months for the AddNeuroMed study and 12, 18, 24 and 36 months for the ADNI study), subjects with MCI were divided into two groups: those who did not progress to AD (stable MCI; MCI-s) and those who did progress to AD (MCI converting to AD; MCI-c).
MRI acquisition protocol
Data acquisition for the AddNeuroMed study was designed to be compatible with that of ADNI [13]. The imaging protocol for both studies included a high-resolution sagittal 3D T1-weighted MPRAGE volume (voxel size 1.1 × 1.1 × 1.2 mm3) and axial proton density/T2-weighted fast spin echo images. The MPRAGE volume was acquired using a customized pulse sequence specifically designed for the ADNI study to ensure compatibility across scanners [13]. Full brain and skull coverage was required and detailed quality control was carried out on all MRI data according to the AddNeuroMed quality control procedure [9, 10].
Post-acquisition image analysis
Volumetric segmentation, cortical surface reconstruction and cortical parcellation, based on the FreeSurfer software package, version 4.5.0 (http://surfer.nmr.mgh.harvard.edu/), were used to quantify the baseline thicknesses and volumes of brain regions, as described in detail previously [14, 15]. The procedure automatically assigns a neuroanatomical label to each voxel in an MRI volume based on probabilistic information automatically estimated from a manually labelled training set. The regional cortical thickness was measured from 34 areas and the regional volumes were measured from 23 areas (see Table 2). Left- and right-sided thicknesses were averaged. Volumetric measures were corrected for differences in head size by dividing each measurement by the estimated total intracranial volume. This segmentation approach has been previously shown to be comparable in accuracy with manual labelling. The atlas-based normalization procedure increases the robustness and accuracy of the segmentation across scanner platforms [16]. This segmentation approach has been used for multivariate classification of patients with AD and healthy CTL subjects [17, 18], neuropsychological image analysis [19], imaging genetic analysis [20] and biomarker discovery [21].
Table 2.
Variables included in OPLS analysis
| Cortical thickness measures | Volumetric measures |
|---|---|
| Banks of superior temporal sulcus | Third ventricle |
| Caudal anterior cingulate | Fourth ventricle |
| Caudal middle frontal gyrus | Brainstem |
| Cuneus cortex | Corpus callosum anterior |
| Entorhinal cortex | Corpus callosum central |
| Fusiform gyrus | Corpus callosum midanterior |
| Inferior parietal cortex | Corpus callosum midposterior |
| Inferior temporal gyrus | Corpus callosum posterior |
| Isthmus of cingulate cortex | Cerebrospinal fluid |
| Lateral occipital cortex | Accumbens |
| Lateral orbitofrontal cortex | Amygdala |
| Lingual gyrus | Caudate |
| Medial orbitalfrontal cortex | Cerebellar cortex |
| Middle temporal gyrus | Cerebellar white matter |
| Parahippocampal gyrus | Hippocampus |
| Paracentral sulcus | Inferior lateral ventricle |
| Frontal operculum | Putamen |
| Orbital operculum | Cerebral cortex |
| Triangular part of inferior frontal gyrus | Cerebral white matter |
| Pericalcarine cortex | Lateral ventricle |
| Postcentral gyrus | Pallidum |
| Posterior cingulate cortex | Thalamus proper |
| Precentral gyrus | Ventral DC |
| Precuneus cortex | |
| Rostral anterior cingulate cortex | |
| Rostral middle frontal gyrus | |
| Superior frontal gyrus | |
| Superior parietal gyrus | |
| Superior temporal gyrus | |
| Supramarginal gyrus | |
| Frontal pole | |
| Temporal pole | |
| Transverse temporal cortex | |
| Insula |
Statistical methods
To determine the sensitivity and specificity for discriminating between AD and healthy CTL subjects, orthogonal partial least squares to latent structures (OPLS), a supervised multivariate data analysis method was employed under SIMCA P+ software package (UMETRICS AB, Umeå, Sweden). FreeSurfer-derived MRI measures were analysed using the OPLS method [22–24].
Pre-processing was performed using unit variance scaling and mean centring. Variables with a high level of variance are more likely to be expressed in modelling than those with a low variance. Therefore, unit variance scaling was selected to scale the data appropriately. This method uses the inverse standard deviation as a scaling weight for each variable. Mean centring improves the interpretability of the data, by subtracting the variable average from the data; thus the dataset is repositioned around the origin. In this study we used seven-fold cross-validation, which means that one-seventh of the data is omitted for each cross-validation round. Balanced groups were maintained during all cross-validation rounds so that further analysis was not affected. The predictive component is given a Q2(Y) value that describes its statistical significance for separating groups. Q2(Y) values >0.05 are regarded as statistically significant (http://www.umetrics.com/Content/Document%20Library/Files/UserGuides-Tutorials/SIMCA-P_12_UG.pdf).
A total of 57 variables were used for OPLS analysis. No feature selection was performed; in other words, all measured variables were included in the analysis. OPLS classifiers were trained on all data (the 57 variables in Table 2) from all subjects in the combined group of AD and CTL subjects and were then applied to data from subjects with MCI. Feature selection was not used, meaning all variables were included. By excluding specific regions, the models might be less representative and structural features measured from a limited set of pre-defined regions might not be able to reflect the complete pattern of structural abnormalities [25]. Furthermore, Cuingnet et al. have shown that feature selection does not improve the classification but does increase computational time [7]. The effect of feature selection was investigated in another recent study [26] and was shown to improve the results for small cohorts but have little effect on larger samples. The cohort size in the present study was much larger than the largest sample used in the latter study.
It is however important to compare new methods with established approaches. Therefore, as well as using the OPLS technique on the 57 regional MRI measures, we created a further three models based on (i) hippocampal volume alone, (ii) the combination of hippocampal volume and lateral ventricles as recently suggested by Heister et al. [27] and (iii) 10 temporal lobe and ventricular measures based on previously reported information about the most affected regions (medial temporal lobe structures: hippocampus, entorhinal cortex, inferior temporal gyrus, medial temporal gyrus, superior temporal gyrus, parahippocampal gyrus, lateral ventricles, inferior lateral ventricles and third and fourth ventricles).
OPLS is a supervised method which means it has both X and Y variables. The X variables are the original variables (volumes and cortical thickness measures) and Y contains the information about group membership. Y is set to 1 for AD cases and 0 for CTL subjects in the AD versus CTL model. The prediction value for a subject to belong to a group is equal to 1 for maximum likelihood and 0 for minimum likelihood, or vice versa depending on the group. The cut-off value for accepting the observation as correctly predicted is 0.5. When the model is generated, each subject receives a predictive Y value while it is omitted from the modeling during the cross-validation rounds and then predicted on to the model. The AD versus CTL model was used as a classifier to investigate how well it could predict conversion from MCI to AD. Each individual MCI subject was predicted on to the AD versus CTL model and this produced a discriminant index (the severity index based on MRI data) for each individual with MCI, reflecting the degree to which the individual’s MR pattern resembled the pattern of AD subjects or the pattern of CTL subjects. MCI subjects demonstrating a more AD-like or CTL-like pattern than the AD and CTL subjects used to generate the OPLS model may be characterised by OPLS scores above 1 or below 0, respectively, as shown by others [28]. A more detailed description of the method has been reported previously [28].
