Previously, Soucek et al. showed the potential of Myc as a therapeutic cancer target. In this study, they show that repeated rounds of Myc inhibition in mice over the course of 1 year with the transcriptional inhibitor Omomyc reversibly blocks expression of all three Myc genes in lung tumors and produces regression of Kras-driven lung tumors. They provide further evidence that Myc inhibition, rather than the targeting of oncogene drivers, could be an effective treatment in human cancers.
Keywords: Myc, cancer therapy, lung cancer, mouse model
Abstract
The principal reason for failure of targeted cancer therapies is the emergence of resistant clones that regenerate the tumor. Therapeutic efficacy therefore depends on not only how effectively a drug inhibits its target, but also the innate or adaptive functional redundancy of that target and its attendant pathway. In this regard, the Myc transcription factors are intriguing therapeutic targets because they serve the unique and irreplaceable role of coordinating expression of the many diverse genes that, together, are required for somatic cell proliferation. Furthermore, Myc expression is deregulated in most—perhaps all—cancers, underscoring its irreplaceable role in proliferation. We previously showed in a preclinical mouse model of non-small-cell lung cancer that systemic Myc inhibition using the dominant-negative Myc mutant Omomyc exerts a dramatic therapeutic impact, triggering rapid regression of tumors with only mild and fully reversible side effects. Using protracted episodic expression of Omomyc, we now demonstrate that metronomic Myc inhibition not only contains Ras-driven lung tumors indefinitely, but also leads to their progressive eradication. Hence, Myc does indeed serve a unique and nondegenerate role in lung tumor maintenance that cannot be complemented by any adaptive mechanism, even in the most aggressive p53-deficient tumors. These data endorse Myc as a compelling cancer drug target.
The ideal cancer drug should target a nonredundant function that is continuously and obligatorily necessary for tumor growth and maintenance throughout all stages of tumor evolution yet dispensable for maintenance and function of normal tissues. To date, the prevailing logic driving the quest for cancer therapy targets has been to identify those gene products that are repeatedly mutated in any cancer type on the basis that such recurring mutations play some inimitable and obligate role in driving the disease. However, two inherent problems confound this otherwise straightforward strategy. First, most solid human cancers pass through episodes of genomic instability, generating extensive mutational noise that can obscure “driver” mutations and their attendant effector pathways. Second, most adult cancers are products of tortuous evolutionary trajectories punctuated by critical bottlenecks through which the proto-neoplasm must pass. Transition through such bottlenecks is likely dependent on specific mutations that, while potentially recurring features of that specific tumor type, may nonetheless be dispensable thereafter and hence not useful as a therapeutic target beyond that point in the tumor evolution.
One possible solution to this conundrum is suggested by the fact that, while a wide range of genes is mutated among the gamut of human cancers, in the main, these mutations serve to deregulate a relatively small number of underlying pathways, each of which channels through a limited cohort of functionally nonredundant, central transducers such as Ras, Myc, and E2F. To this end, we focused our attention on Myc, three broadly isofunctional transcription factors that integrate diverse upstream growth signals and coordinately relay them to the thousands of disparate target genes that, together, underpin somatic cell proliferation (Larsson and Henriksson 2010). Aberrantly high and/or deregulated Myc activity is causally implicated in most cancers and is often associated with aggressive, poorly differentiated, and angiogenic tumors (http://www.myccancergene.org). Interestingly, however, direct mutational activation of Myc genes is relatively uncommon in tumors, at least in their early stages; rather, the aberrantly high levels of expression of Myc are frequently a consequence of its relentless induction by upstream oncogenic signals (Larsson and Henriksson 2010).
Renewed interest in Myc as a therapeutic target has been fostered by recent switchable genetic studies showing that transient systemic inhibition of Myc by the dominant-negative Omomyc has a profound therapeutic effect on diverse tumor types yet elicits surprisingly mild and rapidly reversible side effects on normal tissues (Soucek et al. 2008; Sodir et al. 2011). These startling conclusions have now been endorsed by more recent studies using pharmacological strategies to block Myc expression in the treatment of multiple myeloma (Delmore et al. 2011).
Omomyc is a dominant-negative Myc dimerization domain mutant comprising the basic helix–loop–helix zipper (HLHZip) domain of Myc with four amino acid substitutions in the Myc leucine zipper that confer altered dimerization specificity (Soucek et al. 1998, 2002). Omomyc retains the ability to bind its physiological partner, Max, but also heterodimerizes with wild-type c-Myc, N-Myc, and L-Myc proteins. Since Myc:Omomyc heterodimers can no longer bind to the canonical Myc E-box CACGTG DNA recognition element, Omomyc overexpression inhibits Myc-dependent target gene transactivation (Soucek et al. 2002, 2004; Savino et al. 2011). Omomyc is therefore a competitive inhibitor of Myc transcriptional activation, and we used it to model pharmacological inhibition of Myc by generating mice (TREOmomyc) in which Omomyc expression is driven from a tetracycline-responsive promoter element. When crossed with mice ubiquitously expressing the tetracycline-dependent rtTA transactivator (CMVrtTA), Omomyc expression can be systemically but reversibly induced by administration of doxycycline to the animals (Soucek et al. 2008). Such TREOmomyc;CMVrtTA mice were crossed with the well-established LSL-KrasG12D murine model of lung tumorigenesis (Jackson et al. 2001; Tuveson et al. 2004; Kim et al. 2005), in which inhalation of adenovirus-Cre recombinase triggers sporadic expression of endogenous oncogenic KRasG12D in lung epithelium. This precipitates initiation and evolution of multiple, discrete lung tumors. Focal hyperplasias are evident ∼4 wk post-infection, adenomas by 6–8 wk post-infection, and at ∼18 wk post-infection, adenocarcinomas (Jackson et al. 2001; Tuveson et al. 2004) that closely resemble human non-small-cell lung adenocarcinoma (Sweet-Cordero et al. 2005).
