Smad ubiquitin regulatory factors (Smurfs) play important roles in cell growth and differentiation. Hwang et al. present a new mechanism of Smurf regulation in Xenopus with evidence that Smurf1 and Smurf2 essentially compete for association with ephrin B1 (Smurf2 triggers ephrinB1 degradation, and Smurf1 counteracts it) and so regulate its expression. This unique mechanism for ubiquitin ligases is responsible for regulating the tissue boundaries at the mesoderm/ectoderm border during embryogenesis.
Keywords: Smurf, cell adhesion, ephrinB1, tissue boundaries
Abstract
The formation of tissue boundaries is dependent on the cell–cell adhesion/repulsion system that is required for normal morphogenetic processes during development. The Smad ubiquitin regulatory factors (Smurfs) are E3 ubiquitin ligases with established roles in cell growth and differentiation, but whose roles in regulating cell adhesion and migration are just beginning to emerge. Here, we demonstrate that the Smurfs regulate tissue separation at mesoderm/ectoderm boundaries through antagonistic interactions with ephrinB1, an Eph receptor ligand that has a key role in regulating the separation of embryonic germ layers. EphrinB1 is targeted by Smurf2 for degradation; however, a Smurf1 interaction with ephrinB1 prevents the association with Smurf2 and precludes ephrinB1 from ubiquitination and degradation, since it is a substantially weaker substrate for Smurf1. Inhibition of Smurf1 expression in embryonic mesoderm results in loss of ephrinB1-mediated separation of this tissue from the ectoderm, which can be rescued by the coincident inhibition of Smurf2 expression. This system of differential interactions between Smurfs and ephrinB1 regulates the maintenance of tissue boundaries through the control of ephrinB protein levels.
The creation of the embryonic body and its organization into separate tissues requires the coordinated behavior of cells through adhesive and repulsive mechanisms, which promote and maintain the segregation of different cell populations at tissue boundaries (Nowotschin and Hadjantonakis 2010). Several signaling pathways have been implicated in controlling embryonic morphogenesis, including Wnt, FGF, Eph/ephrin, and bone morphogenetic protein (BMP) pathways (Nowotschin and Hadjantonakis 2010). Regulation of these pathways is critical to proper morphogenesis; for example, the Smad ubiquitin regulatory factors (Smurfs), which are HECT domain E3 ubiquitin ligases, were originally shown to target and regulate the BMP pathway in Xenopus and later found to also affect many morphogenetic pathways, including planar cell polarity and the Par polarity complex (Zhu et al. 1999; Kavsak et al. 2000; Ozdamar et al. 2005; Alexandrova and Thomsen 2006; Osmundson et al. 2008; Narimatsu et al. 2009).
In many cases, Smurf1 and Smurf2 have similar functions within a pathway; for example, both Smurfs target MyD88 protein for ubiquitination and degradation (Lee et al. 2011) and can be recruited by adaptors (i.e., I-Smads) to the TGF-β receptor complex to mediate receptor degradation and down-regulation of TGF-β signaling (Kavsak et al. 2000; Ebisawa et al. 2001; Izzi and Attisano 2004). However, there are reports showing that differences may exist in how the two Smurfs target proteins for ubiquitination. For example, RhoA is specifically targeted by Smurf1 (Wang et al. 2003, 2006; Lu et al. 2011), and Smurf2 is more efficient than Smurf1 in targeting Smad2 (Lin et al. 2000; Bonni et al. 2001; Tang et al. 2011). Par-6 is targeted by Smurf1, but not by Smurf2, when overexpressed in Neuro2a cells, and the target preference of Smurf1 is switched to RhoA upon phosphorylation (Cheng et al. 2011).
Functional redundancy of both Smurfs was implied by the targeted disruption of each gene, which led to viable mice (Yamashita et al. 2005; Tang et al. 2011), while a double knockout caused embryonic lethality with planar cell polarity defects or gastrulation defects (Narimatsu et al. 2009). However, evidence is beginning to emerge that these two proteins are not completely redundant and have some unique functions in embryogenesis (Cao and Zhang 2012). In Xenopus morphogenesis, it has been shown that knockdown of Smurf1 disrupts normal neural tube folding and neural differentiation, while sparing mesoderm differentiation (Alexandrova and Thomsen 2006). A more recent detailed study of Xenopus embryogenesis indicates that Smurf1 and Smurf2 may have redundant functions in the regulation of both neural and tail development but that these proteins also possess unique activities. For example, Smurf1 is important for neural tube closure, and Smurf2 has a critical function in controlling mesoderm induction (Das and Chang 2012). Although it is clear from these studies that Smurf1 and Smurf2 play overlapping but distinct roles in Xenopus embryogenesis, the full extent of their function in this process as well as the additional targets of these ubiquitin ligases remain to be elucidated.
Here we present evidence using biochemical assays and in vivo gain- and loss-of-function experiments that differential interactions with Smurf1 and Smurf2 regulate ephrinB1 stability. EphrinB1, like Smurfs, has functions that intersect with signaling molecules in both the planar cell polarity and Par polarity complex pathways (Tanaka et al. 2003; Lee et al. 2006, 2008, 2009). Eph/ephrin signaling represents a powerful system for regulating tissue separation and morphogenesis (Pasquale 2008), but the mechanisms that regulate the expression levels of these molecules are still poorly understood.
In this study, we propose and test a regulatory model in which ephrinB1 expression is sustained by an interaction with Smurf1, which displays little ubiquitination and degradation activity toward ephrinB1. In contrast, ephrinB1 is degraded when this interaction is lost and supplanted by an interaction with Smurf2, which efficiently targets ephrinB1 for degradation. We used biochemical assays in Xenopus oocytes and embryos to determine that Smurfs interact with ephrinB1 and that Smurf1 and Smurf2 can compete for association with ephrinB1. Using morpholino (MO)-mediated knockdown of individual Smurfs in embryos, we show that the antagonism between Smurf1 and Smurf2 regulates ephrinB1 protein expression in vivo. We further demonstrate that the opposing effects of the Smurfs on ephrinB1 expression affect downstream RhoA activity and play a role in the morphogenetic event of tissue separation at the ectoderm/mesoderm border. MO-mediated inhibition of Smurf1 expression allows ephrinB1 to be targeted for Smurf2-mediated degradation and results in inhibition of ephrinB1-mediated tissue separation.
Results
Smurfs interact with EphrinB1
Several of the ephrinB1-interacting proteins that have been identified are involved in pathways regulating cell movement (Dishevelled, FGFR, RGS3-PDZ, and Grb4) and cell–cell boundaries (Par-6, Cx43, and Claudin) (Bush and Soriano 2012; Daar 2012). To identify additional proteins that might regulate ephrinB1, we used mass spectrometric analysis of proteins that coimmunoprecipitate with ephrinB1 when it is overexpressed in Xenopus embryos (data not shown). From this analysis, Smurf2 was identified as an ephrinB1-interacting protein.
To assess whether endogenous ephrinB1 and Smurf1 or Smurf2 proteins interact, we conducted a coimmunoprecipitation (co-IP) analysis of lysates from HT29 colon carcinoma cells that express abundant levels of all three proteins. Smurf1 and Smurf2 were present in ephrinB1 immune complexes, and, reciprocally, ephrinB1 was found in the Smurf1 and Smurf2 immune complexes but not in the controls (Fig. 1A). These data indicate that an in vivo interaction may exist between ephrinB1 and the Smurf proteins. Other lines of evidence suggest that Smurf1 and Smurf2 may be candidate mediators of ephrinB1 regulation. For example, expression of both Smurf1 and Smurf2 RNA overlaps significantly with ephrinB1 during development (Fig. 1B), and immunoprecipitation analysis of lysates from Xenopus oocytes exogenously expressing ephrinB1 and either Smurf1 or Smurf2 indicates that both proteins can interact with ephrinB1 (Fig. 1C).
Figure 1.
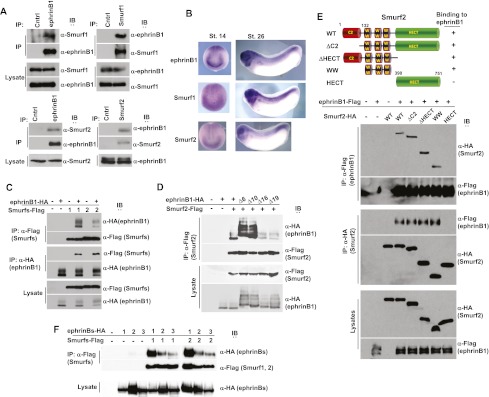
EphrinB1 associates with both Smurf1 and Smurf2. (A) Immunoprecipitations of ephrinB1, Smurf1 or Smurf2, or His (as a control) in HT29 human colon carcinoma cell lysates. (B) Whole-mount in situ hybridizations of ephrinB1, Smurf1, and Smurf2 in stage 14 (dorsal anterior view) and stage 26 embryos (lateral view). (C) Immunoprecipitations of the indicated proteins from embryos exogenously expressing Flag-tagged Smurf1 or Smurf2 and HA-tagged ephrinB1 where indicated, then immunoblotted with the indicated antibodies. (D) Immunoprecipitation and Western blot analysis of lysates from oocytes expressing HA-tagged constructs of wild-type (WT) ephrinB1 or deletion mutants lacking six, 10, 16, or 19 amino acids from the C terminus along with Flag-tagged Smurf2. Note: Deletion of C-terminal 16 amino acids abrogates binding. (E) Immunoprecipitation and Western blot analysis of lysates from oocytes expressing Flag-tagged constructs of wild-type ephrinB1 along with HA-tagged wild-type Smurf2 or Smurf2 mutants as depicted in cartoon. Note: WW domains associate with ephrinB1. (F) Immunoprecipitation and Western blot analysis of lysates from oocytes expressing HA-tagged constructs of wild-type ephrinB1 (1), ephrinB2 (2), and ephrinB3 (3) along with Flag-tagged Smurf1 (1) or Smurf2 (2). Note: More ephrinB1 is detected in immune complexes than ephrinB2 or ephrinB3.