In order to further evaluate the use of the OPLS score for MCI prediction, we created survival curves for the ADNI cohort alone, as follow-up data to 36 months were available.
Receiver operating characteristic (ROC) curves were computed from the resulting scores by using the cross-validated prediction values of the OPLS models and the areas under the ROC curve (AUCs) were computed.
The AUC is similar to the Wilcoxon statistic, which provides a way to approximate the standard error. This enables the comparison of two algorithms based on formal statistical criteria [29–32]. We used the ROCKIT ROC analysis software package (developed at the University of Chicago and part of the Metz ROC software) for statistical comparisons. Values of P<0.05 were considered significant. Bonferroni correction was used to correct for multiple comparisons.
Analysis of variance (ANOVA) was used to compare continuous measures (e.g. FreeSurfer-derived variables, MMSE score and age) between groups.
Four main study groups were used for analyses: CTL, MCI-s, MCI-c and AD.
The severity index is based only on MRI data and characterises the atrophy pattern of each individual subject as AD-like or CTL-like. In order to investigate how demographic factors and cognitive scores (e.g. age, gender, ApoE4 status, baseline MMSE, 1-year change in MMSE, baseline CDR-SOB and 1-year change in CDR-SOB) alter with the severity index, we divided all subjects (CTL, MCI-s, MCI-c and AD) into groups based on AD-like or CTL-like classification and ApoE4 status.
Results
Demographic and clinical characteristics of the study groups are summarized in Table 1. There were no differences in age between the main study groups. Overall the AD subjects had a higher and the MCI-c subjects a lower level of education compared with the MCI-s group.
Of the 434 MCI subjects, 173 progressed to a diagnosis of AD at follow-up (12 months for the AddNeuroMed cohort and 12, 18, 24 and 36 months for the ADNI cohort).
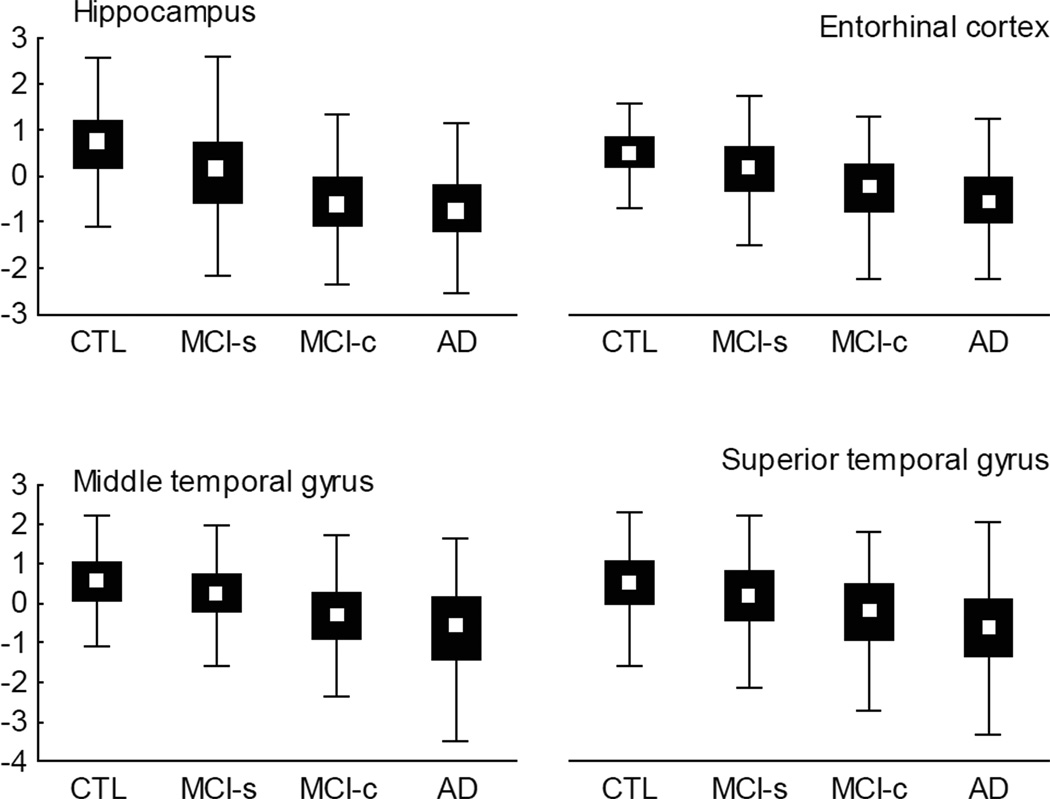
First we assessed whether the FreeSurfer measures replicated previously published differences between the MCI-s and MCI-c groups. We found significant differences between groups for the following structures: banks of superior temporal sulcus, entorhinal cortex, fusiform gyrus, inferior parietal cortex, inferior temporal gyrus, isthmus of cingulate gyrus, lateral occipital cortex, lateral orbitofrontal cortex, medial orbitofrontal cortex, middle temporal gyrus, parahippocampal gyrus, precuneus cortex, rostral middle frontal gyrus, superior parietal gyrus, superior temporal gyrus, supramarginal gyrus, temporal pole, nucleus accumbens, amygdala, hippocampus and inferior lateral ventricle [ANOVA followed by unequal n honestly significant difference (HSD) post hoc analysis]. Figure 1 shows the differences for some of the most AD-specific brain structures (hippocampus, entorhinal cortex, middle temporal gyrus and superior temporal gyrus) in the study groups.
Figure 1.
Box plots of AD-specific brain structures (hippocampal volume and thickness of entorhinal cortex, medial and superior temporal gyruses) that showed significant differences between MCI-s and MCI-c groups using ANOVA with unequal n HSD post hoc test. Data are presented normalised as z-score. Median, percentiles 25–75 (box), range (whiskers). MCI-s, MCI stable; MCI-c, MCI converting to Alzheimer’s disease.
OPLS modelling and quality
An OPLS model was created for CTL versus AD subjects including 57 FreeSurfer measures (34 cortical thickness and 23 volume measures; Table 2). The final model resulted in one predictive and zero orthogonal (1+0) components with cross-validated predictability Q2(Y) = 0.592.
