Differing models of nonsense-mediated mRNA decay (NMD) have been proposed in the literature. In this study, the authors reconcile these models and show that mRNAs bearing premature termination codons (PTCs) are degraded after export to the cytoplasm. In addition, there are two populations of PTC-containing mRNAs—a rapidly degraded one that undergoes NMD and one that degrades with a normal half-life and does not undergo NMD. The evidence presented rules out models that NMD occurs in the nucleus.
Keywords: mRNA decay, single-RNA FISH, mRNA quality control
Abstract
Nonsense-mediated mRNA decay (NMD) is a quality control mechanism responsible for “surveying” mRNAs during translation and degrading those that harbor a premature termination codon (PTC). Currently the intracellular spatial location of NMD and the kinetics of its decay step in mammalian cells are under debate. To address these issues, we used single-RNA fluorescent in situ hybridization (FISH) and measured the NMD of PTC-containing β-globin mRNA in intact single cells after the induction of β-globin gene transcription. This approach preserves temporal and spatial information of the NMD process, both of which would be lost in an ensemble study. We determined that decay of the majority of PTC-containing β-globin mRNA occurs soon after its export into the cytoplasm, with a half-life of <1 min; the remainder is degraded with a half-life of >12 h, similar to the half-life of normal PTC-free β-globin mRNA, indicating that it had evaded NMD. Importantly, NMD does not occur within the nucleoplasm, thus countering the long-debated idea of nuclear degradation of PTC-containing transcripts. We provide a spatial and temporal model for the biphasic decay of NMD targets.
Nonsense-mediated mRNA decay (NMD) is primarily an mRNA surveillance mechanism that targets eukaryotic mRNAs harboring a premature termination codon (PTC) (Hwang et al. 2010; Maquat et al. 2010; Mühlemann and Lykke-Andersen 2010; Nicholson et al. 2010). However, it also regulates either directly or indirectly a substantial number of protein-coding transcripts (for review, see Kervestin and Jacobson 2012). In mammalian cells, PTC recognition during NMD has been suggested to depend on a “pioneer round” of translation when translation terminates sufficiently upstream of a splicing-dependent exon–junction complex (EJC) so that the terminating ribosome can coexist with at least one downstream EJC (Dostie and Dreyfuss 2002; Sato and Maquat 2009).
Despite various attempts to characterize the decay process during NMD, its precise kinetics remain unclear. A short pulse of transcription followed by a cellular fractionation approach has shown that the majority of NMD targets is degraded within an estimated 30 min of formation, while ∼10%–30% escape NMD and undergo decay with a half-life (t1/2) that is indistinguishable from their PTC-free counterparts (Cheng and Maquat 1993; Belgrader et al. 1994; Lejeune et al. 2003). For instance, PTC-containing β-globin (Gl) and triosephosphate isomerase (TPI) mRNAs exhibit bimodal decay kinetics. These measurements, however, have not unequivocally shown whether the two rates of decay occur in single cells rather than in discrete populations of cells. Contrary to these findings are steady-state measurements performed using cultured cells and the transcription inhibitor actinomycin D (ActD). These experiments revealed that the PTC-containing Gl mRNAs decay with a single, 2 h mRNA half-life (Singh et al. 2007). This kinetic model suggests a markedly different mechanism of PTC-containing mRNA decay (and, it follows, a difference in many of the proteins associated with the mRNA substrate) compared with a bimodal model. Thus, in order to understand NMD as a surveillance mechanism, the kinetics of NMD must first be identified.
In addition, whether NMD occurs in the cytoplasm or the nucleoplasm or during nuclear export has been a subject of debate. Cell fractionation experiments suggest that the majority of mammalian NMD occurs “in association” with nuclei (Urlaub et al. 1989; Cheng and Maquat 1993; Belgrader et al. 1994; Lozano et al. 1994; Simpson and Stoltzfus 1994; Carter et al. 1996; Kessler and Chasin 1996; Thermann et al. 1998; Zhang et al. 1998a,b; Brogna 1999). This suggestion is supported by the observation that a fraction of PTC-containing TPI mRNA generated by a transient burst in transcription undergoes decay in what was defined biochemically as the nuclear fraction, whereas decay of the rest of the mRNA occurs in the cytoplasmic fraction at the rate of PTC-free mRNA (Cheng and Maquat 1993; Belgrader et al. 1994). Thus, one model for nucleus-associated NMD stipulates that NMD targets are exported into the cytoplasm and degraded while maintaining an association with nuclear pore complexes (NPCs) after PTC recognition by translationally active cytoplasmic ribosomes (Belgrader et al. 1994; Carter et al. 1996; Maquat 2004).
A second model proposes the existence of nuclear surveillance machinery whereby PTCs are recognized by translating ribosomes within nuclei (Iborra et al. 2001; Bühler et al. 2002; Wilkinson and Shyu 2002). In support of this possibility, it was reported that the inhibition of nuclear export does not decrease the decay of nucleus-associated PTC-containing mRNAs (Bühler et al. 2002); that some ribosome components in yeast accumulate at the site of PTC-containing gene transcription (Brogna et al. 2002); that in mammalian cells, translation can occur in the nucleoplasm and in the nucleolus (David et al. 2012); and that PTC-containing immunoglobulin (Ig)-μ and T-cell receptor-β transcripts, but not their PTC-free counterparts, are retained at their transcription sites (Mühlemann et al. 2001; de Turris et al. 2011). Thus, whether nucleus-associated NMD is a consequence of nuclear or cytoplasmic translation followed by rapid degradation is still questioned (Dahlberg and Lund 2004; Reid and Nicchitta 2012). A third model suggests that the NMD of all PTC-containing mRNAs occurs in the cytoplasm, unassociated with nuclei or the NPCs (Singh et al. 2007), and targets steady-state mRNA (Hogg and Goff 2010).
To clarify the mode and the location of nucleus-associated NMD, we used fluorescent in situ hybridization (FISH) and transiently transfected human osteosarcoma (U2OS) cells in order to localize and count individual newly synthesized PTC-containing or normal (PTC-free) Gl mRNAs in different cellular compartments. This approach allowed us to quantify mRNA abundance as a function of time after mRNA synthesis in intact cells, thereby preserving temporal and spatial information during NMD. By doing so, we obviated errors that arise during ensemble decay measurements using fractionated and chemically perturbed cells (Trcek et al. 2011; Sun et al. 2012).
We found that in individual cells, two PTC-containing mRNA populations coexist that differ in their turnover kinetics: a rapidly decaying population having a t1/2 of <1 min and a population with a t1/2 similar to PTC-free mRNA. The rapid, PTC-dependent decay allows for the time it would take for a translationally active ribosome to reach the PTC and for degradation to occur. Furthermore, we determined that NMD does not occur within nuclei. Rather, our results indicate that NMD abuts nuclei and can, in part, spatially overlap with the cellular location of the NPCs. These results suggest a temporal and spatial model for nucleus-associated NMD.
Results
Ensemble measurements demonstrate bimodal decay of PTC-containing Gl mRNA
To characterize the decay of Gl mRNA that contains a PTC at the 39th codon (termed Gl TER mRNA) and, as a control, PTC-free Gl mRNA (termed Gl NORM mRNA), we initially employed an ensemble approach whereby expression was driven by the serum-inducible c-FOS promoter (Cheng and Maquat 1993; Belgrader et al. 1994). Human osteosarcoma U2OS cells were transiently transfected with a pFOS-Gl TER or pFOS-Gl NORM plasmid and subsequently serum-deprived. After 36 h, serum was added for 30 min to induce a synchronous burst of Gl TER or Gl NORM mRNA transcription, which increased the likelihood of detecting early and rapid decay events. After 30 min of serum addition, ActD was added, RNA was harvested as a function of time, and RT-qPCR was used to quantify the level of Gl TER mRNA or Gl NORM mRNA relative to GAPDH mRNA (Livak and Schmittgen 2001). Data were fitted to a model assuming either one or two exponential decay components weighted by the errors to determine whether TER mRNAs were degraded with a uniform or bimodal t1/2 (Trcek et al. 2011). ActD is a potent transcriptional inhibitor: It acts within minutes of addition (Fig. 4A, see below; de Turris et al. 2011) and maintains its repression for several hours (Supplemental Fig. S2E,F). Thus, in the presence of ActD, the time-dependent change in mRNA levels should reflect mRNA decay with minimal contamination by newly synthesized transcripts due to inefficient transcription inhibition.
