Weber et al. explore the biochemical and structural features of the Aar2p and spliceosomal protein in complex with the two domains of Prp8p during the ordered assembly of the U5 snRNP. They find that Aar2p competes with the helicase Brr2p for binding to Prp8p and also interferes with binding of the U4/U6 di-snRNA to the Prp8p C-terminal fragment. Thus, Aar2p has a role in the biogenesis of U5 snRNP and may also play a role in spliceosome disassembly and/or U5 snRNP recycling.
Keywords: assembly chaperone, pre-mRNA splicing, regulation by phosphorylation, snRNP biogenesis and recycling, X-ray crystallography
Abstract
Yeast U5 small nuclear ribonucleoprotein particle (snRNP) is assembled via a cytoplasmic precursor that contains the U5-specific Prp8 protein but lacks the U5-specific Brr2 helicase. Instead, pre-U5 snRNP includes the Aar2 protein not found in mature U5 snRNP or spliceosomes. Aar2p and Brr2p bind competitively to a C-terminal region of Prp8p that comprises consecutive RNase H-like and Jab1/MPN-like domains. To elucidate the molecular basis for this competition, we determined the crystal structure of Aar2p in complex with the Prp8p RNase H and Jab1/MPN domains. Aar2p binds on one side of the RNase H domain and extends its C terminus to the other side, where the Jab1/MPN domain is docked onto a composite Aar2p–RNase H platform. Known Brr2p interaction sites of the Jab1/MPN domain remain available, suggesting that Aar2p-mediated compaction of the Prp8p domains sterically interferes with Brr2p binding. Moreover, Aar2p occupies known RNA-binding sites of the RNase H domain, and Aar2p interferes with binding of U4/U6 di-snRNA to the Prp8p C-terminal region. Structural and functional analyses of phospho-mimetic mutations reveal how phosphorylation reduces affinity of Aar2p for Prp8p and allows Brr2p and U4/U6 binding. Our results show how Aar2p regulates both protein and RNA binding to Prp8p during U5 snRNP assembly.
Uridine-rich (U) small nuclear ribonucleoprotein particles (snRNPs) are the main subunits of spliceosomes, the large and dynamic RNP machineries required for the removal of noncoding introns from eukaryotic precursor messenger RNAs (pre-mRNAs) and the ligation of neighboring coding exons. The major spliceosome is built from the U1, U2, U4, U5, and U6 snRNPs, each of which contains an individual small nuclear RNA (snRNA), seven common Sm proteins (or like-Sm [LSm] proteins in the case of U6), and a variable set of particle-specific proteins (for review, see Will and Lührmann 2001).
A hallmark of the spliceosome is its stepwise assembly from snRNPs and many non-snRNP factors only in the presence of a substrate pre-mRNA (for review, see Wahl et al. 2009). During this process, the spliceosome is repeatedly remodeled with the help of eight highly conserved DEXD/H-box ATPases/RNA helicases (for review, see Staley and Guthrie 1998). Among these enzymes, the U5-specific Brr2 protein is required for the catalytic activation of an initial inactive spliceosomal assembly and again during the ordered disassembly of the post-splicing complex (for review, see Hahn and Beggs 2010). Brr2p is regulated by two other U5 snRNP proteins: Prp8p, considered the master regulator of the spliceosome (for review, see Grainger and Beggs 2005), and Snu114p, a complex G protein that resembles the ribosomal translocase eEF2 (Bartels et al. 2002, 2003; Brenner and Guthrie 2005; Small et al. 2006).
The Sm-type snRNPs themselves are also assembled in a strictly controlled fashion in vivo (for review, see Patel and Bellini 2008). Their snRNAs are generated by RNA polymerase II, receive a 7-methyl-guanylate cap, and are exported to the cytoplasm. Here, the Sm proteins are attached in a ring-like fashion around a uridine-rich Sm site of the snRNAs (Pomeranz Krummel et al. 2009; Weber et al. 2010; Leung et al. 2011). This process is mediated by two coordinated multifactorial assembly machineries: the protein arginine methyltransferase 5 (PRMT5) complex and the survival of motor neurons (SMN) complex (for reviews, see Battle et al. 2006; Fischer et al. 2011). After assembly of the Sm core RNPs, the snRNA caps are hypermethylated, and the premature particles are returned to the nucleus. Final stages of snRNP assembly take place in the nuclear Cajal bodies, where the particle-specific proteins are thought to be added (Nesic et al. 2004; Schaffert et al. 2004; Stanek and Neugebauer 2004). Furthermore, U4, U5, and U6 snRNPs assemble a U4/U6-U5 tri-snRNP in the Cajal bodies (Schaffert et al. 2004) before incorporation into spliceosomes.
In yeast, a cytoplasmic precursor of U5 snRNP—containing only the U5 snRNA, the Sm proteins, and the U5-specific proteins Prp8p and Snu114p—has been characterized (Gottschalk et al. 2001; Boon et al. 2007). Pre-U5 snRNP lacks the Brr2 helicase and instead includes the Aar2 protein (Gottschalk et al. 2001; Boon et al. 2007), which has been originally characterized as a factor involved in pre-mRNA splicing of the MATa1 transcript in Saccharomyces cerevisiae (Nakazawa et al. 1991). We recently found that Aar2p and Brr2p bind, respectively, to an RNase H-like (RH) domain and a Jab1/MPN-like (Jab1) domain that lie next to each other in the C-terminal region of the Prp8 protein (Weber et al. 2011). Moreover, binding of Aar2p to the Prp8p RH domain sequestered the Jab1 domain and counteracted Brr2p binding to Prp8p (Weber et al. 2011). Furthermore, Aar2p was shown to be phosphorylated at five sites in vivo, and a phospho-mimetic mutation of Aar2p (S253E) diminished binding to the RH domain in vitro and abolished Aar2p–Prp8p interaction in extracts, allowing increased association of Brr2p with Prp8p (Weber et al. 2011).
To further elucidate the molecular basis for the function of Aar2p as a U5 snRNP assembly factor, we determined the crystal structure of Aar2p in complex with the Prp8p RH and Jab1 domains. In the complex, Aar2p packages the two Prp8p domains in a manner incompatible with Brr2p binding. In addition, we show that Aar2p interferes with the binding of U4/U6 di-snRNAs to the Prp8p C-terminal region and demonstrate how reversible phosphorylation can regulate these activities.
Results
Reconstitution and structure analysis of an Aar2pΔloop–RH–Jab1 complex
Our previous crystal structure of subtilisin-treated Aar2p (Aar2pSubt) in complex with the Prp8p RH domain did not reveal how this complex sequesters the Jab1 domain (Weber et al. 2011). We could not use Aar2pSubt to assemble a complex with both Prp8p domains, as this protein lacks a C-terminal expansion that is required for Jab1 sequestration (Weber et al. 2011). We thus generated complexes via two alternative strategies. First, we coproduced and copurified Aar2p with a Prp8p C-terminal fragment (CTF) encompassing both RH and Jab1 domains. Second, we coproduced and copurified Aar2p with the Prp8p RH domain, added a separately purified Jab1 domain, and purified the resulting complex by gel filtration chromatography. In both strategies, we used either wild-type Aar2p or a variant in which a flexible internal loop (residues 153–170) was replaced by five serine residues (Aar2pΔloop) (Supplemental Fig. S1). The same loop was also removed by the subtilisin treatment in Aar2pSubt (Weber et al. 2011).
All attempts to crystallize CTF together with wild-type Aar2p or Aar2pΔloop failed, most likely due to an ∼60-residue flexible linker between the RH and Jab1 domains. In addition, we did not obtain crystals from trials involving wild-type Aar2p and the two separate Prp8p domains. However, the Aar2pΔloop–RH–Jab1 complex (i.e., a “ternary” complex in which the RH and Jab1 domains were separated by removal of the intervening linker) (Supplemental Fig. S2A, lanes 2,3) yielded crystals (Supplemental Fig. S2B) that diffracted to 2.1 Å resolution (Supplemental Table S1). The structure was solved by molecular replacement using the structure coordinates of Aar2pSubt and RH (Protein Data Bank [PBD] ID 3SBT) and refined to low R/Rfree factors with good stereochemistry (Supplemental Table S1).
