Abstract
Circadian rhythms affect olfaction by an unknown molecular mechanism. Independent of the suprachiasmatic nuclei, the mammalian olfactory bulb (OB) has recently been identified as a circadian oscillator. The electrical activity in the OB was reported to be synchronized to a daily rhythm and the clock gene, Period1, was oscillatory in its expression pattern. Because gap junctions composed of connexin36 and α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors (AMPARs) have been reported to work together to synchronize firing of action potentials in the OB, we hypothesized that circadian electrical oscillations could be synchronized by daily changes in the expression of connexins and AMPAR subunits (GluR1–4). We examined the OB for the presence of clock genes by polymerase chain reaction (PCR) and whether Period2, connexins, and AMPARs fluctuated across the light/dark cycle by quantitative PCR or SDS–PAGE/Western blot analysis. We observed significant changes in the messenger RNA and protein expression of our targets across 24 or 48 h. Whereas most targets were rhythmic by some measures, only GluR1 mRNA and protein were both rhythmic by the majority of our tests of rhythmicity across all time scales. Differential expression of these synaptic proteins over the light/dark cycle may underlie circadian synchronization of action potential firing in the OB or modify synaptic interactions that would be predicted to impact olfactory coding, such as alteration of granule cell inhibition, increased number of available AMPARs to bind glutamate, or an increased gap junction conductance between mitral/tufted cells.
Keywords: circadian, gap junction, glutamate receptor, olfaction, Cx36, GluR2
Introduction
The olfactory bulb (OB) is the first site of odor information processing in the brain and sends projections to the piriform cortex, the limbic system, and the lateral hypothalamus (White, 1965). Recently, the OB has been identified as an independent circadian oscillator, as demonstrated by rhythms in Period1 (Per1) expression and synchronized electrical activity (Abe et al., 2002; Granados-Fuentes et al., 2004a; Abraham et al., 2005), but how these rhythms affect OB function is unknown. A subset of clock genes, such as Per1, has been reported in the OB (Abe et al., 2002; Shieh, 2003) and can fluctuate after light exposure (Hamada et al., 2011). OB rhythms are affected by the suprachiasmatic nuclei (Granados-Fuentes et al., 2004b) but the OB does not have a known direct connection to those nuclei. Gap junctions composed of connexin36 (Cx36), working with α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors (AMPARs), a type of ionotropic glutamate receptor, were reported to decrease the current injection that elicited synchronized action potentials from mitral and tufted cells (M/Ts) in the OB, compared to Cx36-knockout mice (Schoppa and Westbrook, 2002; Christie et al., 2005). M/Ts, but not granule cells, have rhythmic expression of Per1 protein (Granados-Fuentes et al., 2004a; Abraham et al., 2005); therefore, diurnal changes in M/T electrical activity may be responsible for the diurnal rhythm of olfactory acuity in rodents (Granados-Fuentes et al., 2006, 2011).
Within the OB, connexins are expressed by neurons, astrocytes, ensheathing cells, and some interneurons (Kosaka and Kosaka, 2003, 2004; Kosaka et al., 2005). Collectively, the literature suggests that only three connexins (Cx) are expressed within the OB: Cx36, Cx43, and Cx45. Early work examining OB primary cell cultures suggested the absence of Cx26, Cx32, Cx40, or Cx45 (Miragall et al., 1997). Cx36 and Cx45 were reported within OB glomeruli (Zhang and Restrepo, 2002, 2003; Rash et al., 2005) and are not expressed by nonneuronal cells (Rash et al., 2005).Rash et al. (2005) were unable to determine whether Cx45 was forming gap junctions between M/Ts or between those cells and neurons within and around glomeruli, called juxtaglomerular cells. M/T-to-juxtaglomerular cell gap junctions were previously reported (Kosaka and Kosaka, 2003, 2004; Kosaka et al., 2005), although the molecular identity of the connexin was not determined. Cx43 is expressed by astrocytes and ensheathing cells in the OB (Rela et al., 2010).
Gap junctions composed of Cx36 help elicit synchronous spiking in the OB, but the required current is generated by AMPARs (Christie et al., 2005). AMPARs (reviewed in Dingledine et al., 1999) are composed of tetramers of the subunits GluR1–4 (also called GluA1–4), and these subunits are present throughout the OB in varying amounts and combinations (Montague and Greer, 1999). AMPARs are involved in synaptic transmission in the OB (Berkowicz et al., 1994; Ennis et al., 1996), glutamate spillover responses (e.g. Pimentel and Margrie, 2008), and astrocytic neurotransmitter release (reviewed in both Volterra and Meldolesi, 2005, and Haydon and Carmignoto, 2006). AMPARs that include an edited GluR2 subunit do not generate Ca2+ flux. Rhythmic changes in the expression of GluR2 throughout the OB would therefore change the Ca2+ permeability of the AMPARs, alter synaptic transmission, lateral excitation, neurotransmitter release, and shift the balance between AMPARs and N-methyl-D-aspartic acid receptors (NMDARs) as a source of intracellular Ca2+ (Blakemore et al., 2006; Ma and Lowe, 2007; Pimentel and Margrie, 2008; Decrock et al., 2011). Therefore, any diurnal modulation in both AMPAR subunit and connexin protein expression could change multiple circuit behaviors in the OB as a function of the time of day.
Changes in the expression of ion channels that function in the millisecond time scale can affect activity over much longer time scales (e.g. diurnal, circadian) by altering the amplitude of synaptic events and the degree of electrical coupling, hence, whether threshold is achieved. Because gap junctions and AMPARs allow for synchronized action potential firing, we hypothesized that expression of gap junctions and AMPARs was rhythmic and would thereby allow synchronized electrical oscillations in the OB. To test our hypothesis, we analyzed OB RNA extracts by quantitative polymerase chain reaction (PCR) and OB membrane proteins by SDS–PAGE sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Western blot analysis to determine whether connexins and AMPAR expression levels changed as a function of the time of day. We also tested for the presence of clock genes and Period2 (Per2) mRNA rhythmicity to establish the validity of our method. Messenger RNA was examined across 48 h in order to establish rhythmicity over two cycles; if mRNA was not rhythmic for two cycles, we could conclude that we observed changes over time but not necessarily rhythmic cycling of that mRNA. We observed significant changes over time in the expression of Cx36, Cx43, Cx45, and GluR1–4, though only GluR1 appeared rhythmic. These changes suggest that OB function is modulated by time- or activity-dependent factors.
Experimental Procedures
Animals
All animals used in this study were Sprague–Dawley rat pups (males and females) ages postnatal day 21 (P21), P22, and P23 (Charles River, Wilmington, MA, USA), or Cx36-knockout mice (Deans et al., 2001). Rats were not weaned before sample collection. Rodents had constant access to food and water ad libitum and were kept on a 12-h-light/12-h-dark cycle. Lights were turned on at 07:00 h EST and lights were turned off at 19:00 h EST. Animals collected in the dark were only briefly exposed to dim red light (approximately 100 lux) while animals collected in the light were exposed to the overhead lights (approximately 440 lux).All animals used in these experiments were anesthetized by isoflurane inhalation and killed by decapitation.
The animals for these experiments were used according to the guidelines of our protocol approved by Florida State University's Animal Care and Use Committee and the National Institutes of Health Guide for the Care and Use of Laboratory Animals (NIH Publications No. 80-23).
RNA extraction
Rat pups of ages P21, P22, and P23 were killed at 3-h intervals over 48 h and their OBs were excised and placed immediately in RNAlater reagent (Qiagen, Valencia, CA, USA) according to manufacturer's instructions. For processing, samples were removed from RNAlater, placed in ice-cold TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and homogenized with a rotor–stator homogenizer (PRO Scientific, Oxford, CT, USA) at 50% amplitude with autoclaved probes washed in diethylpyrocarbonatetreated water. Samples homogenized in TRIzol were processed according to manufacturer's instructions with a repeat of the chloroform-addition step. The resulting pellet was reconstituted in 100 μl of RNase-, DNase-free water (EMD Millipore, Billerica, MA, USA) and processed through the RNEasy Mini Kit (Qiagen) according to the manufacturer's RNA Cleanup protocol with DNase I treatment (Qiagen). Purified RNA samples were spun for 40 min in a vacuum concentrator and stored at −20 °C until use.