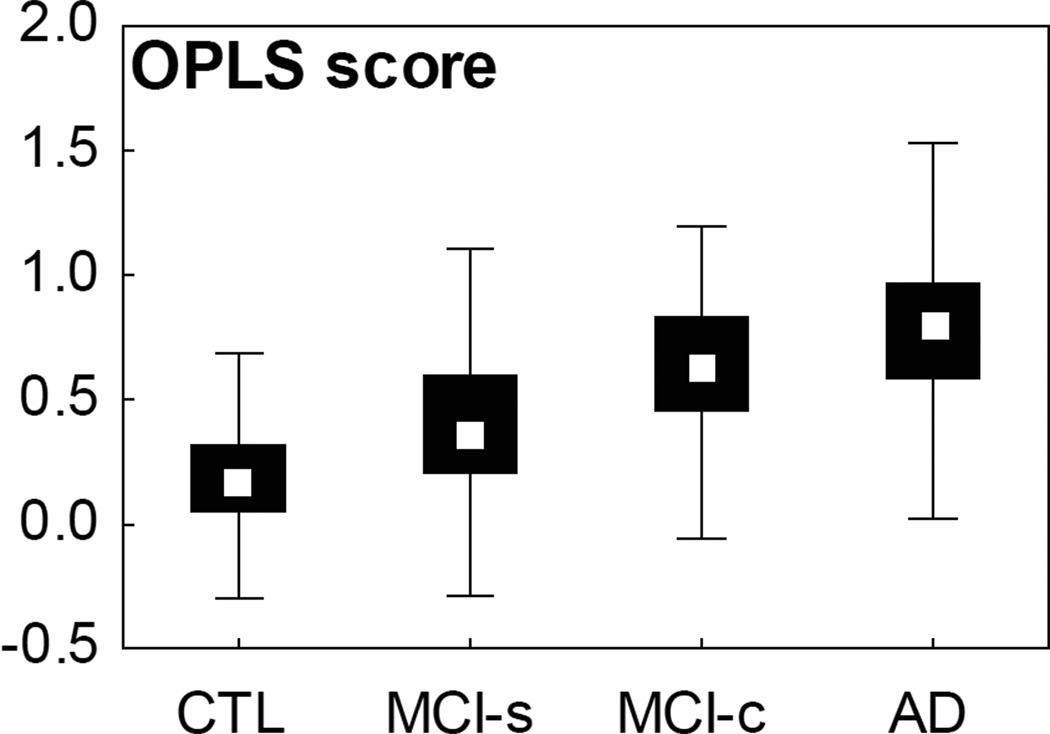
The mean±standard deviation severity indexes were 0.19±0.20 and 0.78±0.28 for the CTL and the AD groups and 0.38±0.28 and 0.65±0.26 for the MCI-s and MCI-c groups, respectively (Figure 2).
Figure 2.
Box plots of the OPLS score for the study groups. Median, percentiles 25–75 (box), range (whiskers). Subjects with a score above 0.5 show a more AD-like pattern of atrophy and below 0.5 a more control-like pattern. MCI-s, MCI stable; MCI-c, MCI converting to AD.
CTL versus AD subjects
The classification results for CTL versus AD subjects are summarized in Table 3. The OPLS classifier enabled the discrimination of subjects with AD from CTLs with high cross-validated sensitivity (86.1%) and specificity (90.5%). Characteristics of the AD and CTL subjects based on their OPLS classification are shown in Table 4. Of 295 AD subjects, 242 were classified as AD-like and 53 as CTL-like. AD subjects classified as CTL-like were younger, although the difference was not statistically significant. This group also had a higher level of education. We found that CTL subjects classified as AD-like were often older than the true negatives, meaning that the oldest controls were more often misclassified, without any influence of the ApoE4 status. Misclassified CTL subjects were also more highly educated.
Table 3.
OPLS modelling: sensitivity, specificity and area under the curve
| Sensitivity | CI | Specificity | CI | Accuracy | CI | AUC | |
|---|---|---|---|---|---|---|---|
| AD vs. CTL | 86.1% | 81.7–89.6 | 90.4% | 86.8–93.2 | 88.4% | 85.7–90.7 | 0.948 |
| MCI-c and MCI-s* | 69.6% | 61.8–76.4 | 66.8% | 61.1–72.0 | 67.7% | 58.0–76.0 | 0.675 |
AD, Alzheimer's disease; MCI, mild cognitive impairment (c, converter; s, stable); CTL, healthy control; CI, confidence interval; AUC, area under the curve.
AD vs. CTL model as training set and MCI subjects as test set. Sensitivity refers to MCI-c classified as AD and specificity to MCI-s classified as CTL.
Table 4.
Characteristics of the AD and CTL subjects based on the OPLS classifier results
| No of subjects |
ApoE 4+/− |
Age (years) |
Baseline MMSE |
Change in MMSE |
Baseline CDR-SOB |
Change in CDR-SOB |
Education (years) |
|
|---|---|---|---|---|---|---|---|---|
| AD | 295 total | |||||||
| AD-like | 242 | 154/83 | 75.56±6.79 | 22.06±3.53* | −2.27±3.99* | 5.38±2.64* | 1.30±3.73* | 11.89±4.67* |
| CTL-like | 53 | 24/27 | 73.82±6.84 | 23.68±3.51* | 0.25±3.48* | 4.33±2.38* | −0.37±3.94* | 12.08±5.23* |
| CTL | 335 total | |||||||
| AD-like | 25 | 5/20 | 78.48±4.81* | 28.84±1.28* | −0.62±1.86 | 0.04±0.14* | 0.26±0.77 | 13.08±6.11* |
| CTL-like | 310 | 83/223 | 74.67±5.72* | 29.15±1.03* | −0.17±2.22 | 0.04±0.14* | 0.03±0.91 | 14.41±4.19* |
Data are mean±standard deviation. AD, Alzheimer's disease; CTL, healthy control; MMSE, Mini Mental State Examination; CDR-SOB, Clinical Dementia Rating – Sum of Boxes. MMSE and CDR-SOB changes are calculated between baseline and 12-month follow-up.
Statistically significant difference between groups.
In order to compare the use of a combination of the 57 measures to use of hippocampal volume alone, hippocampal volume plus lateral ventricle volume, and features selected based on prior knowledge, three additional models were created. The following results were obtained: (i) hippocampus alone: sensitivity 82.7%, specificity 82.1%, accuracy 82.4%, AUC 0.895 [confidence interval (95% CI) for AUC 0.868–0.917]; (ii) hippocampus and lateral ventricles: sensitivity 82.7%, specificity 81.9%, accuracy 82.3%, AUC 0.893 (CI 0.865–0.915); and (iii) hippocampus, entorhinal cortex, inferior temporal gyrus, medial temporal gyrus, superior temporal gyrus, parahippocampal gyrus, lateral ventricles, inferior lateral ventricles and third and fourth ventricles: sensitivity 83.0%, specificity 88.6%, accuracy 85.8%, AUC 0.923 (CI 0.902–0.943). Comparison of the AUC values using the method of Hanley and McNeil [31] showed that each of these three models provided inferior results to the use of the full 57 measures [AUC 0.948 (CI 0.9290–0.9630); P<0.001].
MCI-c versus MCI-s subjects
Using FreeSurfer-derived measures as input to the OPLS model, subjects in the MCI-c group were separated from those in the MCI-s group with 69.6% sensitivity and 66.8% specificity.