Having established that ephrinB1 can interact with Smurf1 and Smurf2, we next determined which region of ephrinB1 is necessary for binding to the Smurfs. We performed an immunoprecipitation analysis of lysates from oocytes coexpressing wild-type or various deletion constructs of ephrinB1 along with wild-type Smurf2. We found that the cytoplasmic domain of ephrinB1 was required for an interaction with Smurf2 (Supplemental Fig. S1A) and that deletion of 16 amino acids from the C terminus of ephrinB1 (ephrinB1Δ16) prevents the interaction with Smurf2 (Fig. 1D). Smurfs can bind to target molecules through an interaction with their WW domains (Rotin and Kumar 2009; Chong et al. 2010; Ogunjimi et al. 2010), and the HECT domain interacts with E2 ligases and ubiquitin (Rotin and Kumar 2009). The N-terminal C2 domain binds phospholipids, and in the case of Smurf1, it can also interact with RhoA (Tian et al. 2011), whereas in Smurf2, this domain can also interact intramolecularly with the HECT domain (Wiesner et al. 2007). Immunoprecipitation analysis was performed with ephrinB1 and Smurf2 deletion constructs, and a construct that consisted of the three WW domains of Smurf2 was able to bind ephrinB1 (Fig. 1E). A co-IP analysis of exogenously expressed ephrinB2 or ephrinB3 showed that both of these ephrins have affinity for Smurf1 and Smurf2 but with a weaker association than observed for ephrinB1 (Fig. 1F).
EphrinB1 is a prominent target for ubiquitination and degradation by Smurf2 but is inadequately targeted by Smurf1 in vivo
Two observations suggested that ephrinB1 may be a target of Smurf2: (1) We observed an interaction between ephrinB1 and the Smurf2 WW domains (Fig. 1E), which are usually associated with substrates of the ubiquitin ligase (Rotin and Kumar 2009). (2) We observed that coexpression of Smurf2 with ephrinB1 in embryos resulted in a marked decrease in ephrinB1 expression (Fig. 1C). This decreased expression was not observed upon similar coexpression of Smurf1 with ephrinB1 (Fig. 1C). Only when Smurf1 was dramatically overexpressed by several fold was a decrease in ephrinB1 expression detected (data not shown), indicating that ephrinB1 is a poor substrate for Smurf1. To test whether the reduction in ephrinB1 expression in embryos was dose-dependent, we coinjected increasing amounts of Smurf2 RNA along with a constant amount of ephrinB1 RNA. Western blot analysis demonstrated a dose-dependent decrease in ephrinB1 expression, indicating that Smurf2 may be targeting ephrinB1 for degradation (Fig. 2A). Of note, we did not observe any effect on ephrinB2 levels when Smurf1 or Smurf2 protein was overexpressed, suggesting that ephrinB2 may not be regulated by the same mechanism as ephrinB1 (Supplemental Fig. S1B).
Figure 2.
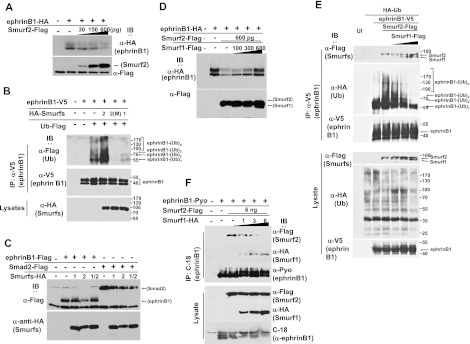
Smurf1 and Smurf2 compete for an interaction with ephrinB1 and regulate its expression in vivo. (A) Western blots showing that the level of exogenously expressed HA-tagged ephrinB1 in stage 16 embryos decreases with increasing expression of Flag-tagged Smurf2. (B) Western analysis showing prominent ubiquitination of V5-tagged ephrinB1 in immunoprecipitations from oocytes exogenously expressing Flag-tagged ubiquitin, V5-tagged ephrinB1, and HA-tagged Smurf2 but markedly less ubiquitination when ephrinB1 is coexpressed with a Smurf2 ligase activity-deficient mutant (M) or Smurf1. (C) Western analysis of lysates from embryos showing that exogenous expression of Flag-tagged ephrinB1 is preserved when it is coexpressed with HA-tagged Smurf1 or both Smurf1 and Smurf2 but not Smurf2 alone. Coexpression of the Flag-tagged Smad2 control with both Smurf1 and Smurf2 shows decreased levels of Smad2 expression. (D) Western analysis showing that HA-tagged ephrinB1 expression is increased with increasing expression of Flag-tagged Smurf1 in the presence of constant expression of Flag-tagged Smurf2. (E) Immunoprecipitation and Western analysis of lysates from oocytes expressing constant amounts of V5-tagged ephrinB1, HA-tagged ubiquitin, and Flag-tagged Smurf2 with increasing amounts of Flag-tagged Smurf1. EphrinB1 immune complexes were probed with anti-HA antibody to reveal ubiquitination. Direct lysates were blotted with the indicated antibodies. Note: The expression of Smurf2 increases the ladder of polyubiquitination of ephrinB1, and with increasing expression of Smurf1, the polyubiquitination is decreased, even below endogenous levels. (F) Immunoprecipitation and Western analysis of lysates from oocytes expressing constant amounts of Pyo-tagged ephrinB1 and Flag-tagged Smurf2 with increasing amounts of HA-tagged Smurf1. Note: With increasing expression of Smurf1, less Smurf2 is found in the ephrinB1 immune complexes.
To assess whether ephrinB1 is a target for Smurf2-mediated ubiquitination, Xenopus oocytes were used for analysis because we observed less background ubiquitination and, most importantly, less degradation of ephrinB1 in the oocyte system when compared with the embryonic system. Oocytes were injected with RNA encoding Flag-tagged ubiquitin along with V5-tagged ephrinB1 and additional RNAs encoding HA-tagged versions of wild-type Smurf1 or wild-type Smurf2 or a ligase activity-deficient mutant of Smurf2 (M) (Fig. 2B). EphrinB1 was immunoprecipitated with an anti-V5 antibody and probed with Flag antibody to reveal the ubiquitinated ephrinB1. We observed background ubiquitination when ephrinB1 and ubiquitin were expressed; however, coexpression of Smurf2 enhanced ephrinB1 ubiquitination (Fig. 2B). Furthermore, coexpression of the ligase activity-deficient mutant of Smurf2 (M) appeared to act in a dominant-negative manner to suppress ubiquitination of ephrinB1 below background levels (Fig. 2B), suggesting that Smurf2 targets ephrinB1. Finally, coexpression of Smurf1 reduced the ubiquitination of ephrinB1 to below background levels as well (Fig. 2B). Collectively, these data indicate that although ephrinB1 can interact with both Smurf1 and Smurf2, it is an accessible target for ubiquitination by Smurf2 but is poorly targeted by Smurf1.
Smurf1 and Smurf2 compete for association with ephrinB1, which regulates ephrinB1 expression in vivo
Since both Smurf1 and Smurf2 can associate with ephrinB1, but only Smurf2 prominently ubiquitinates and targets ephrinB1 for possible degradation, we tested whether coexpression of both Smurf1 and Smurf2 leads to either reduction or protection of ephrinB1 expression. Xenopus embryos were injected with RNA encoding Flag-tagged ephrinB1 alone or in conjunction with HA-tagged Smurf1 or Smurf2 or both. Western blot analysis performed on the embryonic lysates revealed that coexpression of both Smurf1 and Smurf2 led to a sustained (protective) expression of ephrinB1, while expression of only Smurf2 led to reduced ephrinB1 protein levels (Fig. 2C). These data suggest that Smurf1 was able to prevent or markedly inhibit degradation of ephrinB1 mediated by Smurf2 (Fig. 2C). This effect appears to be specific to ephrinB1, since coexpression of both Smurf1 and Smurf2 with Smad2 did not prevent loss of Smad2 expression (Fig. 2C). We tested whether the protective effect of Smurf1 was dose-dependent by introducing a constant amount of ephrinB1 and Smurf2 RNAs into Xenopus embryos, along with increasing amounts of Smurf1 RNA. Western blot analysis shows increasing ephrinB1 protein levels with increasing amounts of Smurf1 expression (Fig. 2D).