Overall structure of an Aar2pΔloop–RH–Jab1 complex
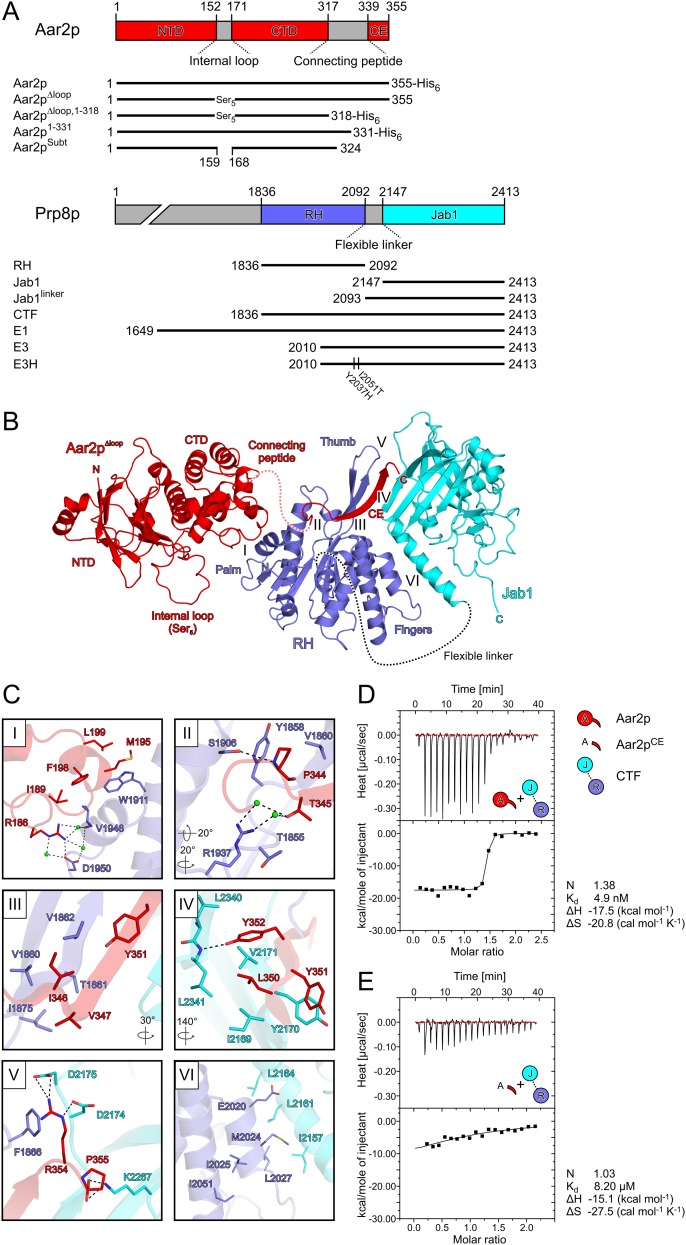
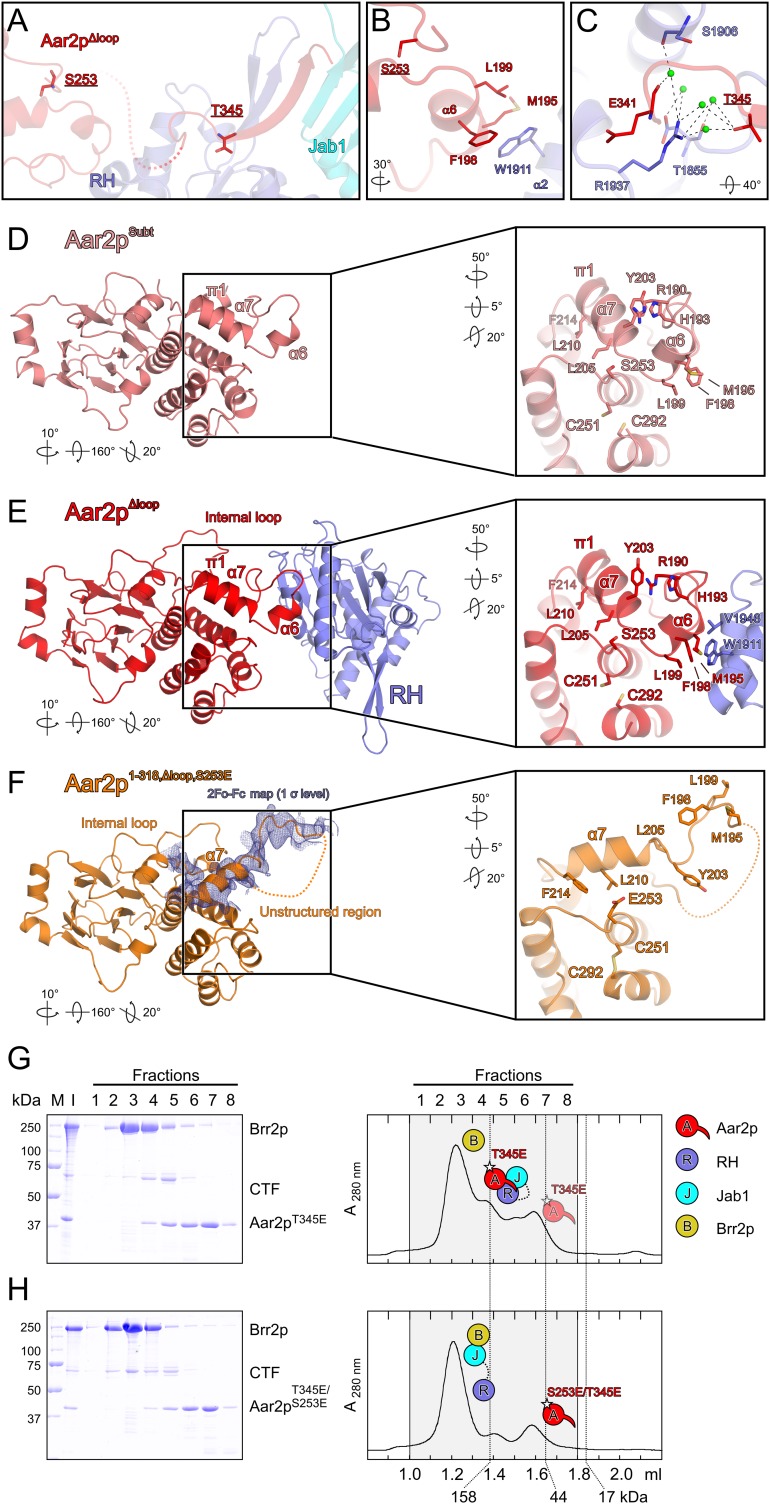
In the structure of the Aar2pΔloop–RH–Jab1 complex, the three molecules form a linear array with the RH domain in the center and the bulk of the other two proteins on either side (Fig. 1A,B). We previously described the structure of the RH domain as a left-handed mitten with the RH-like portion representing the “palm,” an extended β hairpin corresponding to the “thumb,” and an α-helical subdomain forming the “fingers” (Pena et al. 2008). Aar2p is divided into an N-terminal domain with a β-sandwich fold and an α-helical C-terminal domain (Fig. 1A). The Aar2p C-terminal domain interacts with one side of the RH palm, burying 448 Å2 of surface area (Fig. 1C, panel I). This interface includes a number of hydrophobic contacts (residues M195, F198, and L199 in helix α6 of Aar2p interacting with W1911 of RH; residues F198 and L189 of Aar2p contacting V1946 of RH), a salt bridge (Aar2p R186 with RH D1950), and water-mediated interactions (between Aar2p R186 and RH V1946 and D1950).
Figure 1.
Structure of the Aar2pΔloop–RH–Jab1 complex. (A) Scheme showing domains and features of Aar2p and Prp8p constructs used in this study. (NTD) Aar2p N-terminal domain; (CTD) Aar2p C-terminal domain; (CE) Aar2p C-terminal extension. (B) Ribbon plot of the Aar2pΔloop–RH–Jab1 complex. (Red) Aar2pΔloop; (blue) RH; (cyan) Jab1; (dashed red line) flexible Aar2p peptide preceding the C terminus; (internal loop) flexible loop of Aar2p connecting its two domains, which in Aar2pΔloop was replaced by five serines. The N and C termini as well as the RH palm, thumb, and finger regions are labeled. Roman numerals indicate the location of the protein–protein interface regions depicted in C, panels I–VI. (C) Close-up views on protein–protein interface regions of the Aar2pΔloop–RH–Jab1 structure. Interacting residues are shown as sticks and colored by atom type. The color of the carbon is the same as the molecule to which it belongs. Nitrogen is in blue, oxygen is in red, and sulfur is in yellow. Water molecules are shown as green spheres. Dashed lines represent hydrogen bonds or salt bridges. Rotation symbols indicate the views relative to B. (D,E) ITC experiments comparing the binding of wild-type Aar2p (D) and a C-terminal Aar2p extension (residues 331–355) (E) to CTF. Thermodynamic binding parameters are listed. Icons indicate Aar2p variants (red) and CTF variants (RH [blue]; Jab1 [cyan]) used.
The Jab1 domain is located remote from this Aar2p–RH interface on the other side of the RH domain (Fig. 1B). The Aar2p C terminus is preceded by a flexible unstructured peptide (residues 318–338) and extends across the RH palm, where it acts in concert with RH to generate a composite surface to which the Jab1 domain is attached. The C terminus of Aar2p connects the two Prp8p domains via a parallel, intermolecular β sheet, interspersing between one β strand of the RH thumb and the central β barrel of Jab1 (Fig. 1B). The composite binding site for the Jab1 domain is consistent with previous interaction studies, in which neither Aar2p nor RH alone formed stable complexes with Jab1 (Weber et al. 2011). The described Aar2pΔloop–RH–Jab1 complex is the only arrangement in the present crystal structure in which the connecting peptide lacking electron density (21 residues) can easily bridge between the globular part of Aar2pΔloop and the C-terminal extension (21 Å distance compared with 49 Å for the next closest possibility).
Apart from the secondary structure-based backbone hydrogen bonds, the side chains in the C-terminal β strand of Aar2p maintain intricate interactions with both Prp8p domains (Fig. 1C). The interaction between the Aar2p C terminus and the RH palm and thumb regions (733 Å2 of buried surface area) (Fig. 1C, panels II,III) involves a mixture of hydrophobic contacts (Aar2p P344 with RH Y1858 and V1860; Aar2p T345 with RH T1855; Aar2p I346 and V347 with RH V1860, V1862, and I1875; and Aar2p Y351 with RH V1862), a hydrogen bond (Aar2p P344 and RH S1906), a cation–π interaction (Aar2p R354 with RH F1866), and water-mediated interactions (between Aar2p T345 and RH R1937 and T1855). The interface between the C-terminal β strand of Aar2p and Jab1 (508 Å2) (Fig. 1C, panel IV) is dominated by hydrophobic interactions (Aar2p Y351 with Jab1 Y2170 and RH V1862; Aar2p L350 with Jab1 I2169, V2171, and L2341; and Aar2p Y352 with Jab1 V2171 and L2340) with an interspersed hydrophilic contact (Aar2p Y352 with the backbone of Jab1 L2341). The two very C-terminal residues of Aar2p are deeply involved in the interaction network, providing a crucial anchor for RH and Jab1 (Fig. 1C, panel V). The penultimate R354 is positioned by Jab1 residues D2174 and D2175 for stacking on F1866 of RH, while the C-terminal carboxyl group of P355 engages in additional salt bridges with K2267 of Jab1. The interface of RH and Jab1 (481 Å2) (Fig. 1C, panel VI) is formed by a short interdomain α-helical bundle (RH M2024 and E2020 intercalated between Jab1 I2157, L2161, and L2164).
As the C-terminal expansion of Aar2p covers 1195 Å2 of surface area with RH and Jab1, we tested whether it alone binds efficiently to CTF. Isothermal titration calorimetry (ITC) experiments showed that the dissociation constant of a C-terminal Aar2p peptide (residues 331–355) and CTF was >1500 times larger compared with wild-type Aar2p due to a concomitant decrease in reaction enthalpy and increase in reaction entropy (Fig. 1D,E). Therefore, the interaction of the RH domain with the globular part of Aar2p provides a crucial initial anchor for the Aar2p C-terminal expansion to efficiently connect RH and Jab1.