Figure 4.
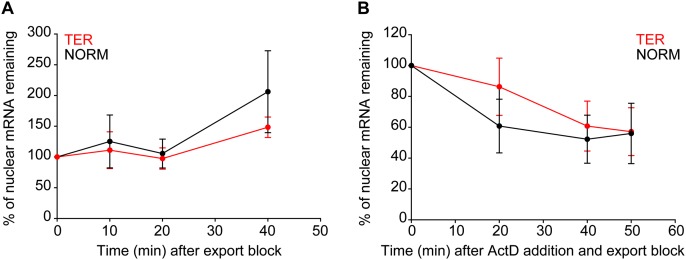
Rapid decay of GI TER mRNAs does not occur within nuclei. (A,B) TER and NORM mRNAs were expressed as described (Fig. 3). Afterward, WGA and DIG were added to block nuclear export while transcription was ongoing (A), or WGA and DIG and ActD were added to block nuclear export and transcription (B). Cells were fixed at various times after the inhibitors were added. The absolute numbers of TER and NORM mRNAs within nuclei were analyzed in the equatorial plane. mRNAs coinciding with the DAPI-defined nuclear boundary were excluded from the analysis. For each time point, the average number of TER mRNAs (red circles and lines) and NORM mRNAs (black circles and lines) per nucleus was determined. The percentage reduction in mRNA levels was determined relative to the level at time 0, which was defined as 100%. An average between two experiments ±SD is shown. For each condition and for each repetition in A and B, between seven and 12 cells were analyzed.
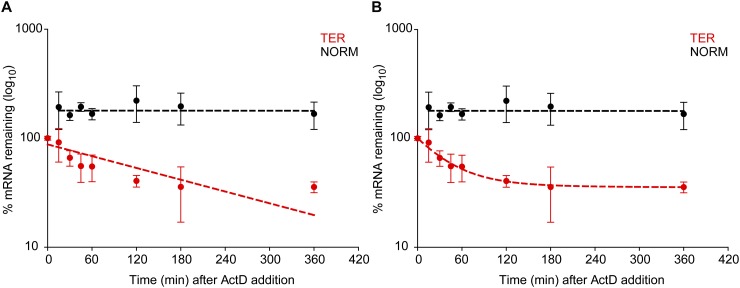
A one-component model yielded GI TER mRNA and GI NORM mRNA populations turning over with, respectively, a t1/2 = 2.75 ± 1.0 h (χ2: 24.6; P < 0.001) (Fig. 1A, red circles and red dashed line) and a t1/2 > 12 h (χ2: 2.4; P > 0. 1) (Fig. 1A, black circles and black dashed line). However, fitting the GI TER mRNA decay data to a two-component model revealed a better alignment than did the one-component model; there were two distinct GI TER populations: one (63.7%) with a t1/2 = 0.5 ± 0.1 h and another (36.3%) with a t1/2 > 12 h (χ2: 0.27; P > 0.1) (Fig. 1B, red circles and red dashed line). The fitting did not reveal two GI NORM mRNA populations with distinct half-lives, but rather a single population having a t1/2 > 12 h (χ2: 2.4; P > 0. 1) (Fig. 1B, black circles and black dashed line).
Figure 1.
(A,B) Whole-cell ensemble measurements demonstrate bimodal decay of PTC-containing Gl mRNA. Human U2OS cells were transiently transfected with a plasmid that produced either PTC-containing (TER) or PTC-free (NORM) Gl mRNA from the serum-inducible c-FOS promoter. Thirty-six hours after serum deprivation, GI TER or GI NORM transcription was induced with the addition of serum to 15%. After 30 min, ActD was added, and RNA was purified from cells as a function of time. Using RT-qPCR, the levels of GI TER and GI NORM mRNAs were normalized to the level of GAPDH mRNA. To determine the half-life (t1/2) of TER and NORM mRNAs, the normalized levels of each mRNA (A,B, red and black circles, respectively) were modeled by a weighted fit by the errors to a decay model having either a single component (A, red and black dashed lines) or two components (B, red and black dashed lines) Results from three independently performed experiments ±SEM are shown.
Based on the goodness of the fit (χ2 < 14.1 P: 0.05), we rejected the one-component model, which demonstrated systematic deviations from the data and thus did not recapitulate the decay kinetics. We therefore conclude that PTC-containing GI mRNAs exhibit two stabilities. Approximately two-thirds decay rapidly, while approximately one-third escapes NMD and decays with a normal half-life. However, this ensemble approach does not provide information as to whether the two Gl TER mRNA populations coexist in a single cell or in different cells. To address this issue, we used intact cells and single-RNA FISH so as to preserve spatial information about the NMD process.
Measuring spatial and temporal constraints of Gl NMD using single-cell, single-RNA FISH
Two-color RNA FISH was employed to simultaneously measure the distribution of a definable number of Gl TER mRNAs and Gl NORM mRNAs in the nucleus, at the nuclear periphery, and in the cytoplasm after the coordinate induction of Gl TER and Gl NORM gene transcription. To distinguish Gl TER and Gl NORM mRNAs, which differ only at codon 39, we replaced 193 nucleotides (nt) in the 3′ untranslated region (UTR) of each mRNA with either of two 330-nt sequences that derive from the open translational reading frame of Escherichia coli β-galactosidase (LacZ) mRNA, termed LacZA or LacZB. Results demonstrated that Gl TER mRNA was subject to NMD and that Gl NORM mRNA was resistant to NMD with either tag (Supplemental Fig. S1A).
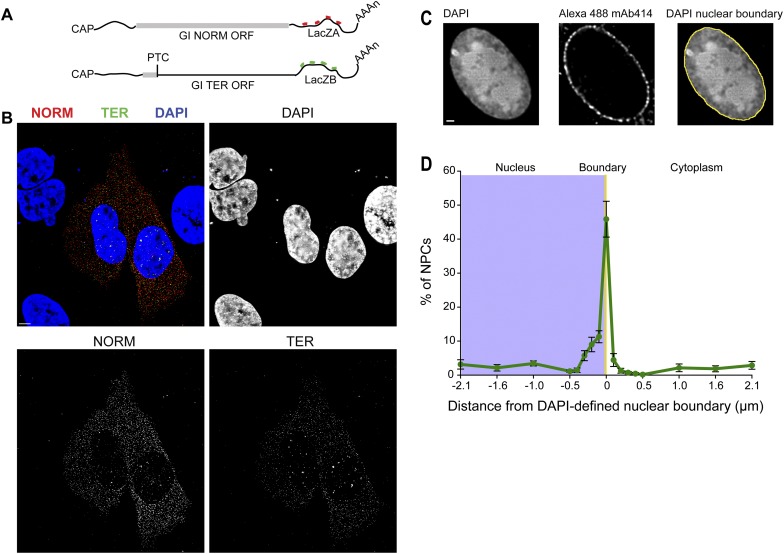
Gl-TER-LacZB (“TER”) and Gl-NORM-LacZA (“NORM”) mRNAs were coexpressed in U2OS Tet-On cells (Darzacq et al. 2006) using a single plasmid containing a doxycycline (Dox)-inducible bidirectional promoter (Supplemental Fig. S1B). This approach allowed the simultaneous expression of both mRNAs under the control of the same tetracycline response element and eliminated gene copy number variability. To detect TER and NORM mRNAs in cells, we used four FISH probes hybridizing to either LacZA or LacZB sequences, each set labeled with the spectrally distinct Cy3 or Cy3.5 fluorophores (Fig. 2A) as described previously (Femino et al. 2003; Zenklusen et al. 2008; Trcek et al. 2012). When coexpressed and detected using the FISH probes, TER mRNAs were represented by green spots in Figure 2B and NORM mRNAs were represented by red spots in Figure 2B, allowing for their distinction within the same cell. Detection of FISH-labeled mRNAs was performed using a computer algorithm (Zenklusen et al. 2008; Trcek et al. 2012) that determined the cellular position of individual FISH-labeled mRNAs at subpixel resolution (Thompson et al. 2002; Larson et al. 2005; Trcek et al. 2012). Hence, the spatial position of an mRNA within a cell relative to other cellular structures could also be assessed.