As a further test for the antagonism between Smurf1 and Smurf2 with regard to targeting ephrinB1 for degradation, we examined whether Smurf2-directed polyubiquitination of ephrinB1 was affected by increasing expression of Smurf1. Oocytes were injected with RNA encoding HA-tagged ubiquitin along with V5-tagged ephrinB1 and additional RNAs encoding Flag-tagged wild-type Smurf2 along with increasing amounts of Flag-tagged wild-type Smurf1 (Fig. 2E). EphrinB1 was immunoprecipitated with V5 antibody and probed with HA antibody to reveal the ubiquitinated ephrinB1. We observed background ubiquitination when ephrinB1 and ubiquitin were expressed, with a significant amount of monoubiquitinated ephrinB1. However, coexpression of Smurf2 enhanced ephrinB1 polyubiquitination, and this decreased as increasing amounts of Smurf1 were coexpressed with Smurf2 (Fig. 2E). These data confirm an antagonistic relationship between Smurf1 and Smurf2, where Smurf2 targets ephrinB1 for ubiquitination.
A previous study indicated that Smurf2 can induce ubiquitin-dependent degradation of Smurf1 in MDA-MB-231 breast cancer cells (Fukunaga et al. 2008). However, if such a mechanism was present in our system, it should lead to a reduction of both Smurf1 and ephrinB1, which was not observed. Moreover, we did not observe a decrease in Smurf2 expression when Smurf1 was coexpressed in embryos (Fig. 2C,D) or oocytes (Fig. 2E), indicating that cross-regulation between Smurfs via ubiquitination and degradation was unlikely to explain the protective effect on ephrinB1 mediated by Smurf1. Thus, we tested whether a possible mechanism of protection was through Smurf1 successfully competing with Smurf2 for binding to ephrinB1. To test such competition, we introduced constant amounts of RNA encoding Polyoma-tagged ephrinB1 and Flag-tagged Smurf2 into Xenopus oocytes and injected increasing amounts of HA-tagged Smurf1. Immunoprecipitation of ephrinB1 was performed and analyzed for the presence of the two Smurfs in the ephrinB1 immune complexes. With increasing amounts of Smurf1 expression in the oocytes, more Smurf1 was detected in the ephrinB1 immune complexes, along with decreasing amounts of associated Smurf2 (Fig. 2F). As expected, neither Smurf affected the expression of the other when coexpressed (Fig. 2F). These data strongly indicate that Smurf1 and Smurf2 can compete for an association with ephrinB1 and that this may represent a mechanism for regulating ephrinB1 expression.
One prediction of a model based on competition between Smurf1 and Smurf2 is that the disruption of Smurf1 expression in vivo would allow Smurf2 to more freely target ephrinB1 for ubiquitination and degradation. To test this model, we used Smurf loss-of-function and gain-of-function strategies in embryos and specifically examined the biochemical effect in the dorsal axial region, where ephrinB1, Smurf1, and Smurf2 expression overlaps (Fig. 3A). To avoid saturating the ability of the endogenous Smurfs to target exogenously introduced ephrinB1, we expressed HA-tagged ephrinB1 at levels normally used to rescue the phenotypes induced by MO-mediated knockdown of ephrinB1. Thus, we either coinjected Smurf1 MO or Smurf2 MO along with ephrinB1 RNA or overexpressed Smurf1 or Smurf2 proteins and removed the dorsal axial region that spans the neural plate and neural fold at the neurula stage for Western blot analysis (Fig. 3B; see also Supplemental Fig. S1C for method). Since in this experiment ephrinB1 is being exogenously expressed in embryos, the protein will be present in areas beyond the region of overlap with the endogenous Smurf proteins. Therefore, we dissected the region where both Smurfs and ephrinB1 normally reside (i.e., dorsal axial tissue) to assess the effect of the endogenous Smurf knockdowns on ephrinB1 expression. Interestingly, a dramatic loss of ephrinB1 expression was found in the tissue containing the Smurf1 MO, suggesting that loss of Smurf1 resulted in a loss of protection of ephrinB1, leading to its degradation by Smurf2 (Fig. 3B). Consistent with this concept, ephrinB1 expression was somewhat increased in the tissue where Smurf2 MO was present, suggesting that Smurf2 may target ephrinB1 for degradation (Fig. 3B). As expected, overexpression of Smurf1 or Smurf2 had opposing effects, where ephrinB1 was somewhat increased in the tissue overexpressing Smurf1 and markedly reduced when Smurf2 was overexpressed (Fig. 3B). These data are also consistent with the similarity in phenotypes observed between ephrinB1 morphants and Smurf1 morphants when compared with Smurf2 morphants (Supplemental Fig. S2).
Figure 3.
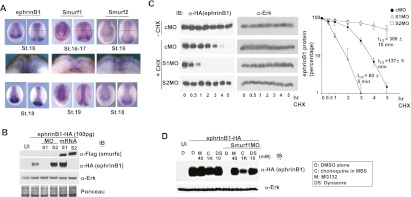
Antagonism between Smurf1 and Smurf2 in the regulation of ephrinB1 degradation. (A) Whole-mount in situ hybridizations of Smurf1, Smurf2, and ephrinB1 in stage 16–19 embryos (for top and bottom panels, left photo is anterior view, and right photo is dorsal view; middle panels represent cross sections of stage 16–17 embryos, and the level of section is represented by a thin horizontal line in the top right panels). (B) Western blot analysis of dorsal axial tissue from stage 16 embryos show that introduction of Smurf1 MO reduces exogenously expressed HA-tagged ephrinB1, while Smurf2 MO increases ephrinB1 in this tissue. Blots were also probed with anti-Erk antibodies and Ponceau-stained as loading controls. (C) Western blot analysis of dorsal axial tissue from stage 14 embryos previously injected with HA-tagged ephrinB1 RNA that was titrated to yield roughly equivalent ephrinB1 protein levels in the presence of the indicated MOs. The explanted tissue was treated with cycloheximide (CHX) for 5 h, and Western analysis shows that introduction of Smurf1 MO reduces exogenously expressed ephrinB1 by half within 60 min, while Smurf2 MO increases ephrinB1 half-life to ∼5 h in this tissue. Blots were also probed with anti-Erk antibodies as loading controls. The quantification of the data from three independent experiments is displayed in the right panel; error bars show SD. (D) One- or two-cell stage embryos were left uninjected (UI) or injected with HA-tagged ephrinB1 (100 pg) RNA alone or along with Smurf1 MO (10 ng). At stages 16–18, dorsal axial tissue was dissected and treated with DMSO carrier (D), chloroquine (C), MG132 (M), or dynasore (DS) at the indicated concentrations for 4–5 h. Samples were analyzed directly by SDS-PAGE, then immunoblotted with anti-HA antibodies or anti-Erk antibodies to reveal exogenous expression levels of ephrinB1 and the endogenous Erk levels (as a loading control), respectively.
We next examined whether the observed reduction in ephrinB1 expression resulting from the presence of the Smurf1 MO was due to a decrease in the half-life of ephrinB1 protein, as would be expected if ephrinB1 were being degraded. For this purpose, we carefully titrated the amount of injected ephrinB1 RNA along with control MO, Smurf1 MO, or Smurf2 MO to yield roughly equivalent ephrinB1 protein levels when embryos reached the neurula stage. We then explanted the neural fold region and cultured the explants in cycloheximide to block further protein synthesis for 5 h (Fig. 3C). Western blot analysis shows that ephrinB1 levels are reduced by half within 137 min (2.3 h) after the addition of cycloheximide to control MO-containing explants. In the Smurf1 MO-containing explants, the half-life of ephrinB1 is reduced to ∼60 min (1.0 h) (Fig. 3C). In contrast, the half-life of ephrinB1 is actually extended to ∼306 min (5.1 h) in the Smurf2 MO-containing explants (Fig. 3C). These data are consistent with Smurf1 hindering ephrinB1 degradation, while Smurf2 targets ephrinB1 for ubiquitination and degradation.
To assess whether the lysosomal pathway and/or the proteasomal pathway may be responsible for Smurf2-induced reduction of ephrinB1 expression, inhibitors of proteasomal degradation (MG132), lysosomal degradation (chloroquine), or dynamin-dependent endocytosis (dynasore) were used to determine the effective pathways used by Smurf2 to regulate ephrinB1. Xenopus dorsal axial tissue was excised after the introduction of HA-tagged ephrinB1 RNA alone or along with Smurf1 MO. This tissue was cultured in the inhibitors for 4 h, and then Western blot analysis was performed. As expected, Smurf1 knockdown mediated by MOs resulted in a dramatic loss of ephrinB1 expression in the dorsal axial tissue. MG132 and dynasore almost equivalently rescued ephrinB1 expression, while chloroquine (at 1 mM) rescued expression but to a lesser degree (Fig. 3D). These data indicate that in the absence of Smurf1, ephrinB1 is targeted for destruction through the lysosomal and proteasomal pathways.