Reassignment of the physiological Aar2p–RH interface
The interface between Aar2p and RH in the present complex differs markedly from the interaction reported for the Aar2pSubt–RH complex (Weber et al. 2011), where Aar2pSubt was described to bind between the thumb and fingers of RH (Supplemental Fig. S3A). The previous assignment of the Aar2pSubt–RH interface was based on a large buried surface area (755 Å2 compared with 440 Å2 for the next largest contact in the crystal lattice) involving 54 contacting residues and a high degree of charge complementarity between the molecules. Crystal contacts with buried surface areas >600 Å2 have a high likelihood of being physiologically relevant (Janin 1997; Krissinel 2009). In addition, Prp8p mutations with enhanced affinity to Brr2p (van Nues and Beggs 2001) and an Aar2p mutation (D281N) that suppressed a slowdown of spliceosome catalytic activation caused by the prp38-1 allele (Pandit et al. 2006) mapped to the presumed Aar2pSubt–RH interface and could be explained by a reduced binding of Aar2p to the Prp8p RH domain. While the present structure still rationalizes the enhanced Brr2 affinity of certain Prp8p mutations (see the Discussion), Aar2p residue D281 is not involved in contacts to RH or Jab1 in our new structure, which therefore does not explain the genetic effects of the D281N mutation.
While there is also no other crystal contact in the present structure that resembles the previously assigned Aar2pSubt–RH complex, an alternative interaction between Aar2pSubt and RH exists in the Aar2pSubt–RH crystal lattice, which closely resembles the interaction observed in the Aar2pΔloop–RH–Jab1 complex (overall root-mean-square deviation [rmsd], 4.16 Å; rmsd Aar2p components, 1.37 Å; rmsd RH components, 1.62 Å) (Supplemental Fig. S3B,C). While the molecular contacts in the two complexes are almost identical (Supplemental Fig. S3B,C), the small size of the interface (∼450 Å2) still allows for small deviations in the relative positioning of the two proteins (Supplemental Fig. S3E).
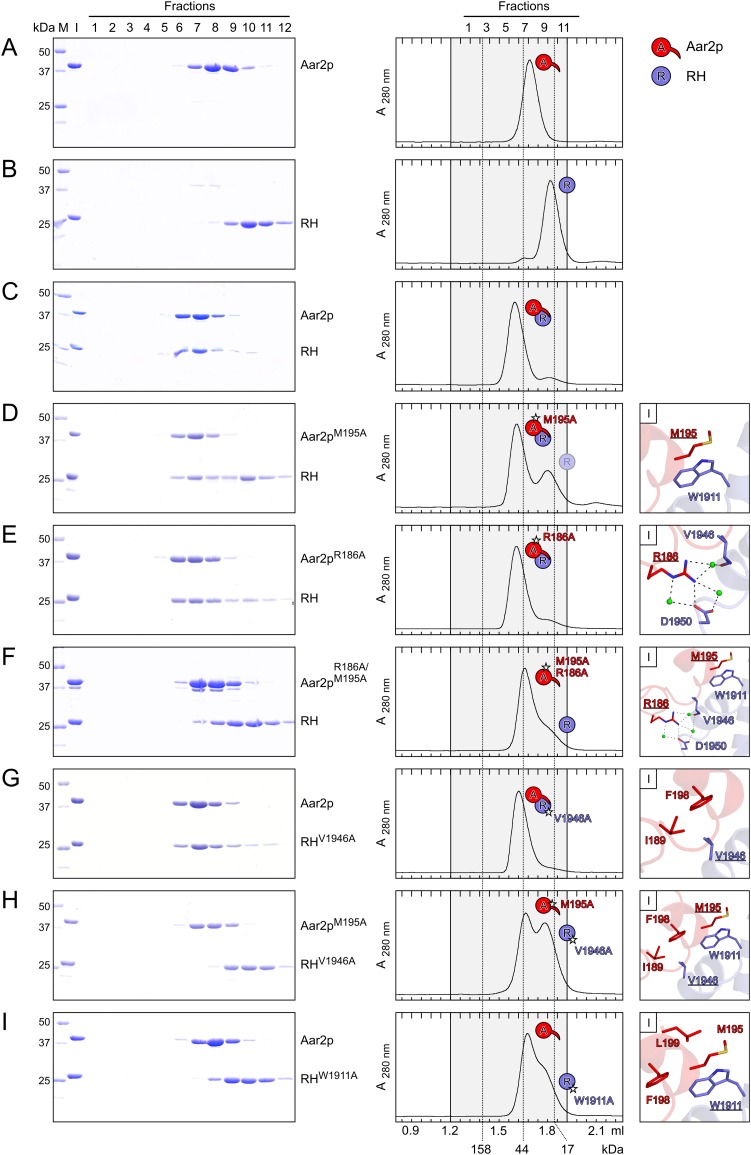
To sort out the physiological Aar2p–RH interaction mode, we mutated residues in either interface and observed their effects on comigration of Aar2p and RH in gel filtration (Fig. 2; Supplemental Fig. S4). Double mutations R55A/I282A in Aar2p (Supplemental Fig. S4D) or V1862A/N2062A in RH (Supplemental Fig. S4E), which introduced changes in residues involved in the previously suggested Aar2pSubt–RH interface, as well as the combination of both double mutants (Supplemental Fig. S4F) still permitted the formation of dimeric Aar2p–RH complexes. Conversely, mutation of residues in the Aar2p–RH interface seen in the present complex had clear effects on complex formation (Fig. 2). Whereas the single residue changes M195A or R186A in Aar2p did not disrupt the complex with RH (Fig. 2D,E), the interaction was abolished with the combined double Aar2p mutation (Fig. 2F). Likewise, the V1946A mutant of RH still bound to Aar2p (Fig. 2G), but the interaction was abolished when we combined this mutation with Aar2pM195A (Fig. 2H). Finally, changing W1911 of RH, which maintains extensive hydrophobic interactions with Aar2p (Fig. 1C, panel I), to alanine also interfered with Aar2p binding (Fig. 2I).
Figure 2.
Alanine-scanning mutagenesis probing the Aar2p–RH interaction. (A–I) Gel filtration analyses assessing the interaction between Aar2p and RH and mutant variants. A–C are identical to Supplemental Figure S4, A–C. All mutated residues contribute to interface I (Fig. 1C). (Left) SDS-PAGE analysis of the eluted gel filtration fractions (indicated at the top of A). The first two lanes show the protein standard (M) and the gel filtration input (I). Molecular mass of the standard proteins in kilodaltons are shown on the left, and protein names are on the right. (Middle) Elution profiles of the respective gel filtration runs. Fractions corresponding to the SDS-PAGE analysis are highlighted in gray, fraction numbers are shown at the top of A, and elution volumes are at the bottom of I. Dashed lines across the elution profiles indicate the elution peaks of molecular mass standards (molecular masses are given at the bottom). Icons denote single proteins or complexes and are defined above the middle panels. Stars denote point mutations. Transparent icons represent an excess of the respective protein running separate from the complex. (Right) Close-up views on the mutated residues (underlined) and their interactions across the Aar2p–RH interface.
All proteins used in this study showed cooperative transitions in a fluorescence-based thermal melting assay (Supplemental Table S2), indicating that introduced mutations did not compromise their overall folds. Furthermore, the observation that several individual and double mutations in Aar2p (M195A, R186A, and R55A/I282A) or RH (V1946A and V1862A/N2062A) still supported complex formation demonstrates that these amino acid exchanges left the overall structures of Aar2p and RH intact. We also crystallized the RHW1911A protein alone and found its structure to be virtually unchanged compared with the wild-type protein (rmsd, 0.55 Å and 1.19 Å for two independent molecules per asymmetric unit) (Supplemental Table S1). We also ascertained that the Aar2p internal loop, which was replaced by five serines in Aar2pΔloop or cleaved in Aar2pSubt, is not involved in the interaction with RH or Jab1 (Supplemental Material; Supplemental Fig. 5). Finally, while this study was being revised, a structure of Aar2p in complex with a larger portion of Prp8p (residue 885-2413) was published (Galej et al. 2013). The corresponding parts of this and our Aar2pΔloop–RH–Jab1 structure are very similar (overall rmsd, 2.64 Å; rmsd Aar2p components, 2.12 Å; rmsd RH components, 1.18 Å; rmsd Jab1 components, 1.34 Å). Taken together, the above analyses clearly show that the contact between Aar2p and RH seen in the present structure represents their physiological interaction mode.
The Aar2pΔloop–RH–Jab1 structure resolves ambiguities in the domain arrangement of a CTF structure
Although CTF did not cocrystallize with Aar2p in our hands, we had previously been able to determine the crystal structure of CTF alone (Weber et al. 2011) but, due to missing electron density for the flexible RH–Jab1 linker, could not reliably assign a physiological contact between the RH and Jab1 domains. We observed that one interaction mode of the two domains in the isolated CTF structure closely corresponds to the RH–Jab1 interaction in the Aar2pΔloop–RH–Jab1 complex (Supplemental Fig. S3D,F). In the CTF structure, the thumb of RH has a different conformation and sustains additional contacts to Jab1 (RH V1862 and V1870 to Jab1 residues 2343–2346). In the Aar2pΔloop–RH–Jab1 complex, the C terminus of Aar2p interjects between these regions, bridging between RH and Jab1 (Fig. 1B). Thus, rather than sequentially interacting first with RH and subsequently with Jab1, Aar2p may bind to a preformed complex between the domains and subsequently stabilize their interaction via its C terminus.
