Abstract
Presently, 2 to 4 days elapse between sampling at infection suspicion and result of microbial diagnostics. This delay for the identification of pathogens causes quite often a late and/or inappropriate initiation of therapy for patients suffering from infections. Bad outcome and high hospitalization costs are the consequences of these currently existing limited pathogen identification possibilities. For this reason, we aimed to apply the innovative method multi-capillary column–ion mobility spectrometry (MCC-IMS) for a fast identification of human pathogenic bacteria by determination of their characteristic volatile metabolomes. We determined volatile organic compound (VOC) patterns in headspace of 15 human pathogenic bacteria, which were grown for 24 h on Columbia blood agar plates. Besides MCC-IMS determination, we also used thermal desorption–gas chromatography–mass spectrometry measurements to confirm and evaluate obtained MCC-IMS data and if possible to assign volatile compounds to unknown MCC-IMS signals. Up to 21 specific signals have been determined by MCC-IMS for Proteus mirabilis possessing the most VOCs of all investigated strains. Of particular importance is the result that all investigated strains showed different VOC patterns by MCC-IMS using positive and negative ion mode for every single strain. Thus, the discrimination of investigated bacteria is possible by detection of their volatile organic compounds in the chosen experimental setup with the fast and cost-effective method MCC-IMS. In a hospital routine, this method could enable the identification of pathogens already after 24 h with the consequence that a specific therapy could be initiated significantly earlier.
Keywords: Pathogen identification, Volatile metabolome, Multi-capillary column (MCC), Ion mobility spectrometry (IMS), Volatile organic compound (VOC)
Introduction
The treatment of suspected bacterial infections like pneumonia or sepsis represents a difficult dilemma for the clinician. Due to a lack of knowledge about the identity of infecting pathogen at the point of infection suspicion, initial treatment mostly starts with broad-spectrum antibiotic therapy followed by narrow-spectrum therapy according to results of microbiological diagnostic. The argument for this strategy is that several studies showed that immediate initiation of an appropriate antibiotic therapy was associated with reduced mortality, reduced length of intensive care unit (ICU) stay and reduced costs (e.g., Kollef 2000; Alvarez-Lerma 1996; Rello et al. 1997; Kumar et al. 2009). Thus, this de-escalating strategy seems to be the preferred approach rather than starting narrow-spectrum therapy and then broadening the spectrum if necessary once culture data are available. In addition, an optimal therapy and thus, the reduction of inappropriate and over treatment may minimize the risk of resistance development seriously (Arias and Murray 2009; Burgess 2009; Chastre 2008).
A fast and efficient determination of pathogens is therefore urgently needed to immediately initiate an appropriate therapy with infection suspicion. Currently, the identity of microorganisms is classically determined using bacterial culture characterization combined with biochemical and susceptibility tests, which takes 2 to 4 days. Consequently, the results are not available early enough to guide antibiotic management in an early stage of the treatment.
Much work has been done in the last decade to overcome the problem of the missing fast and reliable pathogen identification. Different approaches were used for that purpose like proteome based identification by matrix-assisted laser desorption–time-of-flight mass spectrometry (Seng et al. 2009; Barbuddhe et al. 2008) and molecular biological based identification by DNA microarray and PCR (Yoo et al. 2009; Palka-Santini et al. 2009). Besides, first attempts have been made to omit the time intensive cultivation, which is also important for non-cultivatable bacteria (Fenollar and Raoult 2007; Bahrani-Mougeot et al. 2007). But molecular biological methods are with, e.g., EUR 300/test (Lehmann et al. 2010), relatively cost-intensive though.
Next to the proteomic and genomic based pathogen differentiation is a differentiation by metabolic profiling, as metabolism of microorganisms is associated with biodegradation of substrates to products. Some of these products are volatile organic compounds (VOCs) and can therefore be detected in headspace of microbial cultures.
First scientific evidence for microbial VOCs have been presented in 1921 and 1976 already (Zoller and Clark 1921; Stotzky and Schenck 1976) and the role of VOCs for medical application gains more and more interest in nowadays (Phillips et al. 2008; Westhoff et al. 2007; Chaim et al. 2003). Since technology has developed in recent years, more than 120 different VOCs have been detected from Actinomycetes and 80 different VOCs from Streptomyces for example (Schöller et al. 2002; Dickschat et al. 2005). An overview of volatile microbial compounds and their current detection methods has been given in recent publications (Schulz and Dickschat 2007; Kai et al. 2009; Korpi et al. 2009).
A comparatively new approach, which is aimed in this study, is the detection of microbial volatiles by multi-capillary column (MCC)–ion mobility spectrometry (IMS). IMS is well-known for applications like the detection of chemical warfare agents, explosives, and drugs (Eiceman and Karpas 2005; Baumbach and Eiceman 1999; Roehl 1991). New applications in recent years are microbial VOC detections in headspace of microbial cultures (Ruzsanyi et al. 2002; Snyder et al. 1991; Smith et al. 1997; Perl et al. 2011) or even over clinical samples directly (Chaim et al. 2003). Also differential mobility spectrometry, a closely related technology, has been used for that purpose already (Shnayderman et al. 2005). IMS provides a high sensitivity (detection limits down to ng/L-range to pg/L-range, ppbv-range, and pptv-range) combined with high-speed data acquisition and relatively low technical expenditure. By coupling the IMS with a MCC for pre-separation, even the analysis of complex and humid gas samples is possible. The new aspect in this investigation is the application of MCC-IMS for a rapid bacterial discrimination by metabolic profiling. It might be of great importance for several infections and more promising, for the detection of pathogens in breath directly.
The present study describes the use of MCC-IMS as an innovative method to rapidly determine pathogen identities by their emitted VOC patterns. The aim was to prove the principle of differentiation of human pathologic bacteria by metabolic profiling with MCC-IMS. Therefore, 15 clinically relevant human pathogens were grown on Columbia sheep blood agar, and headspace of these cultures were analyzed by MCC-IMS. The results have been validated by help of additional gas chromatography–mass spectrometry (GC/MS) analysis.