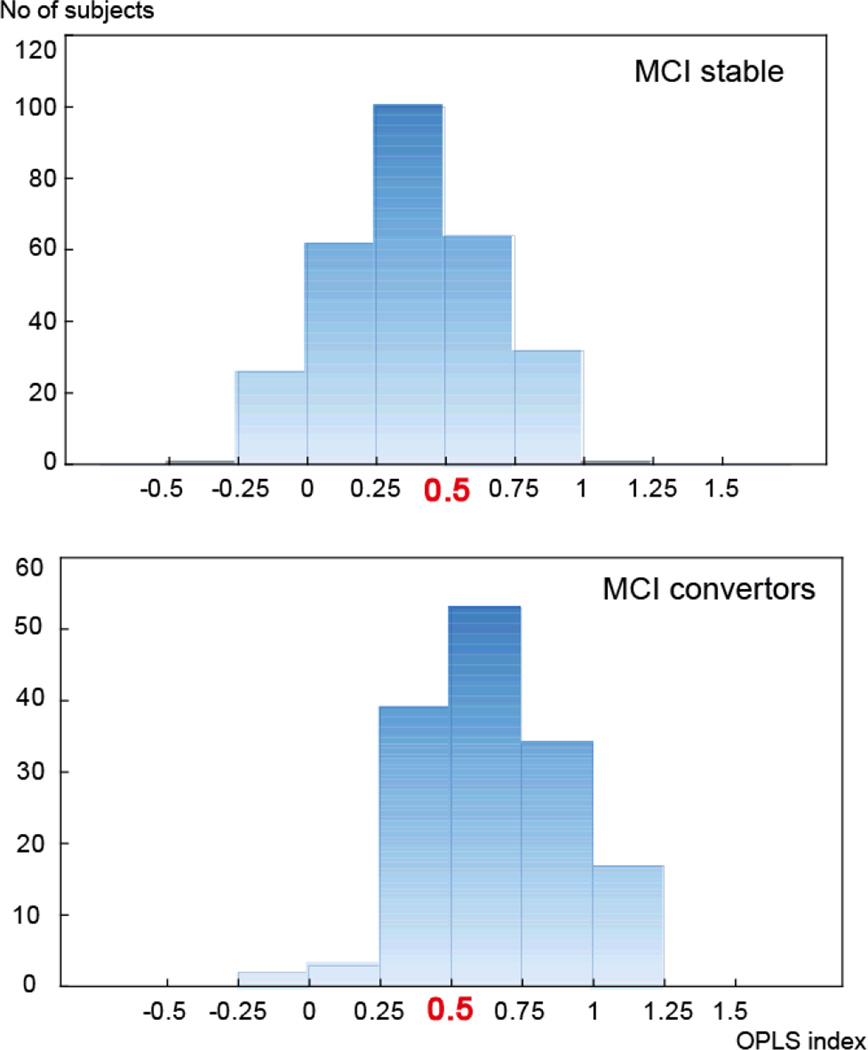
Histograms of the OPLS severity index for both MCI-s and MCI-c groups are presented in Figure 3. Subjects in the MCI-c group were significantly more likely to have the AD atrophy phenotype (112/173, 64.34%) for the OPLS index than the CTL-like phenotype (61/173, 35.26%). ANOVA and post hoc analysis (unequal n HSD method) revealed statistical differences in the severity index between the MCI-s and MCI-c groups (Figure 2). MCI-c subjects classified as CTL-like were younger than those classified as AD-like, but this difference did not reach statistical significance. Of the 173 subjects who progressed to AD (MCI-c), 64.2% were ApoE4 carriers and 28 of these were homozygotes (4/4 genotype). There were no statistically significant differences between the MCI-c AD-like carrier and non-carrier groups nor between the MCI-c CTL-like carrier and non-carrier groups with regard to age, baseline MMSE, 1-year change in MMSE, baseline CDR-SOB, 1-year change in CDR-SOB and years of education (see Table 6).
Figure 3.
Histograms of the severity index for the subjects with MCI (MCI-s, MCI stable; MCI-c, MCI converting to AD). Subjects with a score above 0.5 show a more AD-like pattern of atrophy and below 0.5 a more control-like pattern.
Table 6.
Characteristics of subjects with MCI (stable and convertors) based on the atrophy pattern as detected by the OPLS classifier and ApoE status
| Severity index |
ApoE status | Age (years) | Baseline MMSE |
Change in MMSE |
Baseline CDR-SOB |
Change in CDR-SOB |
Education (years) |
|
|---|---|---|---|---|---|---|---|---|
| MCI-c AD-like | 171 | |||||||
| Carrier | 0.75±0.16 | 75 (66.96%) | 74.72±5.67 | 26.44±1.79 | −1.85±2.47 | 1.87±0.99 | 1.30±1.43 | 14.69±3.57 |
| Non-carrier | 0.84±0.16 | 36 (32.14%) | 75.59±8.83 | 26.69±1.67 | −2.03±2.50 | 1.92±1.08 | 1.06±1.46 | 15.69±3.59 |
| MCI-c CTL-like | 60 | |||||||
| Carrier | 0.39±0.09 | 36 (59.02%) | 71.61±6.82 | 27.00±1.62 | −1.61±2.99 | 1.89±0.95 | 0.85±1.69 | 15.06±3.91 |
| Non-carrier | 0.31±0.18 | 24 (39.34%) | 75.60±6.84 | 26.46±1.61 | −1.14±2.42 | 1.38±0.58 | 0.49±1.62 | 14.25±3.37 |
| MCI-s AD-like | 85 | |||||||
| Carrier | 0.7±0.12 | 34 (39.5%) | 76.74±5.68 | 26.94±1.63 | −0.41±2.60* | 1.44±0.83 | 0.48±1.41 | 12.86±5.15 |
| Non-carrier | 0.68±0.14 | 51 (59.3%) | 75.42±6.07 | 26.91±1.62 | −1.45±2.97* | 1.41±0.83 | 0.57±1.37 | 13.06±5.34 |
| MCI-s CTL-like | 167 | |||||||
| Carrier | 0.24±0.17 | 60 (34.3%) | 73.41±6.80 | 27.23±1.82 | −0.06±2.52 | 1.34±0.75 | 0.39±1.24 | 14.33±4.34 |
| Non-carrier | 0.22±0.18 | 107 (61.1%) | 74.01±6.92 | 27.62±1.66 | 0.29±2.03 | 1.27±0.72 | 0.53±1.74 | 13±4.96 |
Data are mean±standard deviation. AD, Alzheimer's disease; MCI, mild cognitive impairment (c, converter; s, stable); CTL, healthy control; MMSE, Mini Mental State Examination; CDR-SOB, Clinical Dementia Rating – Sum of Boxes. MMSE and CDR-SOB changes are calculated between baseline and 12-month follow-up.
Statistically significant differences.