The composite Aar2p–RH surface is required for the sequestration of Jab1
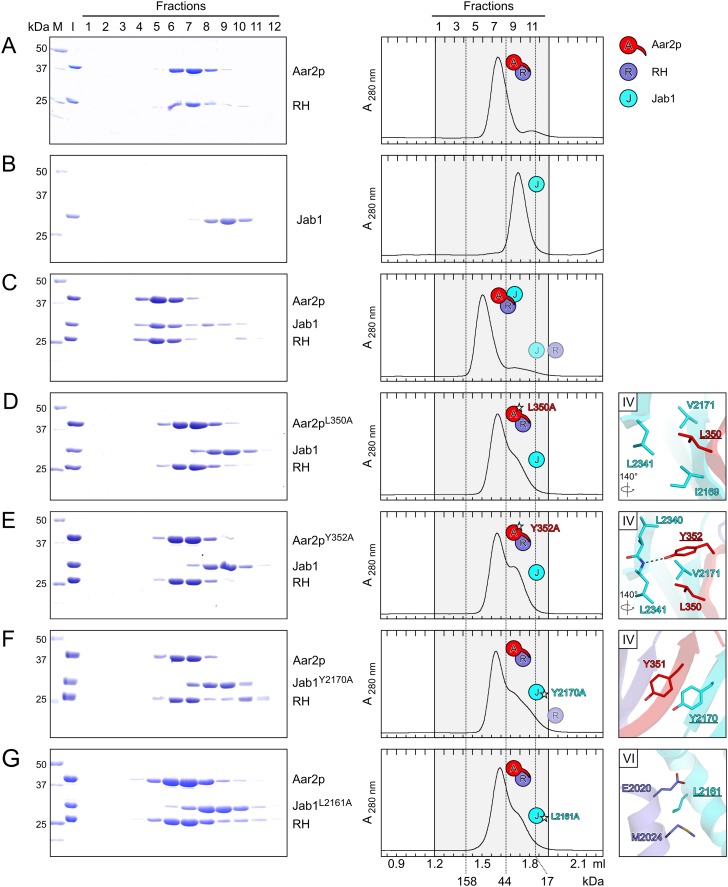
We next tested the physiological relevance of the Aar2p–Jab1 and RH–Jab1 contacts seen in the Aar2pΔloop–RH–Jab1 complex by mutating residues in the respective interfaces. As Jab1 does not stably interact with Aar2p or RH alone and as we could not monitor whether Jab1 was stably sequestered in mutant Aar2p–CTF complexes, we investigated the effects of mutations on binding of Jab1 to preformed Aar2p-–RH complexes by size exclusion chromatography (Fig. 3). Alanine substitution of L350 or Y352 (Fig. 1C, panel IV) at the very C terminus of Aar2p interfered with binding of Jab1 to the mutant Aar2p–RH complexes (Fig. 3D,E). Likewise, the reciprocal Y2170A mutation in Jab1 (Fig. 1C, panel IV) failed to form a complex with Aar2p–RH (Fig. 3F). The same result was obtained with a L2161A variant of Jab1 (Fig. 3G) in which a residue in the center of the RH–Jab1 interface is affected (Fig. 1C, panel VI). These results show that the integrity of both the Aar2p–Jab1 and the RH–Jab1 interfaces is required for efficient binding of Jab1 to Aar2p–RH.
Figure 3.
Alanine-scanning mutagenesis probing the Aar2p–Jab1 and RH–Jab1 interfaces. (A–G) Gel filtration analyses assessing the interaction between the Aar2p–RH complex and Jab1 and variants. A is identical to Figure 2C. Roman numerals in the panels on the right identify the respective interface (Fig. 1C). Other details and labels are as in Figure 2. Icons representing the proteins are defined in the top right. Rotation symbols indicate the views relative to Figure 1B.
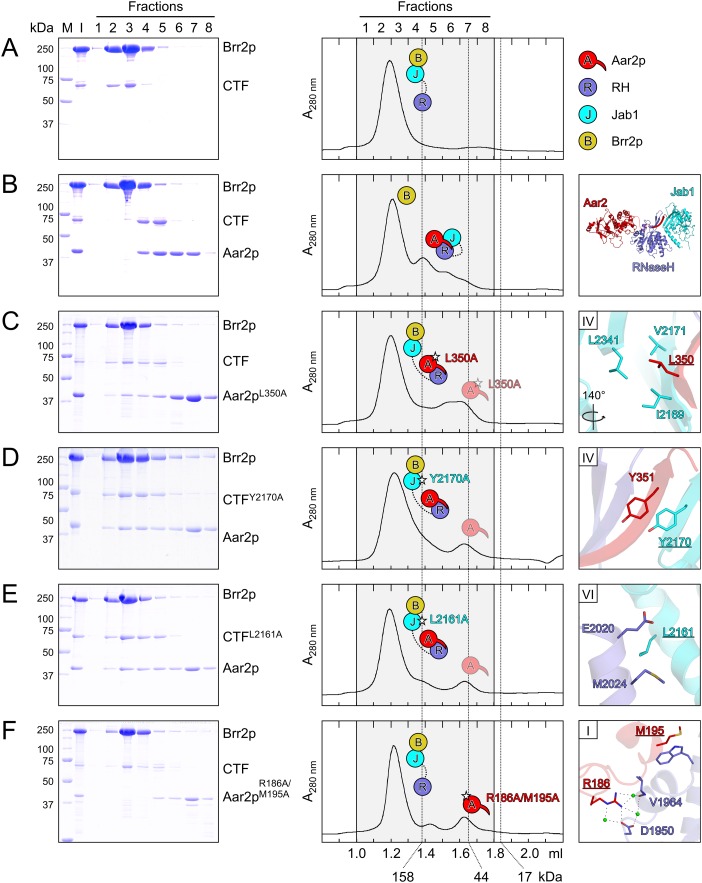
Brr2p interacts with CTF in gel filtration (Fig. 4A), but addition of Aar2p leads to formation of an Aar2p–CTF complex that excludes Brr2p (Weber et al. 2011). To investigate whether stable binding of the Jab1 domain to the composite Aar2p–RH binding site is required for exclusion of Brr2p from CTF, we introduced the above mutations into Aar2p or CTF and investigated the effect on Brr2p binding. We predicted that the mutations would lead to detachment of Jab1 from the Aar2p–RH subcomplex and allow formation of a complex in which Aar2p is bound to RH and Brr2p is bound to Jab1. In line with the observation that Aar2pL350A–RH fails to bind Jab1 (Fig. 3D), the Aar2pL350A mutant allowed formation of an Aar2pL350A–CTF–Brr2p complex (Fig. 4C). Similarly, CTFY2170A bearing a mutation in its Jab1 portion that interfered with Aar2p–RH–Jab1 complex formation (Fig. 3F) allowed concomitant binding of Aar2p and Brr2p to CTF (Fig. 4D). Finally, the same result was obtained with the CTFL2161A variant (Fig. 4E), which carries a mutation in the RH–Jab1 interface detrimental to Aar2p–RH–Jab1 complex formation (Fig. 3G).
Figure 4.
Effect of Aar2p and CTF variants on Brr2p binding. (A–F) Gel filtration analyses probing the interaction between Aar2p and CTF variants and Brr2p. Roman numerals in the panels on the right identify the respective interface (Fig. 1C). Other details and labeling are as in Figure 2. Icons representing the proteins are defined in the top right. (A) Brr2p binds the CTF. Rotation symbols indicate the views relative to Figure 1B. (B) Aar2p sequesters CTF from Brr2p. (C,D) Mutations affecting the interaction of the Aar2p C terminus with Jab1 allow the formation of a Aar2p–CTF–Brr2p complex. (E) Same effect of a mutation in the interface between RH and Jab1. (F) A double mutation in the Aar2p–RH interface does not allow Aar2p binding to the Brr2p–CTF complex.
In contrast to the mutations that affect the Aar2p–Jab1 or RH–Jab1 interfaces, the Aar2pR186A/M195A double mutant, which fails to form a complex with RH (Fig. 2F), was not able to sequester CTF from Brr2p or enter a Brr2p–CTF complex (Fig. 4F). Together, these results indicate that a stable interaction between the globular part of Aar2p and the palm of the Prp8p RH domain is a prerequisite for Aar2p to extend its C terminus across the RH domain and form a composite interface for Jab1 binding. Binding of Jab1 to this composite interface effectively counteracts Brr2p binding to the Jab1 domain.
A covalent connection between RH and Jab1 domains is required for Aar2p-mediated exclusion of Brr2p
While Aar2p was able to bind CTF in a manner that excluded Brr2p (Fig. 4B), Brr2p titrated the isolated Jab1 domain from an Aar2p–RH–Jab1 complex (Supplemental Fig. S6A); i.e., when the RH and Jab1 domains were not connected by the ∼60-residue intervening linker. We therefore investigated whether the linker itself establishes additional contacts that stabilize the Aar2p–CTF complex. To this end, we produced and purified a Jab1 construct that contained the entire linker sequence (Jab1linker; an analogous RHlinker construct was not produced soluble in Escherichia coli). The presence of the linker on Jab1 still allowed Brr2p to titrate the Jab1linker portion from an Aar2p–RH complex (Supplemental Fig. S6B). To investigate the binding thermodynamics of Jab1 and Jab1linker to Aar2p–RH, we conducted ITC experiments. Both Jab1 and Jab1linker bound to Aar2p–RH stoichiometrically (Supplemental Fig. S6C,D). The affinity of Jab1linker to Aar2p–RH was slightly reduced compared with Jab1 due to a larger entropy loss upon binding (Supplemental Fig. S6C,D). Together, these results suggest that the RH–Jab1 linker does not reinforce the Aar2p–CTF interaction by fostering direct contacts. Instead, it acts by tethering the two Prp8p C-terminal domains and thus increasing the local concentration of Jab1 in the neighborhood of RH and Aar2p.
Aar2p blocks U4/U6 di-snRNA binding to CTF
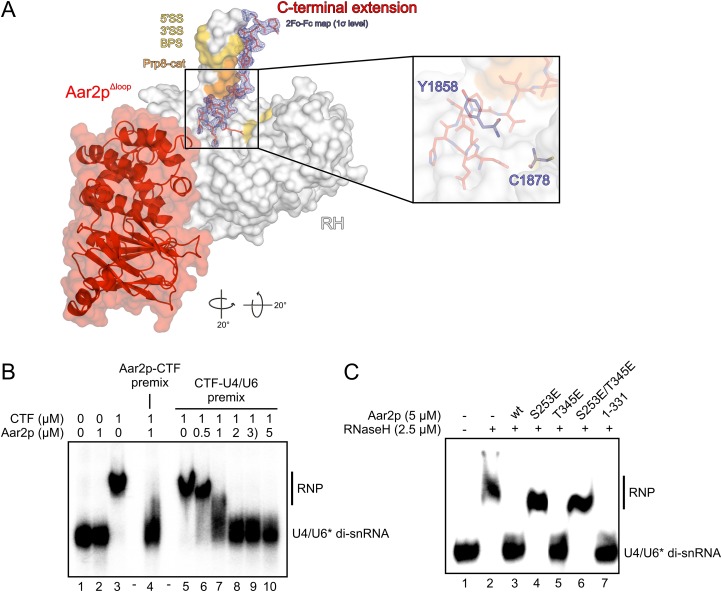
The Prp8p RH domain has been suggested to provide a platform for RNA transactions during spliceosome catalytic activation (Pena et al. 2008). Furthermore, it has been shown to bind several RNAs in vitro (Ritchie et al. 2008; Yang et al. 2008), with U4/U6 di-snRNA representing the strongest interactor known to date (Zhang et al. 2009; Mozaffari-Jovin et al. 2012). Notably, in our present Aar2pΔloop–RH–Jab1 structure, the C terminus of Aar2p occupies several regions on the RH domain that have been implicated in contacting RNA by cross-linking and genetic interactions (Fig. 5A). In particular, the Aar2p C terminus contacts the palm next to an area that could be cross-linked to the 5′ splice site at an early step of spliceosome assembly (Reyes et al. 1996, 1999). It directly interacts with residues V1860, T1861, V1862, and I1875 at the base of the thumb (Fig. 1C, panels II,III). Some prp8-cat alleles (Grainger and Beggs 2005) lead to mutations in these Prp8p residues and suppress u4-cs1 phenotypes originating from a hyperstabilized U4-cs1/U6 duplex (Kuhn et al. 1999; Kuhn and Brow 2000). Aar2p also interacts at the tip of the thumb, where mutations suppress defects in either splice site or the branch point sequence of a pre-mRNA (Umen and Guthrie 1995, 1996; Collins and Guthrie 1999; Siatecka et al. 1999; Query and Konarska 2004). Strikingly, P344 of Aar2p stacks squarely on Y1858, and the following residues run like a lid across C1878 (Figs.1C [panel II], 5A [inset]). These two residues of RH could be UV-cross-linked in vitro to a 3′ single-stranded region of U4 snRNA and a 5′ single-stranded region of U6 snRNA, respectively (Mozaffari-Jovin et al. 2012).