Materials and methods
Bacterial strains
The bacteria Acinetobacter baumannii (DSM 24110), Citrobacter freundii (DSM 24120), Enterobacter cloacae (DSM 30054), Escherichia coli (DSM 1103), Hafnia alvei (DSM 24119), Klebsiella oxytoca (DSM 24121), Klebsiella pneumoniae (DSM 2026), Proteus mirabilis (DSM 4479), Pseudomonas aeruginosa (DSM 46358), Serratia marcescens (DSM 50904), Staphylococcus aureus (DSM 13661), Staphylococcus epidermidis (DSM 18857), Staphylococcus haemolyticus (DSM 24111), Streptococcus agalactiae (DSM 2134), and Streptococcus pneumoniae (DSM 11967) were cultivated on Columbia sheep blood agar (Ref. 43041; BioMérieux, Nürtingen, Germany) for 24 h at 37 °C prior to measurement. Cultures have been stored short term at 4 °C or long term at −80 °C in brain hearth infusion broth (CM1135, OXOID, Wesel, Germany) supplemented with 25% glycerol.
Chemicals and reagents
For MCC-IMS reference measurements, ethanol (CAS 64-17-5), indole (CAS 120-72-9), phenethyl alcohol (CAS 60-12-8), 2-(methylthio)-ethanol (CAS 5271-38-5), 3-methylbutanal (CAS 590-86-3), 3-methyl-1-butanol (CAS 123-51-3), dimethyldisulfide (CAS 624-92-0), 2,3-heptanedione (CAS 96-04-8), 2,5-dimethylpyrazine (CAS 123-32-0), phenol (CAS 108-95-2), dimethyl trisulfide (CAS 3658-80-8), 2-octanone (CAS 111-13-7), 2-acetylthiazole (CAS 24295-03-2), 2-nonanone (CAS 821-55-6), 2-decanone (CAS 693-54-9), N-(phenylmethylene)-1-propanamine (CAS 6852-55-7), 2-undecanone (CAS 112-12-9), S-methyl thiobenzoate (CAS 5925-68-8), benzonitrile (CAS 100-47-0), and 2,3,5-trimethylpyrazine (CAS 1466-55-1) in HPLC grade have been used.
Sampling of bacterial volatiles
In order to detect bacterial VOCs a thermal controlled (37 °C) measurement chamber consistent of stainless steel, a gas inlet and a gas outlet has been constructed (Leibniz-Institute for Analytical Sciences—ISAS—e.V., Dortmund, Germany). Synthetic air (scientific quality, AIR LIQUIDE Deutschland, Düsseldorf) was supplied with 100 mL/min via gas inlet. The gas outlet was either attached to the MCC-IMS measurement system or to an adsorption tube (packed with 200 mg Tenax™ GR, GERSTEL, Mühlheim, Germany) for GC/MS measurement by PTFE tubing (Bohlender, Grünsfeld, Germany). For MCC-IMS analysis, 10 mL, and for GC/MS analysis, 2 L for sample volume have been used, respectively.
Volatile compounds have been detected of the empty measurement chamber and of headspace of Columbia sheep blood agar plates in the measurement chamber to determine the volatile background. After these blank measurements, media with bacterial colonies have been measured after 24 h of cultivation at 37 °C. Headspace of a bacterial culture was continuously rinsed with a synthetic air flow of 100 mL/min. Samples were drawn after 5 and 25 min for MCC-IMS analyses, and after 60 min for GC/MS analyses. Each measurement was performed three times.
Sampling of reference substances
A small aliquot of each substance has been filled into a 1 mL reaction vial (CS-Chromatography Service, Langerwehe) with a screw cap including a gas-permeable membrane to reach a concentration of approximately 1–10 ppb of every reference compound. Afterwards, every reaction vial has been placed in a 250 mL Schott flask with swivelling screw fitting (BOHLENDER, Grünsfeld, Germany). Prior to sampling, the Schott flask containing the reference compound has been incubated for at least 1 h at room temperature under a constant flow with synthetic air of 100 mL/min. For each measurement, the gas outlet of the Schott flask was attached to MCC-IMS measurement system by PTFE tubing (Bohlender, Grünsfeld, Germany).
Multi-capillary column–ion mobility spectrometer
Bacterial volatiles were rinsed from the measurement chamber through the 10 mL sample loop at a flow of 100 mL/min for 5 min, and 10 mL of the sample was afterwards loaded with a carrier gas flow of 150 mL/min via a six-port-valve onto a rapidly pre-separating OV-5 multi-capillary column (20 cm long, consisting of approx. 1,000 parallel glass capillaries, 3 mm total diameter, 43 μm inner diameter of a single capillary; MULTICHROM, Novosibirsk, Russia). The multi-capillary column was operated at a constant temperature of 40 °C and at a flow rate of 150 mL/min using synthetic air as a carrier gas (scientific quality, AIR LIQUIDE Deutschland, Düsseldorf). After chromatographic separation of bacterial volatiles, eluted gas-phase analytes were ionized by a radioactive ionization source (63Ni, 550 MBq) through charge transfer from prior ionized reactant ions and analyzed with a coupled IMS instrument according to Vautz and Baumbach (2008; Leibniz-Institute for Analytical Sciences—ISAS—e.V., Dortmund, Germany) with the following parameters: synthetic air as drift gas, at a flow rate of 100 mL/min, in an electrical field of 330 V/cm, drift tube (length 12 cm, 15 mm diameter), shutter grid opening time of 300 μs, spectra length/sample interval of 100 ms, and spectra resolution of 50 kHz. Under the influence of a positive or negative external electrical field either protonated or negatively charged analyte ions moved towards a detector (Faraday-plate), respectively. During their drift to the detector, ions are being separated through collision with drift gas molecules moving in the opposite direction (100 mL/min). Thus, a constant resulting drift velocity is the consequence depending on size and shape of analyte ions. Ion drift times are measured and ion velocities are calculated for a known drift distance in the following. The so-called ion mobility was then calculated by normalizing drift velocity to the electric field. Finally, reduced ion mobility which is characteristic for an ion and independent on the experimental conditions has been determined by further normalization to temperature and pressure (Vautz et al. 2009).