Reverse transcription, PCR, and quantitative PCR
Purified RNA samples were kept on ice and their concentrations assayed with a Nanodrop 1000 spectrophotometer (Thermo Scientific, Rockford, IL, USA). Sample integrity was assayed by gel electrophoresis with SYBR Safe DNA Dye (Invitrogen). Five micrograms of total RNA was used for the reverse-transcription reaction with a Superscript III Reverse Transcription Supermix Kit (Invitrogen), according to the manufacturer's instructions except for inclusion of both random oligonucleotides and oligodT(20) as primers for the reverse transcription, from the protocol of Resuehr and Spiess (2003). The resulting cDNA was diluted 1:10 in RNase-, DNase-free water.
Polymerase chain reaction (PCR) solutions were composed of HotStarTaq Master Mix (Qiagen), 2 μl of the diluted cDNA, and 500 nM primers, according to manufacturer's instructions. PCR was run on a Veriti thermal cycler (Applied Biosystems, Carlsbad, CA, USA). Reactions were run as follows: 95 °C for 15 min; 40 cycles of 94 °C for 30 s, 55 °C for 1 min, and 72 °C for 1 min; 72 °C for 10min, 4 °C until samples were prepared for gel electrophoresis. PCR products were analyzed using 1.6% agarose gel electrophoresis in Tris-boric acid-EDTA buffer (TBE) and run at 43 V for 2-h, visualized using a Gel Doc XR+ system (Bio-Rad, Hercules, CA, USA). PCRs in which diluted cDNA was replaced with water did not show bands on electrophoresis gels.
The quantitative PCR solution was composed of SYBR Green Master Mix (Applied Biosystems), 2 μl of the diluted cDNA, and 500 nM primers, according to manufacturer's instructions. The quantitative PCR reactions were run on a 7500 Fast Real-Time PCR Thermocycler (Applied Biosystems). All reactions were run as follows: 50 °C for 2 min, 95 °C for 10 min; 40 cycles of 95 °C for 15 s and 60 °C for 1 min; followed by melt curve analysis. Ribosomal protein S28, β-III-tubulin, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNAs were used as reference genes. Each reaction plate also contained a sample from 12 am, P21 as a reference value to allow comparisons between multiple plates. Reactions were compared to each of the reference genes, and line plots for each reference were similar. Reference genes were compared to each other and both β-III-tubulin and GAPDH mRNAs did not show significant fluctuation. Control reactions where cDNA was replaced with water did not reach the critical threshold. Reported results use GAPDH as the reference gene.
Primer design and primer sets used
Primer sets shown in Table 1 were designed with OligoExplorer freeware (http://www.genelink.com/tools/gl-oe.asp), reanalyzed with NetPrimer software (Premier Biosoft, Palo Alto, CA, USA), and ordered from Integrated DNA Technologies (Coralville, IA, USA). Only primer sets that did not form dimers or hairpins in silico were used. The products were then submitted as a BLAST (NCBI, http://blast.ncbi.nlm.nih.gov/Blast.cgi) query against Ref-SeqmRNA in the Rattus norvegicus genome. Any primer sets with nonspecific products were not used. The GAPDH primer set was designed to span between exons 3 and 4 and therefore serve as a check for DNA contamination through the appearance of introns. We tested quantitative PCR products by melt curve, sequencing, and agarose gel electrophoresis to ensure specificity and correct band size. The primer sets for AMPAR subunits GluR1–4 were as published by Santiago et al. (2009), and the primer sets for the clock genes Clock, Bmal1, Period1 (Per1), Period2 (Per2), Period3 (Per3), Cryptochrome1 (Cry1), Cryptochrome2 (Cry2), Rev-erb α, Rev-erb β, and RORα were as published by Kamphuis et al. (2005).
Table 1. Primer sets designed for this study.
| Gene name | Forward primer 5′ to 3′ | Reverse primer 5′ to 3′ | Product (bp) |
|---|---|---|---|
| Connexin36 (Cx36) | TGC TCT GGA GAT TGG GTT TCT GGT | AGA TTG AGC ACC ACA CAA ATG CCG | 186 |
| Connexin43 (Cx43) | GAA CAG GTG GGG ATA AGG GAG GT | GAT GGG GGC AGA GAG AGA AAG C | 164 |
| Connexin45 (Cx45) | GCA GAA CAA AGC CAA TAT CGC CCA | TTC TGG TGA TGG TAG GCC TGG ATT | 144 |
| Ribosomal protein S28 | TTT ATG GAT GAC ACC AGC CGC TCT | TTT CTG ACT CCA ACA GGG TGA GCA | 88 |
| β-III-tubulin | ATG AAG GAG GTG GAT GAG CAG ATG | GCC GAT GAA GGT GGA CGA CA | 141 |
| Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | GGG TGA TGC TGG TGC TGA GTA TGT | CGG AAG GGG CGG AGA TGA TGA | 116 |
Membrane/organelle protein preparation
Rat pups (P21) were killed at 3-h intervals over a 24-h period, and their OBs were dissected and placed immediately in a dry-ice/95%-ethanol slurry for flash-freezing. OBs from the Cx36 −/− mice were harvested similarly but without regard to time. All samples were stored at −80 °C until processing. Before processing, protease inhibitors were added to a homogenization buffer (320 mM sucrose, 1 mM EDTA, 50 mM KCl, 10 mM Tris base, 1 μg/ml leupeptin, 1 μg/ml pepstatin A, 2 μg/ml aprotinin, 1 mg/ml phenylmethylsulfonyl fluoride, pH 7.8; all from Sigma–Aldrich, St. Louis, MO, USA), and then samples were homogenized 50 strokes on ice with a Kontes 20 tissue grinder. Each sample was a pair of OBs from a single rat or mouse. Membrane and organelle proteins were purified by two rounds of low-speed centrifugation (6500g for 10 min at 4 °C) and then one high-speed centrifugation (107,000g for 30 min at 4 °C) with an Optima MAX-XP (Beckman-Coulter, Brea, CA, USA) in a protocol adapted from Guillemin et al. (2005) and Schindler et al. (2006). The pellet was reconstituted by tip sonication (Model 120, Fisher Scientific, Pittsburgh, PA, USA) as previously described (Cook and Fadool, 2002) and stored at −80 °C until use.
SDS–PAGE and Western blot analysis
Protein concentrations were determined using a Bradford assay (Bio-Rad). For each sample, 5–15 μg of membrane/organelle proteins was separated by 10% polyacrylamide gel electrophoresis then electro-transferred to nitrocellulose by semidry transfer (SD TransBlot, Bio-Rad). For samples to be probed with Cx36 antisera, 30 μg of membrane/organelle proteins were separated by 10% polyacrylamide gel electrophoresis and electro-transferred to polyvinylidene fluoride membranes according to standard protocols. Solutions used for electrophoresis and electrotransfer were described by Sambrook and Russell (2001). Target antisera were coapplied with the mouse anti-β-III-tubulin loading control when feasible. Secondary antibodies conjugated to near-infrared dyes were only mixed when dual-color experiments were desired. Nitrocellulose blots were stripped (NewBlot Stripping Buffer, LI-COR Biosciences) and reprobed with a second antibody of interest. Blots for Cx36 were examined by chemiluminescence rather than fluorescence because of the lack of strong fluorescent signal. Blots were visualized by autoradiography with ECL Plus reagents (GE Healthcare, Piscataway, NJ, USA). Polyvinylidene fluoride blots were stripped with the stripping buffer described by Yeung and Stanley (2009) and reprobed with rabbit monoclonal anti-β-III-tubulin (Sigma–Aldrich) as a loading control.