Loss of Smurf1 in vivo impairs RhoA activation, analogous to loss of ephrinB1
Another prediction from the Smurf1/Smurf2 model of ephrinB1 regulation in vivo is that a signal downstream from ephrinB1 should be affected by the loss of Smurf1 and the consequent reduction in ephrinB1 expression. EphB/ephrinB signaling has been shown to be important in tissue separation at the mesoderm/ectoderm border (Medina et al. 2004; Unterseher et al. 2004; Wang 2008; Koster et al. 2010; Rohani et al. 2011), and activated RhoA has been shown to be a downstream effector of EphB/ephrinB signaling (Pasquale 2008) as well as a critical regulator of tissue separation (Wacker et al. 2000; Berger et al. 2009; Koster et al. 2010; Rohani et al. 2011). Therefore, we examined the effect of Smurf1/2 and ephrinB1 knockdowns on RhoA activity at the mesoderm/ectoderm border. Embryos were injected with RNA encoding histone H2B-mRFP (as a cell lineage tracer) along with ephrinB1 MO, Smurf1 MO, Smurf2 MO, or a dominant-negative RhoA as a control.
At the mid-gastrula stage, the dorsal marginal zone (where the ectoderm overlays the involuting mesoderm) was excised, delaminated, fixed, and incubated with a GFP-fused Rhotekin GTPase-binding domain (GFP-RBD) fusion protein to detect localized areas of RhoA activation in situ (for method, see Supplemental Fig. S3; Berger et al. 2009). Confocal analysis revealed the presence of reduced areas of RhoA activity in cells containing the ephrinB1 MO, but this was partially rescued by expression of ephrinB1 through the introduction of MO-resistant ephrinB1 RNA (the relative intensity of the injected cells to uninjected cells is 68% ± 8% for ephrinB1 MO and 85% ± 12% for ephrinB1 MO plus MO-resistant ephrinB1 RNA) (Fig. 4A,B). Having established that loss of ephrinB1 in the assay reduces RhoA activity, we tested whether loss of Smurf1 affected this activity as well. Cells harboring the Smurf1 MO also displayed a reduced level of RhoA activation (76% ± 4%) (Fig. 4A,B), similar to loss of ephrinB1. In contrast, Smurf2 MO-injected cells did not display a reduction in RhoA activation (103% ± 10%) (Fig. 4A,B). As expected, the control cells expressing a dominant-negative RhoA construct also displayed reduced RhoA activity (66% ± 8%) (Fig. 4A,B).
Figure 4.
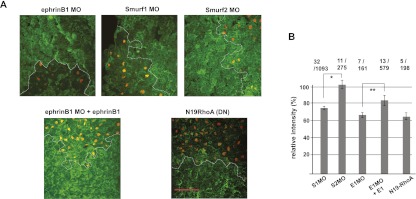
Loss of Smurf1 in vivo impairs RhoA activation, similar to loss of ephrinB1. (A) Embryos unilaterally expressing histone H2B mRFP as a lineage tracer (orange dots) with MO-mediated inhibition of Smurf1 show reduced GFP-RBD binding (green staining) when compared with the control area outside of the dotted line. EphrinB1 MO and Smurf1 MO both cause a similar reduction in GFP-RBD binding at cell contact points. The Smurf2 MO has no observable effect on GFP-RBD binding, and cells exogenously expressing ephrinB1 in the presence of the ephrinB1 MO displayed a partial rescue of this interaction. Bar, 100 μm; all panels are the same scale. See Supplemental Figure S3 for the experimental outline. (B) The green fluorescence intensities representing RhoA activation were measured for the injected and uninjected areas, and the relative intensity of injected areas to the uninjected control areas was calculated and presented as a percentage. The mean relative intensities are shown in bar graphs with error bars (SD). Total tested explant numbers/total measured cell numbers are shown on top. The significance of the difference in comparative pairs was confirmed by Student's t-test as indicated; (*) P = 5.08 × 10−12; (**) P = 0.00186.
To confirm the relationship between Smurf1 inhibition and a reduction in RhoA activity at the mesoderm/ectoderm border in living tissue, RNA encoding a GFP-RBD was coinjected with H2B-mRFP RNA, along with either Smurf1 MO, Smurf2 MO, or control MO in embryos. At stage 16 (neurula), embryos were transversely bisected and cultured in a dish with the sectioned surface contacting a fibronectin-coated coverslip. Time-lapse confocal microscopy revealed that in embryos harboring the Smurf1 MO, the GFP-RBD signal (representing RhoA activation) is attenuated on the somito–ectoderm border and that the border is markedly compromised, allowing mesodermal cells to move into the ectoderm (Supplemental Movie S3). In contrast, uninjected embryos and those with the Smurf2 MO or control MO display prominent GFP-RBD signals (RhoA activation) at the regions of contact between the somite and ectoderm (Supplemental Movies S1, S2, S4). Together, these data indicate that loss of Smurf1, but not Smurf2, leads to a reduction in RhoA activation, which is a key downstream signaling event elicited by ephrinB1. Moreover, these data provide in vivo and ex vivo evidence that loss of Smurf1 expression compromises the tissue boundary between ectoderm and mesoderm.
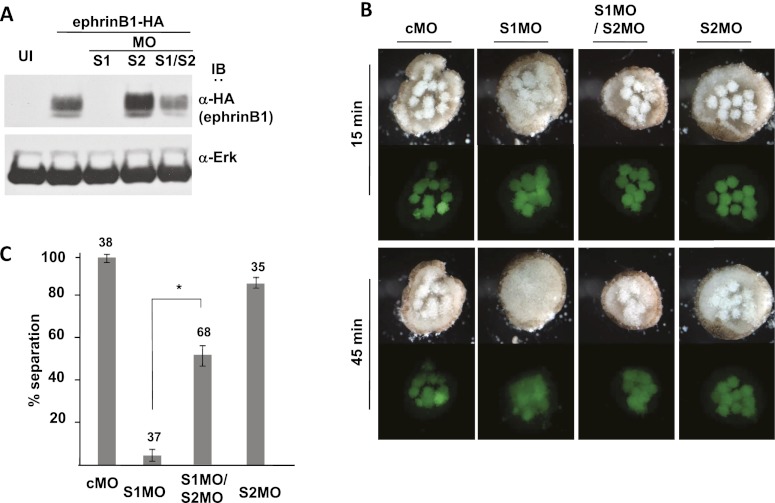
Inhibition of Smurf1 expression results in loss of ephrinB1-mediated tissue separation, which is rescued upon knockdown of Smurf2
As a further test of the opposing roles of Smurf1 and Smurf2 in ephrinB1 regulation, we designed an experiment to determine whether loss of Smurf1 or Smurf2 expression affects tissue separation in living tissue, which represents a functional outcome of ephrinB1 signaling (Rohani et al. 2011). A model for tissue separation in Xenopus is when the internalizing mesendodermal cell mass comes into contact with the blastocoel roof (BCR) during gastrulation (Wacker et al. 2000; Gorny and Steinbeisser 2012). These two tissues remain separated by repulsive forces at Brachet's cleft (Wacker et al. 2000). The in situ hybridizations in Figure 5A show that ephrinB1, Smurf1, and Smurf2 are all expressed in the involuting mesoderm of mid-gastrula stage embryos, where tissue repulsion (separation) is maintained between the mesoderm and overlying ectoderm. EphrinB1 expression is present in both ectoderm and mesoderm and has been shown to be required in both tissues for separation to take place (Rohani et al. 2011). As a functional test of Smurf-mediated regulation of ephrinB1, we used an ex vivo tissue separation assay (Wacker et al. 2000; Gorny and Steinbeisser 2012), where embryos were injected with Smurf1 MO, Smurf2 MO, ephrinB1 MO, or control MO along with fluorescent dextran to label the tissue (Fig. 5B). Ectoderm was removed from the BCR of uninjected late gastrula stage embryos and used as a substrate for excised mesoderm (involuting marginal zone tissue; IMZ) from injected embryos (Fig. 5B,C). As expected, control MO-injected mesoderm explants (IMZ) placed on the ectoderm (BCR) substrate maintained tissue separation (98.3% ± 3.3%), as evidenced by the lack of integration of these mesoderm cells into the BCR (Fig. 5C,D). Strikingly, introduction of the Smurf1 MO into the mesoderm tissue caused a loss of tissue separation (4.2% ± 5%), as evidenced by integration into the BCR (Fig. 5C,D). Mesoderm containing the ephrinB1 MO gave similar results (22.2% ± 3.8%) to the Smurf1 MO, while the mesoderm containing the Smurf2 MO displayed tissue separation (87.5% ± 4.4%), similar to control MO-injected tissue (Fig. 5C,D; see also Supplemental Fig. S4A for statistics).
Figure 5.