The specific reduced ion mobility and retention time related to a particular signal in the MCC-IMS data enables the identification of the corresponding molecule by comparison with a database. For unknown signals which are not available in the MCC-IMS database, identification is possible only by accompanying sampling on adsorption materials for analysis by GC/MS resulting in a proposed analyte, followed by a measurement of the reference analyte by MCC-IMS for validation. Moreover, the MCC-IMS signal intensity is a measure for the concentration of VOCs in gas samples and can be used for quantification under specific conditions by help of an external calibration.
Thermal desorption–gas chromatography–mass spectrometry
Volatile organic compounds were thermally desorbed from Tenax material (Tenax™ GR, GERSTEL, Mühlheim, Germany) using the following temperature program: initial temperature of 25 °C was increased by 30 °C/s to 250 °C, 2.5 min isothermal at 250 °C, splitless for 2 min at a flow rate of 60 mL/min, using helium as carrier gas. Volatile analytes were then transferred at 300 °C to an integrated cooled injection system (CIS; GERSTEL, Mühlheim, Germany), focused at −120 °C, released onto a HP-5MS capillary column (60 m × 0.25 mm × 0.25 μm film thickness; Agilent Technologies, Santa Clara, CA, USA) after the CIS was heated to 250 °C (by 12 °C/min) and temperature was held for 10 min. The GC/MS analysis was performed on an Agilent Technologies 7890A GC system connected with an Agilent Technologies 5975C inert XL mass selective detector (MSD; Agilent Technologies, Santa Clara, CA, USA) at an initial oven temperature of 35 °C which was kept for 2 min, increased by 7 °C/min to 250 °C and held for 7 min. The separation was performed with helium as carrier gas at a constant flow rate of 1.0 mL/min. Electron ionization mode was set at 70 eV and the mass rage of m/z 33–450 was detected.
MCC-IMS data analysis
Detected MCC-IMS signals have been evaluated using BB_IMSAnalyse 1.0 (Leibniz-Institute for Analytical Sciences—ISAS—e.V., Dortmund, Germany). Data are represented as a matrix of signal intensities where the x-axis indicates the inverse ion mobility in volt second per square centimetre (Vs/cm2) and the y-axis indicates the retention time in seconds in a topographic plot (Figs. 1 and 4). In order to normalize all MCC retention times for a better comparison of data, the previously described MCC retention time of 118.3 s for benzothiazole (Perl et al. 2010), a permanent contaminant of synthetic air, has been used. Minor variations in drift and retention time have been corrected by alignment according to Vautz et al. (2009) and Perl et al. (2010). Identification of MCC-IMS signals was performed by comparison of properties of detected signals with information of a database of reference compounds (Leibniz-Institute for Analytical Sciences—ISAS—e.V., Dortmund, Germany) or by own reference measurements. Unidentified detected compounds were named after their position in the 2D topographic plot as p_1000 × inverse ion mobility_retention time (e.g., p_642_1; 0.642 Vs/cm2, 1 s).
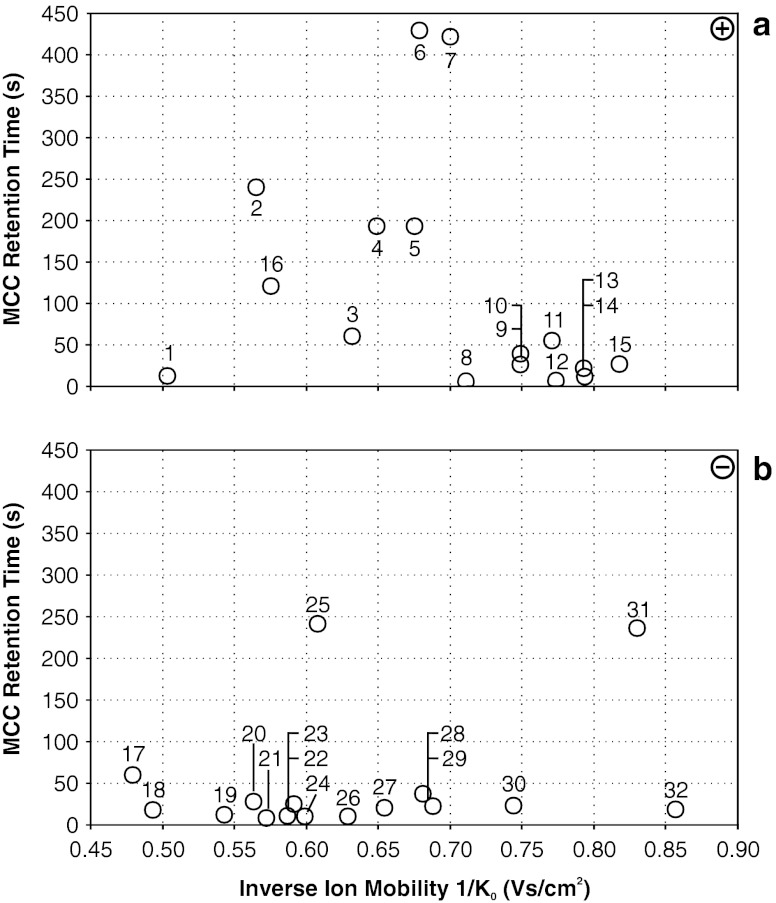
Fig. 1.
Summarized MCC-IMS topographic plot of bacterial volatiles, which can be used for bacterial discrimination. Fifteen (1–15) compounds have been determined in positive (a) and 16 (17–32) compounds have been determined in negative ion detection mode (b). All depicted compounds are listed in Table 1
Fig. 4.
Total ion chromatograms of bacterial headspace samples. For pre-concentration, VOCs of 2 L headspace volume have been adsorbed to Tenax GR. Total ion chromatograms are shown from 7 to 28 min. Identity of VOCs is summarized in Table 2. Cyclohexanol Rt = 12.17 min, styrene Rt = 12.35 min, cyclohexanone Rt = 12.45 min, benzaldehyde Rt = 14.14 min, 2-ethyl-1-hexanol Rt = 15.61 min and acetophenone Rt = 16.68 min have been detected as main volatile compounds over Columbia sheep blood agar. Benzothiazole—a contaminant of the operation gas—has been detected in every headspace sample at Rt = 20.37 min
Thermal desorption–gas chromatography–mass spectrometry data analysis
GC/MS data have been evaluated by MSD ChemStation Data Analysis Application (Agilent Technologies, Santa Clara, CA, USA) and compound mass spectra were identified by AMDIS/NIST (Automated Mass Spectral Deconvolution and Identification System; version 2.62, 2005; NIST version 2.0, 2005).