Antisera
Antisera were diluted in Odyssey Blocking Buffer with 0.2% Tween-20 (OBBT), 5% bovine serum albumin in Tris-buffered saline with 0.2% Tween-20 (TBST*), or different concentrations of nonfat dry milk in Tris-buffered saline with 0.1% Tween-20 (TBST). Primary antisera used in this study were purchased from Cell Signaling Technology (CST; Danvers, MA, USA), Invitrogen, Millipore (Billerica, MA, USA), and Sigma–Aldrich and are as follows, with the epitope, dilution/buffer, and catalog number in parentheses: Invitrogen rabbit anti-Cx36 (cytoplasmic loop between second and third transmembrane domains; 1:250 in 4% milk/TBST; 51-6200), Invitrogen rabbit anti-Cx36 (C-terminus; 1:250 in 1% milk/TBST; 51-6300), Invitrogen mouse anti-Cx36 (C-terminus; 1:500 in 4% milk/TBST; 37-4600), Invitrogen rabbit anti-Cx43 (C-terminus; 1:500 in OBBT; 71-0700), Invitrogen rabbit anti-Cx45 (C-terminus; 1:250/OBBT; 40-7000), Millipore mouse anti-GluR1 (N-terminus; 1:500/OBBT; MAB2263), CST rabbit anti-GluR2 (N-terminus; 1:500/OBBT; 5306S), CST rabbit anti-GluR3 (residues surrounding Pro590 of human GluR3; 1:1500/OBBT; 4676S), CST rabbit anti-GluR4 (residues surrounding Gln890 of human GluR4; 1:1000 in 5% bovine serum albumin/TBST*; 8070S), CST mouse anti-β-III-tubulin (C-terminus; 1:2000/OBBT; 4466S), and Sigma–Aldrich rabbit anti-β-III-tubulin (residues 442–446; 1:5000 in 4% milk/TBST; SAB4300623). Secondary antibodies purchased from LI-COR Biosciences are as follows: goat anti-rabbit conjugated to 800CW dye (1:5000/OBBT; 926-32211), and goat anti-mouse conjugated to 680LT dye (1:20,000/OBBT; 926-68020). Amersham donkey anti-rabbit conjugated to horseradish peroxidase (GE Healthcare) was used in chemiluminescent Western blot experiments (1:5000 in 1 or 4% milk/TBST; NA934).
Quantitative densitometry
Fluorescent Western blots were scanned by a LI-COR Odyssey Imager (LI-COR Biosciences) and densitometry of the fluorescent western signal was performed with the Odyssey's operating software (LI-COR). Fluorescent signals were calculated in comparison to the median background surrounding the band and were normalized to both the β-III-tubulin loading control and a reference sample for standardization of variance across individual gels.
Statistical analysis
Quantitative PCR results and Western blot densitometry results were analyzed by nonparametric ANOVA (Kruskal–Wallis H test) followed by a Dunn's post-hoc test, harmonic regression analysis (Legendre and Legendre, 1998), and JTK_CYCLE analysis (Hughes et al., 2010), all at a 95% confidence level. Real-time PCR data were analyzed as 48-h periods and separate 24-h periods for each analysis, and Western blot data were analyzed as one 24-h period.
Results
Messenger RNA expression of clock genes and changes in mRNA expression of Per2
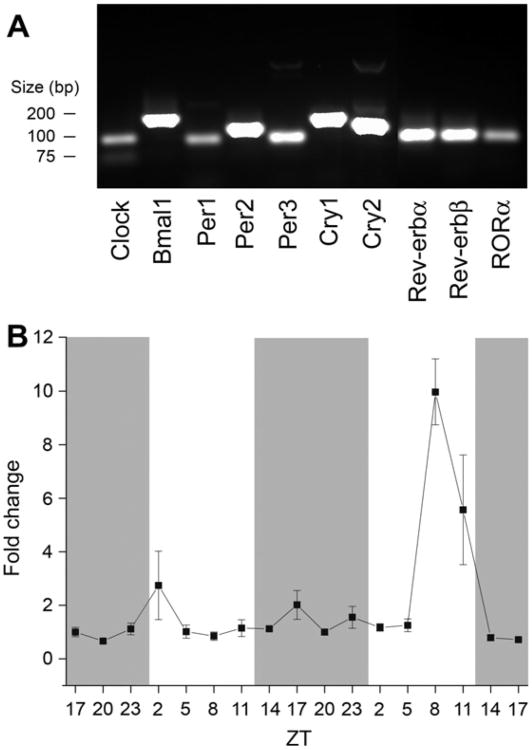
Because only a few clock genes have been found in the OB, we examined total RNA extracts for the presence of canonical clock genes. Clock, Bmal1, Per1, Per2, Per3, Cry1, Cry2, Rev-erb α, Rev-erb β, and RORα mRNAs are present in the OB (Fig. 1A). To test if our experimental procedure could identify mRNA changes in the OB over time, we examined Per2 mRNA over 48-h in rat OB by quantitative PCR (qPCR). Per2 mRNA had two peaks in expression over 48-h (Fig. 1B). Per2 changes were statistically significant (Kruskal–Wallis, P < 0.05) across 48-h, but not necessarily rhythmic (JTK_CYCLE, P > 0.05). When the two days were tested separately, only the second 24-h period was statistically significant (Kruskal– Wallis and JTK_CYCLE, each P <0.05). Neither 24-h period was statistically significant for rhythmicity as assessed by harmonic regression analysis (P < 0.05). Quantitative PCR statistical analyses are summarized in Table 2. Our data were similar at time points close to those previously reported (Hamada et al., 2011). These data establish that all the genes in the core molecular “lock” are present in the OB and that our experimental procedure can identify time-dependent changes in gene expression.
Fig. 1.
Canonical clock genes are present in the rat olfactory bulb (OB). (A) PCR results amplifying cDNA from rat OB and primers as reported by Kamphuis et al. (2005). Messenger RNAs encoding Clock, Bmal1, Period1–3(Per1–3), Cryptochrome1–2(Cry1–2), Reverb α, Rev-erb β, and retinoid-related orphan receptorα (RORα) are present in OB samples. (B) Line plot of Per2 mRNA changes over 48-h. N = 4–5 animals per time point. Points represent mean plus or minus standard error of the mean (SEM) in this and subsequent figures. Gray bars represent dark phases and white bars represent light phases. Per2 mRNA changes were statistically significant across the 48-h and for the second 24-h period (Kruskal–Wallis H test, P < 0.05) but not rhythmic across either 24-h tested (harmonic regression analysis, P > 0.05). Per2 mRNA was statistically significant for JTK_CYCLE analysis only for the second 24-h period (P < 0.05).
Table 2. Statistical results for measured mRNA.
| mRNA | KW48 | KW1–24 | KW24–48 | Dunn's | HR1–24 | HR24–48 | JTK48 | JTK1–24 | JTK24–48 |
|---|---|---|---|---|---|---|---|---|---|
| Cx36 | * | * | NS | * | * | NS | NS | * | NS |
| Cx43 | * | * | NS | * | * | NS | NS | * | NS |
| Cx45 | * | * | NS | * | * | NS | NS | NS | NS |
| GluR1 | * | * | * | * | * | * | * | * | * |
| GluR2 | * | * | * | * | NS | NS | NS | * | NS |
| GluR3 | * | NS | NS | NS | NS | NS | NS | NS | NS |
| GluR4 | NS | * | NS | NS | NS | NS | * | NS | * |
| Per2 | * | NS | * | NS | NS | NS | NS | NS | * |
KW48 is the Kruskal–Wallis H test for 48 h of time points, KW1–24 is the Kruskal–Wallis H test for the first 24 h of time points, KW24–48 is the Kruskal–Wallis H test for the second 24 h of time points. Dunn's is the Dunn's post hoc test that was performed on the Kruskal–Wallis H test results. HR is the harmonic regression analysis, and JTK is the JTK_CYCLE analysis. An “*” indicates that test was considered statistically significant at the 95% confidence level and not significant is “NS”.
Changes in mRNA expression of connexin and AMPA receptor genes
The connexins we tested for diurnal expression patterns in Fig. 2 (Cx36, Cx43, and Cx45) had been previously identified in the OB by freeze-fracture electron microscopy (Rash et al., 2005). Cx36 showed a rhythm of mRNA expression with peaks in the dark phase for both days tested and a trough during the light phase; both dark phase peaks were 5-h after the lights were turned off (ZT17). Cx43 also peaked in the dark phase for the first 24 h, but the second 24 h had large fluctuations in Cx43 expression that did not resemble the previous day's changes. Messenger RNA for Cx45 had peaks during the dark phases over the 48-h. All three connexins had statistically significant mRNA expression changes over 48 h (Kruskal–Wallis, P < 0.05) and for the first, but not the second, 24-h period. Cx36 and Cx43 were not statistically significant across 48 h as judged by JTK_CYCLE analysis but significant for the first 24-h period (P < 0.05). For the three connexins, the first 24-h period was statistically significant for rhythmicity assessed by harmonic regression, (P < 0.05), but the second 24-h period was not.