Figure 2.
Measuring spatial and temporal constraints of GI NMD using single-cell, single-RNA FISH. (A) Schematic of the NORM and TER mRNAs, each hybridized with four Cy3-labeled (red) or Cy3.5-labeled (green) FISH probes. The thick gray lines demarcate each Gl open translational reading frame. The position of the PTC at Gl codon 39 is marked. (B) Maximal projections of transiently transfected U2OS Tet-ON cells coexpressing NORM mRNAs (red) and TER mRNAs (green) using a bidirectional promoter. The DAPI-stained nuclei are shown in blue. The lack of a FISH signal in the neighboring untransfected cells demonstrates the specificity of FISH probe hybridization for target mRNAs. (C,D) Determining the subcellular distribution of NPCs relative to the DAPI-defined nuclear boundary in U2OS Tet-ON cells. NPCs were immunostained using a monoclonal Alexa 488-labeled antibody (mAb414), and the nuclear boundary was determined using DAPI as described (Supplemental Fig. S1C,D). Only the equatorial plane of a three-dimensionally acquired image was analyzed (Supplemental Fig. S1E). For each NPC, its distance in nanometers relative to the DAPI-defined nuclear boundary (0 nm, yellow line) was calculated to obtain the spatial distribution of immunostained NPCs (green line) relative to the DAPI-defined nuclear boundary in the nucleus (blue box in D) and in the cytoplasm, and the spatial NPC distribution was expressed as a percentage. Images in all panels were deconvolved. Bar, 5 μm.
After image acquisition of FISH-hybridized cells, DAPI DNA staining was used to segment each cell into three subcellular compartments—the nucleus, the nuclear boundary, and the cytoplasm (Supplemental Fig. S1C)—where the area of the nuclear boundary spanned 107.5 nm (1 pixel) in both the x and y dimensions. Cells were exposed to DAPI, and the equatorial plane of a three-dimensionally acquired image set was analyzed to distinguish nucleoplasm from cytoplasm. A DAPI fluorescent profile was determined (Fig. 2C; Supplemental Fig. S1C, yellow line), and the half-value between the maximal DAPI intensity (the nuclear interior) and the minimal DAPI intensity (cytoplasmic background) was calculated. Variability in the intensity of DAPI staining within the same nucleus minimally affected the precision of nuclear segmentation; one standard deviation in the threshold intensity resulted in a shift of only 40.6 nm ± 70 nm in the position of the DAPI-defined nuclear boundary (Supplemental Fig. S1D, yellow vs. green bars).
We measured the distance between the NPCs and the DAPI nuclear-defined segmentation line to validate the use of DAPI as a determinant of the nuclear boundary (Fig. 2C,D; Supplemental Fig. S1E). NPCs were immunostained using the monoclonal Alexa 488-labeled antibody mAb414, which reacted with NPC phenylalanine–glycine (FG) repeats to define their cellular locations (Supplemental Fig. S1E). The shortest distance between a detected NPC and the DAPI-defined nuclear boundary was then determined. Since the staining of individual NPCs was diffraction-limited (Rout and Aitchison 2001), their cellular position can be determined at subpixel resolution (Thompson et al. 2002; Larson et al. 2005; Trcek et al. 2012). We found that ∼45% of mAb414 staining spatially coincided with the DAPI-defined nuclear boundary (Fig. 2D, yellow line), with of the rest of staining (consistent with a Gaussian fit) lying within the nucleus (between 0 and 430 nm from the DAPI-defined nuclear boundary) or in the cytoplasm (between 0 and 215 nm from the DAPI-defined nuclear boundary). Thus, the DAPI-defined nuclear boundary serves as a good approximation of the cellular position of the nuclear envelope-bound NPCs.
The steady-state level of cytoplasmic Gl TER mRNA depends on PTC recognition and UPF1 and is resistant to NMD
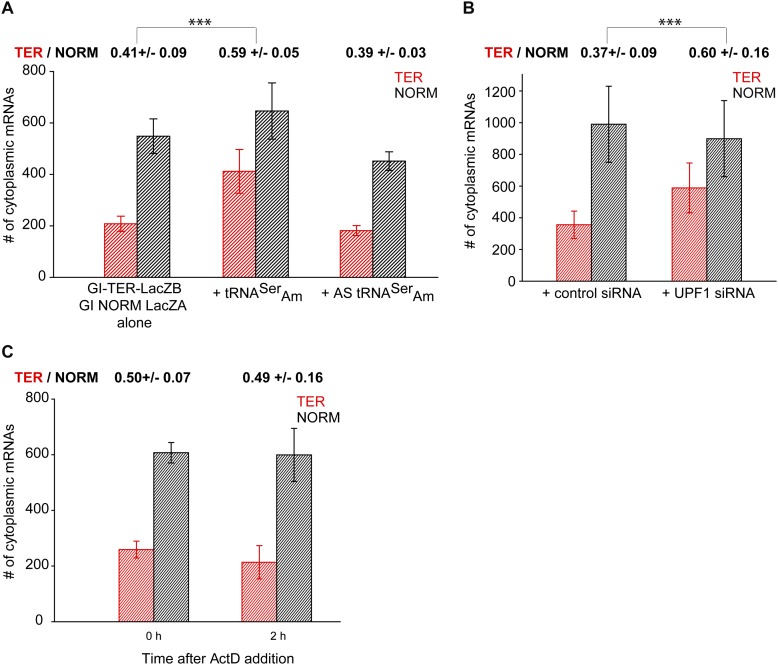
To evaluate whether single-cell measurements recapitulate the essential features of NMD, bidirectional transcription of the TER and NORM genes was induced to reach steady-state expression levels by exposing transfected cells to Dox (Fig. 2B). After 16 h, FISH was used to determine the cytoplasmic abundance of TER and NORM transcripts. The cytoplasmic level of TER mRNA relative to NORM mRNA was 41% (Fig. 3A), which is in the range of anticipated values given the efficiency of NMD (Zhang et al. 1998b; Lejeune et al. 2003). To illustrate that the decrease in the level of TER mRNA abundance was due to the presence of the PTC, we cotransfected the cells with a plasmid producing Amber suppressor tRNA (tRNASerAm) (Belgrader et al. 1993), which directs the incorporation of serine at the PTC in TER mRNA and thus suppresses PTC recognition. A plasmid producing the antisense strand (AS) of tRNASerAm, which fails to suppress the Gl PTC (Belgrader et al. 1993), was used as a control (Supplemental Fig. S2A).
Figure 3.
The steady-state level of cytoplasmic Gl TER mRNA depends on PTC recognition and UPF1 and is resistant to NMD. (A) U2OS Tet-On cells were transfected with the bidirectional promoter plasmid expressing GI TER and NORM mRNAs alone or with a plasmid expressing either tRNASerAm, which suppresses recognition of the PTC codon, or antisense tRNASerAm, which does not suppress recognition of the PTC codon. The nuclear boundary was segmented using the DAPI staining as described (Fig. 2). For each condition, the number of cytoplasmic TER mRNAs (red histograms) and NORM mRNAs (black histograms) was quantified at the equatorial plane of an image stack. DAPI was used for nuclear segmentation. The expression of TER and NORM mRNAs was analyzed in 21 untreated cells, 16 cells coexpressing tRNASerAm and 11 cells coexpressing AS tRNASerAm. (B) Bidirectional promoter plasmid carrying NORM and TER genes was transfected into U2OS Tet-On cells expressing either UPF1 siRNA or a control siRNA (de Turris et al. 2011). At steady state, NORM and TER mRNAs were counted as described above in 17 control siRNA-expressing and 14 UPF1 siRNA-expressing cells. (C) Transcription of TER and NORM mRNAs was induced as described in A. Afterward, cells were fixed (0 h) or treated with ActD for 2 h to stop transcription (2 h). The number of cytoplasmic TER and NORM mRNAs was counted as described above. Twenty and 12 cells were analyzed at 0 h and 2 h, respectively. In A–C, histograms represent the average number of TER and NORM mRNAs per cell ±SEM. The ratio of TER and NORM mRNAs is provided above the histograms. To determine this ratio, the ratio between TER and NORM mRNAs was first determined for each individual cell, after which an average ratio ±SEM for 21, 16, or 11 cells in A, 17 and 14 cells in B, and 20 and 12 cells in C was calculated. (***) t-test with P < 0.001.