Results
Determination of bacterial volatiles by MCC-IMS
All bacterial strains investigated in this study release volatile organic compounds detectable with MCC-IMS (Table 1). By comparison of bacterial VOC patterns with volatiles of Columbia sheep blood agar, several blood agar related compounds have been determined in the positive and negative ion mode by MCC-IMS. These blood agar related compounds were neglected when comparing bacterial strains. The location of single signals in a 2D topographic plot is shown in Fig. 1. All compounds determined by this method are consecutively numbered and in the following either termed by their names or by numbers from 1 to 32 according to Table 1, which is summarizing MCC-IMS results. In addition, the occurrence of the most important VOCs for specification and discrimination in headspace of investigated bacteria is illustrated as region comparison analysis in Fig. 2.
Table 1.
Emitted bacterial VOCs determined by MCC-IMS in a positive and negative IMS mode
| No. | Compound | A. baumannii | C. freundii | E. cloacae | E. coli | H. alvei | K. oxytoca | K. pneumoniae | P. mirabilis | P. aeruginosa | S. marcescens | S. aureus | S. epidermidis | S. haemolyticus | S. agalactiae | S. pneumoniae |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DSM 24110 | DSM 24120 | DSM 30054 | DSM 1103 | DSM 24119 | DSM 24121 | DSM 2026 | DSM 4479 | DSM 46358 | DSM 20904 | DSM 13661 | DSM 18857 | DSM 24111 | DSM 2134 | DSM 11967 | ||
| Positive mode | ||||||||||||||||
| 1 | Ethanol | X | X | XX | X | X | X | XX | XX | X | XX | XX | X | X | X | |
| 2 | Indole | XX | XX | |||||||||||||
| 3 | Phenethyl alcohol | XX | X | X | XX | X | XX | |||||||||
| 4 | p_649_194 | X | ||||||||||||||
| 5 | p_675_194 | XX | ||||||||||||||
| 6 | p_679_429 | X | ||||||||||||||
| 7 | p_700_423 | X | ||||||||||||||
| 8 | p_711_3 | X | X | X | X | X | X | X | X | X | X | |||||
| 9 | p_749_26 | X | ||||||||||||||
| 10 | p_749_37 | X | ||||||||||||||
| 11 | p_771_54 | X | ||||||||||||||
| 12 | p_774_4 | X | X | X | X | X | X | |||||||||
| 13 | p_793_19 | X | ||||||||||||||
| 14 | p_794_12 | X | ||||||||||||||
| 15 | p_818_26 | X | ||||||||||||||
| No. of VOCs | 3 | 3 | 2 | 4 | 4 | 5 | 2 | 14 | 2 | 3 | 1 | 1 | 2 | 2 | 0 | |
| Negative mode | ||||||||||||||||
| 17 | Phenethyl alcohol | X | X | |||||||||||||
| 18 | p_493_17 | X | ||||||||||||||
| 19 | p_543_12 | XX | X | XX | X | X | ||||||||||
| 20 | p_563_24 | XX | ||||||||||||||
| 21 | p_572_8 | X | X | X | XX | X | X | X | X | X | X | X | ||||
| 22 | 2-(Methylthio)-ethanol | X | ||||||||||||||
| 23 | p_591_24 | XX | ||||||||||||||
| 24 | p_599_10 | X | X | X | X | X | X | X | ||||||||
| 25 | Indole | XX | XX | |||||||||||||
| 26 | p_629_10 | X | X | X | ||||||||||||
| 27 | p_654_20 | X | X | |||||||||||||
| 28 | p_681_36 | X | ||||||||||||||
| 29 | p_688_22 | X | ||||||||||||||
| 30 | p_744_22 | X | ||||||||||||||
| 31 | Indole –dimer | X | ||||||||||||||
| 32 | p_857_18 | X | ||||||||||||||
| No. of VOCs | 1 | 6 | 3 | 7 | 4 | 3 | 0 | 8 | 0 | 3 | 2 | 1 | 1 | 1 | 1 | |
| Total no. of VOCs | 4 | 9 | 5 | 11 | 8 | 8 | 2 | 22 | 2 | 6 | 3 | 2 | 3 | 3 | 1 | |
Detected compounds in 10 mL headspace volume of each examined strain are illustrated by an X or XX if occurring in high amounts. 16 benzothiazole has been used for retention time normalization
Fig. 2.