Fig. 2.
Changes in connexin mRNA in the OB across dark and light phases of the diurnal cycle. Line plots of (A) connexin36 (Cx36), (B) connexin43 (Cx43), and (C) connexin45 (Cx45) mRNA expression over 48-h. Sample size and notation as in Fig. 1. Cx36, Cx43, and Cx45 mRNA changes were statistically significant across the 48 h (Kruskal–Wallis H test, P < 0.05). Cx36 was rhythmic for the first 24-h tested and both Cx43 and Cx45 were rhythmic for the second 24-h tested (harmonic regression analysis, P < 0.05). Cx36 and Cx43 mRNAs were statistically significant only for the first 24-h period as tested by JTK_CYCLE analysis (P < 0.05), while Cx45 was not significant (P > 0.05).
We also examined the mRNA expression of the AMPAR subunits (shown in Fig. 3), because Christie et al. (2005) showed that Cx36's effects on neurotransmission required AMPA autoreceptor function. GluR1 had troughs during the light phase for both days and peaks at ZT17, during the dark phase. GluR2 appeared to fluctuate throughout the 48 h without clear, repeated peaks between the two days, although each of the peaks occurred in the dark phase. The data for GluR3 do not appear to show any rhythm but rather show a slight steady increase over the 48 h. GluR4 does not show a well-defined rhythm, although its expression was similar to that of GluR2 in that the peaks occurred in the dark phase. GluR1–3 mRNAs varied significantly over 48 h (Kruskal–Wallis, P < 0.05). When separated, GluR1 and GluR2 varied significantly for both 24-h periods tested (Kruskal–Wallis, P < 0.05), and GluR4 varied significantly for the second 24-h (Kruskal–Wallis). Only GluR1 varied significantly for both 24-h periods for harmonic regression analysis (P < 0.05). According to the JTK_CYCLE analysis, GluR1 and GluR4 were rhythmic over 48 h, GluR1 and GluR2 were rhythmic over the first 24 h, and GluR1 and GluR4 were rhythmic over the second 24 h (P < 0.05).
Fig. 3.
Changes in α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor (AMPAR) subunit mRNA across dark and light phases in rat OB. Line plots of (A) GluR1, (B) GluR2, (C) GluR3, and (D) GluR4 mRNA expression over 48-h. Sample size and notation as in Fig. 1. GluR1, GluR2, and GluR3 mRNA changes were statistically significant across the 48-h (Kruskal–Wallis H test, P < 0.05) but only GluR1 was rhythmic for both 24-h periods (harmonic regression analysis, P < 0.05). GluR1 and GluR4 were rhythmic according to JTK_CYCLE analysis (P < 0.05).
These results indicate that the relative concentrations of Cx36, Cx43, Cx45, GluR1, GluR2, and GluR4 mRNAs fluctuate over time and that GluR1 is likely rhythmic, whereas GluR2, GluR3, and GluR4 are not rhythmic but do change over time.
Changes in connexin and AMPAR subunit proteins expressed in the plasma membrane
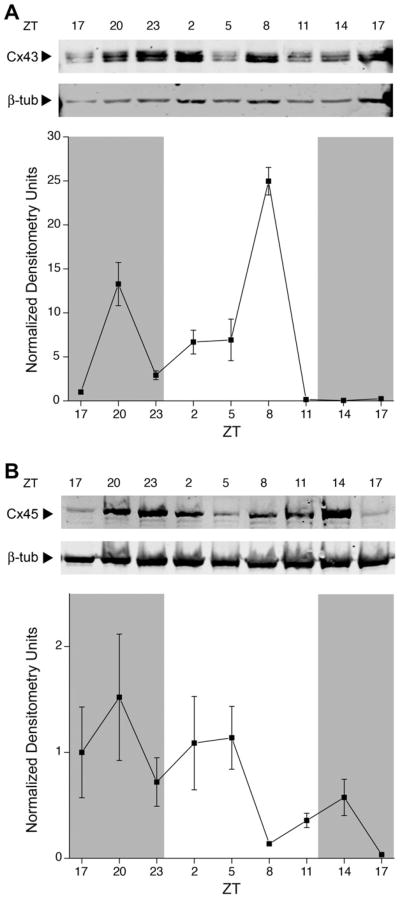
Membrane-bound Cx43 protein expression (Fig. 4A) peaked at ZT20 and ZT8, one peak in the light phase and another peak in the dark. Cx45 (Fig. 4B) peaked at ZT20 and appeared to decrease steadily after that, although the concentration of the protein appeared to remain high until after ZT5, implying that the levels remained high in the dark phase. Differences in membrane protein expression of both connexins were statistically significant (Kruskal–Wallis, P < 0.05), but only Cx43 expression was harmonic (harmonic regression, P < 0.05). Both Cx43 and Cx45 were judged rhythmic by JTK_CYCLE analysis (P < 0.05).
Fig. 4.
Changes in connexin proteins expressed in the rat OB membrane across light and dark phases in the rat OB. (A) Representative membrane proteins harvested over various time points (ZT), separated by SDS–PAGE, and electro-transferred to nitrocellulose. Nitrocellulose blots were probed with anti-Cx43 (1:500) and anti-β-III-tubulin (1:2000). Arrows indicate expected size in kilodaltons (Mr = 43 kDa and 50 kDa, respectively). The corresponding line plot represents the quantitative densitometry of Cx43 in pixel density normalized to β-III-tubulin and the 0 time point. Data are mean +/− SEM for 5–7 animals per time point. Cx43 protein changes were statistically significant across the 24-h (Kruskal–Wallis H test, P < 0.05) and rhythmic (harmonic regression analysis, P < 0.05). (B) Same as (A) but probed with anti-Cx45 (1:250) and anti-β-III-tubulin (1:2000). Arrows indicate expected size in kDa (Mr = 45 kDa and 50 kDa, respectively). Cx45 protein changes were statistically significant across the 24-h (Kruskal–Wallis H test, P < 0.05) but arrhythmic (harmonic regression analysis, P > 0.05). Cx43 and Cx45 protein changes were statistically significant for JTK_CYCLE analysis (P < 0.05).
We also tried to determine if membrane-bound Cx36 protein expression changed over the course of a day, but were unsuccessful in identifying a reliable commercial antibody. We examined Cx36 expression in the rat OB using three different anti-Cx36 antisera. Each antiserum visualized multiple bands (36 and 72 kDa) in rat and wild-type mouse OB and hippocampus; unexpectedly, these antisera also labeled these same bands in the heart, liver, and Cx36-knockout mouse OB and hippocampus. Multiple bandings could not be eliminated with modification in blocker solution or blocking intensity.