We previously showed that systemic induction of Omomyc triggers rapid and dramatic regression of KRasG12D-driven lung tumors within 3 d, and, by 1 wk of sustained Myc inhibition, all animals are free of overt tumors (Soucek et al. 2008). Concurrently, the impact of systemic Myc inhibition on normal tissues was very mild: Proliferation rates were substantially reduced in regenerative tissues such as skin, testis, and intestine, and although this was accompanied by tissue atrophy, mice exhibited no evidence of distress or discomfort and maintained their weights and activity. Moreover, these side effects of Myc inhibition were completely and rapidly reversed upon restoration of Myc function. Hence, Myc inhibition shows remarkable selectivity in killing neoplastic, but not normal, tissues (Soucek et al. 2008). The great potential of Myc as a therapeutic cancer target has recently been further validated through an innovative pharmacological approach in which Myc is indirectly down-regulated, triggering regression of multiple tumor types (Delmore et al. 2011; Zuber et al. 2011). Targeting Myc is now becoming a reality, but three important unanswered questions remain that we now address. Does Myc inhibition eradicate the disease and extend overall survival? Can tumors develop resistance to Myc inhibition by bypassing the requirement for Myc in tumor maintenance? Can ancillary oncogenic mutations, most notably loss of p53, mitigate the therapeutic impact of Myc inhibition?
Results
Myc inhibition dramatically extends overall survival of lung tumor-bearing mice
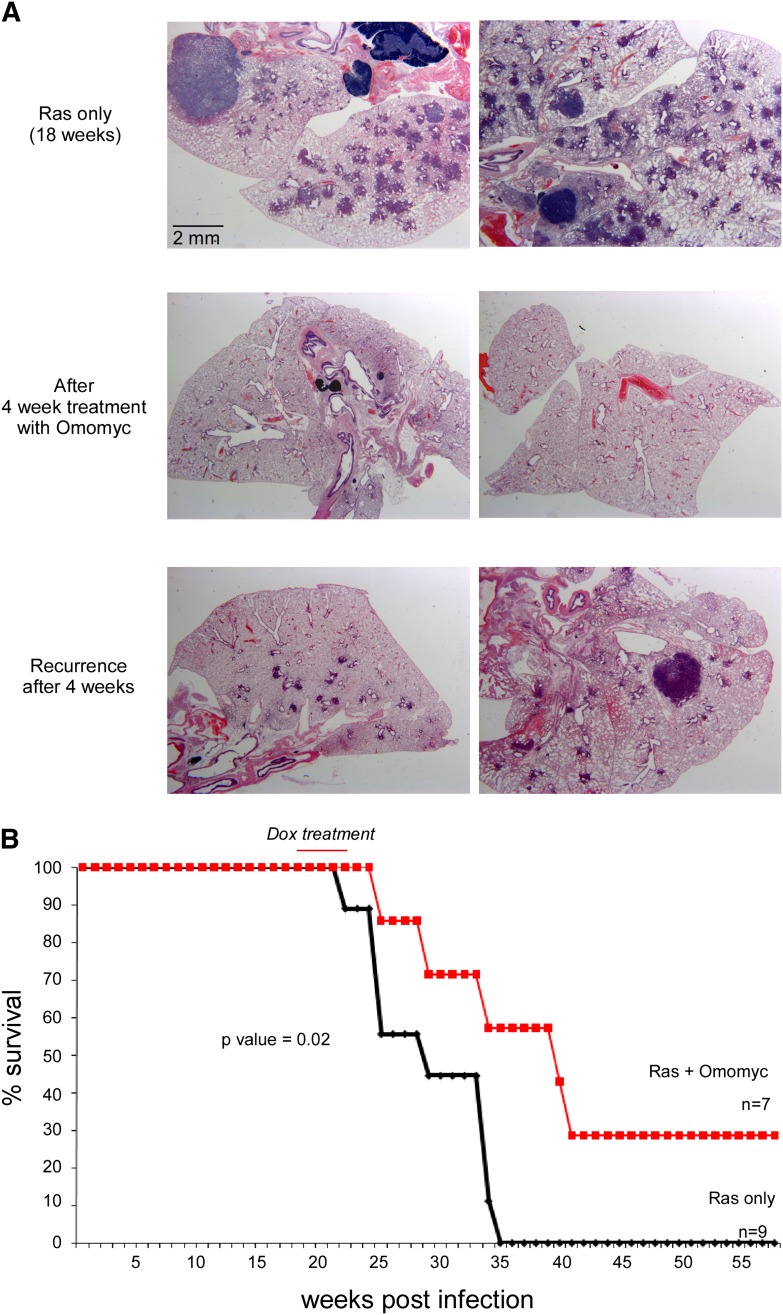
Our previous studies had demonstrated that Myc is required for the maintenance of KRas-driven lung tumors, and its inhibition triggers rapid regression of all tumors, irrespective of stage. To investigate the kinetics and frequency of relapse after surcease of Myc inhibition, lung tumors were induced in TREOmomyc;CMVrtTA;LSL-KrasG12D mice through inhalation of Ad-Cre to activate KRasG12D focally in lung epithelium. At the adenovirus multiplicity of infection (MOI) used, the median number of sporadic tumors generated in each lung section was ∼70, and these were allowed to evolve for 18 wk (Fig. 2B, below). Omomyc was then systemically induced, ostensibly triggering complete regression of all tumors within 7–8 d (Soucek et al. 2008), and then maintained for a total of 4 wk (Fig. 1A; Soucek et al. 2008). Omomyc was then deinduced, and tumor regrowth was monitored. After 4 wk, multiple lesions had reappeared in each lung (Fig. 1A). Histologically, these tumors resemble the more advanced tumors (adenomas and adenocarcinomas) present just prior to Omomyc induction (Supplemental Fig. 1), suggesting that relapse is a consequence of regrowth of the original tumors rather than de novo lesions. Of note, the number of relapsing tumors was, on average, ∼37% lower than the original number prior to Omomyc induction (see Fig. 2B). Even though transient Myc inhibition did not immediately eradicate all lung tumors, the combined impact of initial tumor regression and reduction in tumor multiplicity upon relapse significantly extended overall survival (Fig. 1B).
