Abstract
Background
Cholera is still a significant public health issue in developing countries. The aetiological agent is Vibrio cholerae and only two serogroups, O1 and O139, are known to cause pandemic or epidemic cholera. In contrast, non-O1/non-O139 V. cholerae has only been reported to cause sporadic cholera-like illness and localised outbreaks. The aim of this study was to determine the genetic diversity of non-O1/non-O139 V. cholerae isolates from hospitalised diarrhoeal patients in Zhejiang Province, China.
Results
In an active surveillance of enteric pathogens in hospitalised diarrhoeal patients, nine non-O1/non-O139 V. cholerae isolates were identified from 746 diarrhoeal stool samples at a rate of 1.2%. These isolates and an additional 31 isolates from sporadic cases and three outbreaks were analysed using pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST). PFGE divided the isolates into 25 PFGE types while MLST divided them into 15 sequence types (STs). A single ST, ST80, was predominant which persisted over several years in different cities and caused two outbreaks in recent years. Antibiotic resistance varied with the majority of the isolates resistant to sulphamethoxazole/trimethoprim and nearly all isolates either resistant or intermediate to erythromycin and rifampicin. None of the isolates carried the cholera toxin genes or toxin co-regulated pilus genes but the majority carried a type III secretion system as the key virulence factor.
Conclusions
Non-O1/non-O139 V. cholerae is an important contributor to diarrhoeal infections in China. Resistance to commonly used antibiotics limits treatment options. Continuous surveillance of non-O1/non-O139 V. cholerae is important for control and prevention of diarrhoeal infections.
Keywords: Vibrio cholerae, Non-O1/non-O139 serogroups, Pulsed-field gel electrophoresis, Multilocus sequence typing, Antibiotic resistance, Type III secretion system
Background
Cholera is an acute diarrhoeal disease caused by toxigenic Vibrio cholerae. The two most important serogroups are O1 and O139, which can cause periodic outbreaks reaching epidemic or pandemic proportions [1]. However, non-O1/non-O139 serogroups have been linked with cholera-like-illness sporadically [2-6]. Symptoms may range from mild gastroenteritis to violent diarrhoea, similar to those elicited by the O1 toxigenic strains [7]. However, patients generally suffer a less severe form of the disease than those infected by O1 toxigenic strains [8-10]. Non-O1/non-O139 V. cholerae strains have also caused localised outbreaks in many countries, including India and Thailand [3,11-15]. More recently, an O75 V. cholerae outbreak associated with the consumption of oysters was reported in the USA [5,6].
Non-O1/non-O139 V. cholerae strains are frequently isolated from the environment, particularly from seafood and aquatic sources [11,16,17]. Non-O1/non-O139 V. cholerae strains are highly heterogeneous with considerable serological diversity and vary in virulence properties. The presence of virulence genes amongst some environmental strains is significant, and environmental strains constitute a reservoir of potential pathogenic strains to human diarrhoeal infections [18-21]. Some non-O1/non-O139 strains carry key virulence genes, such as cholera toxin (CT) and toxin co-regulated pili (TCP), which are usually carried by epidemic strains [22]. Some may also carry other virulence factors such as the repeat-like toxin (RtxA) - a cytotoxin and the heat-stable enterotoxin (NAG-ST) [4,18,22-26]. A novel type III secretion system (T3SS) was found in some non-O1/non-O139 strains and appears to be an important virulence factor [27-29]. The T3SS translocates a number of T3SS effectors into the host cell which interfere with host cell signalling [27,28]. Shin et al.[29] showed that T3SS is an essential virulence factor for the non-O1/non-O139 strain AM-19226.
In this study, 40 non-O1/non-O139 V. cholerae isolates from hospitalised diarrhoeal patients in Zhejiang Province, China were analysed using multilocus sequence typing (MLST) and pulsed-field gel electrophoresis (PFGE) to determine their overall genetic relatedness. The presence of key virulence genes including enterotoxins, TCP and T3SS was also analysed.
Results and discussion
Isolation of non-O1/non-O139 V. cholerae isolates from diarrhoeal patients in Zhejiang, China
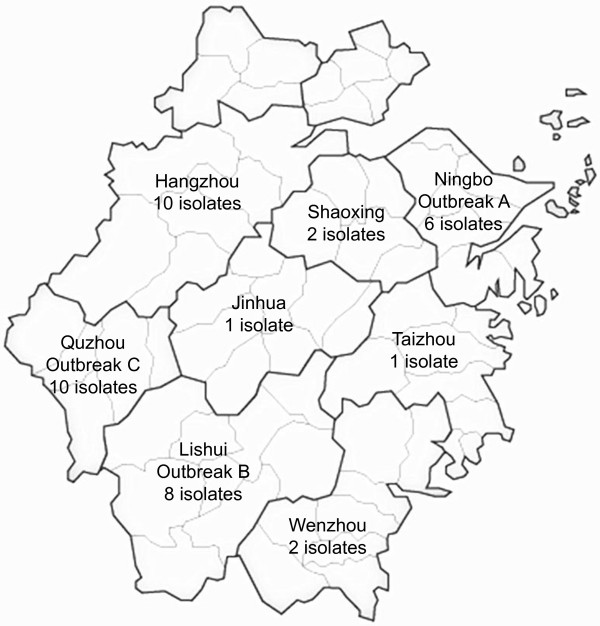
A total of 40 non-O1/non-O139 V. cholerae isolates was retrieved from different cities in Zhejiang Province, China, over a period of six years from 2005 to 2011 (Figure 1, Table 1). Nine isolates were from sporadic cases from seven cities, while 22 isolates were obtained from three outbreaks in three different cities: outbreak A in Ningbo in 2005, outbreak B in Lishui in 2006 and outbreak C in Quzhou in 2011. The three outbreaks were notified as food poisoning events and were investigated. Outbreak A involved 20 cases with symptoms ranging from cholera-like diarrhoea to mild diarrhoea and was initially suspected to be a cholera outbreak. Non-O1/non-O139 V. cholerae was isolated from nine patients. The outbreak occurred in a factory canteen and the food source of the outbreak could not be identified. Outbreak B involved eight cases, all having cholera-like symptoms. Non-O1/non-O139 V. cholerae was isolated from all but one patient. The source of the outbreak was traced to cross contamination of a cold dish from raw cuttlefish. Outbreak C occurred in a family function involving 12 cases with non-O1/non-O139 V. cholerae isolated from nine cases. The source of the outbreak was shrimp.
Figure 1.
Geographical map of Zhejiang Province, China. Cities are demarcated with dark solid lines. City names together with number of non-O1/non-O139 Vibrio cholerae isolates from a given city is shown.
Table 1.