In the presence of tRNASerAm, 59% of TER mRNAs escaped NMD, which represents a statistically significant 18% change relative to untreated cells (Fig. 3A). As expected, tRNASerAm did not completely inhibit PTC recognition (Belgrader et al. 1993). In control experiments, AS tRNASerAm did not change the cytoplasmic ratio of TER and NORM mRNAs.
Additionally, when we inhibited NMD by coexpressing UPF1 siRNA (de Turris et al. 2011), 23% more TER mRNA accumulated in the cytoplasm relative to control cells expressing a random sequence siRNA (Fig. 3B; Supplemental Fig. S2B,C). Therefore, our measurements confirm that the level of cytoplasmic TER mRNAs detected by FISH depends on PTC recognition during NMD surveillance.
To verify that the 41% of cytoplasmic TER mRNAs represented the population that evaded NMD and turned over with a decay rate similar to NORM mRNAs, the bidirectional expression was induced as in Figure 3A, at which point cells were either fixed or treated with ActD for two additional hours before cell fixation (Supplemental Fig. S2D). If the cytoplasmic TER mRNAs were undergoing NMD with a t1/2 of 30 min, as experimentally determined using ensemble measurements (Fig. 1B), then a decay-dependent change in the ratio of cytoplasmic TER and NORM mRNAs should be detected within a 2-h ActD treatment. Conversely, no change in the ratio would indicate that cytoplasmic TER mRNA represents the population of PTC-containing mRNA that is degraded indistinguishably from NORM mRNA.
After treatment with ActD, the ratio of cytoplasmic TER and NORM mRNAs did not change relative to cells that were not treated with ActD (Fig. 3B). Within 2 h, transcription of NORM and TER dropped by 94.2% relative to the initial time point (Supplemental Fig. S2E,F). Thus, the lack of detectable change in the cytoplasmic ratio of TER and NORM mRNAs is not due to ongoing transcription. Rather, we estimate that cytoplasmic TER mRNA is degraded with a t1/2 of ∼7.2 h (Supplemental Fig. S2G), which is similar to the half-life determined for NORM mRNA (Fig. 1B). Furthermore, the 41% of FISH-detected cytoplasmic TER mRNA that escaped NMD was in good agreement with the 36% TER mRNA that escaped NMD, as determined using RT-qPCR (Fig. 1B). Our data demonstrated that the cytoplasmic TER mRNAs detected with FISH in the steady state evaded NMD surveillance and degraded with kinetics similar to NORM mRNAs.
Rapid decay of Gl TER mRNAs does not occur within nuclei
To investigate whether the rapid turnover of TER mRNA detected in Figure 1 occurs in the nucleus, we measured the rates of TER and NORM mRNA decay within nuclei in the presence of wheat germ agglutinin (WGA) and digitonin (DIG), which are general inhibitors of nuclear export (Mor et al. 2010), in either the absence (Fig. 4A) or presence (Fig. 4B) of ActD. Cells were fixed at various time points after WGA, DIG, and, where specified, ActD addition, and the number of TER and NORM mRNAs within nuclei was counted using FISH (Supplemental Fig. S3A–C). Only the nucleoplasm was analyzed so that mRNAs that spatially coincided with the DAPI nuclear boundary—and hence, the NPCs—were excluded from analysis.
When only nuclear export but not transcription was blocked, both TER and NORM mRNAs accumulated over time, suggesting that WGA and DIG were competent inhibitors of mRNA export (Fig. 4A). However, in the presence of WGA, DIG, and ActD, the nuclear levels of TER and NORM mRNAs were depleted during the first 40 min, after which further changes in the level of either mRNA were not detected (Fig. 4B). The initial depletion of mRNAs was likely due to an incomplete block of nuclear export by WGA and DIG. Importantly, the levels of both mRNAs decreased equally, suggesting that TER and NORM mRNAs were subjected to the same nuclear decay and export kinetics. Thus, the rapid NMD-dependent decay of the TER transcripts does not occur within nuclei.
NMD of Gl TER mRNA occurs in the cytoplasm
Thus far, our experiments reveal that (1) using whole cells and ensemble measurements, TER mRNA turned over bimodally; (2) using FISH and individual cells, there was ∼60% less cytoplasmic TER mRNA than cytoplasmic NORM mRNA in the steady state; (3) using suppressor tRNA or UPF1 siRNA, the cytoplasmic abundance of TER mRNA increased when the PTC recognition was blocked; (4) at steady state, a minority of the cytoplasmic TER mRNA turned over with decay kinetics that are indistinguishable from the decay kinetics of NORM mRNA; and (5) the rapid NMD of TER mRNA does not occur within nuclei. It follows that the rapid NMD of TER mRNA occurs in the cytoplasm, possibly during nuclear export or soon thereafter. To study the more rapidly decaying population of TER mRNA that undergoes NMD, we employed a rapidly inducible system that allowed for the detection of newly synthesized TER and NORM mRNAs during and shortly after mRNA export to the cytoplasm. We used an ecdysone-inducible expression vector that relies on the binding of the synthetic insect steroid hormone ponasterone A (PonA) to stimulate transcription (No et al. 1996). We created a plasmid that contains a bidirectional promoter where the enhancer-binding site was oligomerized to create a stretch of 45 PonA repeats, fused on each side to a minimal heat-shock promoter (Supplemental Fig. S4A). The transient transfection of U2OS cells stably expressing a plasma membrane-bound PonA receptor together with the bidirectional PonA-responsive plasmid enabled a rapid, simultaneous, sustained, and calibrated induction of TER and NORM gene transcription within the same cell (Supplemental Fig. S4B).
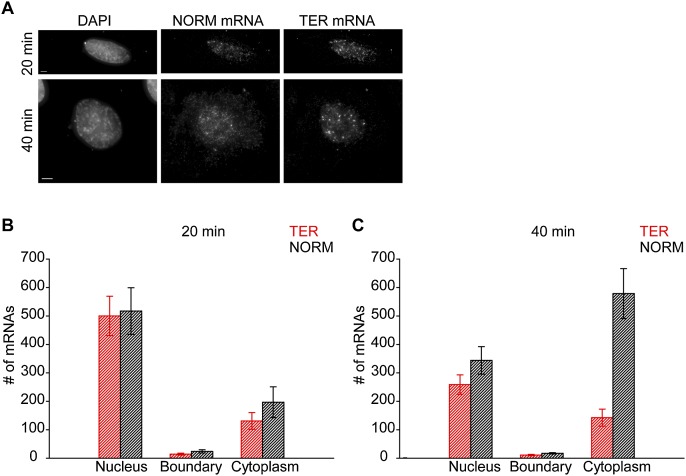
In cell fractionation experiments, the NMD of the PTC-containing mRNAs could be detected ∼15 min after a serum-mediated burst in transcription was over (Belgrader et al. 1994). In our experiments, TER and NORM gene transcription was induced with PonA for 20 or 40 min, after which cells were fixed, and the numbers of TER and NORM mRNAs in the nucleus, the DAPI-defined nuclear boundary, and the cytoplasm were quantified using FISH (Fig. 5A–C).
Figure 5.