Histograms of the severity index for the MCI-s and MCI-c groups are shown in Figure 3. MCI-s subjects were divided into three subgroups based on the OPLS-derived index: <0.25 (MCI-s-1), 0.25–0.75 (MCI-s-2) and >0.75 (MCI-s-3), similar to the reported method of Davatzikos et al. [28]. The characteristics of subjects in the three subgroups are presented in Table 5. Although there were no statistically significant differences between these three groups, a clear trend for all variables was present. Using pattern classification in order to differentiate stable from progressive MCI subjects, the same trend was also observed for the SPARE-AD score [28].
Table 5.
Distribution of the FreeSurfer OPLS score in the MCI cohort
| Severity index |
Age (years) |
Baseline MMSE |
Change in MMSE |
Baseline CDR-SOB |
Change in CDR-SOB |
ADAS1 | Change in ADAS1 |
ApoE4+ | |
|---|---|---|---|---|---|---|---|---|---|
| MCI-c | 0.65±0.26 | 74.42±6.97 | 26.6±1.73 | −2.0±2.53 | 1.81±1.0 | 1.2±1.5 | 5.04±1.27 | −0.50±1.21 | 64.2% |
| MCI-c AD-like | 0.78±0.18 | 74.98±6.80 | 26.50±1.76 | −1.91±2.47 | 1.89±1.01 | 1.19±1.47 | 5.16±1.31 | −0.44±1.20 | 67.0% |
| MCI-c CTL-like | 0.35±0.14 | 73.16±7.00 | 26.82±1.64 | −1.43±2.77 | 1.66±0.86 | 0.88±1.49 | 4.83±1.17 | −0.60±1.24 | 59.0% |
| MCI-s-1 (>0.75) | 0.85±0.07 | 76.5±5.40 | 26.6±1.71 | −0.6±2.88 | 1.60±1.0 | 0.7±1.3 | 5.26±1.25 | −0.40±1.16 | 48.5% |
| MCI-s-2 (0.25–0.75) | 0.46±0.14 | 75.60±6.33 | 27.10±1.67 | −0.2±2.42 | 1.40±0.80 | 0.4±1.6 | 4.71±1.35 | −0.36±1.39 | 42.0% |
| MCI-s-3 (<0.25) | 0.07±0.13 | 72.34±6.86 | 27.77±1.59 | +0.4±1.83 | 1.20±0.60 | 0.4±1.5 | 4.28±1.47 | 0.14±1.67 | 30.3% |
Data are mean±standard deviation. AD, Alzheimer's disease; MCI, mild cognitive impairment (c, converter; s, stable); CTL, healthy control; MMSE, Mini Mental State Examination; CDR-SOB, Clinical Dementia Rating – Sum of Boxes; ADAS1, word list non-learning. The MCI-st group was divided into three subgroups based on the histogram of the OPLS score. MMSE and CDR-SOB changes are calculated between baseline and 12-month follow-up.
Statistically significant difference.
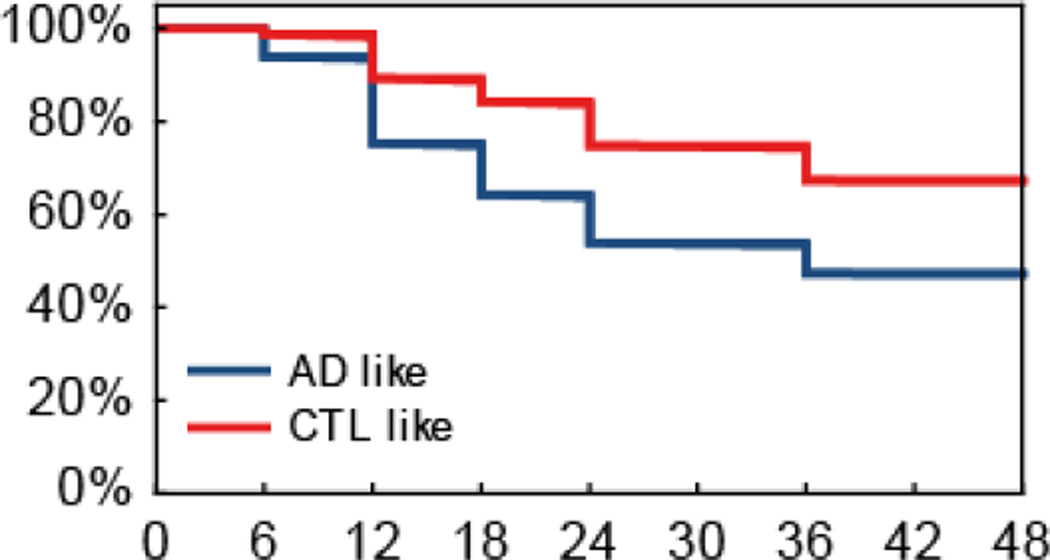
Figure 4 shows survival curves for the AD-like and CTL-like MCI subgroups based on baseline MRI data, illustrating the much higher subsequent conversion rate to AD of the AD-like subgroup.
Figure 4.
Survival curves for the AD-like and CTL-like MCI subgroups based on baseline MRI data for the ADNI cohort. The X axis represents the time in months since entry into the study and the Y axis represents percentage of MCI subjects.
Of the 261 subjects who remained stable (MCI-s), only 36% were ApoE4 carriers (Table 6). Among MCI-s subjects classified as CTL-like, ApoE4 carriers were younger than non-carriers.
Discussion
A challenge in developing informative neuroimaging biomarkers for early AD diagnosis is the need to identify biomarkers that are altered before the onset of clinical symptoms, and which have adequate sensitivity and specificity on an individual patient basis.
We found differences in AD-specific brain structures, which is in line with previous findings of structural MRI as a sensitive biomarker for AD pathology [34]. Volumetric and cortical thickness data from this study extend previously published findings [5, 35]. There were significant differences between the MCI-s and MCI-c groups in hippocampal volume, as well as in the thickness of medial temporal gyrus, superior temporal gyrus and entorhinal cortex. This indicates a widespread pathology for MCI-c subjects; however, whether these findings can be integrated to provide a meaningful clinical intervention remains an open question. Machine learning algorithms and multivariate statistical techniques could have the potential to assist in the early diagnosis of AD. However, manual measures of different brain regions are time consuming and operator dependent and hence are not regularly used in clinical settings. For this reason, in the present study we investigated the applicability of OPLS using only automated regional subcortical volumes and cortical thickness measures. The results show that using all the acquired 57 structural measures and OPLS is superior to using single measures such as hippocampus alone or using feature selection based on prior knowledge (regions known to be affected in AD). Nevertheless, the challenges of translating research techniques into clinical practice should not be underestimated. Jack et al. (36) have highlighted the substantial work required for standardization of hippocampal measures and the significant regulatory hurdles to overcome for even a single measure such as this.