Region comparison image of MCC-IMS analyses of bacterial volatiles. Only monomeric MCC-IMS signals are shown. Red boxes indicate the presence of a VOC, which can be used for bacterial discrimination. Signals from 1 to 14 have been determined in positive ion mode, whereas signals from 17 to 29 have been determined in negative ion mode. An example of headspace VOCs of sheep blood agar and one example of headspace of every bacterial strain is displayed. All strains have been cultivated for 24 h. Signal intensities are illustrated by different colours (white = zero, blue = low, red = medium, yellow = high). The following signals have been used for further analyses: 1 ethanol, 2 indole, 3 phenethyl alcohol, 4 p_649_194, 5 p_675_194, 6 p_679_429, 7 p_700_423, 8 p_711_3, 9 p_749_26, 10 p_749_37, 11 p_771_54, 13 p_793_19, 14 p_794_12, 17 phenethyl alcohol, 18 p_493_17, 19 p_543_12, 20 p_563_24, 21 p_572_8, 22 2-(methylthio)-ethanol, 25 indole, 26 p_629_10, 27 p_654_20, 28 p_681_36 and 29 p_688_22
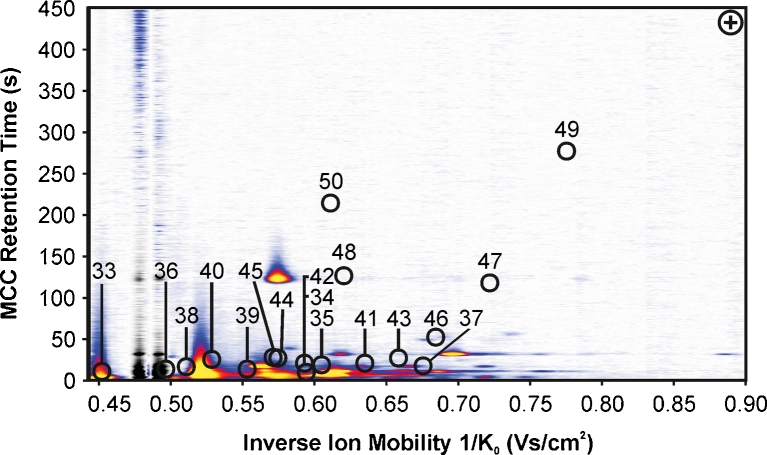
Fifteen significant bacterial volatiles were detected in positive ion mode (1–15) and further 16 significant signals were detected in negative ion mode (17–32) in headspace of all investigated bacteria but not over medium control. These compounds can be expected to enable bacterial differentiation (Table 1). Based on that fact, a specific VOC pattern has been determined for every investigated strain. This includes furthermore that for S. pneumoniae only one emitted VOC has been detected, whereas for P. mirabilis on the other hand 21 generated signals were determined. VOC pattern comparison revealed that all 15 strains are distinguishable unequivocally by MCC-IMS even if the chemical identity of most VOCs has not unambiguously been identified so far. A recently published study by Jünger et al. (2010) suggested an efficient identification strategy of unknown MCC-IMS signals by additionally performed thermal desorption GC/MS. This approach was the secondary aim of this study, which leads to the assignment of ethanol (1), indole (2, 25, 31), phenethyl alcohol (3, 17) and 2-(methylthio)-ethanol (22), variably present over several investigated bacteria. The respective reference compounds ethanol, indole, phenethyl alcohol and 2-(methylthio)-ethanol were measured separately by MCC-IMS and confirmed our assumptions. It has to be emphasized that indole, phenethyl alcohol and 2-(methylthio)-ethanol are detectable in positive as well as in negative ion mode as their proton bound analyte molecules and anions have been formed in the corresponding mode. Due to assign more VOCs, further reference measurements have been performed (Table 2, 33–49), but even if these compounds could definitely be assigned to MCC-IMS signals after the analysis, determination of these compounds in headspace of bacteria gave unsatisfying result. Either the separation from other occurring compounds over investigated strains was not satisfactory or signals could not be detected in the expected area due to amounts present probably below the detection limit, since 10 mL sample volume has been used for MCC-IMS analysis only. Different amounts of ammonia (33, reference data, e.g., Vautz et al. (2010)), also eluting in an area of high background signal density could be determined over all investigated strains using positive ion mode (Fig. 3, Table 2). Consequently, this compound did not provide additional information that can be used for pathogen differentiation purpose.
Table 2.
By MCC-IMS detectable VOCs using positive ion mode which do not possess pathogen differentiation potential by this method due to low amounts in 10 mL headspace or high signal intensity around respective signals
| No. | No. GC/MS | Rt GC (min) | Rt MCC (s) | (+) 1/K0 monomer | (+) 1/K0 dimer | Compound |
|---|---|---|---|---|---|---|
| 33 | >3.00 | 3.0 | 0.451 | – | Ammonia | |
| 34 | 1* | 7.01 | 3.9 | 0.599 | – | 3-Methylbutanal |
| 35 | 2* | 8.48 | 15.0 | 0.614 | 0.742 | 3-Methyl-1-butanol |
| 36 | 3* | 8.77 | 6.3 | 0.488 | 0.559 | Dimethyldisulfide |
| 37 | 7* | 10.95 | 10.0 | 0.671 | – | 2,3-Heptanedione |
| 38 | 8* | 11.57 | 8.8 | 0.515 | 0.583 | 2-(Methylthio)-ethanol |
| 39 | 9* | 12.80 | 14.9 | 0.552 | – | 2,5-Dimethylpyrazine |
| 40 | 10* | 14.41 | 22.8 | 0.529 | – | Phenol |
| 41 | 11* | 14.47 | 19.1 | 0.637 | – | Dimethyl trisulfide |
| 42 | 12* | 14.74 | 18.6 | 0.594 | 0.730 | Benzonitrile |
| 43 | 13* | 14.75 | 23.6 | 0.652 | 0.855 | 2-Octanone |
| 44 | 14* | 15.09 | 23.7 | 0.573 | 0.792 | 2,3,5-Trimethylpyranzine |
| 45 | 16* | 15.55 | 26.0 | 0.556 | 0.726 | 2-Acetylthiazole |
| 46 | 21* | 17.10 | 51.6 | 0.686 | 0.909 | 2-Nonanone |
| 47 | 27* | 19.31 | 119.4 | 0.722 | 0.968 | 2-Decanone |
| 48 | 28* | 19.89 | 121.4 | 0.621 | 0.746 | N-(Phenylmethylene)-1-propanamine |
| 49 | 33* | 21.42 | 276.3 | 0.755 | 1.017 | 2-Undecanone |
| 50 | 35* | 21.75 | 210.0 | 0.610 | 0.815 | S-Methyl thiobenzoate |
Fig. 3.
MCC-IMS topographic plot of volatiles which have been detected in bacterial headspace but have not been chosen for discrimination of bacteria. Because either concentration of VOCs was too low in 10 mL headspace samples or high density of background signals of Columbia sheep blood agar, headspace prevents an unambiguous differentiation. For compound name, see Table 2
Determination of bacterial volatiles by GC/MS
Results of thermal desorption GC/MS analysis confirmed the fact that bacteria can be discriminated by their VOC patterns (Table 3, Fig. 4). As well as for MCC-IMS analysis VOCs determined by GC/MS are either termed by their names and/or by numbers from 1* to 47* according to Table 3. Contrary to MCC-IMS studies, where 10 mL of headspace samples has been analyzed, 2 L of headspace samples has been analyzed by GC/MS. In total 47 different VOCs were detected over investigated strains but not over Columbia sheep blood agar alone. Consequently, an endogenous origin of these compounds can be assumed. In contrast, VOCs, with increased amounts compared to Columbia sheep blood agar background, were not taken in to account like benzaldehyde, which massively increases in its concentration over E. coli (Fig. 3). The elucidation of the origin of those compounds is difficult and has been discussed lately (Schulz and Dickschat 2007). Volatile compounds, which were consumed by microorganisms, were also not investigated further.