Figure 5.
Aar2p interferes with U4/U6 di-snRNA binding to CTF. (A) Binding of the Aar2p C terminus (in sticks) along the thumb of the RH domain (in surface representation). (Gold) Prp8p residues that, upon mutation, suppress defects in either splice site or the branch point sequence of a pre-mRNA (Umen and Guthrie 1995, 1996; Collins and Guthrie 1999; Siatecka et al. 1999; Query and Konarska 2004); (orange) Prp8-cat residues (Grainger and Beggs 2005), which, upon mutation, suppress u4-cs1 phenotypes (Kuhn et al. 1999; Kuhn and Brow 2000). The final 2Fo − Fc electron density contoured at the 1σ level around the Aar2p C terminus is shown as a blue mesh. Rotation symbols indicate the views relative to Figure 1B. (Inset) Position of Prp8p residues Y1858 and C1878 under the Aar2p C terminus, which could be UV-cross-linked in vitro to U4 snRNA and U6 snRNA, respectively (Mozaffari-Jovin et al. 2012). (B) EMSA monitoring binding of U4/U6 di-snRNA in the absence and presence of Aar2p. Protein names and concentrations are given at the top of the gel. (Premix) Preincubation of the indicated components; (U6*) [32P]-labeled U6 snRNA. (C) EMSA monitoring the effects of phospho-mimetic mutations (S253E, T345E, and S253E/T345E) and of a C-terminal deletion in Aar2p (Aar2p1–331) on its ability to compete with U4/U6 di-snRNA for RH.
The above observations suggested that Aar2p might interfere with RNA binding at the Prp8p RH domain. To test this idea, we performed electrophoretic mobility shift assays (EMSAs) with U4/U6 di-snRNA and either CTF (containing RH linked to Jab1) or RH in the absence and presence of Aar2p. Aar2p alone did not interact with U4/U6 di-snRNA (Fig. 5B, lane 2), while CTF bound the RNA complex efficiently (Fig. 5B, lane 3). CTF failed to bind U4/U6 when premixed with an equimolar amount of Aar2p (Fig. 5B, lane 4), and addition of increasing amounts of Aar2p to a preformed CTF–U4/U6 complex led to progressive dissociation of the RNP (Fig. 5B, lanes 5–10). Aar2p also counteracted U4/U6 binding to the RH domain alone (Fig. 5C, lane 3). Surprisingly, the Aar2p C-terminal extension that crosses the RH domain was not required for this effect, as C-terminally truncated Aar2p1–331 also interfered with U4/U6 binding by the RH domain (Fig. 5C, lane 7). Therefore, either stable binding of U4/U6 di-snRNA between the RH thumb and fingers requires surfaces on RH that are occluded by the globular part of Aar2p or this part of Aar2p blocks the path of U4/U6 across the RH domain (Fig. 5A). These data suggest that a second function of Aar2p may be to preclude premature or nonproductive RNA binding to the Prp8p C-terminal region.
Structural and functional consequences of phospho-mimetic mutations in Aar2p
Aar2p can be phosphorylated at five positions in vivo (S253, T274, Y328, S331, and T345) (Supplemental Fig. S1), and its reversible phosphorylation may regulate its ability to compete with Brr2p (Weber et al. 2011). Consistent with this idea, phospho-mimetic S253D or S253E mutations interfered with Aar2p–Prp8p interaction in extracts (Weber et al. 2011). S253 is located next to Aar2p helix α6 residues M195, F198, and L199, which form a hydrophobic pocket for W1911 of RH (Fig. 6A,B). To test whether this RH-binding region of Aar2p could be structurally affected by phosphorylation of S253, we produced and crystallized the Aar21–318,Δloop,S253E protein. This variant contained a C-terminal truncation to allow crystallization in the absence of Prp8p domains, the loop replacement also used for crystallization of the Aar2pΔloop–RH–Jab1 complex, and the phospho-mimetic S253E exchange. The crystal structure at 3.2 Å resolution contained two crystallographically independent Aar2p1–318,Δloop,S253E molecules, in both of which the helices α6/α7 region were dramatically reorganized (Fig. 6D–F). In the conformation seen in Aar2pSubt (Fig. 6D) and the Aar2pΔloop–RH–Jab1 complex (Fig. 6E), the larger E253 would clash with L205 in the N terminus of the α7–π1 helix. To accommodate E253, helix α7–π1 is rotated upwards, the π1 part adopts α-helical conformation, and residues L210 and F214 at the center and C terminus of helix α7 interact with the hydrophobic part of E253, whose carboxyl group is exposed to the solvent (Fig. 6F). As a consequence, the H193–R190–Y203 stacking interaction that anchors the loop preceding helix α6 to the N terminus of helix α7 is disrupted, and helix α6 unfolds completely. In Aar2p1–318,Δloop,S253E, helix α6 residues M195, F198, and L199, which form a key binding site for RH that is preformed in Aar2pSubt, come to lie in a partially disordered loop. The resulting conformation is sterically incompatible with binding to Prp8. We expect that a similar conformational change will be observed upon phosphorylation of S253.
Figure 6.
Effect of phospho-mimetic mutations in Aar2p on Brr2p–CTF complex formation. (A) Position of two Aar2p phosphorylation sites, S253 and T345, in the Aar2pΔloop–RH–Jab1 complex structure. Colors and labels are as in Figure 1C. (B) Close-up view of Aar2p residue S253 (underlined) and its environment. (C) Close-up view of Aar2p residue T345 (underlined) and its environment. (D–F) Comparison of the structures of Aar2pSubt (Weber et al. 2011), the Aar2pΔloop–RH–Jab1 complex, and Aar2p1–318,Δloop,S253E. (Blue mesh in F) 2Fo − Fc electron density covering the α7 region contoured at the 1σ level. Rotation symbols indicate the views relative to Figure 1B. Insets show the α6/α7 region in these structures, which is remodeled upon introducing the S253E mutation. Rotation symbols indicate the views relative to the overviews. (G,H) Gel filtration analysis probing the interaction of Aar2p bearing phospho-mimetic mutations with CTF and Brr2p. Details and labels are as in Figure 2. Icons representing the proteins are defined in the top right. All rotations indicated are relative to Figure 1B. (G) Aar2pT345E still binds CTF and excludes Brr2p. (H) The Aar2pS253E/T345E variant with two phospho-mimetic mutations no longer interferes with Brr2p–CTF complex formation.
Below helix α7 in the Aar2p1–318,Δloop,S253E structure, C251 from helix α9 and C292 from helix α12 reorganized to form a disulfide bridge not seen in any previous structure containing S253 Aar2p variants (Fig. 6F). We believe that this disulfide has formed artificially upon loss of reductive conditions during crystallization, showing an increased flexibility in the region below helix α7 upon introducing the S253E exchange.
Notwithstanding these S253E-triggered conformational changes, Brr2p competition was only partially relieved in Aar2pS253E in vitro (Weber et al. 2011), suggesting that Aar2pS253E can still be driven into a structure that resembles wild-type Aar2p upon contacting Prp8. We therefore investigated whether additional phosphorylation events may contribute to reducing Aar2p affinity for CTF. T274 lies remote from the RH or Jab1 interfaces on the opposite side of Aar2p and is unlikely to exert a direct effect on CTF binding upon phosphorylation. Residues Y328 and S331 map to the flexible peptide connecting the bulk of Aar2p to its C terminus and were not visible in our structure. However, T345 lies in a groove at the base of the RH thumb and engages in water-mediated interactions with RH residues T1855 and R1937 (Fig. 6A,C). Phosphorylation might break this water network and sterically interfere with the interaction. While a T345E version of Aar2p still effectively competed with Brr2p for binding to CTF (Fig. 6G), Brr2p was able to fully titrate CTF from the Aar2pS253E/T345E double mutant (Fig. 6H).
ITC experiments (Supplemental Fig. 6E–H) corroborated these findings. Affinities of Aar2pT345E and Aar2pS253E to CTF were reduced about fivefold and 10-fold, respectively, compared with wild-type Aar2p, while the double Aar2pS253E/T345E mutant showed ∼170-fold weaker binding. Notably, thermodynamic parameters of these interactions are in line with our structural findings. To bind CTF, the disordered loop preceding helix α7 in isolated Aar2pS253E is expected to contract and adopt a structure similar to wild-type Aar2p. This scenario predicts an enthalpic gain (due to the folding upon binding) and an entropic loss (due to compaction and immobilization of the unstructured region) compared with the wild type as indeed seen in ITC (Supplemental Fig. 6E–G). Moreover, the clear effects seen in ITC with freshly produced, reduced Aar2pS253E protein argue that the disulfide bond seen in Aar2p1–318,Δloop,S253E can form as a result of conformational changes due to the S253E mutation rather than causing these conformational changes. Together, these results suggest that phosphorylations at S253 and T345 cooperatively reduce the ability of Aar2p to bind Prp8.
We also tested whether the above phospho-mimetic mutations in Aar2p affect its ability to interfere with the Prp8p–U4/U6 interaction. Gel shift analyses indeed revealed effects similar to those in the modulation of the Prp8p–Brr2p interaction (Fig. 5C). Like C-terminally truncated Aar2p1–331 (Fig. 5C, lane 7), Aar2pT345E still counteracted RH–U4/U6 complex formation, whereas both Aar2pS253E and the double mutant Aar2pS253E/T345E were no longer able to compete with U4/U6 for RH (Fig. 5C, lanes 4–6). These results suggest that in addition to regulating Prp8p–Brr2p interactions, reversible phosphorylation of Aar2p can also control U4/U6 di-snRNA binding at the Prp8p C-terminal region.