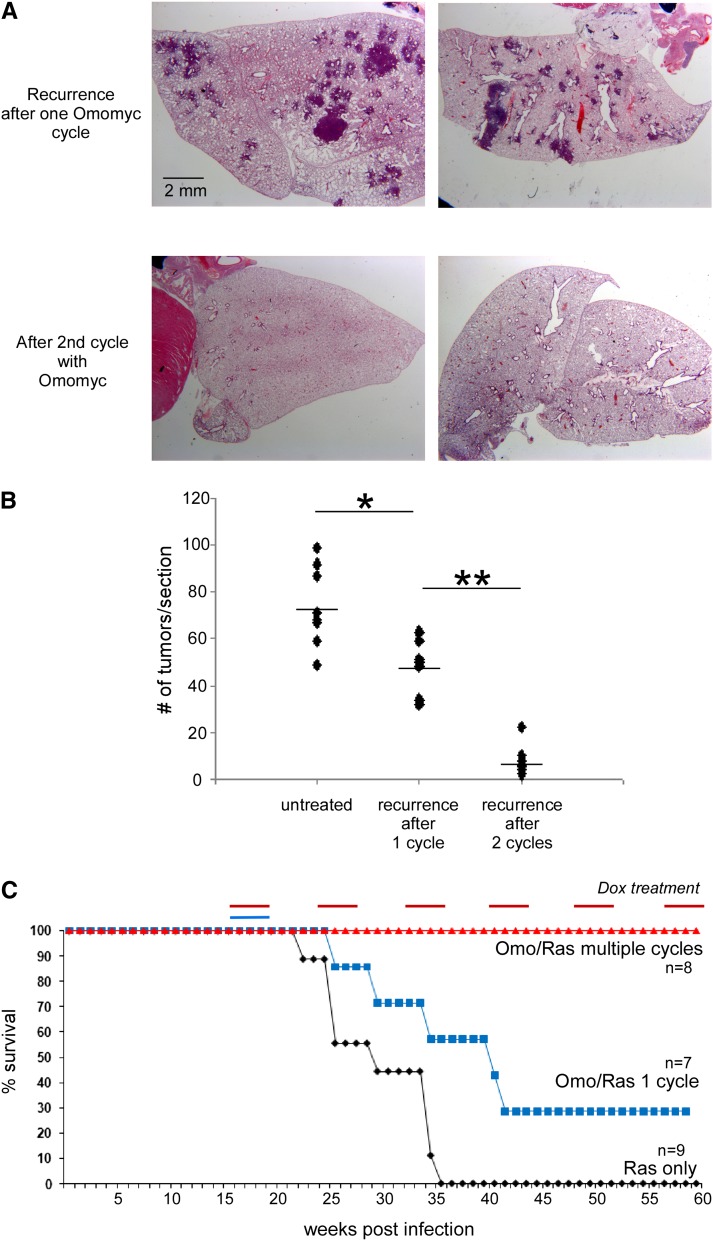
Figure 2.
Recurring tumors remain sensitive to subsequent Myc inhibition, while metronomic Myc inhibition confers indefinite survival. (A) H&E staining of lung tissue samples derived from TREOmomyc;CMVrtTA;LSL-KRasG12D animals after recurrence of the tumors upon discontinuation of Omomyc expression (top panels) and after a second 4-wk round of Omomyc induction (middle panels). (B) Quantification of recurring tumors per tissue section in animals subjected to either one or two 4-wk cycles of Omomyc expression compared with the initial numbers of tumors in untreated animals. The differences indicated by (*) and (**) are both statistically significant (P-value = 0.002 and 0.001, respectively). Each dot corresponds to a tissue section from an independent animal. (C) Survival curve of control TREOmomyc;CMVrtTA;LSL-KRasG12D mice, animals subjected to a single 4-wk period of Omomyc expression, and animals subjected to repeated metronomic Omomyc expression for 4 wk out of every 8 wk (red line). The survival curves of TREOmomyc;CMVrtTA;LSL-KRasG12D subjected to a single 4-wk period of Omomyc expression (blue line) versus no-Omomyc controls (black line) as shown in Figure 1B are repeated here to facilitate comparison.
Figure 1.
Reduced recurrence of tumors and increased survival advantage following one cycle of Myc inhibition. (A) Hematoxylin and eosin (H&E) staining of lung tissue samples derived from TREOmomyc;CMVrtTA;LSL-KRasG12D animals 18 wk after Ad-Cre infection (top panels), after 4 wk induction of Omomyc (middle panels), and 4 wk after surcease of Omomyc expression (bottom panels). (B) Survival curves of TREOmomyc;CMVrtTA;LSLKRasG12D subjected to a single 4-wk period of Omomyc expression (indicated by the red line) versus no-Omomyc controls.
In most solid cancers, relapse after therapy is characterized by emergence of disease that is resistant to the initial therapy, and this is a principal reason for the intractability of many cancers. To ascertain whether the tumors that regrow after surcease of Myc inhibition have acquired resistance to Omomyc, mice were allowed to relapse for 4 wk after cessation of the initial Myc inhibition, and Omomyc was then reinduced. All recurring tumors in all mice rapidly regressed in response to this second round of Myc inhibition at a rate indistinguishable from that in the first round (Fig. 2A). Importantly, even though each animal harbored multiple independent tumors (∼200 per mouse), we saw no evidence of Omomyc refractory tumors in any of the treated animals (Fig. 2A). Hence, lung tumors appear unable to acquire or evolve resistance to Myc inhibition. Remarkably, the multiplicity of tumor regrowth after this second round of Myc inhibition was again markedly reduced, to only ∼11% the original number prior to any Omomyc induction (Fig. 2B).
Although our data indicate that short-term systemic Myc inhibition elicits only mild and reversible side effects, the long-term side effects are likely to be severe, precluding sustained Myc inhibition as a means for tumor containment. However, the rapidity and repeatable efficacy of short-term Myc inhibition in triggering lung tumor regression offered the possibility that repeated, short-term metronomic Myc inhibition might marry minimal side effects with effective disease containment. To test this, LSL-KrasG12D mice were exposed to inhaled Ad-Cre and, 18 wk later, recruited into a regimen of alternating 4-wk periods of Omomyc induction followed by 4-wk deinduction. Remarkably, all mice maintained on this metronomic Myc inhibition remained healthy, retained body weight, and survived indefinitely (Fig. 2C; Supplemental Fig. 2). To assess the tumor status of animals after long-term metronomic Myc inhibition, a cohort of tumor-bearing TREOmomyc;CMVrtTA;LSL-KrasG12D mice was subjected to a total of eight rounds of metronomic Myc inhibition over an ∼1.2-yr period. At the end of the final 4-wk period of Omomyc induction, some of the mice were immediately killed, and their lungs were inspected for any tumors that had failed to regress upon doxycycline treatment. Only two such tumors were found, and staining of both with Omomyc-specific antibody indicated that Omomyc expression was profoundly suppressed, presumably through loss or repression of the Omomyc transgene (Supplemental Fig. 3). Hence, despite strong selection for mechanisms that thwart sensitivity to Myc inhibition, the only observed mechanism of escape in hundreds of individual tumors was repression of Omomyc expression. The remaining mice (n = 4) in the metronomically treated cohort were left for 4 wk without Omomyc, and tumor regrowth was then assessed. Although the initial Ad-Cre infection reproducibly induces tens of independent tumor lesions in each animal, three of the four mice subjected to eight rounds of metronomic Myc inhibition remained tumor-free, while the fourth presented with a single lesion (the Omomyc transgene status of this exceptional lesion is unknown). Taken together, our data strongly support our original contention that Myc is both essential and functionally nonredundant for tumor maintenance, and its inhibition cannot therefore be easily circumvented by an innate adaptive or evolutionary mechanism.