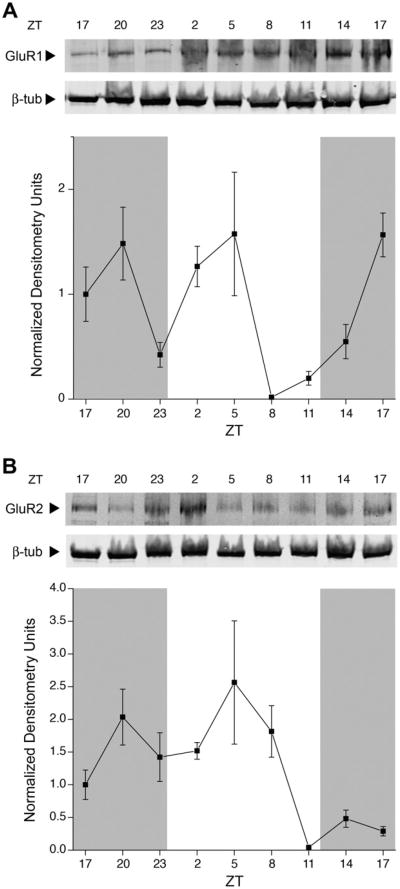
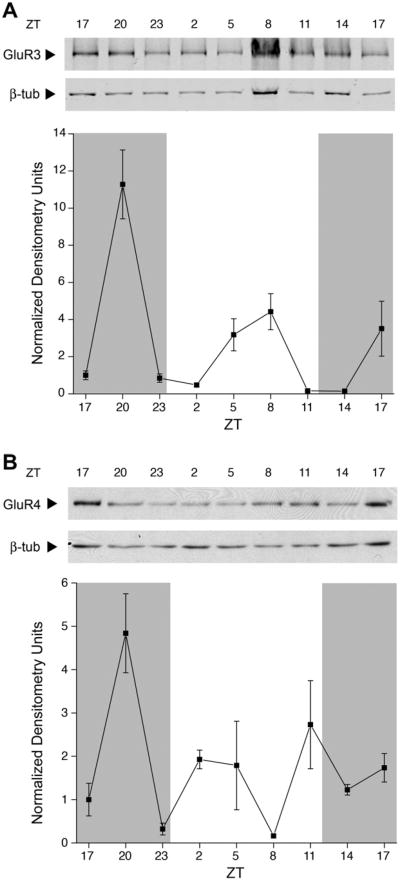
GluR1 protein expression (Fig. 5A) peaked at ZT17. The expression of GluR1 peaked both in the dark phase and the light phase. GluR2 membrane expression (Fig. 5B) peaked at ZT20 and during the light phase; protein expression did not return to the same level of expression at the next ZT17 time point. GluR3 expression (Fig. 6A) peaked in the dark, with a much higher amplitude than was seen at any other time point examined. The lowest point of GluR3 expression was at both light onset and offset. Membrane protein expression of GluR4 (Fig. 6B) peaked both in the dark and the light, without an apparent rhythm. For all 4 AMPAR subunits, membrane protein expression changes were statistically significant (Kruskal–Wallis, P < 0.05), but only GluR2 was harmonic (harmonic regression, P < 0.05). GluR1–3 were judged rhythmic by JTK_CYCLE analysis (P < 0.05).
Fig. 5.
Changes in AMPAR subunit protein expressed in the rat OB membrane across light and dark phases in rat OB. (A) Anti-GluR1 (1:500) and (B) anti-GluR2 (1:500) using experimental design, analysis, notation, sample size, and statistical metric as in Fig. 4. Nitrocellulose blots were stripped and reprobed with anti-β-III-tubulin (1:2000). Arrows indicate expected size in kilodaltons (Mr = 110 kDa for GluR1, 100 kDa for GluR2, and 50 kDa for tubulin). GluR1 and GluR2 protein changes were statistically significant across the 48 h (Kruskal–Wallis H test, P < 0.05) but only GluR2 protein expression was rhythmic in the 24-h tested (harmonic regression analysis, P < 0.05). GluR1 and GluR2 protein expression was statistically significant for JTK_CYCLE analysis (P < 0.05).
Fig. 6.
Changes in AMPAR subunit protein expressed in the rat OB membrane across light and dark phases in the rat OB. (A) Anti-GluR3 (1:1500) and anti-beta-III-tubulin (1:2000) and (B) anti-GluR4 (1:1000) using experimental design, analysis, notation, sample size, and statistical metric as in Fig. 4. Nitrocellulose blots probed with anti-GluR4 were stripped and reprobed with anti-β-III-tubulin (1:2000). Arrows indicate expected size in kilodalton (Mr = 100 kDa for GluR3 and GluR4, and 50 kDa for tubulin). GluR3 and GluR4 protein changes were statistically significant across the 48-h (Kruskal–Wallis H test, P < 0.05) but not rhythmic across either 24-h tested (harmonic regression analysis, P > 0.05). GluR3 protein expression was statistically significant for JTK_CYCLE analysis (P < 0.05).
Taken together, these results demonstrate that the expression patterns of Cx43, Cx45, GluR1, GluR2, GluR3, and GluR4 proteins in the rat OB membrane change over time, and that Cx43, GluR1, and GluR2 may be rhythmic. Statistical analyses of the protein data are summarized in Table 3.
Table 3. Statistical results for measured membrane/organelle protein.
| Protein | KW24 | HR24 | JTK24 |
|---|---|---|---|
| Cx43 | * | * | * |
| Cx45 | * | NS | * |
| GluR1 | * | NS | * |
| GluR2 | * | * | * |
| GluR3 | * | NS | * |
| GluR4 | * | NS | NS |
KW24 is the Kruskal–Wallis H test, HR24 is the harmonic regression analysis, and JTK24 is the JTK_CYCLE analysis. An “*” indicates that test was statistically significant at a 95% confidence level and not significant is “NS”.
Discussion
Although the OB has been demonstrated to be an independent circadian oscillator, little is known about the underlying cellular and molecular mechanisms that generate those oscillations. Our results show that mRNAs for Clock, Bmal1, Per1–3, Cry1–2, Rev-erbα and Rev-erbβ, and RORα are present in the rat OB. Per2 mRNA expression changed over the course of 48 h, but was not rhythmic. A previous report (Hamada et al., 2011) showed that Per2 mRNA changed at ZTs 19, 1, 7, and 13 in the OBs of mice. Our study shows similar changes at time points close to those 4 on the second day (we show ZTs 20, 2, 8, and 14), though our additional time points make it appear that Per2 is not rhythmic in the rat OB. We postulate that this is a difference between mice and rats, and that Per1 (Abraham et al., 2005), but not Per2, expression is rhythmic in the rat OB. We also present evidence that Cx36 and Cx45 mRNA expression in the OB changed across light and dark phases, and the expression of both mRNA and membrane-bound protein of Cx43, Cx45, GluR1, GluR2, GluR3, and GluR4 in the OB changed throughout the course of the day. Our results suggest that changes in the expression of gap junctions and AMPARs alter odor information processing by the OB. Some of the proteins we examined had peaks in expression in both light and dark phases. These peaks may be the results of output of the OB internal clock, input from the suprachiasmatic nuclei, activity-dependent mechanisms in the OB, or developmental changes. Developmental changes in the expression of Cx36 (Song et al., 2012) and AMPARs (Ritter et al., 2002) have been reported in other brain areas.
Cx36 and Cx45 are expressed by neurons in the OB, and gap junctions composed of Cx36 electrically couple these neurons, allowing for synchronous electrical activity. Such activity requires activation of AMPARs (Christie et al., 2005), as well as intraglomerular lateral excitation (Christie and Westbrook, 2006). AMPA receptors are expressed throughout the OB, and changes in the expression of these receptors, as has been shown for other transmitter systems in response to altered sensory experience (Parrish-Aungst et al., 2011), would therefore affect the behavior of many circuits within the bulb by altering responses to excitatory synaptic transmission and glutamate spillover. Because Maher et al. (2009) showed that Cx36-dependent mitral-cell electrical coupling stabilizes at the adult level in P14–P21 mice, we propose that the changes we observed in P21 juveniles persist throughout adulthood.
It is hypothesized that the main rhythmic cells in the OB are M/Ts, because Per1 rhythmicity was specific to M/Ts and not granule cells (Granados-Fuentes et al., 2004a); therefore, any diurnal changes in granule cell activity previously described (Granados-Fuentes et al., 2006) are probably the result of M/T activity on those cells. External tufted cells are known to be endogenous bursters (Hayar et al., 2004) although it is not known whether their patterns change with time of day. The exact mechanism by which M/T input would change granule cell protein expression is unknown; changes in glutamate receptor expression may occur by activation of mGluR5 at mitral cell-granule cell dendrodendritic synapses (Heinbockel et al., 2007).
Cx36
Cx36 is expressed by M/T cells and provides direct electrical connections between their apical dendrites within a glomerulus (Christie et al., 2005; Christie and Westbrook, 2006). The changes in mRNA expression we report could lead to altered behavior of these circuits as a function of the time of day because such connections contribute to synchronous activation of these cells. These alterations could be the molecular basis for the circadian rhythms in olfactory discrimination of mice (Granados-Fuentes et al., 2011). We hypothesize that the changes we saw in Cx36 mRNA would only affect mitral cell–mitral cell, tufted cell–tufted cell, or mitral cell–tufted cell gap junctions, which are composed exclusively of Cx36. Different expression-system studies have shown that Cx36 does not form heteromeric gap junctions with either Cx43 or Cx45 (Li et al., 2008). Cx36 modulation may not occur only at the level of expressed mRNA and protein; in the retina, for example, dopamine has been shown to be able to alter the conductance of Cx36 gap junctions through changes in their phosphorylation state (reviewed by Goodenough and Paul, 2009). The OB has its own population of dopamine neurons, and several studies have shown that dopamine modulates OB circuits (Hsia et al., 1999; Berkowicz and Trombley, 2000; Ennis et al., 2001; Davila et al., 2003). Whether dopamine also alters the function of OB gap junctions is currently under investigation.