Mutations interfering with reversible phosphorylation of Aar2p affect cell viability
To evaluate the cellular consequences of interfering with reversible phosphorylation of Aar2p at specific sites, we systematically replaced the endogenous copy of Aar2p with variants bearing either a nonphosphorylatable alanine or phospho-mimetic aspartic acid or glutamic acid residues at positions 253, 274, 328, 331, or 345. Aar2p variants S253A, S331A, and T274E, when present on the genome, caused a cold-sensitive (cs) growth phenotype (Supplemental Fig. S7A). While our structural and biochemical findings do not provide an explanation for the effects of the S331A and T274E mutations, the growth defect in the case of Aar2pS253A may be explained by enhanced or prolonged binding of the mutant Aar2p to Prp8p, blocking Brr2p binding or activation, or Prp8p binding to U4/U6 di-snRNA, thus resulting in cs U4/U6 unwinding and reduced splicing. Moreover, using the same approach, viable yeast cells bearing double S253A/T345A or S253E/T345E mutations could not be recovered at 30°C, indicating that concomitant phospho-mimetic or nonphosphorylatable mutations at these two residues are synthetic-lethal. This observation is in line with our finding that phospho-mimetic residues at these sites cooperatively weaken the Aar2p–Prp8 interaction in vitro.
We also investigated the effect of Aar2p phospho-mimetic mutations on the growth inhibition caused by co-overproducing Aar2p and Prp8p CTFs (Supplemental Fig. S7B). Prp8 fragment E1 (residues 1649–2413) starts N-terminal of the RH domain and extends to the end of Prp8p. Fragment E3 (residues 2010–2413) includes the C-terminal part of the RH domain and the Jab1 domain. Fragment E3H corresponds to E3 with two point mutations (Y2037H and I2051T) that increase the interaction of the fragment with Brr2p (van Nues and Beggs 2001). The results (Supplemental Fig. S7B) are summarized in Supplemental Table S3. Co-overproduction with E1 had few discernible effects. However, as we reported previously (Weber et al. 2011), overproduction of E3 and, more so, E3H in the presence of Aar2p is detrimental for growth but is counteracted by the S253E mutation.
We now see that substitutions of T274 and T345 in Aar2p have differential effects with E3 and E3H, which are similar for both alanine and aspartate/glutamate substitutions. The E3 and E3H fragments lack most of the RH domain, including portions that build up contact regions I, II, and III with Aar2p in the Aar2pΔloop–RH–Jab1 complex (Fig. 1B). Thus, Aar2p or variants and E3 or E3H fragments should act physically independently of each other. Based on our findings, Aar2p competes with Brr2p for Prp8p. E3 and E3H are expected to disturb this equilibrium by binding Brr2p in competition with endogenous Prp8p. Since E3H has a higher affinity for Brr2p, it is expected to compete more efficiently, consistent with its higher toxicity. Based on our in vitro structural and binding studies, Aar2pT345E exhibits a weaker affinity for Prp8p than wild-type Aar2p. When overproduced, Aar2pT345E will, to some extent, compete with endogenous wild-type Aar2p for Prp8p and provide a Prp8p pool associated with a more easily detached Aar2p variant. In this fashion, Aar2pT345E overproduction could make up for the reduced Prp8p-binding capacity of Brr2p in the presence of E3 or E3H. Again, we cannot explain the T274 effects based on our structural and biochemical findings. Taken together, the growth phenotypes observed upon replacing wild-type Aar2p with phospho-mutants and upon co-overproducing Aar2p phospho-mutants and Prp8p CTF could be explained by the weakening of Aar2p–Prp8p interaction upon phosphorylation of Aar2p residues S253 and T345.
Discussion
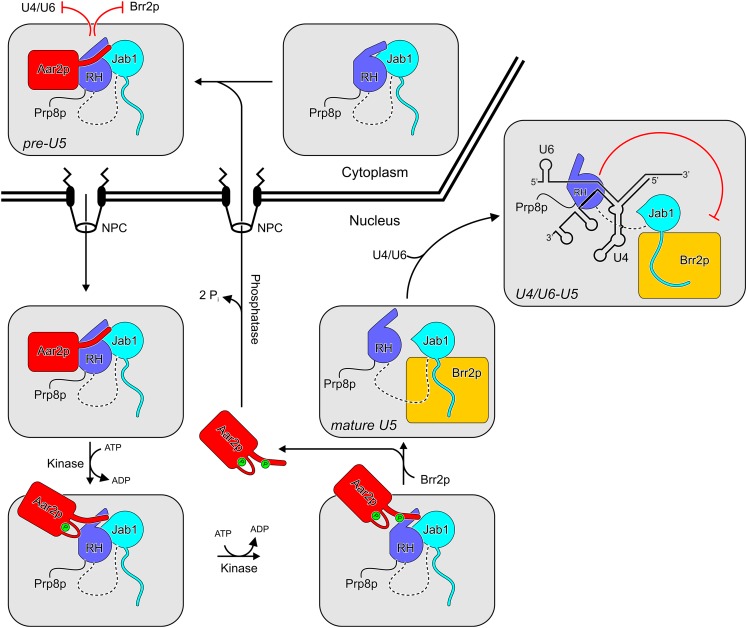
Apart from the PRMT5 and SMN complexes that guide the formation of the Sm core RNPs, only a small number of factors have been characterized that are intermediate subunits of snRNPs but are not integrated into higher-order snRNP assemblies or the spliceosome, representing bona fide assembly factors or chaperones. Perhaps the most prominent example is the Prp24 protein (human p110), which is a subunit of U6 snRNP but is not found in U4/U6 di-snRNP and which facilitates the annealing of U4 and U6 snRNAs during U4/U6 di-snRNP formation (Raghunathan and Guthrie 1998b; Bell et al. 2002). In addition, Lin1p (human U5-52K protein) is specifically associated with U5 snRNP but is released upon U4/U6-U5 tri-snRNP formation (Laggerbauer et al. 2005). The specific function of this protein is presently unknown. Furthermore, several proteins that are associated with spliceosomes have been attributed an additional snRNP assembly function. Both the Lsm2–8 protein complex of U6 snRNP (Rader and Guthrie 2002; Ryan et al. 2002; Verdone et al. 2004) and the Prp19 complex (Chen et al. 2006) were implicated in U4/U6 di-snRNP biogenesis. The Aar2p protein represents the most recent addition to this group of putative or proven snRNP assembly chaperones. Aar2p was the first example of a factor that regulates the incorporation of a snRNP-specific protein, Brr2p. As shown here, it can additionally regulate U4/U6 di-snRNA binding to Prp8p. We employed a combined structural and biochemical approach to investigate the molecular mechanisms underlying these possibly functionally related activities of Aar2p in detail (Fig. 7).
Figure 7.
Scheme illustrating the functions of Aar2p during U5 snRNP assembly. In pre-U5 snRNP, Aar2p blocks RNA (e.g., U4/U6) and Brr2p binding to the Prp8p C-terminal region. Phosphorylation of Aar2p at S253 and T345 in the nucleus (by unknown kinases) leads to its detachment from Prp8p and allows Brr2p binding to form mature U5 snRNP. The same signals allow the RH domain of Prp8p to bind U4/U6 di-snRNA and inhibit their premature unwinding by Brr2p in the U4/U6-U5 tri-snRNP. The precise sequence of phosphorylation events is presently unknown. To participate in further rounds of U5 snRNP assembly, Aar2p has to be dephosphorylated by an unknown phosphatase and shuttled back to the cytoplasm. (NPC) Nuclear pore complex.
Aar2p cements an autoinhibited structure in the C-terminal region of Prp8p
Brr2p has been shown to directly bind the Jab1 domain of Prp8p (Weber et al. 2011). Notably, the unstructured C-terminal tail of the Jab1 domain, which is crucial for Brr2p binding (Boon et al. 2007; Pena et al. 2007; Maeder et al. 2009), is not involved in contacts to Aar2p or RH in our present complex structure. Nevertheless, Aar2p efficiently competes with Brr2p binding to Prp8p CTF (Fig. 4B). While Aar2p or RH could occupy additional essential Brr2p interaction regions on Jab1, we favor the idea that they are attached to Jab1 in a manner that sterically interferes with Brr2p binding. This latter interpretation would provide a straightforward explanation for the enhanced binding of the E3H fragment to Brr2p compared with E3 (van Nues and Beggs 2001). One of the residues mutated in E3H, I2051, stabilizes contact region VI of the present Aar2pΔloop–RH–Jab1 complex (Fig. 1C, panel VI) between the RH and Jab1 domains. Assuming that the RH portion in E3H sterically hinders Brr2p from interacting with Jab1, weakening of the RH–Jab1 interaction should alleviate this inhibition. Attractively, this explanation for the gain-of-function phenotype of E3H does not invoke an unlikely improvement of direct interactions between the mutated residues in E3H and Brr2p.
As essentially the same Jab1 surfaces are covered by RH alone in the structure of the isolated CTF as by RH together with the slender Aar2p C-terminal β strand in the Aar2pΔloop–RH–Jab1 complex (Supplemental Fig. S3C,D), we expect that CTF in its observed closed conformation would also be unable to bind Brr2p and would have to undergo a conformational change to interact with the helicase. As Brr2p efficiently binds CTF in the absence of Aar2p in vitro (Fig. 4A), it can break open the presumed autoinhibitory conformation of CTF. Aar2p reinforces the inhibitory RH–Jab1 interaction and may render this part of Prp8p more rigid so that conformational changes required for Brr2 binding can no longer occur. CTF resembles many other regulatory proteins, which contain multiple domains that interact with different binding partners (Pawson and Nash 2003). Autoinhibition via intramolecular domain–domain interactions is a widespread regulatory principle in such factors (Pufall and Graves 2002; Mackereth et al. 2011; Burmann et al. 2012). In the present case, Aar2p adds an additional level of regulation, representing an assembly factor that counteracts Brr2p binding to Prp8p by reinforcing an autoinhibited conformation in Prp8p CTF.