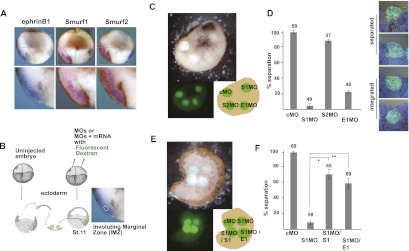
Smurf1 knockdown results in loss of ephrinB1-mediated tissue separation. (A) Whole-mount in situ hybridizations of bisected stage 11 embryos with probes for ephrinB1, Smurf1, and Smurf2. The bottom panel is an enlarged view of the boxed area showing the mesoderm/ectoderm boundary. (B) A schematic depicting the BCR assay. (C) Above view of BCR assay showing mesoderm tissue injected with the indicated MO (i.e., control MO [cMO], Smurf1 MO [S1MO], Smurf2 MO [S2MO], and ephrinB1 MO [E1MO]), and placed on an ectoderm substrate (top panel is light microscopy, and bottom left panel is a reduced image under fluorescent microscopy). (Bottom right) Cartoon depicting the BCR assay samples indicating the location of the explants from the two adjacent panels. (D) Histogram of BCR assay samples as labeled and represented as the percentage of explants displaying tissue separation after 45 min in culture. Error bars represent standard deviation, and the numbers above each bar represent the total number of tested explants. In panels to the right of the bar graph are examples of the categories representing tissue integration (apical surface of mesoderm explant at or below surface of BCR) and tissue separation (apical surface of mesoderm explant above the surface of BCR) of explants as judged by cross-sectional analysis and visual observation. (E) Above view of BCR assay showing mesoderm tissue injected with the indicated MO (as described in C) and RNA (Smurf1 [S1] and ephrinB1 [E1]) and placed on ectoderm substrate (top panel is light microscopy, and bottom left panel is a reduced image under fluorescent microscopy). (Bottom right) Cartoon depicting the BCR assay samples from the two adjacent panels. (F) Histogram of BCR assay samples as indicated and represented as the percentage of explants displaying tissue separation (as described in D). In the MO groups rescued by the introduction of Smurf1 (S1) or ephrinB1 (E1) RNA, the significance was confirmed by Student's t-test ([*] P = 0.00297; [**] P = 0.00289) as indicated. Note: The percentage of each category (G1–G4) and more powerful statistical analysis for D and F are shown in Supplemental Figure S4, A and B.
The above results are consistent with a model in which Smurf1 protects ephrinB1 from being targeted for degradation by Smurf2, and the loss of Smurf1 would compromise ephrinB1 expression that is specifically necessary for tissue separation. Loss of Smurf2 would be expected to maintain tissue separation, since ephrinB1 expression would be preserved or somewhat enhanced (Fig. 3B). One prediction from this model is that in mesoderm, exogenous expression of ephrinB1 in the presence of the Smurf1 MO may rescue tissue repulsion at the mesoderm/ectoderm boundaries. To test this prediction, we introduced the Smurf1 MO along with ephrinB1 RNA at a concentration that was titrated to overcome the dramatic reduction observed in ephrinB1 expression. We found that exogenous expression of ephrinB1 in the presence of the Smurf1 MO rescues tissue separation (59.0% ± 12.2%), while the mesoderm harboring the Smurf1 MO alone (7.9% ± 5.5% separation) integrates into the BCR (Fig. 5E,F; see also Supplemental Fig. S4B for statistics). We also tested whether loss of Smurf1 expression compromises the accompanying tissue separation in vivo by examining intact mid-gastrula stage embryos. Embryos injected with Smurf1 MO displayed a less defined Brachet's cleft (Supplemental Fig. S5A), a visible cleft that separates mesendoderm and ectoderm (Gorny and Steinbeisser 2012). Moreover, the cleft is significantly shorter than either the control MO-injected embryos or the Smurf2 MO-injected embryos (Supplemental Fig. S5A), verifying a role for Smurf1 in tissue separation in vivo. In addition, we determined that exogenous expression of ephrinB1 in the presence of the Smurf1 MO rescues the formation of Brachet's cleft in a dose-dependent manner (Supplemental Fig. S5B). Collectively, the data indicate that ephrinB1 is specifically targeted for degradation in the absence of Smurf1, leading to a loss of tissue separation at the mesoderm/ectoderm boundaries.
Another premise of this model is that the loss of ephrinB1 in the absence of Smurf1 is due to its ubiquitination and degradation by Smurf2. Thus, another testable prediction is that the introduction of Smurf2 MO along with the Smurf1 MO in embryonic mesoderm would rescue the expression of ephrinB1 in the tissue and thus lead to a resumption of tissue separation between mesoderm and the BCR substrate. Western blot analysis was performed on dorsal axial tissue from embryos exogenously expressing HA-tagged ephrinB1 that were also injected with either Smurf1 MO, Smurf2 MO, or both. The results of this analysis show that introduction of both Smurf1 MO and Smurf2 MO caused a marked increase in ephrinB1 expression when compared with the Smurf1 MO-injected embryos (Fig. 6A). Although the ephrinB1 level was not equivalent to that detected in the tissue where ephrinB1 RNA was introduced alone (Fig. 6A), the data indicate that Smurf2 is responsible for a considerable amount of the loss of ephrinB1 expression in the absence of Smurf1. Notably, in the tissue separation assay, mesoderm containing both Smurf1 MO and Smurf2 MO is observed to maintain separation (51.7% ± 9.8%) from the BCR ectoderm (Fig. 6B,C). In contrast, mesoderm explants harboring just the Smurf1 MO (6.1% ± 5.4% separation) integrate into the BCR (Fig. 6B,C). As expected, the mesoderm explants containing the control MO (97.8% ± 3.8%) or the Smurf2 MO (85.6% ± 5.1%) alone displayed tissue separation (Fig. 6B,C; see also Supplemental Fig. S4C for statistics). We further confirmed the above results by performing these experiments using the rigorously tested Smurf2 MO reported by Das and Chang (2012), with similar outcomes (Supplemental Fig. S6). Together, these data indicate that ephrinB1 is targeted for degradation, at least in part, by Smurf2 in the absence of Smurf1 expression and that the coincident loss of Smurf2 rescues tissue separation.
Figure 6.
Disruption of tissue separation due to loss of Smurf1 is rescued by coincident loss of Smurf2 via stabilization of ephrinB1. (A) Western blot analysis of dorsal axial tissue from stage 16 embryos showing that introduction of Smurf1 MO (S1MO) reduces exogenously expressed ephrinB1, but the addition of Smurf2 MO (S1MO/S2MO) increases ephrinB1 expression in this tissue. Blots were also probed with anti-Erk antibody as a loading control. (B) Above view of BCR assay showing mesoderm tissue injected with the indicated MO (10 ng) and placed on an ectoderm substrate (top panels are light microscopy, and bottom panels are fluorescent microscopy) for the indicated times (15 and 45 min). Note that Smurf1 MO (S1MO)-injected mesoderm tissue integrates into the BCR substrate, while addition of a Smurf2 MO (S1MO/S2MO) rescues tissue separation. (C) Histogram of BCR assay samples as labeled and represented as the percentage of explants displaying tissue separation (separation and integration are defined in Fig. 5D). Error bars represent standard deviation, and the numbers above each bar represent the total number of tested explants. P-values are determined by Student's t-test; (*) P = 0.02629. Note: The percentage of each category (G1–G4) and more powerful statistical analysis for C are shown in Supplemental Figure S4C.
Discussion
An understanding of the role Smurfs play in morphogenesis is only beginning to emerge (Zhu et al. 1999; Wang et al. 2003; Ozdamar et al. 2005; Yamashita et al. 2005; Alexandrova and Thomsen 2006; Narimatsu et al. 2009; Huang 2010; Das and Chang 2012). Smurf1 and Smurf2 share 73% identity at the amino acid level, and it is becoming clear that these ubiquitin ligases have both common and distinct functions in Xenopus development, while there appears to be more redundancy in mice.
Smurfs have been shown to ubiquitinate and regulate several molecules involved in cell adhesion and migration (e.g., RhoA, Rap1, talin, hPEM-2, and Pk1) (Schwamborn et al. 2007; Narimatsu et al. 2009; Huang 2010). Smurf1 is localized to lamellipodia and filopodia and can induce protrusions (Wang et al. 2003; Huang et al. 2009), while overexpression or loss of function of Smurf2 promotes or inhibits the migration of some cell lines (Jin et al. 2009; Yang et al. 2009). However, our understanding of the mechanisms and targets of these ligases in specific morphogenetic events is still in its infancy. In this regard, it is now clear that Smurfs can regulate planar cell polarity and convergent extension through a noncanonical Wnt signaling pathway by targeting the core planar cell polarity protein Prickle1 for ubiquitin-mediated degradation (Narimatsu et al. 2009). In mice, it was shown that the epithelial-to-mesenchymal transition and invasion of epicardial cells is dependent on activation of the Par-6/Smurf1/RhoA pathway (Sanchez and Barnett 2012).