Details of Vibrio cholerae strains analysed in this study
| Strain | Place† | Source | Year | Pulse type | ctxAB | toxR | hlyA | tcpA* | zot | NAG-ST |
T3SS |
T3SS |
adk | gyrB | mdh | metE | pntA | purM | pyrC | Sequence type |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ( vcsC2 ) | ( vcsV2 ) | |||||||||||||||||||
| N11192 |
HZ |
sporadic |
2011 |
24 |
- |
+ |
+ |
- |
- |
- |
- |
- |
39 |
45 |
14 |
67 |
50 |
1 |
42 |
93 |
| N10001 |
HZ |
surveillance |
2010 |
13 |
- |
+ |
+ |
- |
- |
- |
+ |
+ |
2 |
44 |
11 |
64 |
6 |
8 |
10 |
94 |
| N10002 |
HZ |
surveillance |
2010 |
N/A‡ |
- |
+ |
+ |
- |
- |
- |
+ |
+ |
2 |
23 |
45 |
48 |
47 |
35 |
5 |
89 |
| N10003 |
HZ |
surveillance |
2010 |
14 |
- |
+ |
+ |
- |
- |
- |
+ |
+ |
2 |
5 |
45 |
56 |
6 |
36 |
51 |
80 |
| N10004 |
HZ |
surveillance |
2010 |
15 |
- |
+ |
+ |
- |
- |
- |
+ |
+ |
2 |
38 |
14 |
56 |
6 |
36 |
51 |
83 |
| N10005 |
HZ |
surveillance |
2010 |
16 |
- |
+ |
+ |
- |
- |
- |
+ |
+ |
2 |
5 |
45 |
56 |
6 |
36 |
51 |
80 |
| N10006 |
HZ |
surveillance |
2010 |
17 |
- |
+ |
+ |
- |
- |
- |
+ |
+ |
2 |
5 |
45 |
56 |
6 |
36 |
51 |
80 |
| N10007 |
HZ |
surveillance |
2010 |
18 |
- |
+ |
+ |
- |
- |
- |
- |
+ |
19 |
46 |
14 |
22 |
48 |
1 |
52 |
86 |
| N10008 |
HZ |
surveillance |
2010 |
19 |
- |
+ |
+ |
- |
- |
- |
- |
- |
38 |
47 |
44 |
65 |
49 |
1 |
53 |
91 |
| N10009 |
HZ |
surveillance |
2010 |
20 |
- |
+ |
+ |
- |
- |
- |
+ |
+ |
2 |
5 |
45 |
56 |
6 |
36 |
51 |
80 |
| N11191 |
JH |
sporadic |
2011 |
23 |
- |
+ |
+ |
- |
- |
- |
- |
- |
24 |
36 |
1 |
50 |
30 |
18 |
45 |
81 |
| N743 |
LS |
outbreak B |
2006 |
8 |
- |
- |
+ |
- |
- |
- |
+ |
+ |
2 |
5 |
45 |
56 |
6 |
36 |
51 |
80 |
| N744 |
LS |
outbreak B |
2006 |
9 |
- |
+ |
+ |
- |
- |
- |
+ |
+ |
2 |
5 |
45 |
56 |
6 |
36 |
51 |
80 |
| N745 |
LS |
outbreak B |
2006 |
10 |
- |
+ |
+ |
- |
- |
- |
+ |
- |
2 |
5 |
45 |
56 |
6 |
36 |
51 |
80 |
| N746 |
LS |
outbreak B |
2006 |
11 |
- |
+ |
+ |
- |
- |
- |
+ |
+ |
2 |
5 |
45 |
56 |
6 |
36 |
51 |
80 |
| N747 |
LS |
outbreak B |
2006 |
9 |
- |
+ |
+ |
- |
- |
- |
+ |
+ |
2 |
5 |
45 |
56 |
6 |
36 |
51 |
80 |
| N748 |
LS |
outbreak B |
2006 |
9 |
- |
+ |
+ |
- |
- |
- |
+ |
+ |
2 |
5 |
45 |
56 |
6 |
36 |
51 |
80 |
| N749 |
LS |
outbreak B |
2006 |
9 |
- |
+ |
+ |
- |
- |
- |
+ |
+ |
2 |
5 |
45 |
56 |
6 |
36 |
51 |
80 |
| N750 |
LS |
sporadic |
2007 |
12 |
- |
+ |
+ |
- |
- |
- |
+ |
+ |
11 |
27 |
11 |
63 |
19 |
1 |
4 |
88 |
| N733 |
NB |
outbreak A |
2005 |
2 |
- |
+ |
+ |
- |
- |
- |
+ |
+ |
2 |
5 |
29 |
19 |
6 |
36 |
51 |
82 |
| N734 |
NB |
outbreak A |
2005 |
2 |
- |
+ |
+ |
- |
- |
- |
+ |
+ |
2 |
5 |
29 |
19 |
6 |
36 |
51 |
82 |
| N735 |
NB |
outbreak A |
2005 |
3 |
- |
+ |
+ |
- |
- |
- |
+ |
+ |
2 |
5 |
29 |
19 |
6 |
36 |
51 |
82 |
| N737 |
NB |
outbreak A |
2005 |
2 |
- |
+ |
+ |
- |
- |
- |
+ |
+ |
2 |
5 |
29 |
19 |
6 |
36 |
51 |
82 |
| N738 |
NB |
outbreak A |
2005 |
2 |
- |
+ |
+ |
- |
- |
- |
+ |
+ |
2 |
5 |
29 |
19 |
6 |
36 |
51 |
82 |
| N739 |
NB |
outbreak A |
2005 |
4 |
- |
+ |
+ |
- |
- |
- |
+ |
+ |
2 |
5 |
29 |
19 |
6 |
36 |
51 |
82 |
| N11193 |
QZ |
outbreak C |
2011 |
17 |
- |
+ |
+ |
- |
- |
- |
+ |
+ |
2 |
5 |
45 |
56 |
6 |
36 |
51 |
80 |
| N11194 |
QZ |
outbreak C |
2011 |
25 |
- |
+ |
+ |
- |
- |
- |
- |
- |
27 |
5 |
14 |
22 |
30 |
18 |
45 |
92 |
| N11196 |
QZ |
outbreak C |
2011 |
17 |
- |
+ |
+ |
- |
- |
- |
+ |
+ |
2 |
5 |
45 |
56 |
6 |
36 |
51 |
80 |
| N11198 |
QZ |
outbreak C |
2011 |
17 |
- |
+ |
+ |
- |
- |
- |
+ |
+ |
2 |
5 |
45 |
56 |
6 |
36 |
51 |
80 |
| N11199 |
QZ |
outbreak C |
2011 |
17 |
- |
+ |
+ |
- |
- |
- |
+ |
+ |
2 |
5 |
45 |
56 |
6 |
36 |
51 |
80 |
| N11200 |
QZ |
outbreak C |
2011 |
17 |
- |
+ |
+ |
- |
- |
- |
+ |
+ |
2 |
5 |
45 |
56 |
6 |
36 |
51 |
80 |
| N11201 |
QZ |
outbreak C |
2011 |
25 |
- |
+ |
+ |
- |
- |
- |
- |
- |
27 |
5 |
14 |
22 |
30 |
18 |
45 |
92 |
| N11202 |
QZ |
outbreak C |
2011 |
17 |
- |
+ |
+ |
- |
- |
- |
+ |
+ |
2 |
5 |
45 |
56 |
6 |
36 |
51 |
80 |
| N11203 |
QZ |
outbreak C |
2011 |
17 |
- |
+ |
+ |
- |
- |
- |
+ |
+ |
2 |
5 |
45 |
56 |
6 |
36 |
51 |
80 |
| N740 |
QZ |
sporadic |
2006 |
6 |
- |
+ |
+ |
- |
- |
- |
- |
+ |
26 |
5 |
46 |
50 |
31 |
14 |
45 |
90 |
| N11041 |
SX |
sporadic |
2011 |
21 |
- |
+ |
+ |
- |
- |
- |
+ |
+ |
19 |
1 |
14 |
66 |
48 |
1 |
52 |
87 |
| N11072 |
SX |
sporadic |
2011 |
22 |
- |
+ |
+ |
- |
- |
- |
+ |
+ |
3 |
41 |
4 |
48 |
47 |
35 |
5 |
84 |
| N742 |
TZ |
sporadic |
2006 |
7 |
- |
+ |
+ |
- |
- |
- |
+ |
+ |
11 |
27 |
11 |
63 |
19 |
1 |
4 |
88 |
| N732 |
WZ |
sporadic |
2005 |
1 |
- |
+ |
+ |
- |
- |
- |
+ |
+ |
3 |
23 |
15 |
48 |
47 |
35 |
5 |
85 |
| N736 | WZ | sporadic | 2005 | 5 | - | + | + | - | - | - | + | + | 3 | 23 | 15 | 48 | 47 | 35 | 5 | 85 |
† Place corresponds to different cities in Zhejiang province: HZ - Hangzhou; JH - Jinhua; LS - Lishui; NB - Ningbo; QZ - Quzhou; SX - Shaoxing; TZ - Taizhou; and WZ – Wenzhou.