Cx43
Expression of Cx43 is limited to astrocytes and ensheathing cells in the OB (Rash et al., 2005), though its expression in the olfactory epithelium can affect synaptic transmission to OB glomeruli (Zhang, 2010). Ensheathing cells do not appear to impact synaptic transmission in the OB; astrocytes, however, can express circadian rhythms (Prolo et al., 2005), and can have effects on synaptic transmission (reviewed in Halassa and Haydon, 2010), though neither has been demonstrated in OB astrocytes. Connexin hexamers, called connexons, can be present on the surface of the cell. The evidence for connexons composed of Cx43 to form a “hemichannel” that allows for a non-junctional membrane conductance is controversial (reviewed in Scemes, 2011). It has not been well explored if Cx36 or Cx45 is capable of forming hemichannels. Gap junction-coupled astrocytes are able to conduct Ca2+ waves that depend on glutamate transmission (Cornell-Bell et al., 1990). In addition, astrocytes are capable of releasing both glutamate and D-serine (reviewed in Haydon and Carmignoto, 2006), which together can activate NMDARs. NMDARs are required for dendrodendritic inhibition within the OB between M/T and granule cells when single electrical pulses are applied to the OB (Schoppa et al., 1998; Christie et al., 2001). Astrocytes are also able to regulate blood flow (Metea and Newman, 2006), which may affect OB neurons by altering local glucose levels (Tucker et al., 2010). We also note that a recent report (Roux et al., 2011) identified connexin30 expression by astrocytes in the OB. This report came out after our study was completed, and we note that connexin30 expression and its effects on astrocyte-to-astrocyte gap junctions may also change across the light/dark cycle and affect olfaction. Further work is required to determine the effects that changes in Cx43 expression have on OB astrocytes and how they may influence circuit behavior.
Cx45
Cx45 is regarded as a neuronal connexin (Rash et al., 2005) and has been hypothesized to connect M/T cells alongside Cx36-containing gap junctions, as happens in the retina (Li et al., 2008). Cx45 has not been well explored in the OB; its main site of study has been in the mammalian heart. Currently, we do not know what effects Cx45 expression has on synaptic transmission because genetic ablation of Cx45 is embryonically lethal and selective connexin pharmacology is currently limited. Little electrical coupling between M/Ts by Cx45-containing gap junctions is apparent in the OB, as Cx36-knockout mice show no electrical coupling between M/Ts despite intact Cx45 (Christie et al., 2005; Christie and Westbrook, 2006). Ultrastructural studies have confirmed that Cx45 is a neuronal connexin (Rash et al., 2005), but because of the lack of electrical coupling of neurons in the Cx36-knockout mouse, Cx45 seems to form junctions with an unknown partner cell type or a minority of M/Ts.
AMPA receptors
Altering the expression of AMPARs in the OB would affect many different circuit behaviors. AMPAR subunits are not segregated to different cell types, as are the connexins, but instead are spread throughout the OB in varying degrees (Montague and Greer, 1999; Horning et al., 2004). OB AMPARs can also have different kinetics and cation selectivity, dependent on subunit composition (Blakemore and Trombley, 2003, 2004). Hypothesizing about the precise effect of changes in GluR1, GluR3, or GluR4 is difficult, but changes in GluR2 expression would affect the populations of Ca2+-permeable AMPARs. If GluR2 in a given AMPAR is not edited posttranscriptionally or not present in the AMPAR, that receptor will allow flux of Ca2+ in addition to Na+ and K+, which can lead to neurotransmitter release, calmodulin/CAM kinase messenger cascades, and even cell death (Trump and Berezesky, 1995), without NMDAR activation. Ca2+-permeable AMPARs are present in the OB (Blakemore et al., 2006; Ma and Lowe, 2007; Pimentel and Margrie, 2008); decreases in GluR2 may allow for more Ca2+-permeable AMPARs, and these receptors could play a greater role in dendrodendritic inhibition, which normally requires NMDAR activity to allow the necessary Ca2+ flux when applying a single electrical pulse to the OB. If one applies repeated pulses, however, AMPARs and kainate receptors are required for dendrodendritic inhibition (Schoppa, 2006b). Genetic deletion of GluR2 increases mitral cell inhibition by granule cells via increased Ca2+ flux (Abraham et al., 2010), which may result in increased synchronous firing of M/Ts (Schoppa, 2006a). We therefore hypothesize that the changes we saw in GluR2 membrane protein expression reflect changes in granule cell electrical activity and subsequently change M/T inhibition.
Conclusion
Our results demonstrate time-dependent changes in ion channels that control the synaptic circuits mediating odor processing by the OB. The OB is a highly laminated structure and odor processing is dependent on the synaptic circuits within each of these layers. Although the whole bulb preparations we use in the present study do not identify changes in specific laminae or cell types, and our approach does not allow precise quantification of surface versus cytoplasmic proteins, we have demonstrated temporal changes in the OB as a whole, some of which appear to be diurnal. We hypothesize that these temporal changes affect synaptic transmission and thus olfaction. The rhythms reported here may affect intercellular communication through gap junctions and glutamatergic transmission. These changes may be part of the molecular makeup of the electrical rhythms in the OB reported by Granados-Fuentes et al., 2004a,b and the circadian patterns of olfactory discrimination in rodents (Granados-Fuentes et al., 2011).
Acknowledgments
The authors thank J. Olcese, C. DeHeer, B. Lynn, B.K. Washburn, C. Fitch-Pye, and S.H. Miller for technical advice, K. Kornacker for donating the JTK_CYCLE script, J. Travis for statistical advice, L.J. Blakemore for help with the manuscript, and B.W. Connors and D.L. Paul for tissue from Cx36-knockout mice. We thank Ms. Stephanie Zych and Mr. Christopher Kovach for technical assistance. This work was supported by Grants from the US National Institutes of Health (NIH) [R01 DC003387 and T32 DC00044] from the National Institute on Deafness and Other Communication Disorders (NIDCD) and intramural funding from the Program in Neuroscience at Florida State University.