Our study reveals an interesting antagonistic mechanism by which Smurf1 and Smurf2 can regulate the morphogenetic process of tissue separation by their differential activity toward ephrinB1, an Eph ligand. There is persuasive evidence that Eph/ephrinB signaling has a crucial role in the control of cell–cell adhesion complexes and morphogenetic processes that rely on Rho family GTPase activity, leading to repulsion or separation between tissues or cell types (Pasquale 2005, 2008; Rohani et al. 2011). A recent study provided evidence that both EphB ligands and receptors can be found on both sides of the mesoderm/ectoderm boundary in the gastrulating Xenopus embryo and that tissue separation depends on forward signaling across this boundary in both directions (Rohani et al. 2011). Here, using overexpression studies in Xenopus oocytes, we show that both Smurf1 and Smurf2 interact with ephrinB1 and that they compete for this interaction. This mechanism differs from the reported cross-regulation of the Smurfs through ubiquitination and degradation, where Smurf2 was shown to target Smurf1, but Smurf1 did not target Smurf2 in a breast cancer cell line (Fukunaga et al. 2008). This mechanism also differs from the recently revealed interaction between the E3 ligase Cdh1 and Smurf1 that can either target Smurf1 for degradation (Kannan et al. 2012) or activate it (Wan et al. 2011).
Our in vivo experiments with the Smurfs reveal a regulatory mechanism that relies on differential activity of Smurf1 and Smurf2 toward ephrinB1, where Smurf2 is able to target ephrinB1 for ubiquitination, while Smurf1 is greatly diminished in this capacity. This may be due to the recently reported autoinhibitory activity conferred by the C2 domain of Smurf1 (Wan et al. 2011). In that study, Smurf1 was shown to form an autoinhibited homodimer (unlike Smurf2, which forms an autoinhibited monomer) with the C2 domain of one monomer binding to the HECT domain of another. In this case, the Cdh1 protein binds to the C2 domain of Smurf1 and frees it from the autoinhibited dimer state (Wan et al. 2011). In our study, it may also be possible that the C2 domain plays a role in the selection of ephrinB1 as a substrate or to localize Smurf1 in proximity to ephrinB1. A recent report found that the C2 domain of Smurf1 confers differential activity toward RhoA rather than Smad5 and Runx2 and identifies a role for the Smurf1 C2 domain in substrate selection and cell localization (Lu et al. 2011). Alternatively, it may be possible that the C2 domain interacts with a protein responsible for reducing the activity of Smurf1 toward ephrinB1.
The competition model for the binding and targeting of ephrinB1 was tested in vivo and ex vivo by Smurf1 or Smurf2 MO-mediated knockdown experiments in Xenopus embryos and explants. A dramatic loss of ephrinB1 expression was found in the dorsal axial tissue, where Smurf1 protein expression was suppressed by a Smurf1 MO, and increased expression of ephrinB1 was observed in the tissue where Smurf2 MO was present, suggesting that loss of Smurf1 resulted in a loss of protection of ephrinB1, leading to its degradation by Smurf2. This model was further tested by specifically knocking down Smurf2 in the presence of the Smurf1 MO, resulting in a substantial rescue of ephrinB1 expression. Since signaling elicited by ephrinB2 is also critical for proper tissue separation during embryogenesis (Rohani et al. 2011), it too may be subject to regulation by the Smurf1/Smurf2 system. However, unlike ephrinB1, we did not observe any effect on ephrinB2 expression levels in dorsal axial tissue when Smurf1 or Smurf2 proteins were overexpressed (Supplemental Fig. S1B). These data strongly suggest that the Smurf competition model may be specific to ephrinB1; however, it may also be possible that ephrinB2 is subject to regulation by Smurf1 and/or Smurf2 in a different context. Moreover, MO-mediated suppression of Smurf1 expression results in the loss of tissue separation, and this is specifically rescued by re-expression of ephrinB1 or MO-mediated coincident loss of Smurf2, leading to reaccumulation of ephrinB1 protein. This reveals a system whereby balancing the association of ephrinB1 with either Smurf1 or Smurf2 E3 ubiquitin ligases allows regulation of ephrinB1 levels and, consequently, its signaling through Eph receptors to control tissue separation (Rohani et al. 2011). Understanding how the ephrin/Smurf interactions at tissue boundaries are coordinated to control cell–cell adhesion and repusion is likely to have implications in morphogenesis as well as metastatic disease.
Materials and methods
Plasmids and reagents
cDNA clones encoding full-length Xenopus Smurf1 and Smurf2 were obtained from OPEN Biosystems (Clone ID 5084177 and 7976230; GenBank accession BC073111.1 and BC110749.1), and N-terminal HA-tagged Smurf1 was a gift from Dr. Gerald Thomsen (State University of New York at Stony Brook, NY). The ephrinB1 MO was described previously (Moore et al. 2004), as was the Smurf1 MO (Alexandrova and Thomsen 2006). The Smurf2 MO was 25 nucleotides long with the base composition 5′-GGATCCCTGATTAGACATGGCACCG-3′ (Gene Tools) (see also the Supplemental Material). Various HA-tagged, Polyoma-tagged, and Flag-tagged wild-type and deletion mutants of ephrinB1 (Δ6, Δ16, Δ19, Δ60, and T/C) and HA-tagged Smurf2 (ΔC2, ΔHECT, HECT, and WW) were generated by PCR and subcloned into pCS2+, while the ligase-deficient version of Smurf2 (M) was generated by substituting an alanine in place of a cysteine at amino acid position 719, also by PCR. The Smurf2 9MT (MO-resistant construct) was created by site-directed mutagenesis using the Quik-Change method (Agilent Technologies) in the wobble positions of codons 2–6. H2B-mRFP (Kieserman et al. 2008), H10GFP-RBD (Goulimari et al. 2005), and N19RhoA (Wunnenberg-Stapleton et al. 1999) were provided as gifts and originally constructed as reported previously.
Cryosections
Cryosections of IMZ/BCR hybrid explants were generated. Explants were fixed in MEMFA (0.1 M MOPS at pH 7.4, 2 mM EGTA,1 mM MgSO4, 3.7% formaldehyde) containing 0.25% glutaraldehyde for 30 min at room temperature, then washed in PBS three times. The embryos were soaked in 15% fish gelatin until they submerged (4 h), then in 25% fish gelatin overnight. Explants were frozen on dry ice in 25% fish gelatin blocks and kept frozen at −80°C. Ten-micrometer sections were cut at −29°C (metal body) and −19°C (air) and collected on precoated glass slides (Fisher Scientific). Slides were dried in a fume hood for 1 h and refixed with acetone for 2 min followed by complete evaporation and three PBS washes, then mounted with DAPI-containing solution (H-1500, Vector Laboratories).
Embryo and oocyte injections
Xenopus embryos and oocytes were obtained by standard methods (Moore et al. 2004). For injections, synthetic capped mRNAs were made using the SP6 mMessage mMachine kit (Ambion) and microinjected into embryos or oocytes as previously described (Chong et al. 2000). Western analysis of embryonic (or oocyte) lysates used the same number of embryos (or oocytes) in all samples, and one and a half embryo (or oocyte) equivalents of total protein were loaded. For rescue of MO effects in embryos, MO-resistant mRNAs encoding ephrinB1ΔUTR and Smurf1ΔUTR, which lack the 5′ untranslated region (UTR), were synthesized. For the tissue separation assay, MOs and/or mRNAs were microinjected into embryos along with fluorescent dextran (25 ng; D22910, Invitrogen).
Tissue separation assay
The assay was performed largely as described (Wacker et al. 2000), where intermediate mesoderm was precisely dissected from the dorsal lip region using an eyebrow knife. These mesoderm explants were derived from uninjected control or MO- or RNA-injected stage 11 embryos. The tissue was placed on ectodermal explants (animal caps) from uninjected embryos and incubated in 1× Steinberg solution for 45 min, followed by fixation. For each experiment, 10–30 explants were tested for a single group. Each experiment was scored for the percentage of explants that were separated or integrated into the BCR substrate (as judged by cross-sectional analysis and visual observation). G3 and G4 categories were considered “separated” due to the observed protrusion above the substrate under light microscopy, while G1 and G2 were considered integrated (not separated), since they do not protrude above the substrate. Each experiment was scored for the percentage of explants that were separated from the BCR substrate. A total of three to four repetitions of an individual experiment are represented as a bar graph with the standard deviation (error bar) indicated. Additional statistical analysis was performed (see the statistics section of the Materials and Methods; Supplemental Fig. S4).