‡ The isolate was unable to be typed by PFGE.
*Two primer pairs of tcpA (see Table 2) were used. Both were negative.
Nine other non-O1/non-O139 V. cholerae isolates were obtained during an active surveillance of enteric bacterial pathogens conducted by Zhejiang Provincial CDC in two Provincial hospitals in Hangzhou between May and December in 2010. These nine cases of non-O1/non-O139 V. cholerae infections were identified from a total of 746 diarrhoeal stool samples screened. All samples were screened for Salmonella, Shigella, Campylobacter, Yersinia enterocolitica, pathogenic Vibrio spp., pathogenic E. coli, Aeromonas hydrophila, Plesiomonas shigelloides, rotavirus, enteric adenovirus, norovirus, sapovirus, and astrovirus. There were no other enteric pathogens isolated from these nine cases. This data gave a non-O1/non-O139 V. cholerae infection rate of 1.2 per 100 diarrhoeal patients. Thus, non-O1/non-O139 V. cholerae is an important pathogen in this population and has been neglected as a pathogen generally.
The prevalence of non-O1/non-O139 V. cholerae in clinical samples varied in other countries. In Thailand, the proportion of non-O1/non-O139 V. cholerae isolated from diarrhoeal patients was between 1.0 and 1.3% [3], which is comparable to our study. In Italy, two non-O1/non-O139 V. cholerae infections (3.4%) were identified among 58 hospitalized patients with acute diarrhoea and both were associated with seafood consumption [30]. In cholera endemic regions, isolation of non-O1/non-O139 V. cholerae seems to be higher. In a 2003 survey in Kolkata, India, non-O1/non-O139 V. cholerae constituted 27.4% of the total V. cholerae isolations from hospitalised patients with acute diarrhoea [16], although estimates based on the number of diarrhoeal cases were not available.
Molecular typing of non-O1/non-O139 V. cholerae isolates
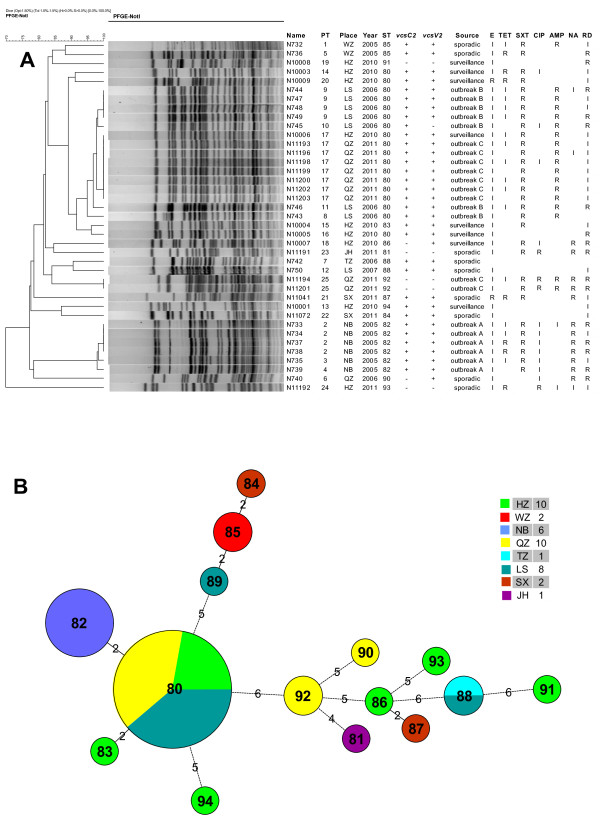
In order to determine the genetic and epidemiological relatedness among the isolates, we first performed PFGE analysis using the PulseNet standardised PFGE protocol for V. cholerae. PFGE is the gold standard of epidemiological typing as it offers high discriminatory power [31] and is routinely used for epidemiological typing of food-borne pathogens by the Zhejiang Provincial Center for Disease Control and Prevention. Thirty nine of the 40 isolates were typed using PFGE and were divided into 25 PFGE types (PTs) (Figure 2A). Of the six outbreak A isolates, four belonged to the same PFGE pattern (PT2), while the other two had two different patterns (PT3 and PT4) with only one band difference to PT2. Four outbreak B isolates had the same PFGE pattern (PT9) and three others had a unique pattern (PT8, PT10 and PT11). PT9 and PT10 were very similar to each other while PT11 and PT8 differed by three and four bands from PT9 respectively. The nine outbreak C isolates were separated into two distinctive patterns (PT17 with seven isolates and PT25 with two isolates). The similarity in PFGE patterns among the isolates was estimated using dice coefficient and was represented by a dendrogram in Figure 2A. The dendrogram showed that outbreak C was most likely caused by two different strains since PT17 and PT25 were well separated in the dendrogram. Interestingly, one isolate (N10006) obtained in the 2010 active surveillance in Hangzhou shared the same PFGE pattern (PT17) with seven outbreak C isolates from Quzhou. It seems that the PT17 strain causing the 2011 outbreak in Quzhou has been circulating in the neighbouring Hangzhou city a year earlier.