NMD of GI TER mRNAs occurs in the cytoplasm. (A–C) U2OS PonA cells were transiently transfected with a plasmid producing TER and NORM mRNAs from a bidirectional PonA-regulated promoter that allowed for a calibrated coexpression of both mRNAs within individual cells (Supplemental Fig. S4A,B). mRNA expression was induced using PonA for 20 or 40 min, after which cells were fixed. (A) Maximal projections of cells coexpressing TER and NORM mRNAs after 20 or 40 min of PonA addition. Bar, 5 μm. For each time point (20 min [B] and 40 min [C]), the number of TER mRNAs (B,C; red histograms) and NORM mRNAs (B,C; black histograms) was quantified in the equatorial plane of acquired images in the nucleus, at the DAPI-defined nuclear boundary, and in the cytoplasm. An average number of mRNAs in each compartment for seven (A) and 10 (B) cells ±SEM is shown (see also Supplemental Table S1).
After 20 min of PonA treatment, the number of TER and NORM mRNAs was similar in the nucleus, indicating that the initial rate of transcription for the corresponding genes was the same. At the DAPI-defined nuclear boundary and in the cytoplasm, there were, respectively, 35% and 28% fewer TER mRNAs than NORM mRNAs (Fig. 5B; Supplemental Table S1). Forty minutes after induction, the nuclear abundance of both mRNAs was depleted to roughly the same extent (to ∼250 mRNAs for each over the 20-min period), indicating that the export of both mRNAs was equally efficient (Fig. 5C). It is unlikely that the initial buildup of NORM and TER mRNAs in the nucleus was due to a short burst of transcription, since synthesis of both genes using PonA promoters was sustained over several hours (Supplemental Fig. S4B). Rather, this transient accumulation could be a consequence of a rapid synthesis of a magnitude of mRNAs in a short period of time. However, while NORM mRNAs accumulated in the cytoplasm by roughly the same amount as the number of NORM mRNAs that were depleted in the nucleus, TER mRNAs did not accumulate in the cytoplasm (Fig. 5B,C). Similar to the 20-min time point, we did not detect an accumulation of either TER or NORM mRNA at the DAPI nuclear boundary at 40 min after the PonA-mediated induction (Fig. 5C; Supplemental Table S1).
Since our data exclude the possibility of NMD within nuclei (Fig. 4) and the rates of TER and NORM gene transcription were similar (Fig. 5B), we attribute the lack of cytoplasmic TER mRNA accumulation to cytoplasmic NMD. Additionally, during a 20-min window, little change in the levels of cytoplasmic TER mRNAs was detected at 40 min relative to 20 min after induction (Fig. 5B,C; Supplemental Table S1). The depletion of nuclear TER mRNA and the lack of a corresponding cytoplasmic accumulation between the 20-min and 40-min time points allowed us to estimate the rapid t1/2 of TER mRNA during NMD. The nuclear abundance of TER mRNAs was depleted by 243 mRNAs between the 20- and 40-min time points (Fig. 5B,C; Supplemental Table S1). Assuming that 243 TER mRNAs that were transcribed during the first 20 min of transcription induction with PonA (Fig. 5B; Supplemental Table S1) are released into the cytoplasm either at once or sequentially and degrade following a unimodal exponential decay within the next 20 min after PonA addition (to reach 40 min time point), we estimated the rapid TER mRNA half-life to be between 2.9 and 56 sec. These are upper limits of TER mRNA half-life, as our simplified calculation does not account for ongoing transcription (Supplemental Fig. S4B). The estimated half-life was also several orders of magnitude faster than that determined by an ensemble approach using fractionated cells or cell lysates treated with ActD (Fig. 1B; Zhang et al. 1998b; Lejeune et al. 2003; Singh et al. 2007). The short t1/2 presumably includes the time it would take for translating ribosomes to reach the PTC at codon 39. It is therefore likely that TER mRNA does not undergo multiple rounds of translation before it undergoes NMD.
Furthermore, we detected a precipitous drop in the amount of TER mRNAs relative to NORM mRNAs in the cytoplasm proximal to the DAPI-defined nuclear boundary 40 min after transcriptional induction with PonA. No change in the ratio could be detected either in the nucleus or distally in the cytoplasm (Supplemental Fig. S4C–E). Thus, changes in the cellular distribution of TER mRNA that are attributable to NMD occur in the cytoplasm in close proximity to the nuclear envelope, while TER mRNA in the nucleoplasm and located in the cytoplasm distally from DAPI staining exhibit a decay rate that is similar to NORM mRNA, in agreement with the data obtained from the steady-state experiments (Figs. 1, 4, 5). The steep gradient, which levels off ∼430 nm away from the DAPI-defined nuclear boundary, supports our estimates of a short TER mRNA half-life, indicating that TER mRNAs likely do not diffuse far away from the nucleus before they are degraded. Thus, TER mRNA exhibits bimodal decay kinetics within individual cells. We therefore conclude that NMD is a rapid process that eliminates the majority of newly synthesized Gl TER mRNAs as soon as they are exported to the cytoplasm.
Discussion
Using single-RNA FISH to analyze single cells transiently expressing PTC-containing and PTC-free Gl mRNAs from bidirectional, inducible promoters, we characterized the temporal and spatial decay features of individual mRNAs during NMD. Our results demonstrate that ∼60% fewer PTC-containing Gl mRNAs accumulate in the cytoplasm relative to their PTC-free counterparts. We also show that two populations of PTC-containing mRNAs with two distinct lifetimes coexist within the same cell: one population undergoing rapid, cytoplasmic decay with a half-life of <1 min proximal to the nucleus and a second population with a half-life >12 h throughout the cytoplasm, approximating that of the corresponding PTC-free mRNA. We further show that the rapid decay does not occur within nuclei. We conclude that the majority (∼60% in our experiments) of all PTC-containing mRNAs is interdicted in the cytoplasm for the purposes of quality control by NMD, and the remainder (∼40%) escapes NMD and is degraded by the cell as if it were normal mRNA. Our data further show that TER mRNAs undergoing rapid decay likely do not undergo multiple rounds of translation before they are degraded. The fact that a fraction of PTC-containing mRNAs escapes NMD suggests that there exists a temporally and spatially defined checkpoint that, once passed, leads to immunity from further surveillance.
It makes mechanistic sense that NMD would be quick and restricted to the first rounds of translation, which can involve more than one ribosome depending on the translation initiation efficiency and length of the translational reading frame (Chiu et al. 2004; Isken et al. 2008). Thus, it is likely that every newly synthesized mRNA, even those without a PTC, initiates translation while associated with the nucleus or immediately thereafter unless the efficiency of the pioneer round of translation was too low—a case that typifies PTC-containing glutathione peroxidase 1 mRNA, which undergoes largely cytoplasmic NMD (Sato et al. 2008). The efficiency of mRNA export would not be impeded by the myriad of mRNAs exiting the nucleus: We calculate that an mRNA exits an NPC once per minute based on measurements of 3000 NPCs per nucleus (Maul 1977) and ∼1 million mRNAs per cell [based on 0.2 pg of poly(A) per cell] (Holland et al. 1980) and an average mRNA half-life that is equal to the cell division time of 18 h (Singer and Penman 1973). This is ample time for a surveillance mechanism to translate every exporting mRNA without creating a bottleneck during export. Notably, our upper estimate of 56 sec for the half-life of PTC-containing Gl mRNA is compatible with this frequency; i.e., the time necessary to scan to a PTC at codon 39. Assuming a translation rate of approximately five codons per sec (Ingolia et al. 2011), translation of TER mRNA would only take 8 sec, which is consistent with a t1/2 of 2.9 sec for PTC-containing Gl mRNA.