Aar2p coordinates the binding of Brr2p and U4/U6 di-snRNA to the Prp8p C-terminal region
Our results show that apart from competing with Brr2p for binding to Prp8p, Aar2p also interferes with binding of U4/U6 di-snRNA to Prp8p CTF (Fig. 5B,C). The latter observation may simply indicate an additional role of Aar2p in safeguarding against premature or nonspecific RNA interactions at the Prp8p C-terminal region during U5 snRNP assembly. However, the two seemingly disparate activities of Aar2p may also be functionally closely interrelated. During spliceosome catalytic activation, Brr2p unwinds the U4/U6 duplex and displaces all U4/U6-associated proteins (Laggerbauer et al. 1998; Raghunathan and Guthrie 1998a). This activity of Brr2p needs to be tightly controlled because the enzyme already encounters its U4/U6 substrate in the U4/U6–U5 tri-snRNP before incorporation into the spliceosome. We recently found that the Prp8p RH domain acts as an inhibitor of Brr2p by occluding regions of U4/U6 di-snRNA also required for Brr2p binding (Mozaffari-Jovin et al. 2012). Our present competition assays (Fig. 5B,C) show that the RH domain is released from Aar2p by the same signals that make the Jab1 available to Brr2p. Thus, with the entry of Brr2p into U5 snRNP, the Prp8p RH domain is ready to dock the U4/U6 di-snRNA (Fig. 7). This assembly strategy would thus be suitable to ensure that mature U5 snRNP is incorporated into U4/U6–U5 tri-snRNP in a way that precludes premature unwinding of U4/U6.
It is possible that the subnuclear sites of tri-snRNP assembly coincide with the locations where U5 snRNP is matured; e.g., the Cajal bodies in higher eukaryotes. On the other hand, a free pool of mature U5 snRNP exists in yeast (Stevens et al. 2001) and other eukaryotes (Bringmann and Lührmann 1986). Therefore, further work is required to determine whether Aar2p only functions as a U5 snRNP assembly factor or additionally coordinates or physically links U5 snRNP and U4/U6-U5 tri-snRNP assembly.
Strategy for Aar2p release
During the assembly of macromolecular machines, assembly factors may engage in interactions that have to be released again during later stages of assembly or have to unleash biological activity (Chari and Fischer 2010). Obviously, the affinities of such factors for their assembly intermediates have to be finely tuned to avoid terminal stalling of the assembly process. In the case of Aar2p, the sizeable Aar2p–CTF interaction surface (2125 Å2 of buried surface area) is distributed among several smaller interfaces (Fig. 1C, panels I–VI). Each of these interfaces alone is easier to disrupt than a continuous protein–protein interaction surface of the combined size (Fig. 7). A similar strategy has been suggested for the σ factor that guides bacterial RNA polymerase to its promoters but has to be released upon entry of the elongation phase (Murakami and Darst 2003).
Our observations with phospho-mimetic mutations suggest that the affinity of Aar2p for CTF is regulated by reversible phosphorylation (Weber et al. 2011). We identified two putative regulatory residues: S253 and T345. Phospho-mimetic mutations at these residues cooperatively reduced Aar2p interaction with Prp8p CTF (Fig. 6; Supplemental Fig. S6E–H), and double nonphosphorylatable S253A/T345A or phospho-mimetic S253E/T345E mutations were synthetic-lethal. These observations suggest that phosphorylation of S253 and T345 of Aar2p affect the same process; i.e., release of Aar2p from Prp8p during U5 snRNP assembly. The distribution of the two residues among the Aar2p–Prp8p interfaces suggests that down-regulation of Aar2p affinity by phosphorylation is efficiently combined with facilitation of Aar2p release based on multiple small contact surfaces—phosphorylation of S253 interferes with the contact region I, and phosphorylation of T345 interferes with contact region II (Figs. 1C, 7).
The above principles are corroborated by our observation that the interaction of the Aar2p C-terminal expansion alone with CTF is very weak (Fig. 1E). Although this interaction is highly specific due to the large buried surface area (1195 Å2), the intrinsically unstructured nature of the Aar2p C-terminal expansion (and the concomitant loss in entropy associated with its immobilization) renders the interaction only marginally stable and dependent on the anchoring of the globular part of Aar2p on RH (Fig. 1D,E) and allows it to be easily undone; e.g., via a phosphorylation event. Thus, Aar2p combines several strategies (dissection of the total interaction surface, multiple phosphorylation events that affect different interfaces, and entropy-enthalpy compensation) to act as a reversible inhibitor of macromolecular interactions during U5 snRNP assembly.
Possible roles of Aar2p in spliceosome disassembly and U5 snRNP recycling
During the splicing process, several snRNPs are profoundly remodeled (Staley and Guthrie 1998; Wahl et al. 2009). Thus, for continued pre-mRNA splicing, the snRNPs have to be regenerated after each round of splicing. Interestingly, it has been observed that Aar2p-depleted nuclear extract did not permit consecutive rounds of splicing (Gottschalk et al. 2001). Thus, Aar2p not only may be involved in the de novo biogenesis of U5 snRNP, but could also play a role in spliceosome disassembly and/or U5 snRNP recycling.
Spliceosome disassembly is the least understood phase of the splicing reaction. While proteomics analyses of purified spliceosomal complexes have not detected Aar2p in spliceosome assembly or catalysis intermediates, the complete proteome of post-splicing complexes is presently unknown. The Prp8p RH domain is thought to act as an assembly platform for the catalytic RNA network of the spliceosome (Pena et al. 2008; Ritchie et al. 2008; Yang et al. 2008). However, UV cross-links between components of this network and other regions of Prp8p during later stages of the splicing reaction (Turner et al. 2006) suggest that catalytic core RNAs are handed over to other binding sites after initial assembly on the RH domain. Thus, late in the splicing cycle, the Prp8p RH domain may be available for rebinding of Aar2p. Whether rebinding of Aar2p at RH actually occurs, whether it would also affect Prp8p–Brr2p interactions during spliceosome disassembly, and whether U5 snRNP recycling resembles its de novo biogenesis pathway remain to be investigated.
Materials and methods
Protein production and crystallographic analyses
All yeast proteins for biochemical, biophysical, and structural studies, except Brr2p, were produced in E. coli and purified to near homogeneity by chromatographic techniques. Brr2p was produced in insect cell culture (Santos et al. 2012). Crystallization was performed by the sitting drop vapor diffusion method, and diffraction data were collected on beamline 14.2 of the BESSY II storage ring. The structures were solved by molecular replacement and refined with standard protocols. Details for these and other methods are given in the Supplemental Material.
ITC and EMSA
ITC measurements were conducted on a MicroCal iTC200 system (GE Healthcare) using Jab1 and Jab1linker as a sample and Aar2p–RH complex as the titrant. U4 and U6 snRNAs were in vitro transcribed by T7 polymerase. The U6 snRNA was 5′-end-radiolabeled and annealed to the U4 snRNA, followed by purification of the U4/U6 di-snRNA by 6% nondenaturing PAGE. Complex formation was analyzed on a native polyacrylamide gel and visualized by autoradiography.
Database deposition
Structure coordinates and diffraction data were deposited with the Protein Data Bank (http://www.pdb.org) under accession codes 4ILG (Aar2pΔloop-RH-Jab1), 4ILJ (RHW1911A), 4ILH (Aar2p1–331-RH), and 4ILI (Aar2p1–318,Δloop,S253E) and will be released upon publication.
Acknowledgments
We thank Claudia Kipar, Claudia Alings, and Traudy Wandersleben (Freie Universität Berlin) for help with cloning, expression, protein preparation, in vitro interaction analysis, and crystallization, and Amit Gautam (University of Edinburgh) for aar2 double mutant analysis in vivo. We acknowledge access to beamline BL14.2 of the BESSY II storage ring (Berlin, Germany) via the Joint Berlin MX-Laboratory sponsored by the Helmholtz Zentrum Berlin für Materialien und Energie, the Freie Universität Berlin, the Humboldt-Universität zu Berlin, the Max-Delbrück Centrum, and the Leibniz-Institut für Molekulare Pharmakologie. This work was supported by the Deutsche Forschungsgemeinschaft (grants SFB 740/2 and WA 1126/4-1 to M.C.W.) and the Wellcome Trust (grant 087551 to J.D.B.). J.D.B. holds the Royal Society Darwin Trust Research Professorship. G.W., V.F.C., K.F.S., S.M.J., A.C.H., and N.H. performed the experiments. All authors contributed to the analysis of the data and the interpretation of the results. G.W., R.L., J.D.B., and M.C.W. designed and coordinated the study. G.W., J.D.B., and M.C.W. wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.213207.113.