Table 3.
Emitted bacterial VOCs determined by thermal desorption GC/MS of 2 L headspace samples
| No. | Rt GC (min) | Compound | E. cloacae | E. coli | K. pneumoniae | P. mirabilis | P. aeruginosa | S. marcescens | S. aureus | S. epidermidis | S. agalactiae | S. pneumoniae |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DSM 30054 | DSM 1103 | DSM 2026 | DSM 4479 | DSM 46358 | DSM 50904 | DSM 13661 | DSM 18857 | DSM 2134 | DSM 11967 | |||
| 1* | 7.01 | 3-Methylbutanal | X | X | X | X | X | X | X | X | ||
| 2* | 8.48 | 3-Methyl-1-butanol | XX | XX | X | X | XX | X | ||||
| 3* | 8.77 | Dimethyldisulfide | X | X | XX | X | X | |||||
| 4* | 9.48 | 3-Methyl-2-buten-1-ol | X | |||||||||
| 5* | 9.73 | 3-Methyl-2-butenal | X | |||||||||
| 6* | 10.66 | Methylpyrazine | X | |||||||||
| 7* | 10.95 | 2,3-Heptanedione | X | |||||||||
| 8* | 11.57 | 2-(Methylthio)-ethanol | X | |||||||||
| 9* | 12.80 | 2-(Methylthio)-ethanol | X | X | X | X | X | X | X | |||
| 10* | 14.41 | Phenol | X | X | X | |||||||
| 11* | 14.47 | Dimethyl trisulfide | X | X | X | |||||||
| 12* | 14.74 | Benzonitrile | X | X | X | X | X | X | X | |||
| 13* | 14.75 | 2-Octanone | X | |||||||||
| 14* | 15.09 | 2,3,5-Trimethylpyrazine | X | X | X | |||||||
| 15* | 15.25 | 3-Methylbutyl 2-methylpropanoate | X | |||||||||
| 16* | 15.55 | 2-Acetylthiazole | X | |||||||||
| 17* | 15.57 | 1-Methoxy-4-methylbenzene | X | |||||||||
| 18* | 15.98 | 3-Methyl-N-(3-methylbutylidene)-1-butanamine | X | |||||||||
| 19* | 16.19 | N-(Phenylmethylene)-methanamine | X | |||||||||
| 20* | 17.08 | 1-Undecene | X | |||||||||
| 21* | 17.10 | 2-Nonanone | X | X | X | X | X | X | ||||
| 22* | 17.33 | Methyl benzoate | X | |||||||||
| 23* | 17.72 | Phenethyl alcohol | X | X | ||||||||
| 24* | 17.81 | N,N′-Dibenzylideneethylenediamine | X | |||||||||
| 25* | 18,32 | Benzyl nitrile | X | X | X | |||||||
| 26* | 19.01 | Benzyl methyl sulfide | X | |||||||||
| 27* | 19.31 | 2-Decanone | X | X | X | |||||||
| 28* | 19.89 | N-(Phenylmethylene)-1-propanamine | X | |||||||||
| 29* | 20.50 | Ethyl phenylacetate | X | |||||||||
| 30* | 20.78 | 2-Phenyl ethyl acetate | X | X | ||||||||
| 31* | 21.03 | N-(Phenylmethylene)-1-butanamine | X | X | ||||||||
| 32* | 21.20 | N-Butyl-benzenamine | X | |||||||||
| 33* | 21.42 | 2-Undecanone | X | X | X | X | ||||||
| 34* | 21.62 | Indole | XX | |||||||||
| 35* | 21.75 | S-Methyl thiobenzoate | X | |||||||||
| 36* | 21.79 | 1-Methyl-naphthalene | X | X | ||||||||
| 37* | 21.94 | 2-(3-Methylbutyl)-3,5-dimethylpyrazine | X | X | X | X | ||||||
| 38* | 22.21 | 2-Methyl-naphthalene | X | X | ||||||||
| 39* | 22.45 | Phenylacetic acid propylester | X | |||||||||
| 40* | 23.22 | 3-Methyl-N-(2-phenylethylidene)-1-butanamine | X | |||||||||
| 41* | 23.49 | p-Pentylaniline | X | |||||||||
| 42* | 23.51 | 3-Methyl-1H-indole | X | |||||||||
| 43* | 23.53 | Phenylethyl butyrate | X | X | ||||||||
| 44* | 24.34 | Isoamyl benzoate | X | |||||||||
| 45* | 26.18 | 4-Chloro-1H-indole | X | |||||||||
| 46* | 26.66 | N-(1,1-Dimethylethyl)-benzamide | X | |||||||||
| 47* | 27.59 | N-n-Butylphthalimide | X | |||||||||
| No. of VOCs | 14 | 22 | 4 | 24 | 7 | 11 | 3 | 2 | 11 | 4 | ||
An X indicates the detection of a compound, whereas XX indicates compounds occurring in high amounts
Discussion
Currently, the identification of human pathogenic bacteria takes, with 2 to 4 days after infection suspicion, too much time using routine diagnostics. An innovative tool to possibly overcome bacterial identification problems is the MCC-IMS. The results of this study demonstrate that it is indeed possible to differentiate all 15 bacterial strains by their different VOC patterns via the fast and powerful tool MCC-IMS (Table 1). Analysis in positive or negative ion mode only would lead to less discriminating power and cannot be recommended. Both ionization modes used in this investigation are important for pathogen differentiation.
The differentiability of pathogens per se could also be confirmed by corresponding GC/MS analyses of several strains (Table 3).