However in contrast to these calculations, our data in Figure 5 indicate that PTC-containing or PTC-free Gl mRNAs do not accumulate at NPCs. These measurements are supported by live-cell single-RNA imaging that indicates that mRNAs do not associate with the NPCs for extended periods of time during export. High-speed imaging of a PTC-free β-actin mRNA transiting the NPC revealed that the total transport time lasted ∼170 msec, and mRNA remained associated with NPCs for ∼80 msec once on the cytoplasmic side. Nonetheless, a fraction of mRNAs (∼10%) were retained at the NPC for up to 6 sec (Grünwald and Singer 2010). Studies of an artificial 8-kb mRNA revealed that its nuclear export time took ∼500 msec. Significant accumulations of the mRNAs at the NPCs were not observed on either side of the nuclear envelope, indicating that export is not a rate-limiting step, at least not for PTC-free mRNAs (Mor et al. 2010). However, PTC-containing mRNAs may exhibit different kinetics, and although we did not observe accumulation of PTC-containing mRNA at the envelope, our study was not designed to test this rigorously. Further work using live-cell imaging will be necessary to determine whether PTC-containing mRNAs show, for instance, an increase in release times from an NPC at the cytoplasmic side similar to the slow component measured previously (Grünwald and Singer 2010). This is possible, considering that the nature of a translation termination complex at a PTC differs mechanistically and possibly compositionally from the nature of a termination complex at a codon that does not trigger NMD (Amrani et al. 2004; Kashima et al. 2006).
Our data demonstrate that the NMD of PTC-containing mRNA is precipitous and is spatially proximal to the nuclear envelope. Since cytoplasmic ribosomes are not limiting (Warner 1999), a rapid association of mRNA during or soon after export with the ribosome is expected, which can expedite the recognition of the PTC and the eventual degradation of the mRNA. This leads us to postulate several models for NMD. One is that the mRNA diffuses away from the nuclear envelope while undergoing the first round of translation, which, as estimated above, takes place within 8 sec. If a diffusion rate of between 0.09 μm2/sec and 1 μm2/sec is assumed (Shav-Tal et al. 2004; Grünwald and Singer 2010), a PTC-containing mRNA could diffuse ∼0.72–8 μm2 away from the NPCs during this time period—in either case, farther than the observed gradient in Supplemental Figure S4 would predict. This implies that diffusion of the NMD complex may be impeded by association with a cellular structure or is a large complex with significantly slower diffusion than mRNA alone. It is thus possible that the NMD of PTC-containing Gl mRNA begins immediately after export, where a fraction of these mRNAs degrades while still associated with the outer nuclear envelope, possibly even while associated with the NPC (for review, see Maquat 2005). Some mRNA may dissociate from the NPC or the envelope but finish NMD rapidly thereafter such that a gradient results proximal to the DAPI-defined nuclear boundary.
The NMD-resistant population turns over with a half-life >7 h. The decay kinetics of this older population of mRNA resembles the decay kinetics of PTC-free Gl mRNA, indicating that this population is immune to NMD. Our data thus support previous measurements of a bimodal behavior for PTC-containing mRNAs during and after NMD (Cheng and Maquat 1993; Belgrader et al. 1994). The model suggesting that association with the nuclear envelope provides a mechanism for NMD may also partly explain the observed immunity to NMD, since a lack of association would allow the mRNA to diffuse away from a site of degradation faster than degradation factors could assemble on the mRNA. Increasing the efficiency of a newly synthesized mRNA to engage in translation would be expected to decrease the time to NMD and hence decrease the distance from the envelope where NMD occurs (Sato et al. 2008).
One final model should be considered. It has been shown that mRNA degradation factors can be acquired upon gene transcription (Enssle et al. 1993; Bregman et al. 2011; Trcek et al. 2011) and that PTC-containing transcripts can be recognized at the site of transcription (Mühlemann et al. 2001; de Turris et al. 2011). It is possible that the first PTC-containing mRNAs to undergo NMD set up a recognition process at the site of the gene that allows degradation factors to be assembled there so that NMD can be activated upon arrival in the cytoplasm. In support of this, there is a measureable delay of ∼20 min from the time of transcription until NMD is apparent (Fig. 5; Supplemental Fig. S4C,D). Further work using techniques similar to those described here should distinguish among these models.
Materials and methods
Mammalian cell lines and plasmids
L-M(TK−) mouse fibroblast cells (American Type Culture Collection accession no. CCL-1.3) were grown at 37°C and 5% CO2 in DMEM containing 4.5 g/L glucose, 10% FBS, and 1% penicillin–streptomycin, as were human U2OS Tet-On cells (Darzacq et al. 2006) and U2OS PonA cells, except the concentration of glucose was 1 g/L. Plasmids used in this study are provided in Supplemental Table S2.
Transient transfections
For NMD efficiency experiments using RT–PCR, L-M(TK−) cells seeded at ∼95% confluency in a 10-cm dish were transiently transfected with 3 μg of the pFOS-Gl NORM, pFOS-Gl TER, pFOS-Gl NORM-LacZA, pFOS-Gl NORM-LacZB, pFOS-Gl TER- LacZA, or pFOS-Gl TER-LacZB test plasmid and 1 μg of the phCMV-MUP reference plasmid using Lipofectamine 2000. Cells were harvested after 48 h, and total cell RNA was extracted using Trizol as described previously (Sato and Maquat 2009).
For experiments using Dox, U2OS Tet-On cells were seeded onto tissue culture-treated coverslips (Fisherbrand, 12-545-84) at ∼25% confluency, placed in 12-well tissue culture plates, supplemented with 1 mL of growth medium, and allowed to attach overnight. The next day, cells were transfected with pTRE-Tight-BI-Gl NORM-LacZA TER-LacZB at a 6:1 ratio of Fugene to DNA, following Fugene specifications. For the suppression of PTC recognition in Figure 3A, U2OS Tet-On cells were cotransfected with pTRE-Tight-BI-Gl NORM-LacZA TER-LacZB and p53TS+A encoding an tRNASerAm or with p53TS+B encoding an AS tRNASerAm (Capone et al. 1985), while maintaining the 6:1 Fugene-to-DNA ratio.
To induce transcription of NORM LacZA and TER LacZB with Dox, the old medium was replaced by 1 mL of fresh, 37°C prewarmed medium containing 1 μg/mL Dox (Janicki et al. 2004). Transcription was induced for 16 h to reach steady state. When steady state was reached, cells were fixed in 4% paraformaldehyde for 20 min, washed once with 1× PBS, and stored in 70% ethanol at 4°C for the subsequent FISH analysis.
For mRNA half-life experiments in Figure 3B, transfection and induction of transcription of pTRE-Tight-BI-Gl NORM-LacZA TER-LacZB in U2OS Tet-ON cells was carried out as described above. After 16 h of Dox addition, cells were fixed in 4% paraformaldehyde as described above, which defined the 0 time point (0 h). Alternatively, after 16 h of transcription induction using Dox, the medium containing Dox was removed and replaced by 1 mL of fresh, 37°C prewarmed medium containing 4μg/mL ActD to inhibit NORM LacZA and TER LacZB transcription. Cells were then incubated in the presence of ActD for 2 h, after which cells were fixed in 4% paraformaldehyde, which defined the 2-h time point. Expression of NORM LacZA and TER LacZB mRNAs at t0 and t2 was subsequently analyzed using FISH.
For experiments using PonA in Figure 5, U2OS PonA cells were propagated and seeded onto coverslips as described above. Cells were transfected with pPonA-BI-Gl NORM-LacZA TER-LacZB at a 6:1 ratio of Fugene to DNA. Four hours after transfection, the transfection medium was replaced by 1 mL of fresh, 37°C prewarmed medium, and cells were incubated overnight. The next day, PonA was added to a final concentration of 100 μM for 20 or 40 min to induce expression of pPonA-BI-Gl-NORM LacZA TER LacZB (No et al. 1996). Cells were then fixed and prepared for FISH analysis as described above.
Single-RNA FISH
Mammalian cell single-RNA FISH was performed as previously described. FISH probes, each ∼50 nt long, containing five modified nucleotides that were coupled with fluorescent dyes were used. Labeling efficiency of each probe with fluorescent dyes was ≥90% efficient (Supplemental Table S3; Femino et al. 2003; Zenklusen et al. 2008; Trcek et al. 2012). Images were acquired using a microscope setup previously described (Trcek et al. 2012). Cells were optically sectioned using a 200-nm Z step, spanning a 3.0-μm Z depth. Eight-hundred-millisecond exposure times were used to acquire each plane in the Cy3 and Cy3.5 channel, and an ∼25-msec exposure was used for the DAPI channel.