p53 facilitates, but is not essential for, the therapeutic effect of Myc inhibition
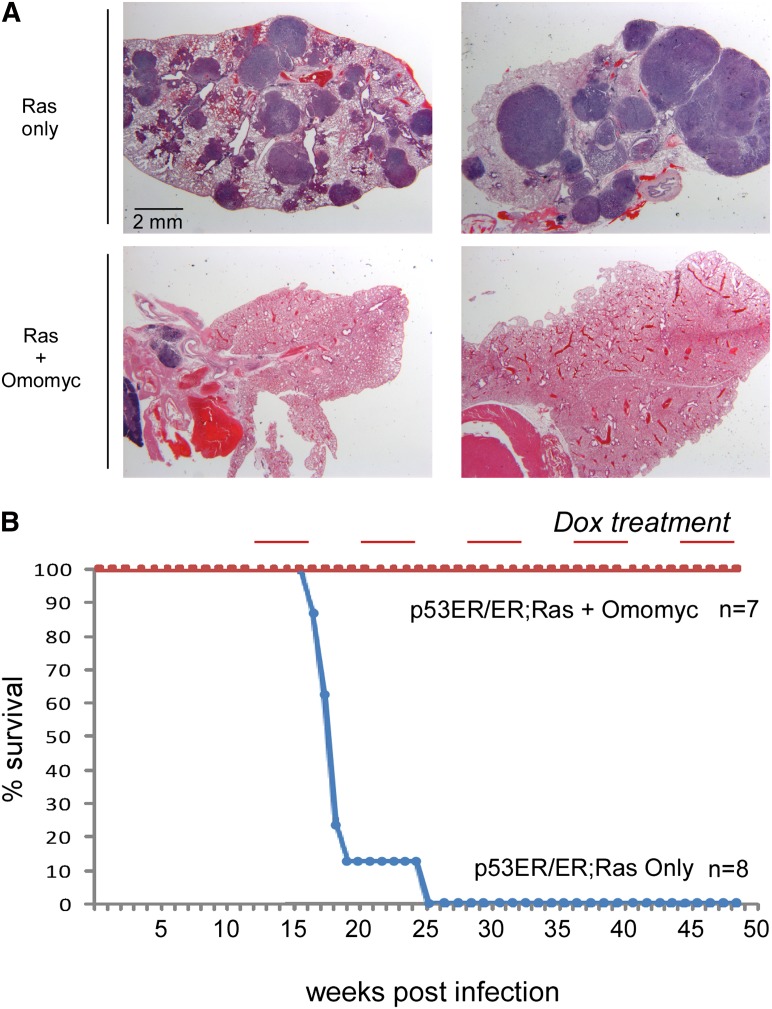
Compared with lung tumors arising in p53 wild-type mice, lung tumors in p53-defective LSL-KrasG12D mice develop with a significantly shorter latency and exhibit accelerated evolution into adenocarcinomas (Jackson et al. 2001) that closely resemble human nonsquamous non-small-cell lung cancer (NSCLC) in their pathology and gene expression (Sweet-Cordero et al. 2005). Moreover, absence of functional p53 also makes the lung tumors arising in such p53-deficient mice very resistant to chemotherapy, again mimicking the therapeutic intractability of their human counterparts (Oliver et al. 2010; Singh et al. 2010). Since Myc inhibition triggered quantitative regression of advanced lung tumors, many of which harbor cells that have sporadically lost p53, it seemed likely that p53 inactivation would not compromise the therapeutic impact of Myc inhibition. Nonetheless, to test this unequivocally, we crossed the TREOmomyc;CMVrtTA;LSL-KrasG12D mice into the p53ER/ER background in which the endogenous p53 gene has been replaced by one encoding p53ERTAM, a 4-hydroxytamoxifen (4OHT)-dependent conditional allele of p53 (Christophorou et al. 2005). In the absence of 4OHT, such TREOmomyc;CMVrtTA;LSL-KrasG12D;p53ER/ER mice are de facto p53-null but can be reversibly toggled to p53 wild-type status by systemic administration of 4OHT (Christophorou et al. 2005; Junttila et al. 2010), allowing for direct comparison of the therapeutic efficacy of Myc inhibition in the presence or absence of functional p53. As expected, KRas-driven lung tumors progress far more quickly in TREOmomyc;CMVrtTA;LSL-KrasG12D;p53ER/ER mice never given 4OHT relative to those in p53wt mice (Jackson et al. 2005; Junttila et al. 2010), exhibiting large, high-grade lesions by 15 wk after KRasG12D activation (Fig. 3A). At that time point, Omomyc was induced, and its effect on the tumors was assessed. Myc inhibition once again elicited profound tumor regression, clearing the lungs of all overt tumors (Fig. 3A) and rapidly ameliorating any respiratory distress the animals might be experiencing (data not shown), albeit with delayed kinetics relative to the regression of lung tumors in p53wt mice: Whereas Myc inhibition clears the tumors from lungs of p53-competent mice within 1 wk, complete tumor clearance took up to 4 wk in p53-negative animals. Of note, clearance of such large tumors was accompanied by substantial inflammatory cell infiltration in the main macrophages (data not shown), presumably reflecting the extensive tissue remodeling such rapid regression elicits. Just as for the tumor-bearing p53wt mice, metronomic Myc inhibition in TREOmomyc;CMVrtTA;LSL-KrasG12D;p53ER/ER animals maintained their disease-free viability indefinitely (Fig. 3B). Occasional tumors resistant to the doxycycline regimen did eventually emerge, at ∼2.5-fold higher frequency than in p53wt animals (overall, 0.5% of all p53-deficient tumors compared with 0.2% in p53 wild-type mice). However, just as with p53-competent tumors, resistance was always associated with repression of Omomyc expression (data not shown).