Actually, the identification of bacterial species by MCC-IMS is a phenotypic method. Using this approach, different VOC patterns have been detected for 15 investigated bacterial strains. As we focused on bacterial discrimination by the innovative tool MCC-IMS primarily as a proof of principle, the time-consuming GC/MS analyses have not been performed for all bacteria investigated. Identification of unknown MCC-IMS signals was our secondary aim only and has to be given more emphasise in the future. Due to different properties of both analytical methods, not all compounds detected by thermal desorption (TD)–GC/MS could be verified by MCC-IMS and vice versa. Most important properties of the analytical systems are highlighted in Table 4. The MCC-IMS offers a powerful analysis of smaller molecules in complex gas mixtures. The high separation power of small molecules is based on a doubled separation, primary the chromatographic by MCC and secondary by differences in drift time in the IMS. However, only volatile compounds which are ionisable can be accelerated in the IMS drift region and thus are detectable. That is one reason for the gap between compounds detected with GC/MS and MCC-IMS. Further on, even if some of the signals detected with MCC-IMS have been assigned to volatile compounds by comparison to GC/MS measurements and reference measurements, not all signals have been elucidated up to now. Therefore, further investigations are necessary to assign unknown MCC-IMS signals and to develop a microbiological reference database for MCC-IMS data. Hence, it perfectly complements and improves already existing methods like Gram staining and susceptibility tests. But even if not all MCC-IMS signals could be assigned to volatile compounds so far, the MCC-IMS is a promising tool to be used for clinical samples in the near future.
Table 4.
Comparison of methodical properties
| Property | MCC-IMS | TD-GC/MS |
|---|---|---|
| Sample volume | 10 mL | 2 L |
| Sample preparation | No pre-concentration needed | Pre-concentration on, e.g., Tenax |
| Molecular weight of analytes | Approx. 17–180 g/mol | Approx. 85–200 g/mol |
| Duration of sampling | 5 min | 20 min |
| Duration of sample analysis | 10 min | 60 min |
| Data analysis (reference databases) | Approx. 120 compounds | More than 190,000 compounds NIST standard reference database |
| ISAS customized database | ||
| Temperature program during analysis | Isothermally at 40 °C | Gradient from 35 °C to 250 °C |
| Necessity of vacuum for separation | Not needed | Necessary |
Some of the MCC-IMS signals have been assigned to corresponding GC/MS signals (Table 2). For these compounds, even quantitative measurements after calibration of the device to these compounds are possible. Thus, the MCC-IMS demonstrates its capacity beyond plain phenotypic tools as the electronic noses (Pavlou et al. 2002).
A further aspect, also important for the current investigation, is the detection of compound dimers. As previously described 63Ni as ionization source leads to the formation of proton bound monomers and can also lead under certain conditions (e.g., high concentrations) to the formation of proton bound dimers using positive ion mode (Borsdorf and Eiceman 2006). The formation of proton bound dimers might be present for detected compounds in this investigation as well. Therefore, p_818_26 (15) could be the proton bound dimer of p_749_26 (9) and p_774_4 (12) could be the proton bound dimer of p_771_3 (8). This phenomenon can be observed in this study for measurements in negative ion mode too and has also been previously reported (Borsdorf and Eiceman 2006). Accordingly, indole has been detected as negatively charged monomer (25) in K. oxytoca and E. coli but the negatively charged dimer (31) has been determined over E. coli only. The occurrence of the indole dimer was associated with high signal intensities for an indole monomer. Further on the monomer p_563_24 (20) and the corresponding dimer p_591_24 (23) were detected over C. freundii. Likewise it seems to be the case for the signals p_688_22 (29) and p_744_22 (30) from C. freundii although their relations need to be investigated further. In addition the compounds p_572_8 (21) and p_599_10 (24), widely distributed over the investigated strains, seem to be the negatively charged monomer and dimer as well.
In this context, we made no attempts to distinguish between different isolates of one species, since the present investigation was designed as a proof of principle. Furthermore, we did not quantify VOC amounts and bacterial cultures, we did not determine detection limits and we did not check different growth media although it is known that medium and growth conditions strongly influence VOC compositions (Wagner et al. 2003; Schulz and Dickschat 2007). We rather used a medium which is routinely used in clinical laboratories (Columbia sheep blood agar). In addition, we made no attempts to optimize the setup as par example variation of sample gas flow. We used a sample gas flow of 100 mL/min to rinse the headspace of the measurement chamber and the sample loop to sample 10 mL afterwards. A reduction of sample gas flow will lead to a higher concentration of volatile compounds and therefore differences might occur in lower bacterial count.
Moreover, we focused in this investigation on compounds which have apparently been formed by pathogens and which are not present over Columbia blood agar alone. The origin of benzaldehyde for example, which is massively released by E. coli and is also present in moderate amounts over the used medium alone, has to be studied in further investigations. Benzaldehyde has been published as VOC of several bacteria though (Schulz and Dickschat 2007).
The major advantage of the MCC-IMS system is the feasibility of rapid measurements without pre-concentration or preparation of samples and analysis of complex gas mixtures regardless of the water vapour content in an online setup with relatively low technical expenditure. Beside the discrimination of different pathogens by volatile metabolic profiles grown on agar plates, the detection of respective VOCs in patients’ breath suffering from infections or with infection suspicion might be the most promising approach. But since VOC patterns of bacteria depend on growth conditions and growth phase (O’Hara and Mayhew 2009), pathogen determination by breath sampling could lead to different VOC patterns. Also infection related endogenous compounds could be detected in patient’s breath. Consequently, an earlier diagnosis of infections and identification of pathogens by MCC-IMS leading to an early and appropriate therapy could be achieved compared to current strategies.
In summary, the application of multi-capillary column–ion mobility spectrometry with respect to a rather fast identification of human pathogenic bacteria could be realised by determination of their specific volatile metabolomes. It is possible to determine volatile organic compound pattern of 15 human pathogenic bacteria directly, which were grown on Columbia blood agar plates. Besides MCC-IMS determination TD-GC/MS was used to confirm and evaluate the MCC-IMS data obtained. In addition, GC/MS helps to assign volatile compounds to so far unknown MCC-IMS signals. All investigated strains showed different VOC patterns by MCC-IMS using positive and negative ion mode for every single strain. Thus, the discrimination of investigated bacteria is possible by detection of their volatile organic compounds in the chosen experimental setup with the fast and cost-effective method MCC-IMS leading to a possible reduction of microbial identification time from 2 to 4 days by traditional methods to only 24 h by using the MCC-IMS.