Immunostaining NPCs
NPCs were immunostained using the monoclonal antibody mAb414 labeled with the Alexa 488 dye (Covance, A488-120L). Immunostaining was carried out as previously detailed (Darzacq et al. 2006). Cells were imaged as described above using 100 msec in the FITC channel.
Inhibition of nuclear export by WGA and DIG
Production of NORM LacZA and TER LacZB was induced with Dox in U2OS cells to reach steady state as described above. At steady state, WGA (5 μg/mL), DIG (30 μg/mL) (Mor et al. 2010), and/or ActD (4 μg/mL) were added, which defined the 0 time point, or incubated in the presence of inhibitors for various time points afterward. Cells were fixed in 4% paraformaldehyde and subsequently analyzed using FISH.
RT–PCR
RNA isolation and RT–PCR analysis were performed as described before (Ishigaki et al. 2001). mRNAs were detected using RT–PCR and the primer pair 5′-GCAGCGAGCAACTGAGAAGC-3′ (sense) and 5′-GGGTTTAGTGGTACTTGTGAGC-3′ (antisense) in the case of Gl mRNA or 5′-CTGATGGGGCTCTATG-3′ (sense) and 5′-TCCTGGTGAGAAGTCTCC-3′ (antisense) in the case of MUP mRNA. Individual RT–PCR products were quantified using ImageQuant.
RT-qPCR and mRNA decay modeling
To characterize the decay of Gl TER and Gl NORM mRNAs in Figure 1, we employed an ensemble approach whereby expression was driven by the serum-inducible c-FOS promoter (Cheng and Maquat 1993; Belgrader et al. 1994). U2OS cells were transiently transfected with a pFOS-Gl TER or pFOS-Gl NORM plasmid and subsequently serum-deprived. After 36 h, serum was added for 30 min to induce Gl transcription. After 30 min, 4μg/mL ActD was added to inhibit transcription, and RNA was harvested as a function of time. RT-qPCR was used to quantify the level of Gl TER mRNA or Gl NORM mRNA relative to GAPDH mRNA (Livak and Schmittgen 2001) using the following primer pairs: 5′-CCTGGACAACCTCAAGGG-3′ (sense) and 5′-CGATCATATTGCCCAGGAGCCTG-3′ (antisense) for Gl NORM and Gl TER mRNAs and 5′-TGCACCACCAACTGCTTAGC-3′ (sense) and 5′-GGCATGGACTGTGGTCATGAG-3′ (antisense) for GAPDH mRNA. To determine whether GI NORM mRNA or GI TER mRNA was degraded with a uniform or a bimodal t1/2, data were modeled using SigmaPlot software and a weighted fit by the errors assuming either one (y = ae−bk) or two (y = ae−bk + ce−dx) exponential decay components (Trcek et al. 2011).
To measure induction of NORM LacZA and TER LacZB gene expression using the bidirectional PonA promoter plasmid, U2OS PonA cells were transfected with pPonA-BI-Gl-NORM LacZA TER LacZB and NORM LacZA, and TER LacZB expression was induced with PonA as described above. RNA was harvested at various time points after gene expression induction, and NORM LacZA and TER LacZB mRNA levels were measured using RT-qPCR, as described above. The following primer pairs were used for RT-qPCR: 5′-GATGGCGTTAACTCGGCG-3′ (sense) and 5′-GCGAGGCGGTTTTCTCC-3′ (antisense) for NORM LacZA and 5′-GCTGTGCCGAAATGGTC-3′ (sense) and 5′-CCTGCCAGTATTTAGCG-3′ (antisense) for TER LacZB transcripts.
siRNA-mediated UPF1 down-regulation
U2OS Tet-ON cells were transfected with pSUPERpuro-shRNA-UPF1 or control pSUPERpuro-shRNA-scr coding a scrambled sequence as described previously (de Turris et al. 2011). Down-regulation of UPF1 was induced for 48 h, after which cells were reseeded in medium containing 5 μg/mL puromycin and transfected with pTRE-Tight-BI-Gl NORM-LacZA TER-LacZB. Four hours after transfection, the transfection medium was replaced by 1 mL of fresh, 37°C prewarmed medium containing 1ug/mL Dox, and cells were incubated overnight to reach steady-state NORM LacZA and TER LacZB expression as described above. Anti-RENT1 UPF1 antibody (Bethyl Laboratories, A300-036A) was used for Western Blot (de Turris et al. 2011). β-Actin, detected by rabbit anti-actin antibody (Sigma, A2103) was used as a loading control.
Acknowledgments
We thank Sean P. Burke for providing the algorithm that calculated the shortest distance between an mRNA and the DAPI-defined nuclear boundary. We thank Dr. Daniel R. Larson for providing the nuclear segmentation algorithm. H.S. was supported by a Fellowship from the Japan Society for the Promotion of Science. This work was supported by NIH R01 GM84364 and GM86217 to R.H.S., and NIH R01GM59614 to L.E.M.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.209635.112.
References
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A 2004. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432: 112–118 [DOI] [PubMed] [Google Scholar]
- Belgrader P, Cheng J, Maquat LE 1993. Evidence to implicate translation by ribosomes in the mechanism by which nonsense codons reduce the nuclear level of human triosephosphate isomerase mRNA. Proc Natl Acad Sci 90: 482–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgrader P, Cheng J, Zhou X, Stephenson LS, Maquat LE 1994. Mammalian nonsense codons can be cis effectors of nuclear mRNA half-life. Mol Cell Biol 14: 8219–8228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregman A, Avraham-Kelbert M, Barkai O, Duek L, Guterman A, Choder M 2011. Promoter elements regulate cytoplasmic mRNA decay. Cell 147: 1473–1483 [DOI] [PubMed] [Google Scholar]
- Brogna S 1999. Nonsense mutations in the alcohol dehydrogenase gene of Drosophila melanogaster correlate with an abnormal 3′ end processing of the corresponding pre-mRNA. RNA 5: 562–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogna S, Sato TA, Rosbash M 2002. Ribosome components are associated with sites of transcription. Mol Cell 10: 93–104 [PubMed] [Google Scholar]
- Bühler M, Wilkinson MF, Mühlemann O 2002. Intranuclear degradation of nonsense codon-containing mRNA. EMBO Rep 3: 646–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capone JP, Sharp PA, RajBhandary UL 1985. Amber, ochre and opal suppressor tRNA genes derived from a human serine tRNA gene. EMBO J 4: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter MS, Li S, Wilkinson MF 1996. A splicing-dependent regulatory mechanism that detects translation signals. EMBO J 15: 5965–5975 [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Maquat LE 1993. Nonsense codons can reduce the abundance of nuclear mRNA without affecting the abundance of pre-mRNA or the half-life of cytoplasmic mRNA. Mol Cell Biol 13: 1892–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu SY, Lejeune F, Ranganathan AC, Maquat LE 2004. The pioneer translation initiation complex is functionally distinct from but structurally overlaps with the steady-state translation initiation complex. Genes Dev 18: 745–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlberg JE, Lund E 2004. Does protein synthesis occur in the nucleus? Curr Opin Cell Biol 16: 335–338 [DOI] [PubMed] [Google Scholar]
- Darzacq X, Kittur N, Roy S, Shav-Tal Y, Singer RH, Meier UT 2006. Stepwise RNP assembly at the site of H/ACA RNA transcription in human cells. J Cell Biol 173: 207–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David A, Dolan BP, Hickman HD, Knowlton JJ, Clavarino G, Pierre P, Bennink JR, Yewdell JW 2012. Nuclear translation visualized by ribosome-bound nascent chain puromycylation. J Cell Biol 197: 45–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Turris V, Nicholson P, Orozco RZ, Singer RH, Mühlemann O 2011. Cotranscriptional effect of a premature termination codon revealed by live-cell imaging. RNA 17: 2094–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J, Dreyfuss G 2002. Translation is required to remove Y14 from mRNAs in the cytoplasm. Curr Biol 12: 1060–1067 [DOI] [PubMed] [Google Scholar]
- Enssle J, Kugler W, Hentze MW, Kulozik AE 1993. Determination of mRNA fate by different RNA polymerase II promoters. Proc Natl Acad Sci 90: 10091–10095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Femino AM, Fogarty K, Lifshitz LM, Carrington W, Singer RH 2003. Visualization of single molecules of mRNA in situ. Methods Enzymol 361: 245–304 [DOI] [PubMed] [Google Scholar]