Here, we investigated whether structural brain measures (both volume and thickness) combined under the form of a severity index can be used to differentiate between clinically relevant groups. The OPLS model resulted in a prediction accuracy which was significantly better than chance for the discrimination of patients with AD from normal ageing (88.4%), with high sensitivity (86.1%) and specificity (90.4%). Both misclassified AD and CTL subjects were younger and more educated than correctly classified subjects. This finding is in line with previously published results [7]. The severity index was higher than 0.5 in most MCI-c subjects. As expected, those in the MCI-s group presented a more heterogeneous pattern for the severity index.
The performance of our classifier is comparable to that of previously published methods and suggests similar conclusions [7, 25]. Cuingnet et al. compared several different methods for the separation of MCI-c and MCI-s using subjects from the ADNI study and confirmed that discrimination is difficult, even with different analysis approaches [7]. Similarly, Davatzikos et al. [28] derived a classification index which they compared with clinical assessment and prognosis, with a performance similar to the one used in the current study (66.6% sensitivity) when applied to a subset of the ADNI cohort. We used the same MCI-c subgroups as Davatzikos and colleagues [28] to allow direct comparison between the two methods. However, a strength of the present study is that we used a much larger cohort than in the latter study. McEvoy et al. used rigid regularized quadratic discriminant analysis on a subsample of the ADNI cohort [37]. Fifty-eight FreeSurfer measurements were used as input into the analysis and, as in our study, an atrophy score for each MCI subject was generated that was used to compute an average risk of conversion. The authors concluded that individuals with atrophy scores in the highest percentile had a greater than two-fold increase in risk of conversion to AD, whereas those with atrophy scores in the lowest percentile had a five-fold decreased risk.
The issue of non-converter MCI subjects cannot be solved without long-term follow-up, although MRI changes can be detected at least 3 years before the diagnosis of AD [38]. Longitudinal studies have shown that the majority of subjects with MCI progress to AD in the first 2 years [39]. Thus, a 1-year follow-up period (as in the AddNeuroMed cohort) may be insufficient to ensure adequate clinical separation between the MCI-c and MCI-s groups. This may explain the lower accuracy achieved for the MCI predictions. The survival curves shown in Figure 4 however do illustrate that a substantially higher proportion of the MCI subjects who demonstrate an AD-like MRI pattern at baseline convert to AD in the following 36 months compared to those with a CTL-like pattern.
These results further support the notion that sophisticated statistical methods, such as multivariate analysis, are necessary to capture complex patterns of brain atrophy. Such methods are more informative for predicting clinical course, compared to the use of a limited number of predefined regions known to be affected early in the disease course. Using a limited set of predefined regions may not reflect completely the spatial and temporal pattern of structural and physiological abnormalities [25]. Most existing pattern classification methods usually use only one individual modality of biomarker that may affect the overall classification performance, as is the case in this study. In addition to neuroimaging biomarkers (structural MRI and FDG-PET), biological and genetic biomarkers are available. Different biomarkers provide complementary information, which may be useful for the diagnosis of AD and MCI when used together. Recently, Zhang et al. proposed a new multimodal data fusion and classification method based on kernel combination for AD and MCI subjects [25]. We have also previously combined MRI measures and cerebrospinal fluid (CSF) markers to predict conversion at several future time points using OPLS in a subsample of the ADNI cohort. We found that the addition of CSF markers further improved the predictions [30].
Multiple follow-up diagnostic visits were available for the subjects in the ADNI cohort, but not those in the AddNeuroMed study. Although the clinical evolution of MCI remains poorly understood, our study is one of the largest to date to investigate an MRI-based severity index. As the annual conversion rate for the MCI subjects is 10–15%, it is anticipated that many in the MCI-s group will convert to AD in the near future. The distribution of the severity index suggests that a subgroup of MCI-s subjects has normal brain structures. However, a large subgroup has a distinct AD-like pattern, which we believe reflects the underlying AD pathology.
Subjects with MCI who did not progress to AD within the follow-up period of this study are characterized by differences in characteristics (e.g. age, MMSE score and ApoE4 status) depending on severity index subgroup. The subgroup of MCI-s with the highest severity index (MCI-s-1) was associated with a faster decline in MMSE score and older age. By contrast, subjects belonging to the MCI-s-2 and MCI-s-3 groups, although showing relatively similar decline in MMSE scores, had statistically significant differences in severity index. Among MCI-s subjects, the subgroup with the highest severity index had a different proportion of APOE4 carriers compared to the other two subgroups.
Dividing the MCI-s group into three subgroups revealed that gradual brain changes over long periods of time might eventually lead to clinical progression. These results are important because they demonstrate the robustness of the structural MRI dementia measure that we have used to detect structural brain differences between groups. In a previous study by Davatzikos et al., the MCI-s group was categorized in a similar way [28]. The present results confirm their findings in a much larger cohort. Although not statistically significant, we observed a trend towards lower MMSE scores, higher CDR-SOB and ADAS1 scores and a higher percentage of ApoE4 carriers with a higher severity index.
MRI is a promising adjunct to the clinical diagnosis of AD and is useful for assessing the early stages of disease. However, there are several obstacles to the widespread use of volumetric MRI in the clinical setting: variation in imaging protocols, spatial distortion of MRI data, the absence of normative values and labour-intensive methods have all reduced the potential impact of MRI measures. Developments are needed to allow consistency in acquisition of MRI data across sites and fully automated image segmentation before being able to introduce the clinical use of volumetric MRI. Large, multicentre trials, such as ADNI and AddNeuroMed, are important for facilitating greater use of MRI in clinical settings by providing image standardization, correction for spatial distortion, improved data throughput, and on-site quality control procedures.
These results confirm that joint evaluation of brain regions is beneficial for increasing the accuracy of predicting progression to AD. Future studies with longer follow-up periods will improve our estimates of specificity. In addition, information such as age, ApoE4 status and level of education should be used as co-factors when deciding cut-off values for severity indices similar to the index proposed in this study.
Supplementary Material
Acknowledgements
This study was supported by InnoMed (Innovative Medicines in Europe), an Integrated Project funded by the European Union of the Sixth Framework programme priority FP6-2004-LIFESCIHEALTH-5, Life Sciences, Genomics and Biotechnology for Health.
Data collection and sharing in this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott; Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Amorfix Life Sciences Ltd.; AstraZeneca; Bayer HealthCare; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129, K01 AG030514, and the Dana Foundation.
The authors thanks Swedish Brain Power, the Strategic Research Programme in Neuroscience at Karolinska Institutet (StratNeuro), Hjärnfonden, the Gamla Tjänarinnor foundation, the Swedish Alzheimer’s Association, the regional agreement on medical training and clinical research (ALF) between Stockholm County Council and Karolinska Institutet, Health Research Council of Academy of Finland, University of Eastern Finland UEFBRAIN, EVO funding from Kuopio University Hospital and Stockholm Medical Image Laboratory and Education (SMILE). AS and SL were supported by funds from NIHR Biomedical Research Centre for Mental Health at the South London and Maudsley NHS Foundation Trust and Institute of Psychiatry, Kings College London.