Figure 3.
Myc inhibition triggers lung tumor regression in p53-deficient mice. (A) H&E staining of lung tissue samples derived from TREOmomyc;CMVrtTA;LSL-KRasG12D;p53ER/ER animals 15 wk after Ad-Cre infection (top panels) and after 4 wk of treatment with Omomyc (bottom panels). (B) Survival curve of control (not doxycycline-treated) TREOmomyc;CMVrtTA;LSL-KRasG12D;p53ER/ER mice versus counterparts subjected to repeated metronomic Omomyc induction as in Figure 2C.
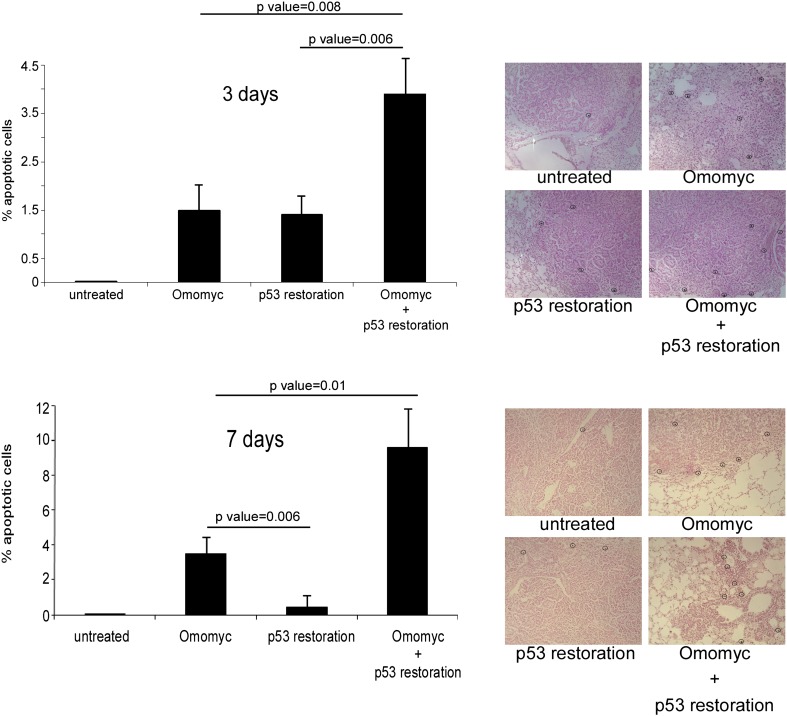
We recently showed that p53 restoration alone has only a limited therapeutic effect in lung tumors of LSL-KrasG12D mice because only the more aggressive, high-grade cells within such tumors harbor the preternaturally elevated oncogenic KRas signal strength necessary to trigger p53-induced apoptosis. Since the less aggressive cells in each tumor are spared, p53 restoration alone cannot eradicate such tumors (Junttila et al. 2010). While our data indicate that Myc inhibition is lethal to lung tumor cells irrespective of p53 status, the greater rapidity of tumor regression in p53-competent animals suggests that p53, when present, senses some aberrant state induced by Myc inhibition and then relays this to downstream cell death machineries. If so, then restoration of p53 in p53-deficient tumors should synergize therapeutically with Myc inhibition. To ascertain directly whether Myc inhibition synergizes therapeutically with p53 restoration in p53-deficient LSL-KrasG12D lung cancer, tumors were initiated in TREOmomyc;CMVrtTA;LSL-KrasG12D;p53ER/ER mice by Ad-Cre inhalation, and, 15 wk later, mice were treated for either 3 or 7 d with doxycycline alone (to inhibit Myc), tamoxifen alone (to restore p53 function), or doxycycline and tamoxifen together. Apoptosis was then quantified histologically (Fig. 4). Three days of sustained p53 restoration alone induced apoptosis in ∼1.5% of tumor cells, but this was restricted only to high-grade regions of tumors, as previously reported (Fig. 4, top graph; Junttila et al. 2010). Three days of Myc inhibition induced a similar level (∼1.5%) of tumor cell apoptosis, but unlike p53 restoration, this was broadly distributed irrespective of tumor grade. When p53 was restored concurrently with Myc inhibition, the two effects were additive (∼3.5% of tumor cells in both low- and high-grade regions). More illuminating was the situation after 7 d (Fig. 4, bottom graph). At this time, Myc inhibition alone is still killing tumor cells (∼3.8% apoptosis), but p53-dependent apoptosis has fallen to baseline (∼0.4%) because all of the high-grade tumor cells sensitive to p53 restoration have been killed. However, the combination of p53 restoration and Myc inhibition still exhibits significant synergy, eliciting widespread and tumor-grade-independent apoptosis (∼9.8% apoptosis). These observations intimate that inhibition of endogenous Myc does indeed elicit a p53-activating signal in KRasG12D-driven lung tumor cells irrespective of tumor grade, and this then engages p53-dependent apoptosis. To test this directly, we assayed both the disposition and extent of p53 activation in lung tumors immunohistochemically using expression of p21cip1 (encoded by the p53 target gene cdkn1a) following p53 restoration alone and in combination with Myc inhibition. As expected, restoration of p53 alone elicited p21cip1 expression only in high-grade tumor cells that harbor sufficiently elevated levels of KRasG12D signaling to engage p19ARF (Fig. 5; as previously described in Junttila et al. 2010). In contrast, Myc inhibition triggered widespread activation of restored p53 throughout all tumors and irrespective of tumor grade (Fig. 5). No p21cip1 was induced in tumors when Myc was inhibited in the absence of p53 function. Our data confirm the notion that Myc inhibition induces a p53-activating signal in lung tumor cells, triggering expeditious tumor cell apoptosis. However, since Myc inhibition also kills in the absence of p53, it must nonetheless elicit a dysfunctional state in tumor cells that is intrinsically lethal.
Figure 4.
Myc inhibition in lung tumors cooperates with p53 in inducing apoptosis. Quantification of TUNEL-positive cells in tumor sections derived from TREOmomyc;CMVrtTA;LSL-KrasG12D;p53ER/ER animals after 3 d (top) or 7 d (bottom) of sustained doxycycline administration (to induce Omomyc), 4OHT (to restore p53), or both. Quantification was made by counting cells in at least six tumors in each of three mice. Error bars indicate SD.