Figure 2.
Relationships of the non-O1/non-O139 Vibrio cholerae isolates. A. Dendrogram analysis generated using the unweighted pair group method with arithmetic based on pulsed field gel electrophoresis (PFGE) patterns. Place corresponds to different cities in Zhejiang province: HZ - Hangzhou; JH - Jinhua; LS - Lishui; NB - Ningbo; QZ - Quzhou; SX - Shaoxing; TZ - Taizhou; and WZ – Wenzhou. The classification of the PFGE type (PT), sequence type (ST); presence (+) or absence (−) of the two T3SS genes (vcsC2 and vcsV2); and resistance (R) or intermediate (I) to antibiotics (E - erythromycin, TET - tetracycline, SXT - sulphamethoxazole/trimethoprim, CIP – ciprofloxacin, AMP – ampicillin, NA - nalidixic acid and RD – rifampicin) is shown. B. Minimum spanning tree based on MLST data. The number in the circle indicates the ST and the size of the circle corresponds the total number of isolates belonging to that ST. Different localities are indicated in colour and specified in the colour legend together with the total number of isolates from each city in brackets. City name abbreviations are the same as in A above. The number of allelic difference between STs is indicated on the branches. Nodes were connected by a dashed line if the difference is more than two alleles.
All ST80 outbreak C isolates (PT17) were grouped together but were placed within outbreak B PTs and were closest to PT9 and PT10 (Figure 2A). It should be noted that PT17 looked nearly identical to PT9 in Figure 2A. However, closer examination of the PFGE patterns showed that the two bands in PT17 clearly were not identical to those in PT9. Since the two outbreaks were separated by time and locality, it is interesting to note such a close relationship of the isolates, which also shows that epidemiological information must be considered in addition to PFGE patterns in detecting outbreaks.
We further used multilocus sequence typing (MLST) to determine the relationships of and genetic heterogeneity among the isolates. Seven housekeeping genes (adk, gyrB, metE, mdh, pntA, purM and pyrC) selected based on a previous study [32] were used for the MLST (Octavia et al. manuscript in preparation). MLST divided the 40 isolates into 15 sequence types (STs) (Figure 2B). ST80 was predominant which consisted of 18 isolates. eBURST [33] analysis showed none of the STs formed a clonal complex. A minimum spanning tree was constructed (Figure 2B) which showed three sets of closely related STs: ST82, ST83 and ST80; ST84, ST85 and ST89; and ST86 and ST87, all differing by two genes. Two STs (ST80 and ST88) were isolated over two or more years and from different cities, suggesting that these two STs had a wide geographical distribution. For the three outbreaks, outbreak A was caused by ST82 while outbreaks B and C were caused by ST80. However, the ST80 isolates from outbreaks B and C can be separated by one band difference by PFGE. Additionally, two of the nine outbreak C isolates belonged to ST92. Therefore, outbreak C was caused by two STs and possibly due to contamination of the source (shrimp) by two different strains. There was also heterogeneity in isolates from the same city. The nine isolates from the 2010 active surveillance in Hangzhou were separated into six STs. Thus, our MLST analysis showed that these non-O1/non-O139 isolates were genetically diverse and some strains such as those belonging to ST80 can predominate across the regions.
We compared the relationships of isolates based on MLST (Figure 2B) with those based on PFGE. For the five STs (ST80, ST82, ST85, ST88 and ST92) with two or more isolates, each individual ST is associated with distinct PFGE nodes with all isolates of the same ST contained within the same node (Figure 2A). Additionally, two isolates of different STs, N10004 of ST83 and N10005 of ST80 were grouped together by PFGE with a three-band difference and a 95% similarity (Figure 2A). This was consistent with the MLST relationship as ST83 was linked with ST80 with a two-allele difference (Figure 2B). The two alleles differed between ST83 and ST80 were gyrB and mdh with 5 bp and 4 bp differences, respectively. The differences in these genes may be due to recombination as V. cholerae undergoes recombination quite frequently [32]. Therefore, relationships of isolates with high similarity in PFGE patterns are consistent between PFGE and MLST.
In contrast, the relationships of isolates with less similar PFGE patterns were inconsistent with those based on MLST. For example, the ST86 isolate N10007 was grouped together with the ST81 isolate N11191 by PFGE, while by MLST ST81 and ST86 were not linked together on the MST (Figure 2B). These two isolates differed substantially in their banding patterns (Figure 2B) and also differed in all seven alleles by MLST. Similarly the grouping together of ST84 and ST94 by PFGE was also inconsistent with their relationship based on MLST (Figure 2B).
As measured by the index of diversity (D), the discriminatory power of PFGE (D = 0.945) was clearly higher than MLST (D = 0.781) for characterisation of non-O1/non-O139 V. cholerae. PFGE further divided isolates within an ST for all STs except ST92 in which there were only two isolates and both were from the same outbreak.
Antibiotic resistance patterns amongst non-O1/non-O139 V. cholerae isolates
Antimicrobial susceptibility testing was carried out using disk diffusion assay for 13 antibiotics and all or nearly all isolates were susceptible to cephalothin, cefotaxime, gentamicin, amikacin, doxycycline and norfloxacin. Resistance to other antibiotics varied with 80% of the isolates resistant to sulphamethoxazole/trimethoprim (SXT), 47.5% to ampicillin, 42.5% to rifampicin, 30% to nalidixic acid, 15% to tetracycline, 5% to ciprofloxacin and 5% to erythromycin. Additionally, for rifampicin, erythromycin and tetracycline, the majority or nearly all of the remaining isolates were intermediate to the respective antibiotics (Figure 2A). Isolates obtained from the same outbreak may also vary in antibiotic resistance. However, most of these variations were due to intermediate resistance (Figure 2A). The use of antimicrobial agents is generally regarded as an effective method to reduce the duration and symptoms of diarrhoea. Tetracycline, erythromycin, SXT and ciprofloxacin have all been generally considered as the drug of choice for the treatment of cholera. However, the resistance profiles indicate that these antibiotics will not be or less effective for treating non-O1/non-O139 V. cholerae infections.
Antibiotic resistance profiles were also correlated with PFGE or MLST relationships. All ST82 isolates and all except one ST80 isolate were resistant to SXT. The only SXT susceptible ST80 isolate was grouped away from the other ST80 isolates. All ST80 isolates associated with outbreaks (either outbreak B or outbreak C) were resistant to ampicillin. Nalidixic acid resistance also has a restricted distribution. With the exception of the nalidixic acid resistant ST90 isolate (N740) and the nalidixic acid resistant ST87 isolate (N11041) which are unrelated, nalidixic acid resistance was present only in the two ST92 outbreak C isolates, all ST82 outbreak A isolates and the two related ST86 and ST81 isolates. The two ST92 isolates were the most drug resistant and shared the same resistance profile with resistance or intermediate to six antibiotics (erythromycin, SXT, ciprofloxacin, ampicillin, nalidixic acid and rifampicin). The ST86 and ST81 isolates (N10007 and N11191, respectively) grouped together by PFGE shared a similar resistance profile with resistance or intermediate to five antibiotics (erythromycin, SXT, ciprofloxacin, nalidixic acid and rifampicin).