- Grünwald D, Singer RH 2010. In vivo imaging of labelled endogenous β-actin mRNA during nucleocytoplasmic transport. Nature 467: 604–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg JR, Goff SP 2010. Upf1 senses 3′UTR length to potentiate mRNA decay. Cell 143: 379–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland CA, Mayrand S, Pederson T 1980. Sequence complexity of nuclear and messenger RNA in HeLa cells. J Mol Biol 138: 755–778 [DOI] [PubMed] [Google Scholar]
- Hwang J, Sato H, Tang Y, Matsuda D, Maquat LE 2010. UPF1 association with the cap-binding protein, CBP80, promotes nonsense-mediated mRNA decay at two distinct steps. Mol Cell 39: 396–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iborra FJ, Jackson DA, Cook PR 2001. Coupled transcription and translation within nuclei of mammalian cells. Science 293: 1139–1142 [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishigaki Y, Li X, Serin G, Maquat LE 2001. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106: 607–617 [DOI] [PubMed] [Google Scholar]
- Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JW, Maquat LE 2008. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell 133: 314–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, Prasanth KV, Ried T, Shav-Tal Y, Bertrand E, Singer RH, et al. 2004. From silencing to gene expression: Real-time analysis in single cells. Cell 116: 683–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S 2006. Binding of a novel SMG-1–Upf1–eRF1–eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev 20: 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kervestin S, Jacobson A 2012. NMD: A multifaceted response to premature translational termination. Nat Rev Mol Cell Biol 13: 700–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler O, Chasin LA 1996. Effects of nonsense mutations on nuclear and cytoplasmic adenine phosphoribosyltransferase RNA. Mol Cell Biol 16: 4426–4435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DR, Johnson MC, Webb WW, Vogt VM 2005. Visualization of retrovirus budding with correlated light and electron microscopy. Proc Natl Acad Sci 102: 15453–15458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F, Li X, Maquat LE 2003. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol Cell 12: 675–687 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−Δ Δ CT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lozano F, Maertzdorf B, Pannell R, Milstein C 1994. Low cytoplasmic mRNA levels of immunoglobulin κ light chain genes containing nonsense codons correlate with inefficient splicing. EMBO J 13: 4617–4622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE 2004. Nonsense-mediated mRNA decay: Splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol 5: 89–99 [DOI] [PubMed] [Google Scholar]
- Maquat LE 2005. Nonsense-mediated mRNA decay in mammals. J Cell Sci 118: 1773–1776 [DOI] [PubMed] [Google Scholar]
- Maquat LE, Tarn WY, Isken O 2010. The pioneer round of translation: Features and functions. Cell 142: 368–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul GG 1977. The nuclear and the cytoplasmic pore complex: Structure, dynamics, distribution, and evolution. Int Rev Cytol Suppl 1977: 75–186 [PubMed] [Google Scholar]
- Mor A, Suliman S, Ben-Yishay R, Yunger S, Brody Y, Shav-Tal Y 2010. Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nat Cell Biol 12: 543–552 [DOI] [PubMed] [Google Scholar]
- Mühlemann O, Lykke-Andersen J 2010. How and where are nonsense mRNAs degraded in mammalian cells? RNA Biol 7: 28–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlemann O, Mock-Casagrande CS, Wang J, Li S, Custodio N, Carmo-Fonseca M, Wilkinson MF, Moore MJ 2001. Precursor RNAs harboring nonsense codons accumulate near the site of transcription. Mol Cell 8: 33–43 [DOI] [PubMed] [Google Scholar]
- Nicholson P, Yepiskoposyan H, Metze S, Zamudio Orozco R, Kleinschmidt N, Mühlemann O 2010. Nonsense-mediated mRNA decay in human cells: Mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell Mol Life Sci 67: 677–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- No D, Yao TP, Evans RM 1996. Ecdysone-inducible gene expression in mammalian cells and transgenic mice. Proc Natl Acad Sci 93: 3346–3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid DW, Nicchitta CV 2012. The enduring enigma of nuclear translation. J Cell Biol 197: 7–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD 2001. The nuclear pore complex as a transport machine. J Biol Chem 276: 16593–16596 [DOI] [PubMed] [Google Scholar]
- Sato H, Maquat LE 2009. Remodeling of the pioneer translation initiation complex involves translation and the karyopherin importin β. Genes Dev 23: 2537–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H, Hosoda N, Maquat LE 2008. Efficiency of the pioneer round of translation affects the cellular site of nonsense-mediated mRNA decay. Mol Cell 29: 255–262 [DOI] [PubMed] [Google Scholar]
- Shav-Tal Y, Darzacq X, Shenoy SM, Fusco D, Janicki SM, Spector DL, Singer RH 2004. Dynamics of single mRNPs in nuclei of living cells. Science 304: 1797–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson SB, Stoltzfus CM 1994. Frameshift mutations in the v-src gene of avian sarcoma virus act in cis to specifically reduce v-src mRNA levels. Mol Cell Biol 14: 1835–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer RH, Penman S 1973. Messenger RNA in HeLa cells: Kinetics of formation and decay. J Mol Biol 78: 321–334 [DOI] [PubMed] [Google Scholar]
- Singh G, Jakob S, Kleedehn MG, Lykke-Andersen J 2007. Communication with the exon–junction complex and activation of nonsense-mediated decay by human Upf proteins occur in the cytoplasm. Mol Cell 27: 780–792 [DOI] [PubMed] [Google Scholar]
- Sun M, Schwalb B, Schulz D, Pirkl N, Etzold S, Lariviere L, Maier KC, Seizl M, Tresch A, Cramer P 2012. Comparative dynamic transcriptome analysis (cDTA) reveals mutual feedback between mRNA synthesis and degradation. Genome Res 22: 1350–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thermann R, Neu-Yilik G, Deters A, Frede U, Wehr K, Hagemeier C, Hentze MW, Kulozik AE 1998. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J 17: 3484–3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RE, Larson DR, Webb WW 2002. Precise nanometer localization analysis for individual fluorescent probes. Biophys J 82: 2775–2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trcek T, Larson DR, Moldon A, Query CC, Singer RH 2011. Single-molecule mRNA decay measurements reveal promoter-regulated mRNA stability in yeast. Cell 147: 1484–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trcek T, Chao JA, Larson DR, Park HY, Zenklusen D, Shenoy SM, Singer RH 2012. Single-mRNA counting using fluorescent in situ hybridization in budding yeast. Nat Protoc 7: 408–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlaub G, Mitchell PJ, Ciudad CJ, Chasin LA 1989. Nonsense mutations in the dihydrofolate reductase gene affect RNA processing. Mol Cell Biol 9: 2868–2880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci 24: 437–440 [DOI] [PubMed] [Google Scholar]
- Wilkinson MF, Shyu AB 2002. RNA surveillance by nuclear scanning? Nat Cell Biol 4: E144–E147 [DOI] [PubMed] [Google Scholar]
- Zenklusen D, Larson DR, Singer RH 2008. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat Struct Mol Biol 15: 1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Sun X, Qian Y, LaDuca JP, Maquat LE 1998a. At least one intron is required for the nonsense-mediated decay of triosephosphate isomerase mRNA: A possible link between nuclear splicing and cytoplasmic translation. Mol Cell Biol 18: 5272–5283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Sun X, Qian Y, Maquat LE 1998b. Intron function in the nonsense-mediated decay of β-globin mRNA: Indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA 4: 801–815 [DOI] [PMC free article] [PubMed] [Google Scholar]