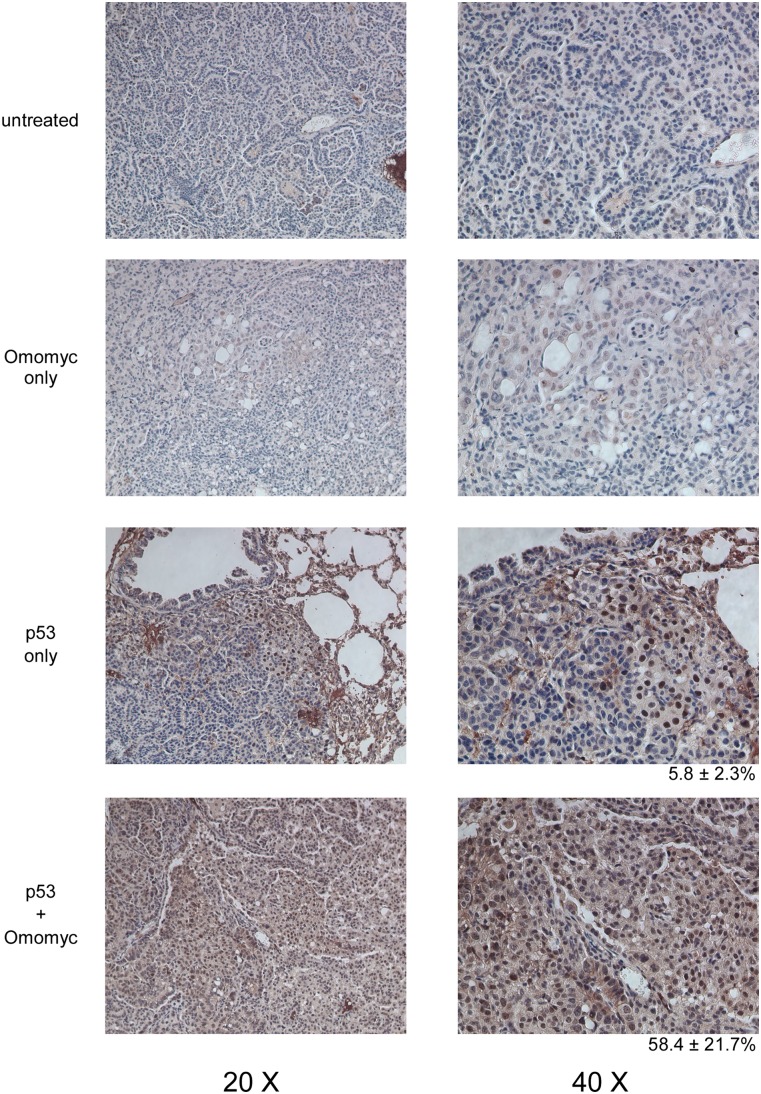
Figure 5.
Myc inhibition induces expression of p21cip1. Immunohistochemical staining for p21cip1 in tissue samples derived from TREOmomyc;CMVrtTA;LSL-KrasG12D;p53ER/ER animals after 7 d of sustained p53 restoration (daily tamoxifen) or sustained Omomyc induction (doxycycline) together with p53 restoration.
Discussion
Evolution of drug resistance is the major factor confounding durable cancer treatment. Hence, the efficacy of a targeted therapeutic depends on not only potent inhibition of the target's activity, but also how redundant such a target is and the likelihood of evolutionary compensation for its function.
Most classical and targeted cancer drugs are directed at highly plastic and adaptive signaling networks or cellular processes, and, as a consequence, their therapeutic effectiveness is relatively rapidly eroded, and the patient relapses with disease that is both drug-resistant and often more aggressive as a consequence of the selective pressure imposed by the therapy.
The Myc proteins comprise three broadly isofunctional transcription factors: c-Myc, N-Myc, and L-Myc. All share a similar HLHZip domain that mediates dimerization with their common obligate partner protein, Max. Such dimerization, which is essential for Myc proliferative and oncogenic activities, is blocked by the dominant-negative Omomyc mutant. As a competitive inhibitor of c-Myc, N-Myc and L-Myc dimerization that specifically blocks Myc transcriptional activation, Omomyc more closely resembles a generic Myc inhibitor than do more classical Myc gene deletion studies, making it a useful tool with which to model pharmacological inhibition of Myc.
Given its unique role as a broad integrator of expression of so many different genes, it is no surprise that Myc function cannot be replaced or substituted by any other pathway or mechanism. This makes Myc an excellent candidate cancer drug target, at least in principle, since tumor cells should not be able to adapt around, or evolve independence from, Myc inhibition. There is no better way of directly testing this than by subjecting hundreds of individual tumors to repeated rounds of Myc inhibition interspersed with periods of recovery and tumor regrowth. However, very few tumors survived our metronomic Myc inhibition regimen, and those that did all had inactivated expression of the Omomyc transgene. Our data therefore support the notion that targeting a functionally nondegenerate signaling node like Myc is an effective strategy for mitigating the evolution of therapeutic resistance.
Surprisingly, while Myc inhibition elicits profound growth inhibition in normal regenerative tissues (Soucek et al. 2008), the side effects of short-term systemic Myc inhibition are remarkably mild. However, the situation is very different in KRasG12D-driven lung tumor cells, where Myc inhibition-induced growth arrest is accompanied by widespread tumor cell death and tumor regression irrespective of tumor size or stage of tumor evolution. Quite why Myc inhibition exhibits this fortunate therapeutic proclivity to kill tumor cells remains unclear. However, since Myc inhibition activates p53, this necessarily indicates that it provokes a form of intracellular dysfunction to which p53 has evolved to respond.