The distribution of SXT resistance on the tree (Figure 2A) revealed an interesting evolutionary history. SXT resistance in V. cholerae is carried by a conjugative, self-transmissible and integrative element (SXT element) that also provides resistance to chloramphenicol and streptomycin [18,34,35]. The wide distribution of SXT resistance along the tree suggests that the SXT element is widespread, although previous studies mostly analysed V. cholerae O1 and O139 toxigenic strains for the presence of SXT element [35-37]. Interestingly, one of the ST80 isolates which was obtained from the 2010 active surveillance was susceptible to SXT and presumably has lost the SXT element since every other isolate in the large PFGE clusters was resistant to SXT. In contrast, the four SXT susceptible isolates (two ST88 isolates, one ST84 isolate and one ST94 isolate) were grouped together as two pairs of isolates on different branches of the tree and are likely to have not gained the SXT element. Resistance to the other antibiotics may be due to chromosomal mutations, plasmids or other mobile elements [38] and are more difficult to make any evolutionary inference of the observed resistance patterns.
Detection and distribution of virulence factors genes
PCR assays (Table 2) were used for the detection of the ctxAB[39], tcpA[40], zot[41], NAG-ST [16], T3SS (vcsC2 and vcsV2) [16,28], ompW[42], toxR[42] and hlyA genes [43]. All isolates were positive for V. cholerae specific gene ompW by PCR, but were negative for ctxAB, zot, tcpA and NAG-ST. All isolates were positive for toxR (Table 1), except for N743 which was toxR negative. Interestingly, N743 also differed from other ST80 isolates in its PFGE pattern. toxR codes for the transcriptional regulatory protein ToxR [44] and is expected to be present in all V. cholerae isolates. Negative PCR amplification of toxR from N743 may be due to sequence divergence in primer binding regions. Similarly, all isolates were positive for the haemolysin gene hlyA (Table 1). In contrast, the absence of ctxAB, zot, tcpA and NAG-ST suggests that these non-O1/non-O139 isolates caused diarrhoea by a different mechanism from that used by toxigenic V. cholerae O1 and O139.
Table 2.
PCR primers used in this study
| Gene target |
Primer sequence (5’-3’) |
Probe | Ta* | Amplicon size (bp) | Reference | |
|---|---|---|---|---|---|---|
| Forward | Reverse | |||||
|
ompW |
TCCTCAACGCTTCTGTGTGGTAT |
ATTGATTTCAACATCCGTGGATT |
FAM-TGAAACAACGGCAACCTACAAAGCAGG-BHQ1 |
55 |
92 |
This study |
|
hlyA |
AGTGGTCAACCGATGCGATT |
TTCAGGATCTGCGCTTTATTGTT |
ROX-CCCAAGATTATCGCTTCGTGTTTAACGCA- BHQ2 |
47-55 |
76 |
This study |
|
toxR |
GATTCGACAAAGTCCCCACAA |
TCGGGCGATCAATTGGTAA |
HEX-CGTCAAAACGGTTCCGAAACGCG-BHQ1 |
47-55 |
66 |
This study |
|
ctxAB |
CTCAGACGGGATTTGTTAGGCACG |
TCTATCTCTGTAGCCCCTATTACG |
- |
55 |
303 |
[39] |
|
tcpA (1)# |
GTGACTGAAAGTCATCTCTTC |
AATCCGACACCTTGTTGGTA |
- |
55 |
1248 |
[40] |
|
tcpA (2)# |
ATATGCAATTATTAAAACAGC |
TTATTATTACCCGTTGTCGG |
- |
55 |
1052 |
[40] |
|
ace |
AGAGCGCTGCATTTATCCTTATTG |
AACTCGGTCTCGGCCTCTCGTATC |
- |
55 |
655 |
[41] |
|
zot |
GCTATCGATATGCTGTCTCCTCAA |
AAAGCCGACCAATACAAAAACCAA |
- |
55 |
1000 |
[41] |
| T3SS (vcsC2) |
GGAAAGATCTATGCGTCGACGTTACCGATGCTATGGGT |
CATATGGAATTCCCGGGATCCATGCTCT AGAAGTCGGTTGTTTCGGTAA |
- |
47-60 |
535 |
[16] |
| T3SS (vcsV2) |
ATGCAGATCTTTTGGCTCACTTGATGGG |
ATGCGTCGACGCCACATCATTGCTTGCT |
- |
47-55 |
742 |
[16] |
| NAG-ST | CCTATTCATTAGCATAATG | CCAAAGCAAGCTGGATTGC | - | 47-55 | 215 | [16] |
*Ta – Annealing temperature.
# Two primer pairs of tcpA primers were used. These two primer pairs have been used previously to amplify divergent tcpA alleles [24].
Recent reports suggest that T3SS is present in some non-O1/non-O139 isolates and plays an important role in virulence [16,28]. We tested for the presence of T3SS using two T3SS genes (vcsC2 and vcsV2). Thirty two carried both genes and presumably have a functional T3SS while five were negative for both genes. Three isolates were negative for one of the genes, two isolates negative for vcsC2 and one isolate negative for vcsV2. The primer binding regions in the genes of these isolates may be divergent leading to non-amplification, but it is also possible that the genes are deleted. It seemed that the pathogenicity of the majority of the isolates was due to the presence of the T3SS since 35 isolates possessed one or both T3SS genes (87.5%),which is different from that reported in Bangladesh (38.9%) [45] and in India (31.5%) [16]. The varying presence of virulence factors among different non-O1/non-O139 strains may be associated with their ability to cause disease. Further studies are warranted.
Conclusion
Our study is the first report which showed that non-O1/non-O139 V. cholerae was an important pathogen in China, causing diarrhoeal infections with an isolation rate of 1.2%. MLST revealed that a single ST, ST80, was predominant in Zhejiang Province. ST80 persisted over several years and appeared in different cities. It caused two outbreaks in recent years. Since the majority of the isolates were positive for T3SS but negative for any other virulence factors tested, the T3SS was likely to be the key virulence factor for these isolates. Resistance to commonly used antibiotics limits choice of drugs for treating non-O1/non-O139 V. cholerae infections. Our study highlights that non-O1/non-O139 V. cholerae has been neglected as an important cause of diarrhoea in China and may be the same in other developing countries. Close monitoring of non-O1/non-O139 V. cholerae capable of causing outbreaks in China is necessary to reduce the health burden of diarrhoeal infections caused by this pathogen.