Immunoprecipitation and Western blot analysis
HT29 cells, oocytes, embryos, or ectodermal explants were prepared with ice-cold lysis buffer as previously described (Rohani et al. 2011). Immunoprecipitations were performed for 1 h on HT29 cell extracts or 15 oocyte (embryo) equivalents with antibodies raised against HA (Applied Biological Materials), Flag (Applied Biological Materials), ephrinB1 (C-18, Santa Cruz Biotechnology) and Smurf1 (F-20, Santa Cruz Biotechnology), or Smurf2 (P-18, Santa Cruz Biotechnology) and protein-A/G agarose (Santa Cruz Biotechnology). Washes and immunoblots were performed as previously described (Chong et al. 2000) using HRP-conjugated anti-Flag (Sigma), HRP-conjugated anti-HA (Roche), anti-ephrinB1 (extracellular domain; AF-473, R&D Systems), or anti-His (SC-803 or SC-803G, Santa Cruz Biotechnology) antibodies. Lysates of dorsal axial explants were immunoblotted with anti-ERK2 (C-14, Santa Cruz Biotechnology) antibodies for loading normalization. Note: Stage V/VI oocytes display substantially less degradation of ephrinB1 than embryos, and thus oocytes were used for several protein–protein interaction experiments where stable amounts of ephrinB1 were required.
Immunofluorescence
Xenopus embryos were collected at stage 11, explants were prepared as previously described (Rohani et al. 2011), and immunofluorescence was visualized as previously described (Berger et al. 2009) using a Zeiss LSM510 confocal microscope with a Plan-Neofluar 40× oil objective for active RhoA detection. The GFP-RBD fusion protein was prepared as described previously (Goulimari et al. 2005). The GFP-RBD signal intensity was measured for the injected and uninjected regions within a sample using ImageJ (Smurf1 MO: N = 32 explants/1093 injected cells; Smurf2 MO: N = 11 explants/275 injected cells; ephrinB1 MO: N = 7/ explants/161 injected cells; ephrinB1 MO plus ephrinB1-MO-resistant RNA: N = 13 explants/579 injected cells; RhoA N19 RNA: N = 5 explants/198 injected cells), and the signal intensity of the injected region was reported as a percentage of the uninjected region. For the BCR assay, we used a Zeiss LSM510 confocal microscope with a Plan-Neofluar 20× objective for visualization.
Statistics
The degree to which integration or separation occurs is scored with a four-level ordered categorical variable: 1 and 2 integrated, and 3 and 4 separated. A cumulative logit model is appropriate for analyzing ordinal categorical responses. The generalized estimating equations (GEE) method was used to account for the correlation of multiple measurements made on tissue taken from a single group of embryos. Pairwise treatment comparisons of the degree of separation were summarized by odds ratios, multiplicative constants by which the odds of high separation in one treatment group differs from the odds of high separation in another treatment group. The parameters of the cumulative logit models were estimated using the SAS GENMOD procedure (SAS version 9.1; SAS Institute, Inc.). The consistency of the separation score over the different groups of embryos was measured using Piccarreta's τ, a nominal-ordinal measure of association (Berry et al. 2009).
In vivo ubiquitination assay
RNAs encoding Flag-tagged Smurfs (Smurf2, 10 ng per oocyte; Smurf1, 2, 4, 8 ng per oocyte) and HA-Ub (4 ng per oocyte) were injected into oocytes followed by overnight culture at 16°C in oocyte medium. The following day, oocytes were injected with 10 nL of MG132 and dynasore (both 100 μM solution in water containing 1% DMSO) and cultured in oocyte medium containing 10 μM each inhibitor and also injected with V5-tagged ephrinB1 RNA (2 ng per oocyte). Oocytes were harvested after 8 h and lysed in TNSG buffer (20 mM Tris at pH 8.0, 137 mM NaCl, 10% glycerol, 1% NP-40) containing protease inhibitor cocktail (Roche Diagnostics), sodium vanadate (100 mM), PMSF (100 mM), β-glycerol phosphate and ubiquitin-aldehyde (5 μM; Boston Biochem). Samples were incubated with anti-V5 (Invitrogen) for 1 h at 4°C and after addition of protein A/G beads for 4 h at 4°C. Immune complexes were collected and washed three times in TNSG before SDS-PAGE and Western blotting.
Acknowledgments
We thank Dr. Robert Grosse for H10GFP-RBD and the H10GFP-spacer-MCS construct, Dr. Ken W. Cho for the Rho N19 construct, Dr. Gerald Thomsen for the HA-Smurf1 construct, and Dr. John Wallingford for the H2B-mRFP construct. We also thank Dr. Robert Stephens for generating the Xenopus database, Dr. Ming Zhou and Dr. Tim Veenstra for mass spectrometric analysis, Dr. Robert Leighty for statistical analysis, and the Cancer Genetics and Signaling Group for helpful suggestions and discussions. Also, we thank Dr. Renping Zhou, Dr. Jairaj Acharya, and Dr. Shyam Sharan for critical reading of this manuscript. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, and a grant of the Korean Health Technology R&D Project, Ministry for Health, Welfare, and Family Affairs, Republic of Korea (A100335). H.L., Y.H., and I.D. designed the experiments, analyzed the data, and wrote the manuscript. H.L., Y.H., K.M., H.C., E.W., and T.K. performed the experiments. A.S. and Y.J. assisted in the design of the experiments.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.208355.112.
References
- Alexandrova EM, Thomsen GH 2006. Smurf1 regulates neural patterning and folding in Xenopus embryos by antagonizing the BMP/Smad1 pathway. Dev Biol 299: 398–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger CD, Marz M, Kitzing TM, Grosse R, Steinbeisser H 2009. Detection of activated Rho in fixed Xenopus tissue. Dev Dyn 238: 1407–1411 [DOI] [PubMed] [Google Scholar]
- Berry KJ, Johnston JE, Mielke PW Jr 2009. Nominal-ordinal measures of association: A comparison of two measures. Percept Mot Skills 109: 285–294 [DOI] [PubMed] [Google Scholar]
- Bonni S, Wang HR, Causing CG, Kavsak P, Stroschein SL, Luo K, Wrana JL 2001. TGF-β induces assembly of a Smad2–Smurf2 ubiquitin ligase complex that targets SnoN for degradation. Nat Cell Biol 3: 587–595 [DOI] [PubMed] [Google Scholar]
- Bush JO, Soriano P 2012. Eph/ephrin signaling: Genetic, phosphoproteomic, and transcriptomic approaches. Semin Cell Dev Biol 23: 26–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Zhang L 2012. A Smurf1 tale: Function and regulation of an ubiquitin ligase in multiple cellular networks. Cell Mol Life Sci doi: 10.1007/s00018-012-1170-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng PL, Lu H, Shelly M, Gao H, Poo MM 2011. Phosphorylation of E3 ligase Smurf1 switches its substrate preference in support of axon development. Neuron 69: 231–243 [DOI] [PubMed] [Google Scholar]
- Chong LD, Park EK, Latimer E, Friesel R, Daar IO 2000. Fibroblast growth factor receptor-mediated rescue of x-ephrin B1-induced cell dissociation in Xenopus embryos. Mol Cell Biol 20: 724–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong PA, Lin H, Wrana JL, Forman-Kay JD 2010. Coupling of tandem Smad ubiquitination regulatory factor (Smurf) WW domains modulates target specificity. Proc Natl Acad Sci 107: 18404–18409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daar IO 2012. Non-SH2/PDZ reverse signaling by ephrins. Semin Cell Dev Biol 23: 65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Chang C 2012. Regulation of early Xenopus embryogenesis by Smad ubiquitination regulatory factor 2. Dev Dyn 241: 1260–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebisawa T, Fukuchi M, Murakami G, Chiba T, Tanaka K, Imamura T, Miyazono K 2001. Smurf1 interacts with transforming growth factor-β type I receptor through Smad7 and induces receptor degradation. J Biol Chem 276: 12477–12480 [DOI] [PubMed] [Google Scholar]
- Fukunaga E, Inoue Y, Komiya S, Horiguchi K, Goto K, Saitoh M, Miyazawa K, Koinuma D, Hanyu A, Imamura T 2008. Smurf2 induces ubiquitin-dependent degradation of Smurf1 to prevent migration of breast cancer cells. J Biol Chem 283: 35660–35667 [DOI] [PubMed] [Google Scholar]
- Gorny A-K, Steinbeisser H 2012. Brachet's cleft: A model for the analysis of tissue separation in Xenopus. Wiley Interdiscip Rev Dev Biol 1: 294–300 [DOI] [PubMed] [Google Scholar]
- Goulimari P, Kitzing TM, Knieling H, Brandt DT, Offermanns S, Grosse R 2005. Gα12/13 is essential for directed cell migration and localized Rho-Dia1 function. J Biol Chem 280: 42242–42251 [DOI] [PubMed] [Google Scholar]