Abbreviations
- AMPA
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- AMPAR
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor
- Cx36
connexin36
- Cx43
connexin43
- Cx45
connexin45
- EDTA
ethylenediaminetetraacetic acid
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- M/T
mitral/tufted
- NMDAR
N-methyl-D-aspartic acid receptor
- OB
olfactory bulb
- OBBT
Odyssey Blocking Buffer with 0.2% Tween-20
- PCR
polymerase chain reaction
- TBST
Tris-buffered saline with 0.1% Tween-20
- TBST*
Tris-buffered saline with 0.2% Tween-20
References
- Abe M, Herzog ED, Yamazaki S, Straume M, Tei H, Sakaki Y, Menaker M, Block GD. Circadian rhythms in isolated brain regions. J Neurosci. 2002;22:350–356. doi: 10.1523/JNEUROSCI.22-01-00350.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham NM, Egger V, Shimshek DR, Renden R, Fukunaga I, Sprengel R, Seeburg PH, Klugmann M, Margrie TW, Schaefer AT, Kuner T. Synaptic inhibition in the olfactory bulb accelerates odor discrimination in mice. Neuron. 2010;65:399–411. doi: 10.1016/j.neuron.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham U, Prior JL, Granados-Fuentes D, Piwnica-Worms DR, Herzog ED. Independent circadian oscillations of Period1 in specific brain areas in vivo and in vitro. J Neurosci. 2005;25:8620–8626. doi: 10.1523/JNEUROSCI.2225-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkowicz DA, Trombley PQ. Dopaminergic modulation at the olfactory nerve synapse. Brain Res. 2000;855:90–99. doi: 10.1016/s0006-8993(99)02342-2. [DOI] [PubMed] [Google Scholar]
- Berkowicz DA, Trombley PQ, Shepherd GM. Evidence for glutamate as the olfactory receptor cell neurotransmitter. J Neurophysiol. 1994;71:2557–2561. doi: 10.1152/jn.1994.71.6.2557. [DOI] [PubMed] [Google Scholar]
- Blakemore LJ, Trombley PQ. Kinetic variability of AMPA receptors among olfactory bulb neurons in culture. Neuroreport. 2003;14:965–970. doi: 10.1097/01.wnr.0000070826.57864.00. [DOI] [PubMed] [Google Scholar]
- Blakemore LJ, Trombley PQ. Diverse modulation of olfactory bulb AMPA receptors by zinc. Neuroreport. 2004;15:919–923. doi: 10.1097/00001756-200404090-00037. [DOI] [PubMed] [Google Scholar]
- Blakemore LJ, Resasco M, Mercado MA, Trombley PQ. Evidence for Ca2+ -permeable AMPA receptors in the olfactory bulb. Am J Physiol Cell Physiol. 2006;290:C925–C935. doi: 10.1152/ajpcell.00392.2005. [DOI] [PubMed] [Google Scholar]
- Christie JM, Westbrook GL. Lateral excitation within the olfactory bulb. J Neurosci. 2006;26:2269–2277. doi: 10.1523/JNEUROSCI.4791-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie JM, Bark C, Hormuzdi SG, Helbig I, Monyer H, Westbrook GL. Connexin36 mediates spike synchrony in olfactory bulb glomeruli. Neuron. 2005;46:761–772. doi: 10.1016/j.neuron.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Christie JM, Schoppa NE, Westbrook GL. Tufted cell dendrodendritic inhibition in the olfactory bulb is dependent on NMDA receptor activity. J Neurophysiol. 2001;85:169–173. doi: 10.1152/jn.2001.85.1.169. [DOI] [PubMed] [Google Scholar]
- Cook KK, Fadool DA. Two adaptor proteins differentially modulate the phosphorylation and biophysics of Kv1.3 ion channel by Src kinase. J Biol Chem. 2002;277:13268–13280. doi: 10.1074/jbc.M108898200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell-Bell AH, Finkbeiner SM, Cooper MS, Smith SJ. Glutamate induces calcium waves in cultured astrocytes: long-range glial signaling. Science. 1990;247:470–473. doi: 10.1126/science.1967852. [DOI] [PubMed] [Google Scholar]
- Davila NG, Blakemore LJ, Trombley PQ. Dopamine modulates synaptic transmission between rat olfactory bulb neurons in culture. J Neurophysiol. 2003;90:395–404. doi: 10.1152/jn.01058.2002. [DOI] [PubMed] [Google Scholar]
- Deans MR, Gibson JR, Sellitto C, Connors BW, Paul DL. Synchronous activity of inhibitory networks in neocortex requires electrical synapses containing connexin36. Neuron. 2001;31:477–485. doi: 10.1016/s0896-6273(01)00373-7. [DOI] [PubMed] [Google Scholar]
- Decrock E, Vinken M, Bol M, D'Herde K, Rogiers V, Vandenabeele P, Krysko DV, Bultynck G, Leybaert L. Calcium and connexin-based intercellular communication, a deadly catch? Cell Calcium. 2011;50:310–321. doi: 10.1016/j.ceca.2011.05.007. [DOI] [PubMed] [Google Scholar]
- Dingledine R, Borges K, Bowie D, Traynelis SF. The glutamate receptor ion channels. Pharmacol Rev. 1999;51:7–61. [PubMed] [Google Scholar]
- Ennis M, Zhou FM, Ciombor KJ, Aroniadou-Anderjaska V, Hayar A, Borrelli E, Zimmer LA, Margolis F, Shipley MT. Dopamine D2 receptor-mediated presynaptic inhibition of olfactory nerve terminals. J Neurophysiol. 2001;86:2986–2997. doi: 10.1152/jn.2001.86.6.2986. [DOI] [PubMed] [Google Scholar]
- Ennis M, Zimmer LA, Shipley MT. Olfactory nerve stimulation activates rat mitral cells via NMDA and non-NMDA receptors in vitro. Neuroreport. 1996;7:989–992. doi: 10.1097/00001756-199604100-00007. [DOI] [PubMed] [Google Scholar]
- Goodenough DA, Paul DL. Gap junctions. Cold Spring Harb Perspect Biol. 2009;1:a002576. doi: 10.1101/cshperspect.a002576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granados-Fuentes D, Ben-Josef G, Perry G, Wilson DA, Sullivan-Wilson A, Herzog ED. Daily rhythms in olfactory discrimination depend on clock genes but not the suprachiasmatic nucleus. J Biol Rhythms. 2011;26:552–560. doi: 10.1177/0748730411420247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granados-Fuentes D, Prolo LM, Abraham U, Herzog ED. The suprachiasmatic nucleus entrains, but does not sustain, circadian rhythmicity in the olfactory bulb. J Neurosci. 2004a;24:615–619. doi: 10.1523/JNEUROSCI.4002-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granados-Fuentes D, Saxena MT, Prolo LM, Aton SJ, Herzog ED. Olfactory bulb neurons express functional, entrainable circadian rhythms. Eur J Neurosci. 2004b;19:898–906. doi: 10.1111/j.0953-816x.2004.03117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granados-Fuentes D, Tseng A, Herzog ED. A circadian clock in the olfactory bulb controls olfactory responsivity. J Neurosci. 2006;26:12219–12225. doi: 10.1523/JNEUROSCI.3445-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemin I, Becker M, Ociepka K, Friauf E, Nothwang HG. A subcellular prefractionation protocol for minute amounts of mammalian cell cultures and tissue. Proteomics. 2005;5:35–45. doi: 10.1002/pmic.200400892. [DOI] [PubMed] [Google Scholar]
- Halassa MM, Haydon PG. Integrated brain circuits: astrocytic networks modulate neuronal activity and behavior. Annu Rev Physiol. 2010;72:335–355. doi: 10.1146/annurev-physiol-021909-135843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada T, Honma S, Honma K. Light responsiveness of clock genes, Per1 and Per2, in the olfactory bulb of mice. Biochem Biophys Res Comm. 2011;409:727–731. doi: 10.1016/j.bbrc.2011.05.076. [DOI] [PubMed] [Google Scholar]
- Hayar A, Karnup S, Shipley MT, Ennis M. Olfactory bulb glomeruli: external tufted cells intrinsically burst at theta frequency and are entrained by patterned olfactory input. J Neurosci. 2004;24(5):1190–1199. doi: 10.1523/JNEUROSCI.4714-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydon PG, Carmignoto G. Astrocyte control of synaptic transmission and neurovascular coupling. Physiol Rev. 2006;86:1009–1031. doi: 10.1152/physrev.00049.2005. [DOI] [PubMed] [Google Scholar]
- Heinbockel T, Laaris N, Ennis M. Metabotropic glutamate receptors in the main olfactory bulb drive granule cell-mediated inhibition. J Neurophysiol. 2007;97:858–870. doi: 10.1152/jn.00884.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horning MS, Kwon B, Blakemore LJ, Spencer CM, Goltz M, Houpt TA, Trombley PQ. Alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionate receptor subunit expression in rat olfactory bulb. Neurosci Lett. 2004;372:230–234. doi: 10.1016/j.neulet.2004.09.044. [DOI] [PubMed] [Google Scholar]
- Hsia AY, Vincent JD, Lledo PM. Dopamine depresses synaptic inputs into the olfactory bulb. J Neurophysiol. 1999;82:1082–1085. doi: 10.1152/jn.1999.82.2.1082. [DOI] [PubMed] [Google Scholar]
- Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms. 2010;25:372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphuis W, Cailotto C, Dijk F, Bergen A, Buijs RM. Circadian expression of clock genes and clock-controlled genes in the rat retina. Biochem Biophys Res Comm. 2005;330:18–26. doi: 10.1016/j.bbrc.2005.02.118. [DOI] [PubMed] [Google Scholar]
- Kosaka T, Kosaka K. Neuronal gap junctions in the rat main olfactory bulb, with special reference to intraglomerular gap junctions. Neurosci Res. 2003;45:189–209. doi: 10.1016/s0168-0102(02)00222-5. [DOI] [PubMed] [Google Scholar]
- Kosaka T, Kosaka K. Neuronal gap junctions between intraglomerular mitral/tufted cell dendrites in the mouse main olfactory bulb. Neurosci Res. 2004;49:373–378. doi: 10.1016/j.neures.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Kosaka T, Deans MR, Paul DL, Kosaka K. Neuronal gap junctions in the mouse olfactory bulb: morphological analyses on transgenic mice. Neurosci. 2005;134:757–769. doi: 10.1016/j.neuroscience.2005.04.057. [DOI] [PubMed] [Google Scholar]
- Legendre P, Legendre L. Numerical Ecology. 2nd English. Amsterdam: Elsevier Publishing; 1998. [Google Scholar]
- Li X, Kamasawa N, Ciolofan C, Olson CO, Lu S, Davidson KGV, Yasumura T, Shigemoto R, Rash JE, Nagy JI. Connexin45-containing neuronal gap junctions in rodent retina also contain connexin36 in both apposing hemiplaques, forming bihomotypic gap junctions, with scaffolding contributed by zonula occludens-1. J Neurosci. 2008;28:9769–9789. doi: 10.1523/JNEUROSCI.2137-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Lowe G. Calcium permeable AMPA receptors and autoreceptors in external tufted cells of rat olfactory bulb. Neurosci. 2007;144:1094–1108. doi: 10.1016/j.neuroscience.2006.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher BJ, McGinley MJ, Westbrook GL. Experience-dependent maturation of the glomerular microcircuit. PNAS. 2009;106:16865–16870. doi: 10.1073/pnas.0808946106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metea MR, Newman EA. Glial cells dilate and constrict blood vessels: a mechanism of neurovascular coupling. J Neurosci. 2006;26:2862–2870. doi: 10.1523/JNEUROSCI.4048-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miragall F, Simbürger E, Dermietzel R. Mitral and tufted cells of the mouse olfactory bulb possess gap junctions and express connexin43 mRNA. Neurosci Lett. 1997;216:199–202. doi: 10.1016/0304-3940(96)13042-1. [DOI] [PubMed] [Google Scholar]
- Montague AA, Greer CA. Differential distribution of ionotropic glutamate receptor subunits in the rat olfactory bulb. J Compar Neurol. 1999;405:233–246. doi: 10.1002/(sici)1096-9861(19990308)405:2<233::aid-cne7>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Parrish-Aungst S, Kiyokage E, Szabo G, Yanagawa Y, Shipley MT, Puche AC. Sensory experience selectively regulates transmitter synthesis enzymes in interglomerular circuits. Brain Res. 2011;1382:70–76. doi: 10.1016/j.brainres.2011.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel DO, Margrie TW. Glutamatergic transmission and plasticity between olfactory bulb mitral cells. J Physiol. 2008;586(8):2107–2119. doi: 10.1113/jphysiol.2007.149575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prolo LM, Takahashi JS, Herzog ED. Circadian rhythm generation and entrainment in astrocytes. J Neurosci. 2005;25:404–408. doi: 10.1523/JNEUROSCI.4133-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rash JE, Davidson KGV, Kamasawa G, Yasumura T, Kamasawa M, Zhang C, Michaels R, Restrepo D, Ottersen OP, Olson CO, Nagy JI. Ultrastructural localization of connexins (Cx36, Cx43, Cx45), glutamate receptors and aquaporin-4 in rodent olfactory mucosa, olfactory nerve and olfactory bulb. J Neurocytol. 2005;34:307–341. doi: 10.1007/s11068-005-8360-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rela L, Bordey A, Greer CA. Olfactory ensheathing cell membrane properties are shaped by connectivity. Glia. 2010;58:665–678. doi: 10.1002/glia.20953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resuehr D, Spiess AN. A real-time polymerase chain reaction-based evaluation of cDNA synthesis priming methods. Anal Biochem. 2003;322:287–291. doi: 10.1016/j.ab.2003.07.017. [DOI] [PubMed] [Google Scholar]
- Ritter LM, Vazquez DM, Meador-Woodruff JH. Ontogeny of ionotropic glutamate receptor subunit expression in the rat hippocampus. Dev Brain Res. 2002;139:227–236. doi: 10.1016/s0165-3806(02)00572-2. [DOI] [PubMed] [Google Scholar]
- Roux L, Benchenane K, Rothstein JD, Bonvento G, Giaume C. Plasticity of astroglial networks in olfactory glomeruli. PNAS. 2011;108:18442–18446. doi: 10.1073/pnas.1107386108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell D. Molecular Cloning. 3rd. Long Island: Cold Spring Harbor Press; 2001. [Google Scholar]
- Santiago AR, Gaspar JM, Baptista FI, Cristóvão AJ, Santos PF, Kamphuis W, Ambrósio AF. Diabetes changes the levels of ionotropic glutamate receptors in the rat retina. Mol Vis. 2009;15:1620–1630. [PMC free article] [PubMed] [Google Scholar]
- Scemes E. Nature of plasmalemmal functional “hemichannels”. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbamem.2011.06.005. http://dx.doi.org/10.1016/j.bbamem.2011.06.005. [DOI] [PMC free article] [PubMed]
- Schindler J, Jung S, Niedner-Schatteburg G, Friauf E, Nothwang HG. Enrichment of integral membrane proteins from small amounts of brain tissue. J Neural Transm. 2006;113:995–1013. doi: 10.1007/s00702-006-0508-4. [DOI] [PubMed] [Google Scholar]
- Schoppa NE. Synchronization of olfactory bulb mitral cells by precisely timed inhibitory inputs. Neuron. 2006a;49:271–283. doi: 10.1016/j.neuron.2005.11.038. [DOI] [PubMed] [Google Scholar]
- Schoppa NE. AMPA/kainate receptors drive rapid output and precise synchrony in olfactory bulb granule cells. J Neurosci. 2006b;26:12996–13006. doi: 10.1523/JNEUROSCI.3503-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoppa NE, Kinzie JM, Sahara Y, Segerson TP, Westbrook GL. Dendrodendritic inhibition in the olfactory bulb is driven by NMDA receptors. J Neurosci. 1998;18:6790–6802. doi: 10.1523/JNEUROSCI.18-17-06790.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoppa NE, Westbrook GL. AMPA autoreceptors drive correlated spiking in olfactory bulb glomeruli. Nat Neurosci. 2002;5:1194–1202. doi: 10.1038/nn953. [DOI] [PubMed] [Google Scholar]
- Shieh KR. Distribution of the rhythm-related genes rPeriod1, rPeriod2, and rClock, in the rat brain. Neurosci. 2003;118:831–843. doi: 10.1016/s0306-4522(03)00004-6. [DOI] [PubMed] [Google Scholar]
- Song JH, Wang Y, Fontes JD, Belousov AB. Regulation of connexin 36 expression during development. Neurosci Lett. 2012;513:17–19. doi: 10.1016/j.neulet.2012.01.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trump BF, Berezesky IK. Calcium-mediated cell injury and death. FASEB J. 1995;9:219–228. doi: 10.1096/fasebj.9.2.7781924. [DOI] [PubMed] [Google Scholar]
- Tucker K, Cavallin MA, Jean-Baptiste P, Biju KC, Overton JM, Pedarzani P, Fadool DA. The olfactory bulb: a metabolic sensor of brain insulin and glucose concentrations via a voltage-gated potassium channel. Results Probs Cell Differ. 2010;52:147–157. doi: 10.1007/978-3-642-14426-4_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volterra A, Meldolesi J. Astrocytes, from brain glue to communication elements: the revolution continues. Nat Rev Neurosci. 2005;6:626–640. doi: 10.1038/nrn1722. [DOI] [PubMed] [Google Scholar]
- White LE. Olfactory bulb projections of the rat. Anat Rec. 1965;152:465–479. [Google Scholar]
- Yeung YG, Stanley ER. A solution for stripping antibodies from polyvinylidene fluoride immunoblots for multiple reprobing. Anal Biochem. 2009;389:89–91. doi: 10.1016/j.ab.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C. Gap junctions in olfactory neurons modulate olfactory sensitivity. BMC Neurosci. 2010;11:108–123. doi: 10.1186/1471-2202-11-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Restrepo D. Expression of connexin 45 in the olfactory system. Brain Res. 2002;929:37–47. doi: 10.1016/s0006-8993(01)03372-8. [DOI] [PubMed] [Google Scholar]
- Zhang C, Restrepo D. Heterogeneous expression of connexin 36 in the olfactory epithelium and glomerular layer of the olfactory bulb. J Comp Neurol. 2003;459:426–439. doi: 10.1002/cne.10617. [DOI] [PubMed] [Google Scholar]