Methods
Bacterial isolates
Faecal samples from sporadic and outbreak cases were collected by local hospitals as part of standard patients care over a five year period from diarrhoeal patients at local hospitals in Zhejiang Province, China, and were sent to Zhejiang Provincial CDC laboratory for isolation of V. cholerae. Potential V. cholerae isolates from the faecal samples were grown onto No. 4 Agar (1% sodium citrate, 0.5% pig gall powder, 0.003% rivano powder, 0.2% sodium sulphite, 0.1% sodium lauryl sulphate, 0.001% potassium tellurite, and 500 μg/L gentamicin). All retrieved isolates were serologically tested for agglutination of O1 or O139 antisera (Denka Seiken, Japan) and all were shown to be negative.
V. cholerae isolates were also obtained from an active surveillance program of enteric bacterial pathogens which was coordinated by Zhejiang Provincial CDC and was conducted in two Provincial hospitals in Hangzhou between May and December in 2010. Faecal specimens were obtained with written informed consent of the patients and with the approval of the Zhejiang Provincial CDC ethics committee, according to the medical research regulations of Ministry of Health, China.
Molecular techniques
DNA was prepared using DNeasy Blood & Tissue kit (QIAGEN, Inc., Valencia, CA). Seven housekeeping genes (adk, gyrB, metE, mdh, pntA, purM and pyrC) selected based on a previous study [32] were used for the MLST (Octavia et al. manuscript in preparation). The amplified products were sequenced commercially by Beijing Genomics Institute. PFGE was performed according to the US CDC PulseNet standardised PFGE protocol for V. cholerae[31]. Simplex PCR assays (Table 2) were used for the detection of ctxAB[39], tcpA[40], zot[41], NAG-ST [16], T3SS (vcsC2 and vcsV2) [16,28], and performed in a Mastercycler (Eppendorf, Hamburg, Germany). The reactions were carried out as follows: 5 min at 94°C; followed by 30 cycles of 30 s at 94°C, 30 s at the annealing temperature specified in Table 2, and 30 s at 72°C; followed by a final 5 min at 72°C. For detection of ompW[42], toxR[42] and hlyA[43] genes, new primer pairs (Table 2) were designed to be used in a multiplex real time PCR assay. The reaction was performed in an ABI7500 fast real-time PCR system (Applied Biosystems, CA, USA). The cycling conditions were as follows: 2 min at 95°C, followed by 40 cycles of 15 s at 95°C, and 45 s at the annealing temperature specified in Table 2. Isolate N10002 was typed by MLST and PCR but not typed by PFGE nor tested for antibiotic sensitivity as only DNA was available.
Bioinformatics
Sequence alignments were done using ClustalW [46]. The PFGE dendrogram was constructed using the unweighted pair group method with arithmetic mean algorithm and Dice coefficient of two patterns at 0.5% pattern optimisation and 1.5% band position tolerance, available from Bionumerics (Applied Math). Note that one band in PT17 (band 16 from higher molecular weight end, Figure 2A) was recognised as two bands by the software to which manual correction was applied to become one band as this affected the placement of PT17. Sequence types were numbered from ST80 onwards. ST1 to ST79 were pre-assigned to isolates of another study (Octavia et al. manuscript in preparation). eBURST [33] was used to identify clonal complexes which are defined using the difference of one out of the seven genes typed. Minimum spanning tree using the allelic difference between isolates of the seven housekeeping genes was constructed using Bionumerics (Applied Math). The Simpson’s index of diversity (D value) [47] was calculated using an in-house program, MLEECOMP package [48].
Antibiotic resistance
Antimicrobial susceptibility testing for 13 antibiotics including amikacin, ampicillin, cephalothin, cefotaxime, ciprofloxacin, doxycycline, erythromycin, gentamicin, nalidixic acid, norfloxacin, rifampicin, SXT and tetracycline, was carried out using disk diffusion assay according to the protocol of the Clinical and Laboratory Standards Institute [49]. Antibiotic discs were purchased from Oxoid (Hampshire, UK). Results were analysed using WHONET 5.4 software (WHO Collaborating Centre for the Surveillance of Antibiotics Resistance, Geneva, Switzerland). Isolates were classified as susceptible, intermediate or resistant based on the guidelines for each antibiotic.
GenBank accession numbers
The sequences obtained in this study have been submitted to GenBank with accession numbers JX905826-JX05848.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Experimental work and data collection were carried out by YL, JY, DJ, GD, ZZ, LM. YL, RL and SO contributed to data analysis and interpretation. The study was conceived and designed by YL and RL. The manuscript was drafted by YL, RL and SO. All authors have read and approved the final manuscript.
Contributor Information
Yun Luo, Email: yluo@cdc.zj.cn.
Julian Ye, Email: jlye@cdc.zj.cn.
Dazhi Jin, Email: dzjin@cdc.zj.cn.
Gangqiang Ding, Email: gqding@cdc.zj.cn.
Zheng Zhang, Email: zhzhang@cdc.zj.cn.
Lingling Mei, Email: llmei@cdc.zj.cn.
Sophie Octavia, Email: s.octavia@unsw.edu.au.
Ruiting Lan, Email: r.lan@unsw.edu.au.
Acknowledgements
We thank our colleagues Xiaofei Fang and Linna Han for isolating the strains and PCR detections. We are grateful to Junhang Pan for providing epidemiological data. We thank Junchao Wei for coordinating the active surveillance program. We thank the anonymous reviewers for helpful suggestions to improve the manuscript.