To gain some insight into the possible mechanism by which Omomyc induces death of tumor cells, we interrogated expression microarrays from 15-wk-old TREOmomyc;CMVrtTA;LSL-KrasG12D;p53ER/ER lung tumors either untreated or treated with Omomyc in vivo for 3 d. Intriguingly, the two most significantly up-regulated networks elicited by Omomyc expression are both potentially related to tumor cell death (Supplemental Fig. 4A,B) and focused on several major nodes; namely, PPARG, FOXO1, and NFE2L2. PPARG is a member of the peroxisome proliferator-activated receptor family known to be involved in inflammation and metabolism and to induce apoptosis in various cancer cell lines (Tachibana et al. 2008; Grabacka et al. 2010). FOXO1 loss has been shown to cooperate with Myc in inducing tumorigenesis, and its expression induces the p19ARF tumor suppressor (Bouchard et al. 2007). Finally, NFE2L2, also known as Nrf2, is a modulator of NfkB signaling, and its loss has been shown to predispose mice to carcinogen-induced tumorigenesis, while its expression could have an anti-inflammatory or proapoptotic effect on damaged cells (Li et al. 2008). When the same expression data were interrogated specifically for cell death-related networks, three major nodes appeared as the most significantly regulated by Omomyc treatment: p53, HTT, and ZFP36 (Supplemental Fig. 5). A role for p53 is consistent with its therapeutic synergy with Omomyc (as shown in Fig. 4), but any potential roles for HTT and ZFP36 remain unclear. HTT, also known as huntingtin, is a protein associated with Huntington's disease (Nance et al. 1999). Intriguingly, Huntington's disease patients are resistant to a variety of cancers (Ji et al. 2012). The reason for such resistance is currently unknown. Finally, loss of ZFP36 (also known as Tristetraprolin [TTP]) is typical of cancers with Myc involvement, and its restoration impairs Myc-induced lymphomagenesis and abolishes maintenance of the malignant state (Rounbehler et al. 2012).
Another likely mechanism contributing to the peculiar lethality of Omomyc in tumor cells is disruption of the cross-talk between tumor and stromal cells that maintains the tumor microenvironment. We recently showed in a model of pancreatic β-cell carcinoma that Myc inhibition profoundly blocks the production by tumor cells of angiogenic and proinflammatory signals (Sodir et al. 2011), precipitating collapse of tumor vasculature and stromal maintenance. Similarly, in p53-deficient KRasG12D-driven lung tumors, Myc inhibition abruptly perturbs the panoply of chemokines, cytokines, angiogenic factors, and cognate receptors underpinning the lung tumor microenvironment (Supplemental Fig. 6).
Despite the ostensibly complete tumor regression induced by a single round of Myc inhibition, all of the animals eventually relapse, albeit with reduced tumor multiplicity. Since the recurring tumors bear the hallmarks of more advanced lesions, such relapse is most probably due to regeneration of original tumors rather than evolution of new ones. Furthermore, while it is conceivable that these residual surviving cells represent a specific, resistant subpopulation of tumor-regenerating cells, their progressive eradication over repeated rounds of Myc inhibition argues against their harboring inherent resistance to Myc inhibition.
Recent pharmacological studies that target Myc indirectly by blocking its expression appear to offer great promise (Delmore et al. 2011; Zuber et al. 2011). The switchable genetic data using Omomyc described here reinforce the promise of Myc as a drug target whose inhibition would elicit durable therapeutic responses in a wide range of cancer types.
Materials and methods
Generation and maintenance of genetically engineered mice
TRE-Omomyc;CMVrtTA;LSLKRas [Tg(TetO-MYC*)4Gev;Tg(rtTAhCMV)4Bjd/J; Krastm4Tyj] and p53ER/ER (Trp53tm1Gev) mice have been described previously (Christophorou et al. 2005; Soucek et al. 2008). Omomyc expression was systemically induced in TRE-Omomyc;CMVrtTA mice by addition of doxycycline (2 mg/mL, plus 5% sucrose) to their drinking water. Sucrose only was administered to control mice. p53 restoration was achieved by daily i.p. injection with tamoxifen (1 mg per mouse dissolved in peanut oil). Animals were maintained and treated in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) at the University of California at San Francisco (UCSF). For each experiment, cohorts of at least four mice were used for each data point.
Tissue preparation and histology
Mice were killed, and the lungs were inflated and perfused with zinc-buffered formalin. Each lung was divided in two: Half (two or three lobes) was embedded in paraffin, and half was embedded in optimal cutting temperature (OCT) compound. For paraffin embedding, tissues were fixed overnight in zinc-buffered formalin and processed. Tissue sections (5 μM) were stained with hematoxylin and eosin (H&E) using standard reagents and protocols. For frozen sections, lungs were embedded in OCT, frozen on dry ice, and stored at −80°C. Tumor quantification and grading was performed on H&E-stained paraffin sections. Adenomas and adenocarcinomas were scored, while atypical adenomatous hyperplasias were excluded from the analysis.
Immunohistochemistry
For immunohistochemical analysis, sections were deparaffinized, rehydrated, and subjected to high-temperature antigen retrieval in 10 mM citrate buffer (pH 6.0). Primary antibodies were as follows: rabbit polyclonal anti-Omomyc [prepared in-house, Evan laboratory (Soucek et al. 2004)] and mouse monoclonal anti-p21 (clones SX118 and SXM30, BD Pharmingen). Primary antibodies were applied for 2 h in blocking buffer (2.5% BSA, 5% goat serum, 0.3% Triton X-100 in PBS), sections were washed, and species-appropriate secondary Alexa fluor 488 dye-conjugated antibodies (Amersham) or Vectastain ABC kit and DAB reagents (Vector Laboratories) were applied. Fluorescence antibody-labeled slides were mounted in DAKO fluorescent mounting medium containing 1 μg/mL Hoechst counterstain. HRP-conjugated secondary antibodies were visualized by DAB staining (Vector Laboratories). Images were collected with a Lumar V12 microscope equipped with an Axiocam HRC, an Axiovert 5100 TV inverted fluorescence microscope (Zeiss) and Open Lab 3.5.1 software, or an Axiovert 100 inverted microscope (Zeiss) equipped with a Hamamatsu Orca digital camera.
Microarray analyses
For expression profiling, TREOmomyc;CMVrtTA;LSL-KrasG12D;p53ER/ER mice were treated with either doxycycline (2 mg/mL, plus 5% sucrose; referred to as “treated”) or 5% sucrose (referred to as “untreated”) in their drinking water for 3 d, 15 wk after Ad-Cre infection. Tumors were dissected, and total RNA was isolated using TRIzol RNA isolation protocol and processed for hybridization on Affymetrix mouse set ST1.0 at the Genomics Core of the Gladstone Institute. Robust multiarray (RMA) analysis using Affymetrix Power Tool “APT” software was used to normalize and summarize probe level intensities, and any transcript clusters that, after normalization, had a log-base-2 score <3.0 were discarded. The fold change in expression for each gene was calculated from the ratio of median expression of treated samples versus untreated.