Footnotes
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.ucla.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.ucla.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf
Conflict of Interest Statement
No conflict of interest was declared.
References
- 1.Albert MS, DeKosky ST, Dickson D, Dubois B, Feldman HH, Fox NC, Gamst A, Holtzman DM, Jagust WJ, Petersen RC, Snyder PJ, Carrillo MC, Thies B, Phelps CH. The diagnosis of mild cognitive impairment due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7:270–279. doi: 10.1016/j.jalz.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mariani E, Monastero R, Mecocci P. Mild cognitive impairment: a systematic review. J Alzheimers Dis. 2007;12:23–35. doi: 10.3233/jad-2007-12104. [DOI] [PubMed] [Google Scholar]
- 3.Gauthier S, Reisberg B, Zaudig M, Petersen RC, Ritchie K, Broich K, Belleville S, Brodaty H, Bennett D, Chertkow H, Cummings JL, de Leon M, Feldman H, Ganguli M, Hampel H, Scheltens P, Tierney MC, Whitehouse P, Winblad B. Mild cognitive impairment. Lancet. 2006;367:1262–1270. doi: 10.1016/S0140-6736(06)68542-5. [DOI] [PubMed] [Google Scholar]
- 4.Ewers M, Sperling RA, Klunk WE, Weiner MW, Hampel H. Neuroimaging markers for the prediction and early diagnosis of Alzheimer's disease dementia. Trends Neurosci. 2011;34:430–442. doi: 10.1016/j.tins.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fennema-Notestine C, Hagler DJJ, McEvoy LK, Fleisher AS, Wu EH, Karow DS, Dale AM. Structural MRI biomarkers for preclinical and mild Alzheimer's disease. Hum Brain Mapp. 2009;30:3238–3253. doi: 10.1002/hbm.20744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bylesjö M, Rantalainen M, Nicholson JK, Holmes E, Trygg J. K-OPLS package: kernel-based orthogonal projections to latent structures for prediction and interpretation in feature space. BMC Bioinformatics. 2008;9:106. doi: 10.1186/1471-2105-9-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cuingnet R, Gerardin E, Tessieras J, Auzias G, Lehéricy S, Habert M, Chupin M, Benali H, Colliot O. Automatic classification of patients with Alzheimer's disease from structural MRI: a comparison of ten methods using the ADNI database. Neuroimage. 2011;56:766–781. doi: 10.1016/j.neuroimage.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 8.Westman E, Simmons A, Muehlboeck J, Mecocci P, Vellas B, Tsolaki M, Kłoszewska I, Soininen H, Weiner MW, Lovestone S, Spenger C, Wahlund L. AddNeuroMed and ADNI: similar patterns of Alzheimer's atrophy and automated MRI classification accuracy in Europe and North America. Neuroimage. 2011b;58:818–828. doi: 10.1016/j.neuroimage.2011.06.065. [DOI] [PubMed] [Google Scholar]
- 9.Simmons A, Westman E, Muehlboeck S, Mecocci P, Vellas B, Tsolaki M, Kłoszewska I, Wahlund L, Soininen H, Lovestone S, Evans A, Spenger C. MRI measures of Alzheimer's disease and the AddNeuroMed study. Ann N Y Acad Sci. 2009;1180:47–55. doi: 10.1111/j.1749-6632.2009.05063.x. [DOI] [PubMed] [Google Scholar]
- 10.Simmons A, Westman E, Muehlboeck S, Mecocci P, Vellas B, Tsolaki M, Kłoszewska I, Wahlund L, Soininen H, Lovestone S, Evans A, Spenger C. The AddNeuroMed framework for multi-centre MRI assessment of Alzheimer's disease: experience from the first 24 months. Int J Geriatr Psychiatry. 2011;26:75–82. doi: 10.1002/gps.2491. [DOI] [PubMed] [Google Scholar]
- 11.American Psychiatric Association Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition, Text Revision. 2000. [Google Scholar]
- 12.Rosen WG, Mohs RC, Davis KL. A new rating scale for Alzheimer's disease. Am J Psychiatry. 1984;141:1356–1364. doi: 10.1176/ajp.141.11.1356. [DOI] [PubMed] [Google Scholar]
- 13.Jack CRJ, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, Britson PJ, L Whitwell J, Ward C, Dale AM, Felmlee JP, Gunter JL, Hill DLG, Killiany R, Schuff N, Fox-Bosetti S, Lin C, Studholme C, DeCarli CS, Krueger G, Ward HA, Metzger GJ, Scott KT, Mallozzi R, Blezek D, Levy J, Debbins JP, Fleisher AS, Albert M, Green R, Bartzokis G, Glover G, Mugler J, Weiner MW. The Alzheimer's Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008;27:685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- 15.Fischl B, van der Kouwe A, Destrieux C, Halgren E, Ségonne F, Salat DH, Busa E, Seidman LJ, Goldstein J, Kennedy D, Caviness V, Makris N, Rosen B, Dale AM. Automatically parcellating the human cerebral cortex. Cereb Cortex. 2004;14:11–22. doi: 10.1093/cercor/bhg087. [DOI] [PubMed] [Google Scholar]
- 16.Han X, Fischl B. Atlas renormalization for improved brain MR image segmentation across scanner platforms. IEEE Trans Med Imaging. 2007;26:479–486. doi: 10.1109/TMI.2007.893282. [DOI] [PubMed] [Google Scholar]
- 17.Westman E, Wahlund L, Foy C, Poppe M, Cooper A, Murphy D, Spenger C, Lovestone S, Simmons A. Magnetic resonance imaging and magnetic resonance spectroscopy for detection of early Alzheimer's disease. J Alzheimers Dis. 2011;26(Suppl 3):307–319. doi: 10.3233/JAD-2011-0028. [DOI] [PubMed] [Google Scholar]
- 18.Westman E, Cavallin L, Muehlboeck J, Zhang Y, Mecocci P, Vellas B, Tsolaki M, Kłoszewska I, Soininen H, Spenger C, Lovestone S, Simmons A, Wahlund L. Sensitivity and specificity of medial temporal lobe visual ratings and multivariate regional MRI classification in Alzheimer's disease. PLoS One. 2011a;6:e22506. doi: 10.1371/journal.pone.0022506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, Paajanen T, Zhang Y, Westman E, Wahlund L, Simmons A, Tunnard C, Sobow T, Mecocci P, Tsolaki M, Vellas B, Muehlboeck S, Evans A, Spenger C, Lovestone S, Soininen H. Combination analysis of neuropsychological tests and structural MRI measures in differentiating AD, MCI and control groups--the AddNeuroMed study. Neurobiol Aging. 2011;32:1198–1206. doi: 10.1016/j.neurobiolaging.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y, Paajanen T, Westman E, Wahlund L, Simmons A, Tunnard C, Sobow T, Proitsi P, Powell J, Mecocci P, Tsolaki M, Vellas B, Muehlboeck S, Evans A, Spenger C, Lovestone S, Soininen H. Effect of APOE ε4 allele on cortical thicknesses and volumes: the AddNeuroMed study. J Alzheimers Dis. 2010;21:947–966. doi: 10.3233/JAD-2010-100201. [DOI] [PubMed] [Google Scholar]