References
- Faruque SM, Albert MJ, Mekalanos JJ. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol Mol Biol Rev. 1998;62:1301–1314. doi: 10.1128/mmbr.62.4.1301-1314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalsgaard A, Albert MJ, Taylor DN, Shimada T, Meza R, Serichantalergs O, Echeverria P. Characterization of Vibrio cholerae non-O1 serogroups obtained from an outbreak of diarrhea in Lima, Peru. J Clin Microbiol. 1995;33:2715–2722. doi: 10.1128/jcm.33.10.2715-2722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalsgaard A, Forslund A, Bodhidatta L, Serichantalergs O, Pitarangsi C, Pang L, Shimada T, Echeverria P. A high proportion of Vibrio cholerae strains isolated from children with diarrhoea in Bangkok, Thailand are multiple antibiotic resistant and belong to heterogenous non-O1, non-O139 O-serotypes. Epidemiol Infect. 1999;122:217–226. doi: 10.1017/S0950268899002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalsgaard A, Serichantalergs O, Forslund A, Lin W, Mekalanos J, Mintz E, Shimada T, Wells JG. Clinical and environmental isolates of Vibrio cholerae serogroup O141 carry the CTX phage and the genes encoding the toxin-coregulated pili. J Clin Microbiol. 2001;39:4086–4092. doi: 10.1128/JCM.39.11.4086-4092.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onifade TJ, Hutchinson R, Van Zile K, Bodager D, Baker R, Blackmore C. Toxin producing Vibrio cholerae O75 outbreak, United States, March to April 2011. Eurosurveillance. 2011;16:19870. [PubMed] [Google Scholar]
- Tobin-D'Angelo M, Smith AR, Bulens SN, Thomas S, Hodel M, Izumiya H, Arakawa E, Morita M, Watanabe H, Marin C. Severe diarrhea caused by cholera toxin-producing Vibrio cholerae serogroup O75 infections acquired in the Southeastern United States. Clin Infect Dis. 2008;47:1035–1040. doi: 10.1086/591973. [DOI] [PubMed] [Google Scholar]
- Cariri FA, Costa AP, Melo CC, Theophilo GN, Hofer E, de Melo Neto OP, Leal NC. Characterization of potentially virulent non-O1/non-O139 Vibrio cholerae strains isolated from human patients. Clin Microbiol Infect. 2010;16:62–67. doi: 10.1111/j.1469-0691.2009.02763.x. [DOI] [PubMed] [Google Scholar]
- Ko WC, Chuang YC, Huang GC, Hsu SY. Infections due to non-O1 Vibrio cholerae in southern Taiwan: predominance in cirrhotic patients. Clin Infect Dis: an official publication of the Infectious Diseases Society of America. 1998;27:774–780. doi: 10.1086/514947. [DOI] [PubMed] [Google Scholar]
- Blake PA, Allegra DT, Snyder JD, Barrett TJ, McFarland L, Caraway CT, Feeley JC, Craig JP, Lee JV, Puhr ND. Cholera- a possible endemic focus in the United States. New Engl J Med. 1980;302:305–309. doi: 10.1056/NEJM198002073020601. [DOI] [PubMed] [Google Scholar]
- Morris JM. , Jr. In: Vibrio cholerae and cholera: molecular to global perspectives. Wachsmuth IK, Blake PA, Olsvik O, editor. Washington: American Society for Microbiology; 1994. Non-O1 group 1 Vibrio cholerae strains not associated with epidemic disease. [Google Scholar]
- Bagchi K, Echeverria P, Arthur JD, Sethabutr O, Serichantalergs O, Hoge CW. Epidemic of diarrhea caused by Vibrio cholerae non-O1 that produced heat-stable toxin among Khmers in a camp in Thailand. J Clin Microbiol. 1993;31:1315–1317. doi: 10.1128/jcm.31.5.1315-1317.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthy T, Bag PK, Pal A, Bhattacharya SK, Bhattacharya MK, Shimada T, Takeda T, Karasawa T, Kurazono H, Takeda Y. Virulence patterns of Vibrio cholerae non-O1 strains isolated from hospitalised patients with acute diarrhoea in Calcutta, India. J Med Microbiol. 1993;39:310–317. doi: 10.1099/00222615-39-4-310. [DOI] [PubMed] [Google Scholar]
- Rudra S, Mahajan R, Mathur M, Kathuria K, Talwar V. Cluster of cases of clinical cholera due to Vibrio cholerae O10 in east Delhi. Indian J Med Res. 1996;103:71–73. [PubMed] [Google Scholar]
- Sharma C, Thungapathra M, Ghosh A, Mukhopadhyay AK, Basu A, Mitra R, Basu I, Bhattacharya SK, Shimada T, Ramamurthy T. Molecular analysis of non-O1, non-O139 Vibrio cholerae associated with an unusual upsurge in the incidence of cholera-like disease in Calcutta, India. J Clin Microbiol. 1998;36:756–763. doi: 10.1128/jcm.36.3.756-763.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya MK, Dutta D, Bhattacharya SK, Deb A, Mukhopadhyay AK, Nair GB, Shimada T, Takeda Y, Chowdhury A, Mahalanabis D. Association of a disease approximating cholera caused by Vibrio cholerae of serogroups other than O1 and O139. Epidemiol Infect. 1998;120:1–5. doi: 10.1017/S0950268897008352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Ghosh K, Raychoudhuri A, Chowdhury G, Bhattacharya MK, Mukhopadhyay AK, Ramamurthy T, Bhattacharya SK, Klose KE, Nandy RK. Incidence, virulence factors, and clonality among clinical strains of non-O1, non-O139 Vibrio cholerae isolates from hospitalized diarrheal patients in Kolkata, India. J Clin Microbiol. 2009;47:1087–1095. doi: 10.1128/JCM.02026-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh CS, Chua KH, Thong KL. Genetic variation analysis of Vibrio cholerae using multilocus sequencing typing and multi-virulence locus sequencing typing. Infect Genet Evol. 2011;11:1121–1128. doi: 10.1016/j.meegid.2011.04.005. [DOI] [PubMed] [Google Scholar]
- Rivera IN, Chun J, Huq A, Sack RB, Colwell RR. Genotypes associated with virulence in environmental isolates of Vibrio cholerae. Appl Environ Microbiol. 2001;67:2421–2429. doi: 10.1128/AEM.67.6.2421-2429.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh DV, Matte MH, Matte GR, Jiang S, Sabeena F, Shukla BN, Sanyal SC, Huq A, Colwell RR. Molecular analysis of Vibrio cholerae O1, O139, non-O1, and non-O139 strains: clonal relationships between clinical and environmental isolates. Appl Environ Microbiol. 2001;67:910–921. doi: 10.1128/AEM.67.2.910-921.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faruque SM, Mekalanos JJ. Pathogenicity islands and phages in Vibrio cholerae evolution. Trends Microbiol. 2003;11:505–510. doi: 10.1016/j.tim.2003.09.003. [DOI] [PubMed] [Google Scholar]