Additional filtering was applied to select genes analyzed using Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems, http://www.ingenuity.com), and genes with fold change between −2-fold and 1.5-fold change were excluded. Each gene symbol was mapped in the Ingenuity Pathways Knowledge Base. Networks of these genes were algorithmically generated based on their connectivity and assigned a score. The score was generated using a P-value calculation and displayed as the negative log of that P-value. This score indicates the likelihood that the assembly of a set of focus genes in a network could be explained by random chance alone. Further explanation regarding gene symbols and functional relationships can be found at http://www.ingenuity.com.
Acknowledgments
We are indebted to Fanya Rostker for her technical assistance, and to our colleagues in the Evan laboratory for their invaluable criticism and advice. We also thank Roderik Kortlever for his assistance with animal care, and Josep Villanueva for his advice on the use of IPA software. This work was supported by grants NIH/NCI 2R01 CA98018, Cancer Research UK A12077, and AICR 09-0649 to G.I.E., and the Miguel Servet Program, the FERO Foundation, and the Bear Necessities Pediatric Cancer Foundation to L.S.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.205542.112.
References
- Bouchard C, Lee S, Paulus-Hock V, Loddenkemper C, Eilers M, Schmitt CA 2007. FoxO transcription factors suppress Myc-driven lymphomagenesis via direct activation of Arf. Genes Dev 21: 2775–2787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophorou MA, Martin-Zanca D, Soucek L, Lawlor ER, Brown-Swigart L, Verschuren EW, Evan GI 2005. Temporal dissection of p53 function in vitro and in vivo. Nat Genet 37: 718–726 [DOI] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. 2011. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146: 904–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabacka M, Pierzchalska M, Reiss K 2010. Peroxisome proliferator activated receptor α ligands as anti-cancer drugs targeting mitochondrial metabolism. Curr Pharm Biotechnol PMID: 21133850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA 2001. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes & Dev 15: 3243–3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M, Jacks T 2005. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res 65: 10280–10288 [DOI] [PubMed] [Google Scholar]
- Ji J, Sundquist K, Sundquist J 2012. Cancer incidence in patients with polyglutamine diseases: A population-based study in Sweden. Lancet Oncol 13: 642–648 [DOI] [PubMed] [Google Scholar]
- Junttila MR, Karnezis AN, Garcia D, Madriles F, Kortlever RM, Rostker F, Brown Swigart L, Pham DM, Seo Y, Evan GI, et al. 2010. Selective activation of p53-mediated tumour suppression in high-grade tumours. Nature 468: 567–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CF, Jackson EL, Kirsch DG, Grimm J, Shaw AT, Lane K, Kissil J, Olive KP, Sweet-Cordero A, Weissleder R, et al. 2005. Mouse models of human non-small-cell lung cancer: Raising the bar. Cold Spring Harb Symp Quant Biol 70: 241–250 [DOI] [PubMed] [Google Scholar]
- Larsson LG, Henriksson MA 2010. The Yin and Yang functions of the Myc oncoprotein in cancer development and as targets for therapy. Exp Cell Res 316: 1429–1437 [DOI] [PubMed] [Google Scholar]
- Li W, Khor TO, Xu C, Shen G, Jeong WS, Yu S, Kong AN 2008. Activation of Nrf2-antioxidant signaling attenuates NFκB-inflammatory response and elicits apoptosis. Biochem Pharmacol 76: 1485–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nance MA, Mathias-Hagen V, Breningstall G, Wick MJ, McGlennen RC 1999. Analysis of a very large trinucleotide repeat in a patient with juvenile Huntington's disease. Neurology 52: 392–394 [DOI] [PubMed] [Google Scholar]
- Oliver TG, Mercer KL, Sayles LC, Burke JR, Mendus D, Lovejoy KS, Cheng MH, Subramanian A, Mu D, Powers S, et al. 2010. Chronic cisplatin treatment promotes enhanced damage repair and tumor progression in a mouse model of lung cancer. Genes Dev 24: 837–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounbehler RJ, Fallahi M, Yang C, Steeves MA, Li W, Doherty JR, Schaub FX, Sanduja S, Dixon DA, Blackshear PJ, et al. 2012. Tristetraprolin impairs myc-induced lymphoma and abolishes the malignant state. Cell 150: 563–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savino M, Annibali D, Carucci N, Favuzzi E, Cole MD, Evan GI, Soucek L, Nasi S 2011. The action mechanism of the Myc inhibitor termed Omomyc may give clues on how to target Myc for cancer therapy. PLoS ONE 6: e22284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Lima A, Molina R, Hamilton P, Clermont AC, Devasthali V, Thompson JD, Cheng JH, Bou Reslan H, Ho CC, et al. 2010. Assessing therapeutic responses in Kras mutant cancers using genetically engineered mouse models. Nat Biotechnol 28: 585–593 [DOI] [PubMed] [Google Scholar]
- Sodir NM, Swigart LB, Karnezis AN, Hanahan D, Evan GI, Soucek L 2011. Endogenous Myc maintains the tumor microenvironment. Genes Dev 25: 907–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek L, Helmer-Citterich M, Sacco A, Jucker R, Cesareni G, Nasi S 1998. Design and properties of a Myc derivative that efficiently homodimerizes. Oncogene 17: 2463–2472 [DOI] [PubMed] [Google Scholar]
- Soucek L, Jucker R, Panacchia L, Ricordy R, Tato F, Nasi S 2002. Omomyc, a potential Myc dominant negative, enhances Myc-induced apoptosis. Cancer Res 62: 3507–3510 [PubMed] [Google Scholar]
- Soucek L, Nasi S, Evan GI 2004. Omomyc expression in skin prevents Myc-induced papillomatosis. Cell Death Differ 11: 1038–1045 [DOI] [PubMed] [Google Scholar]
- Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, Karnezis AN, Swigart LB, Nasi S, Evan GI 2008. Modelling Myc inhibition as a cancer therapy. Nature 455: 679–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet-Cordero A, Mukherjee S, Subramanian A, You H, Roix JJ, Ladd-Acosta C, Mesirov J, Golub TR, Jacks T 2005. An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nat Genet 37: 48–55 [DOI] [PubMed] [Google Scholar]
- Tachibana K, Yamasaki D, Ishimoto K, Doi T 2008. The role of PPARs in cancer. PPAR Res 2008: 102737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuveson DA, Shaw AT, Willis NA, Silver DP, Jackson EL, Chang S, Mercer KL, Grochow R, Hock H, Crowley D, et al. 2004. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell 5: 375–387 [DOI] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. 2011. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478: 524–528 [DOI] [PMC free article] [PubMed] [Google Scholar]