- 21.Thambisetty M, Simmons A, Velayudhan L, Hye A, Campbell J, Zhang Y, Wahlund L, Westman E, Kinsey A, Güntert A, Proitsi P, Powell J, Causevic M, Killick R, Lunnon K, Lynham S, Broadstock M, Choudhry F, Howlett DR, Williams RJ, Sharp SI, Mitchelmore C, Tunnard C, Leung R, Foy C, O'Brien D, Breen G, Furney SJ, Ward M, Kloszewska I, Mecocci P, Soininen H, Tsolaki M, Vellas B, Hodges A, Murphy DGM, Parkins S, Richardson JC, Resnick SM, Ferrucci L, Wong DF, Zhou Y, Muehlboeck S, Evans A, Francis PT, Spenger C, Lovestone S. Association of plasma clusterin concentration with severity, pathology, and progression in Alzheimer disease. Arch Gen Psychiatry. 2010;67:739–748. doi: 10.1001/archgenpsychiatry.2010.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eriksson L, Johansson E, Kettaneh-Wold N, Trygg J, Wiksröm C. Wold S Multi- and Megavariate Data Analysis (Part I -Basics and Principles and Applications) 2nd ed. 2006. [Google Scholar]
- 23.Stenlund H, Gorzsás A, Persson P, Sundberg B, Trygg J. Orthogonal projections to latent structures discriminant analysis modeling on in situ FT-IR spectral imaging of liver tissue for identifying sources of variability. Anal Chem. 2008;80:6898–6906. doi: 10.1021/ac8005318. [DOI] [PubMed] [Google Scholar]
- 24.Bylesjö M, Eriksson D, Kusano M, Moritz T, Trygg J. Data integration in plant biology: the O2PLS method for combined modeling of transcript and metabolite data. Plant J. 2007;52:1181–1191. doi: 10.1111/j.1365-313X.2007.03293.x. [DOI] [PubMed] [Google Scholar]
- 25.Zhang D, Wang Y, Zhou L, Yuan H, Shen D. Multimodal classification of Alzheimer's disease and mild cognitive impairment. Neuroimage. 2011;55:856–867. doi: 10.1016/j.neuroimage.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chu C, Hsu A, Chou K, Bandettini P, Lin C. Does feature selection improve classification accuracy? Impact of sample size and feature selection on classification using anatomical magnetic resonance images. Neuroimage. 2012;60:59–70. doi: 10.1016/j.neuroimage.2011.11.066. [DOI] [PubMed] [Google Scholar]
- 27.Heister D, Brewer JB, Magda S, Blennow K, McEvoy LK. Predicting MCI outcome with clinically available MRI and CSF biomarkers. Neurology. 2011;77:1619–1628. doi: 10.1212/WNL.0b013e3182343314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davatzikos C, Bhatt P, Shaw LM, Batmanghelich KN, Trojanowski JQ. Prediction of MCI to AD conversion, via MRI, CSF biomarkers, and pattern classification. Neurobiol Aging. 2011;32:2322.e19–2322.e27. doi: 10.1016/j.neurobiolaging.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Westman E, Muehlboeck J, Simmons A. Combining MRI and CSF measures for classification of Alzheimer's disease and prediction of mild cognitive impairment conversion. Neuroimage. 2012;62:229–238. doi: 10.1016/j.neuroimage.2012.04.056. [DOI] [PubMed] [Google Scholar]
- 30.Hanley JA, McNeil BJ. A method of comparing the areas under receiver operating characteristic curves derived from the same cases. Radiology. 1983;148:839–843. doi: 10.1148/radiology.148.3.6878708. [DOI] [PubMed] [Google Scholar]
- 31.Metz C, Herman B, Roe C. Statistical comparison of two roc-curve estimates obtained from partially-paired datasets. Medical Decision Making. 1998;18:110–121. doi: 10.1177/0272989X9801800118. [DOI] [PubMed] [Google Scholar]
- 32.Metz C, Kronman H. Statistical signicance tests for binormal roc-curves. Journal of Mathematical Psychology. 1980;22:218–243. [Google Scholar]
- 33.Metz C, Pan X. Proper binormal roc curves: Theory and maximum-likelihood estimation. Journal of mathematical psychology. 1999;43:1–33. doi: 10.1006/jmps.1998.1218. [DOI] [PubMed] [Google Scholar]
- 34.Reiman EM, Jagust WJ. Brain imaging in the study of Alzheimer's disease. Neuroimage. 2012;61(2):505–516. doi: 10.1016/j.neuroimage.2011.11.075. doi: 10.1016/j.neuroimage.2011.11.075. Epub 2011 Dec 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, Paajanen T, Westman E, Zhang Y, Wahlund L, Simmons A, Tunnard C, Sobow T, Proitsi P, Powell J, Mecocci P, Tsolaki M, Vellas B, Muehlboeck S, Evans A, Spenger C, Lovestone S, Soininen H. APOE ε2 allele is associated with larger regional cortical thicknesses and volumes. Dement Geriatr Cogn Disord. 2010;30:229–237. doi: 10.1159/000320136. [DOI] [PubMed] [Google Scholar]
- 36.Jack CR, Jr, Barkhof F, Bernstein MA, Cantillon M, Cole PE, Decarli C, Dubois B, Duchesne S, Fox NC, Frisoni GB, Hampel H, Hill DL, Johnson K, Mangin JF, Scheltens P, Schwarz AJ, Sperling R, Suhy J, Thompson PM, Weiner M, Foster NL. Steps to standardization and validation of hippocampal volumetry as a biomarker in clinical trials and diagnostic criterion for Alzheimer's disease. Alzheimers Dementia. 2011 Jul 7;(4):474–485.e4. doi: 10.1016/j.jalz.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McEvoy LK, Holland D, Hagler DJJ, Fennema-Notestine C, Brewer JB, Dale AM. Mild cognitive impairment: baseline and longitudinal structural MR imaging measures improve predictive prognosis. Radiology. 2011;259:834–843. doi: 10.1148/radiol.11101975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitwell JL, Przybelski SA, Weigand SD, Knopman DS, Boeve BF, Petersen RC, Jack CRJ. 3D maps from multiple MRI illustrate changing atrophy patterns as subjects progress from mild cognitive impairment to Alzheimer's disease. Brain. 2007;130:1777–1786. doi: 10.1093/brain/awm112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson RS, Bennett DA, Bienias JL, Aggarwal NT, Mendes De Leon CF, Morris MC, Schneider JA, Evans DA. Cognitive activity and incident AD in a population-based sample of older persons. Neurology. 2002;59:1910–1914. doi: 10.1212/01.wnl.0000036905.59156.a1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.