- Faruque SM, Naser IB, Islam MJ, Faruque AS, Ghosh AN, Nair GB, Sack DA, Mekalanos JJ. Seasonal epidemics of cholera inversely correlate with the prevalence of environmental cholera phages. Proc Natl Acad Sci USA. 2005;102:1702–1707. doi: 10.1073/pnas.0408992102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faruque SM, Chowdhury N, Kamruzzaman M, Dziejman M, Rahman MH, Sack DA, Nair GB, Mekalanos JJ. Genetic diversity and virulence potential of environmental Vibrio cholerae population in a cholera-endemic area. Proc Natl Acad Sci USA. 2004;101:2123–2128. doi: 10.1073/pnas.0308485100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Fullner KJ, Clayton R, Sexton JA, Rogers MB, Calia KE, Calderwood SB, Fraser C, Mekalanos JJ. Identification of a Vibrio cholerae RTX toxin gene cluster that is tightly linked to the cholera toxin prophage. Proc Natl Acad Sci U S A. 1999;96:1071–1076. doi: 10.1073/pnas.96.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arita M, Takeda T, Honda T, Miwatani T. Purification and characterization of Vibrio cholerae non-O1 heat-stable enterotoxin. Infect Immun. 1986;52:45–49. doi: 10.1128/iai.52.1.45-49.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa A, Kato J, Watanabe H, Nair BG, Takeda T. Cloning and nucleotide sequence of a heat-stable enterotoxin gene from Vibrio cholerae non-O1 isolated from a patient with traveler's diarrhea. Infect Immun. 1990;58:3325–3329. doi: 10.1128/iai.58.10.3325-3329.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theophilo GN, Rodrigues Ddos P, Leal NC, Hofer E. Distribution of virulence markers in clinical and environmental Vibrio cholerae non-O1/non-O139 strains isolated in Brazil from 1991 to 2000. Rev Inst Med Trop Sao Paulo. 2006;48:65–70. doi: 10.1590/S0036-46652006000200002. [DOI] [PubMed] [Google Scholar]
- Alam A, Miller KA, Chaand M, Butler JS, Dziejman M. Identification of Vibrio cholerae type III secretion system effector proteins. Infect Immun. 2011;79:1728–1740. doi: 10.1128/IAI.01194-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziejman M, Serruto D, Tam VC, Sturtevant D, Diraphat P, Faruque SM, Rahman MH, Heidelberg JF, Decker J, Li L. Genomic characterization of non-O1, non-O139 Vibrio cholerae reveals genes for a type III secretion system. Proc Natl Acad Sci USA. 2005;102:3465–3470. doi: 10.1073/pnas.0409918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin OS, Tam VC, Suzuki M, Ritchie JM, Bronson RT, Waldor MK, Mekalanos JJ. Type III secretion is essential for the rapidly fatal diarrheal disease caused by non-O1, non-O139 Vibrio cholerae. MBio. 2011;2:e00106–00111. doi: 10.1128/mBio.00106-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottaviani D, Leoni F, Rocchegiani E, Santarelli S, Masini L, Di Trani V, Canonico C, Pianetti A, Tega L, Carraturo A. Prevalence and virulence properties of non-O1 non-O139 Vibrio cholerae strains from seafood and clinical samples collected in Italy. Int J Food Microbiol. 2009;132:47–53. doi: 10.1016/j.ijfoodmicro.2009.03.014. [DOI] [PubMed] [Google Scholar]
- Cooper KL, Luey CK, Bird M, Terajima J, Nair GB, Kam KM, Arakawa E, Safa A, Cheung DT, Law CP. Development and validation of a PulseNet standardized pulsed-field gel electrophoresis protocol for subtyping of Vibrio cholerae. Foodborne Pathog Dis. 2006;3:51–58. doi: 10.1089/fpd.2006.3.51. [DOI] [PubMed] [Google Scholar]
- Salim A, Lan R, Reeves PR. Vibrio cholerae pathogenic clones. Emerg Infect Dis. 2005;11:1758–1760. doi: 10.3201/eid1111.041170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004;186:1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaber JW, Hochhut B, Waldor MK. Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J Bacteriol. 2002;184:4259–4269. doi: 10.1128/JB.184.15.4259-4269.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldor MK, Tschape H, Mekalanos JJ. A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. J Bacteriol. 1996;178:4157–4165. doi: 10.1128/jb.178.14.4157-4165.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochhut B, Lotfi Y, Mazel D, Faruque SM, Woodgate R, Waldor MK. Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 SXT constins. Antimicrob Agents Chemother. 2001;45:2991–3000. doi: 10.1128/AAC.45.11.2991-3000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Zhou Y, Wang R, Lou J, Zhang L, Li J, Bi Z, Kan B. Multiple antibiotic resistance of Vibrio cholerae serogroup O139 in China from 1993 to 2009. PLoS One. 2012;7:e38633. doi: 10.1371/journal.pone.0038633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitaoka M, Miyata ST, Unterweger D, Pukatzki S. Antibiotic resistance mechanisms of Vibrio cholerae. J Med Microbiol. 2011;60:397–407. doi: 10.1099/jmm.0.023051-0. [DOI] [PubMed] [Google Scholar]
- Shirai H, Nishibuchi M, Ramamurthy T, Bhattacharya SK, Pal SC, Takeda Y. Polymerase chain reaction for detection of the cholera enterotoxin operon of Vibrio cholerae. J Clin Microbiol. 1991;29:2517–2521. doi: 10.1128/jcm.29.11.2517-2521.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay CY, Reeves PR, Lan R. Importation of the major pilin TcpA gene and frequent recombination drive the divergence of the Vibrio pathogenicity island in Vibrio cholerae. FEMS Microbiol Lett. 2008;289:210–218. doi: 10.1111/j.1574-6968.2008.01385.x. [DOI] [PubMed] [Google Scholar]
- Leal NC, Sobreira M, Leal-Balbino TC, de Almeida AM, de Silva MJ, Mello DM, Seki LM, Hofer E. Evaluation of a RAPD-based typing scheme in a molecular epidemiology study of Vibrio cholerae O1, Brazil. J Appl Microbiol. 2004;96:447–454. doi: 10.1111/j.1365-2672.2004.02090.x. [DOI] [PubMed] [Google Scholar]
- Nandi B, Nandy RK, Mukhopadhyay S, Nair GB, Shimada T, Ghose AC. Rapid method for species-specific identification of Vibrio cholerae using primers targeted to the gene of outer membrane protein OmpW. J Clin Microbiol. 2000;38:4145–4151. doi: 10.1128/jcm.38.11.4145-4151.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun R, Elbourne LD, Lan R, Reeves PR. Evolutionary relationships of pathogenic clones of Vibrio cholerae by sequence analysis of four housekeeping genes. Infect Immun. 1999;67:1116–1124. doi: 10.1128/iai.67.3.1116-1124.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skorupski K, Taylor RK. Control of the ToxR virulence regulon in Vibrio cholerae by environmental stimuli. Mol Microbiol. 1997;25:1003–1009. doi: 10.1046/j.1365-2958.1997.5481909.x. [DOI] [PubMed] [Google Scholar]
- Dziejman M, Balon E, Boyd D, Fraser CM, Heidelberg JF, Mekalanos JJ. Comparative genomic analysis of Vibrio cholerae: genes that correlate with cholera endemic and pandemic disease. Proc Natl Acad Sci USA. 2002;99:1556–1561. doi: 10.1073/pnas.042667999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W- Impoving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specifc gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pupo GM, Lan R, Reeves PR, Baverstock P. Population Genetics of Escherichia coli in a Natural Population of Native Australian Rats. Environ Microbiol. 2000;2:594–610. doi: 10.1046/j.1462-2920.2000.00142.x. [DOI] [PubMed] [Google Scholar]
- Anonymous. Performance Standards for Antimicrobial Disk Susceptibility Tests;Approved Standard-Ninth Edition. ninth. 940 West Valley Road, Suite 1400, Wayne, Pennsylvania 19087–1898 USA: Clinical And Laboratory Standards Institute; 2006. Clinical and Laboratory Standards Institute.Clinical and Laboratory Standards Institute document M2-A9. ISBN 1-56238-586-0. [Google Scholar]