- Huang C 2010. Roles of E3 ubiquitin ligases in cell adhesion and migration. Cell Adhes Migr 4: 10–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Rajfur Z, Yousefi N, Chen Z, Jacobson K, Ginsberg MH 2009. Talin phosphorylation by Cdk5 regulates Smurf1-mediated talin head ubiquitylation and cell migration. Nat Cell Biol 11: 624–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzi L, Attisano L 2004. Regulation of the TGFβ signalling pathway by ubiquitin-mediated degradation. Oncogene 23: 2071–2078 [DOI] [PubMed] [Google Scholar]
- Jin C, Yang YA, Anver MR, Morris N, Wang X, Zhang YE 2009. Smad ubiquitination regulatory factor 2 promotes metastasis of breast cancer cells by enhancing migration and invasiveness. Cancer Res 69: 735–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan M, Lee SJ, Schwedhelm-Domeyer N, Stegmuller J 2012. The E3 ligase Cdh1-anaphase promoting complex operates upstream of the E3 ligase Smurf1 in the control of axon growth. Development 139: 3600–3612 [DOI] [PubMed] [Google Scholar]
- Kavsak P, Rasmussen RK, Causing CG, Bonni S, Zhu H, Thomsen GH, Wrana JL 2000. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF β receptor for degradation. Mol Cell 6: 1365–1375 [DOI] [PubMed] [Google Scholar]
- Kieserman EK, Glotzer M, Wallingford JB 2008. Developmental regulation of central spindle assembly and cytokinesis during vertebrate embryogenesis. Curr Biol 18: 116–123 [DOI] [PubMed] [Google Scholar]
- Koster I, Jungwirth MS, Steinbeisser H 2010. xGit2 and xRhoGAP 11A regulate convergent extension and tissue separation in Xenopus gastrulation. Dev Biol 344: 26–35 [DOI] [PubMed] [Google Scholar]
- Lee HS, Bong YS, Moore KB, Soria K, Moody SA, Daar IO 2006. Dishevelled mediates ephrinB1 signalling in the eye field through the planar cell polarity pathway. Nat Cell Biol 8: 55–63 [DOI] [PubMed] [Google Scholar]
- Lee HS, Nishanian TG, Mood K, Bong YS, Daar IO 2008. EphrinB1 controls cell–cell junctions through the Par polarity complex. Nat Cell Biol 10: 979–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Mood K, Battu G, Ji YJ, Singh A, Daar IO 2009. Fibroblast growth factor receptor-induced phosphorylation of ephrinB1 modulates its interaction with Dishevelled. Mol Biol Cell 20: 124–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Park JS, Kim JH, Jung SM, Lee JY, Kim SJ, Park SH 2011. Smad6-specific recruitment of Smurf E3 ligases mediates TGF-β1-induced degradation of MyD88 in TLR4 signalling. Nat Commun 2: 460. [DOI] [PubMed] [Google Scholar]
- Lin X, Liang M, Feng XH 2000. Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-β signaling. J Biol Chem 275: 36818–36822 [DOI] [PubMed] [Google Scholar]
- Lu K, Li P, Zhang M, Xing G, Li X, Zhou W, Bartlam M, Zhang L, Rao Z, He F 2011. Pivotal role of the C2 domain of the Smurf1 ubiquitin ligase in substrate selection. J Biol Chem 286: 16861–16870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina A, Swain RK, Kuerner KM, Steinbeisser H 2004. Xenopus paraxial protocadherin has signaling functions and is involved in tissue separation. EMBO J 23: 3249–3258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore KB, Mood K, Daar IO, Moody SA 2004. Morphogenetic movements underlying eye field formation require interactions between the FGF and ephrinB1 signaling pathways. Dev Cell 6: 55–67 [DOI] [PubMed] [Google Scholar]
- Narimatsu M, Bose R, Pye M, Zhang L, Miller B, Ching P, Sakuma R, Luga V, Roncari L, Attisano L, et al. 2009. Regulation of planar cell polarity by Smurf ubiquitin ligases. Cell 137: 295–307 [DOI] [PubMed] [Google Scholar]
- Nowotschin S, Hadjantonakis AK 2010. Cellular dynamics in the early mouse embryo: From axis formation to gastrulation. Curr Opin Genet Dev 20: 420–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogunjimi AA, Wiesner S, Briant DJ, Varelas X, Sicheri F, Forman-Kay J, Wrana JL 2010. The ubiquitin binding region of the Smurf HECT domain facilitates polyubiquitylation and binding of ubiquitylated substrates. J Biol Chem 285: 6308–6315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmundson EC, Ray D, Moore FE, Gao Q, Thomsen GH, Kiyokawa H 2008. The HECT E3 ligase Smurf2 is required for Mad2-dependent spindle assembly checkpoint. J Cell Biol 183: 267–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdamar B, Bose R, Barrios-Rodiles M, Wang HR, Zhang Y, Wrana JL 2005. Regulation of the polarity protein Par6 by TGFβ receptors controls epithelial cell plasticity. Science 307: 1603–1609 [DOI] [PubMed] [Google Scholar]
- Pasquale EB 2005. Eph receptor signalling casts a wide net on cell behaviour. Nat Rev Mol Cell Biol 6: 462–475 [DOI] [PubMed] [Google Scholar]
- Pasquale EB 2008. Eph-ephrin bidirectional signaling in physiology and disease. Cell 133: 38–52 [DOI] [PubMed] [Google Scholar]
- Rohani N, Canty L, Luu O, Fagotto F, Winklbauer R 2011. EphrinB/EphB signaling controls embryonic germ layer separation by contact-induced cell detachment. PLoS Biol 9: e1000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotin D, Kumar S 2009. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol 10: 398–409 [DOI] [PubMed] [Google Scholar]
- Sanchez NS, Barnett JV 2012. TGFβ and BMP-2 regulate epicardial cell invasion via TGFβR3 activation of the Par6/Smurf1/RhoA pathway. Cell Signal 24: 539–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwamborn JC, Muller M, Becker AH, Puschel AW 2007. Ubiquitination of the GTPase Rap1B by the ubiquitin ligase Smurf2 is required for the establishment of neuronal polarity. EMBO J 26: 1410–1422 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tanaka M, Kamo T, Ota S, Sugimura H 2003. Association of Dishevelled with Eph tyrosine kinase receptor and ephrin mediates cell repulsion. EMBO J 22: 847–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang LY, Yamashita M, Coussens NP, Tang Y, Wang X, Li C, Deng CX, Cheng SY, Zhang YE 2011. Ablation of Smurf2 reveals an inhibition in TGF-β signalling through multiple mono-ubiquitination of Smad3. EMBO J 30: 4777–4789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Bai C, Lin Q, Lin H, Liu M, Ding F, Wang HR 2011. Binding of RhoA by the C2 domain of E3 ligase Smurf1 is essential for Smurf1-regulated RhoA ubiquitination and cell protrusive activity. FEBS Lett 585: 2199–2204 [DOI] [PubMed] [Google Scholar]
- Unterseher F, Hefele JA, Giehl K, De Robertis EM, Wedlich D, Schambony A 2004. Paraxial protocadherin coordinates cell polarity during convergent extension via RhoA and JNK. EMBO J 23: 3259–3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker S, Grimm K, Joos T, Winklbauer R 2000. Development and control of tissue separation at gastrulation in Xenopus. Dev Biol 224: 428–439 [DOI] [PubMed] [Google Scholar]
- Wan L, Zou W, Gao D, Inuzuka H, Fukushima H, Berg AH, Drapp R, Shaik S, Hu D, Lester C, et al. 2011. Cdh1 regulates osteoblast function through an APC/C-independent modulation of Smurf1. Mol Cell 44: 721–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BC 2008. PTPases: ‘Eph’ ective arbitrators of attraction. Blood 112: 455–456 [DOI] [PubMed] [Google Scholar]
- Wang HR, Zhang Y, Ozdamar B, Ogunjimi AA, Alexandrova E, Thomsen GH, Wrana JL 2003. Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science 302: 1775–1779 [DOI] [PubMed] [Google Scholar]
- Wang HR, Ogunjimi AA, Zhang Y, Ozdamar B, Bose R, Wrana JL 2006. Degradation of RhoA by Smurf1 ubiquitin ligase. Methods Enzymol 406: 437–447 [DOI] [PubMed] [Google Scholar]
- Wiesner S, Ogunjimi AA, Wang HR, Rotin D, Sicheri F, Wrana JL, Forman-Kay JD 2007. Autoinhibition of the HECT-type ubiquitin ligase Smurf2 through its C2 domain. Cell 130: 651–662 [DOI] [PubMed] [Google Scholar]
- Wunnenberg-Stapleton K, Blitz IL, Hashimoto C, Cho KW 1999. Involvement of the small GTPases XRhoA and XRnd1 in cell adhesion and head formation in early Xenopus development. Development 126: 5339–5351 [DOI] [PubMed] [Google Scholar]
- Yamashita M, Ying SX, Zhang GM, Li C, Cheng SY, Deng CX, Zhang YE 2005. Ubiquitin ligase Smurf1 controls osteoblast activity and bone homeostasis by targeting MEKK2 for degradation. Cell 121: 101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Chen SP, Zhang XP, Wang H, Zhu C, Lin HY 2009. Smurf2 participates in human trophoblast cell invasion by inhibiting TGF-β type I receptor. J Histochem Cytochem 57: 605–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Kavsak P, Abdollah S, Wrana JL, Thomsen GH 1999. A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature 400: 687–693 [DOI] [PubMed] [Google Scholar]