References
- Bartels C, Klatt C, Lührmann R, Fabrizio P 2002. The ribosomal translocase homologue Snu114p is involved in unwinding U4/U6 RNA during activation of the spliceosome. EMBO Rep 3: 875–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels C, Urlaub H, Lührmann R, Fabrizio P 2003. Mutagenesis suggests several roles of Snu114p in pre-mRNA splicing. J Biol Chem 278: 28324–28334 [DOI] [PubMed] [Google Scholar]
- Battle DJ, Kasim M, Yong J, Lotti F, Lau CK, Mouaikel J, Zhang Z, Han K, Wan L, Dreyfuss G 2006. The SMN complex: An assembly machine for RNPs. Cold Spring Harb Symp Quant Biol 71: 313–320 [DOI] [PubMed] [Google Scholar]
- Bell M, Schreiner S, Damianov A, Reddy R, Bindereif A 2002. p110, a novel human U6 snRNP protein and U4/U6 snRNP recycling factor. EMBO J 21: 2724–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon KL, Grainger RJ, Ehsani P, Barrass JD, Auchynnikava T, Inglehearn CF, Beggs JD 2007. prp8 mutations that cause human retinitis pigmentosa lead to a U5 snRNP maturation defect in yeast. Nat Struct Mol Biol 14: 1077–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner TJ, Guthrie C 2005. Genetic analysis reveals a role for the C terminus of the Saccharomyces cerevisiae GTPase Snu114 during spliceosome activation. Genetics 170: 1063–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann P, Lührmann R 1986. Purification of the individual snRNPs U1, U2, U5 and U4/U6 from HeLa cells and characterization of their protein constituents. EMBO J 5: 3509–3516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmann BM, Knauer SH, Sevostyanova A, Schweimer K, Mooney RA, Landick R, Artsimovitch I, Rösch P 2012. An α helix to β barrel domain switch transforms the transcription factor RfaH into a translation factor. Cell 150: 291–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chari A, Fischer U 2010. Cellular strategies for the assembly of molecular machines. Trends Biochem Sci 35: 676–683 [DOI] [PubMed] [Google Scholar]
- Chen CH, Kao DI, Chan SP, Kao TC, Lin JY, Cheng SC 2006. Functional links between the Prp19-associated complex, U4/U6 biogenesis, and spliceosome recycling. RNA 12: 765–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins CA, Guthrie C 1999. Allele-specific genetic interactions between Prp8 and RNA active site residues suggest a function for Prp8 at the catalytic core of the spliceosome. Genes Dev 13: 1970–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U, Englbrecht C, Chari A 2011. Biogenesis of spliceosomal small nuclear ribonucleoproteins. Wiley Interdiscip Rev RNA 2: 718–731 [DOI] [PubMed] [Google Scholar]
- Galej WP, Oubridge C, Newman AJ, Nagai K 2013. Crystal structure of Prp8 reveals active site cavity of the spliceosome. Nature 493: 638–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk A, Kastner B, Lührmann R, Fabrizio P 2001. The yeast U5 snRNP coisolated with the U1 snRNP has an unexpected protein composition and includes the splicing factor Aar2p. RNA 7: 1554–1565 [PMC free article] [PubMed] [Google Scholar]
- Grainger RJ, Beggs JD 2005. Prp8 protein: At the heart of the spliceosome. RNA 11: 533–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn D, Beggs JD 2010. Brr2p RNA helicase with a split personality: Insights into structure and function. Biochem Soc Trans 38: 1105–1109 [DOI] [PubMed] [Google Scholar]
- Janin J 1997. Specific versus non-specific contacts in protein crystals. Nat Struct Biol 4: 973–974 [DOI] [PubMed] [Google Scholar]
- Krissinel E 2009. Crystal contacts as nature's docking solutions. J Comput Chem 31: 133–143 [DOI] [PubMed] [Google Scholar]
- Kuhn AN, Brow DA 2000. Suppressors of a cold-sensitive mutation in yeast U4 RNA define five domains in the splicing factor Prp8 that influence spliceosome activation. Genetics 155: 1667–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn AN, Li Z, Brow DA 1999. Splicing factor Prp8 governs U4/U6 RNA unwinding during activation of the spliceosome. Mol Cell 3: 65–75 [DOI] [PubMed] [Google Scholar]
- Laggerbauer B, Achsel T, Lührmann R 1998. The human U5-200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc Natl Acad Sci 95: 4188–4192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laggerbauer B, Liu S, Makarov E, Vornlocher HP, Makarova O, Ingelfinger D, Achsel T, Lührmann R 2005. The human U5 snRNP 52K protein (CD2BP2) interacts with U5-102K (hPrp6), a U4/U6.U5 tri-snRNP bridging protein, but dissociates upon tri-snRNP formation. RNA 11: 598–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Nagai K, Li J 2011. Structure of the spliceosomal U4 snRNP core domain and its implication for snRNP biogenesis. Nature 473: 536–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackereth CD, Madl T, Bonnal S, Simon B, Zanier K, Gasch A, Rybin V, Valcarcel J, Sattler M 2011. Multi-domain conformational selection underlies pre-mRNA splicing regulation by U2AF. Nature 475: 408–411 [DOI] [PubMed] [Google Scholar]
- Maeder C, Kutach AK, Guthrie C 2009. ATP-dependent unwinding of U4/U6 snRNAs by the Brr2 helicase requires the C terminus of Prp8. Nat Struct Mol Biol 16: 42–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozaffari-Jovin S, Santos KF, Hsiao HH, Will CL, Urlaub H, Wahl MC, Lührmann R 2012. The Prp8 RNase H-like domain inhibits Brr2-mediated U4/U6 snRNA unwinding by blocking Brr2 loading onto the U4 snRNA. Genes Dev 26: 2422–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami KS, Darst SA 2003. Bacterial RNA polymerases: The wholo story. Curr Opin Struct Biol 13: 31–39 [DOI] [PubMed] [Google Scholar]
- Nakazawa N, Harashima S, Oshima Y 1991. AAR2, a gene for splicing pre-mRNA of the MATa1 cistron in cell type control of Saccharomyces cerevisiae. Mol Cell Biol 11: 5693–5700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesic D, Tanackovic G, Krämer A 2004. A role for Cajal bodies in the final steps of U2 snRNP biogenesis. J Cell Sci 117: 4423–4433 [DOI] [PubMed] [Google Scholar]
- Pandit S, Lynn B, Rymond BC 2006. Inhibition of a spliceosome turnover pathway suppresses splicing defects. Proc Natl Acad Sci 103: 13700–13705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SB, Bellini M 2008. The assembly of a spliceosomal small nuclear ribonucleoprotein particle. Nucleic Acids Res 36: 6482–6493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T, Nash P 2003. Assembly of cell regulatory systems through protein interaction domains. Science 300: 445–452 [DOI] [PubMed] [Google Scholar]
- Pena V, Liu S, Bujnicki JM, Lührmann R, Wahl MC 2007. Structure of a multipartite protein-protein interaction domain in splicing factor prp8 and its link to retinitis pigmentosa. Mol Cell 25: 615–624 [DOI] [PubMed] [Google Scholar]
- Pena V, Rozov A, Fabrizio P, Lührmann R, Wahl MC 2008. Structure and function of an RNase H domain at the heart of the spliceosome. EMBO J 27: 2929–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeranz Krummel DA, Oubridge C, Leung AK, Li J, Nagai K 2009. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature 458: 475–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pufall MA, Graves BJ 2002. Autoinhibitory domains: Modular effectors of cellular regulation. Annu Rev Cell Dev Biol 18: 421–462 [DOI] [PubMed] [Google Scholar]
- Query CC, Konarska MM 2004. Suppression of multiple substrate mutations by spliceosomal prp8 alleles suggests functional correlations with ribosomal ambiguity mutants. Mol Cell 14: 343–354 [DOI] [PubMed] [Google Scholar]
- Rader SD, Guthrie C 2002. A conserved Lsm-interaction motif in Prp24 required for efficient U4/U6 di-snRNP formation. RNA 8: 1378–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunathan PL, Guthrie C 1998a. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr Biol 8: 847–855 [DOI] [PubMed] [Google Scholar]
- Raghunathan PL, Guthrie C 1998b. A spliceosomal recycling factor that reanneals U4 and U6 small nuclear ribonucleoprotein particles. Science 279: 857–860 [DOI] [PubMed] [Google Scholar]
- Reyes JL, Kois P, Konforti BB, Konarska MM 1996. The canonical GU dinucleotide at the 5′ splice site is recognized by p220 of the U5 snRNP within the spliceosome. RNA 2: 213–225 [PMC free article] [PubMed] [Google Scholar]
- Reyes JL, Gustafson EH, Luo HR, Moore MJ, Konarska MM 1999. The C-terminal region of hPrp8 interacts with the conserved GU dinucleotide at the 5′ splice site. RNA 5: 167–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie DB, Schellenberg MJ, Gesner EM, Raithatha SA, Stuart DT, Macmillan AM 2008. Structural elucidation of a PRP8 core domain from the heart of the spliceosome. Nat Struct Mol Biol 15: 1199–1205 [DOI] [PubMed] [Google Scholar]
- Ryan DE, Stevens SW, Abelson J 2002. The 5′ and 3′ domains of yeast U6 snRNA: Lsm proteins facilitate binding of Prp24 protein to the U6 telestem region. RNA 8: 1011–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos KF, Jovin SM, Weber G, Pena V, Lührmann R, Wahl MC 2012. Structural basis for functional cooperation between tandem helicase cassettes in Brr2-mediated remodeling of the spliceosome. Proc Natl Acad Sci 109: 17418–17423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffert N, Hossbach M, Heintzmann R, Achsel T, Lührmann R 2004. RNAi knockdown of hPrp31 leads to an accumulation of U4/U6 di-snRNPs in Cajal bodies. EMBO J 23: 3000–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siatecka M, Reyes JL, Konarska MM 1999. Functional interactions of Prp8 with both splice sites at the spliceosomal catalytic center. Genes Dev 13: 1983–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small EC, Leggett SR, Winans AA, Staley JP 2006. The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H box ATPase. Mol Cell 23: 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C 1998. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell 92: 315–326 [DOI] [PubMed] [Google Scholar]
- Stanek D, Neugebauer KM 2004. Detection of snRNP assembly intermediates in Cajal bodies by fluorescence resonance energy transfer. J Cell Biol 166: 1015–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens SW, Barta I, Ge HY, Moore RE, Young MK, Lee TD, Abelson J 2001. Biochemical and genetic analyses of the U5, U6, and U4/U6 x U5 small nuclear ribonucleoproteins from Saccharomyces cerevisiae. RNA 7: 1543–1553 [PMC free article] [PubMed] [Google Scholar]
- Turner IA, Norman CM, Churcher MJ, Newman AJ 2006. Dissection of Prp8 protein defines multiple interactions with crucial RNA sequences in the catalytic core of the spliceosome. RNA 12: 375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen JG, Guthrie C 1995. A novel role for a U5 snRNP protein in 3′ splice site selection. Genes Dev 9: 855–868 [DOI] [PubMed] [Google Scholar]
- Umen JG, Guthrie C 1996. Mutagenesis of the yeast gene PRP8 reveals domains governing the specificity and fidelity of 3′ splice site selection. Genetics 143: 723–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nues RW, Beggs JD 2001. Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes of Saccharomyces cerevisiae. Genetics 157: 1451–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdone L, Galardi S, Page D, Beggs JD 2004. Lsm proteins promote regeneration of pre-mRNA splicing activity. Curr Biol 14: 1487–1491 [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R 2009. The spliceosome: Design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Weber G, Trowitzsch S, Kastner B, Lührmann R, Wahl MC 2010. Functional organization of the Sm core in the crystal structure of human U1 snRNP. EMBO J 29: 4172–4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber G, Cristão VF, de L Alves F, Santos KF, Holton N, Rappsilber J, Beggs JD, Wahl MC 2011. Mechanism for Aar2p function as a U5 snRNP assembly factor. Genes Dev 25: 1601–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Lührmann R 2001. Spliceosomal UsnRNP biogenesis, structure and function. Curr Opin Cell Biol 13: 290–301 [DOI] [PubMed] [Google Scholar]
- Yang K, Zhang L, Xu T, Heroux A, Zhao R 2008. Crystal structure of the β-finger domain of Prp8 reveals analogy to ribosomal proteins. Proc Natl Acad Sci 105: 13817–13822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Xu T, Maeder C, Bud LO, Shanks J, Nix J, Guthrie C, Pleiss JA, Zhao R 2009. Structural evidence for consecutive Hel308-like modules in the spliceosomal ATPase Brr2. Nat Struct Mol Biol 16: 731–739 [DOI] [PMC free article] [PubMed] [Google Scholar]