Acknowledgements
The dedicated work of Rita Fobbe, Luzia Seifert, Stefanie Sina, Rieke Bufe, Nadine Lange and Jacqueline Friedrich, technicians at ISAS, was indispensable for the success of the investigations. The financial support of the Bundesministerium für Bildung und Forschung and the Ministerium für Wissenschaft und Forschung des Landes Nordrhein-Westfalen is gratefully acknowledged. The work was funded partly by the high-tech strategy funds of the Federal Republic of Germany (Project Metabolit—01SF0716).
References
- Alvarez-Lerma F. Modification of empiric antibiotic treatment in patients with pneumonia acquired in the intensive care unit. ICU-Acquired Pneumonia Study Group. Intensive Care Med. 1996;22:387–394. doi: 10.1007/BF01712153. [DOI] [PubMed] [Google Scholar]
- Arias CA, Murray BE. Antibiotic-resistant bugs in the 21st century—a clinical super-challenge. N Engl J Med. 2009;360:439–443. doi: 10.1056/NEJMp0804651. [DOI] [PubMed] [Google Scholar]
- Bahrani-Mougeot FK, Paster BJ, Coleman S, Barbuto S, Brennan MT, Noll J, Kennedy T, Fox PC, Lockhart PB. Molecular analysis of oral and respiratory bacterial species associated with ventilator-associated pneumonia. J Clin Microbiol. 2007;45:1588–1593. doi: 10.1128/JCM.01963-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbuddhe SB, Maier T, Schwarz G, Kostrzewa M, Hof H, Domann E, Chakraborty T, Hain T. Rapid identification and typing of Listeria species by matrix-assisted laser desorption ionization–time of flight mass spectrometry. Appl Environ Microbiol. 2008;74:5402–5407. doi: 10.1128/AEM.02689-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumbach JI, Eiceman GA. Ion mobility spectrometry: arriving on-site and moving beyond a low profile. Appl Spectrosc. 1999;53:338A–355A. doi: 10.1366/0003702991947847. [DOI] [PubMed] [Google Scholar]
- Borsdorf H, Eiceman GA. Ion mobility spectrometry: principles and applications. Appl Spectrosc Rev. 2006;41:323–375. doi: 10.1080/05704920600663469. [DOI] [Google Scholar]
- Burgess DS. Curbing resistance development: maximizing the utility of available agents. J Manag Care Pharm. 2009;15(5 Suppl):S5–S9. doi: 10.18553/jmcp.2009.15.s5.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaim W, Karpas Z, Lorber A. New technology for diagnosis of bacterial vaginosis. Eur J Obstet Gynecol Reprod Biol. 2003;111:83–87. doi: 10.1016/S0301-2115(03)00210-0. [DOI] [PubMed] [Google Scholar]
- Chastre J. Evolving problems with resistant pathogens. Clin Microbiol Infect. 2008;14(Suppl 3):3–14. doi: 10.1111/j.1469-0691.2008.01958.x. [DOI] [PubMed] [Google Scholar]
- Dickschat JS, Martens T, Brinkhoff T, Simon M, Schulz S. Volatiles released by a Streptomyces species isolated from the North Sea. Chem Biodivers. 2005;2:837–865. doi: 10.1002/cbdv.200590062. [DOI] [PubMed] [Google Scholar]
- Eiceman GA, Karpas Z. Ion mobility spectrometry. Boca Raton, Ann Arbor, London, Tokyo: CRC Press; 2005. pp. 1–228. [Google Scholar]
- Fenollar F, Raoult D. Molecular diagnosis of bloodstream infections caused by non-cultivable bacteria. Int J Antimicrob Agents. 2007;30:S7–S15. doi: 10.1016/j.ijantimicag.2007.06.024. [DOI] [PubMed] [Google Scholar]
- Jünger M, Bödeker B, Baumbach JI. Peak assignment in multi-capillary column-ion mobility spectrometry using comparative studies with gas chromatography–mass spectrometry for VOC analysis. Anal Bioanal Chem. 2010;396:471–482. doi: 10.1007/s00216-009-3168-z. [DOI] [PubMed] [Google Scholar]
- Kai M, Haustein M, Molina F, Petri A, Scholz B, Piechulla B. Bacterial volatiles and their action potential. Appl Microbiol Biotechnol. 2009;81:1001–1012. doi: 10.1007/s00253-008-1760-3. [DOI] [PubMed] [Google Scholar]
- Kollef MH. Inadequate antimicrobial treatment: an important determinant of outcome for hospitalized patients. Clin Infect Dis. 2000;31(Suppl 4):S131–S138. doi: 10.1086/314079. [DOI] [PubMed] [Google Scholar]
- Korpi A, Järnberg J, Pasanen AL. Microbial volatile organic compounds. Crit Rev Toxicol. 2009;39:139–193. doi: 10.1080/10408440802291497. [DOI] [PubMed] [Google Scholar]
- Kumar A, Ellis P, Arabi Y, Roberts D, Light B, Parrillo JE, Dodek P, Wood G, Kumar A, Simon D, Peters C, Ahsan M, Chateau D, Cooperative Antimicrobial Therapy of Septic Shock Database Research Group Initiation of inappropriate antimicrobial therapy results in a fivefold reduction of survival in human septic shock. Chest. 2009;136:1237–1248. doi: 10.1378/chest.09-0087. [DOI] [PubMed] [Google Scholar]
- Lehmann LE, Herpichboehm B, Kost GJ, Kollef MH, Stuber F (2010) Cost and mortality prediction using polymerase chain reaction pathogen detection in sepsis: evidence from three observational trials. Crit Care 14:R186 [DOI] [PMC free article] [PubMed]
- O’Hara M, Mayhew CA. A preliminary comparison of volatile organic compounds in the headspace of cultures of Staphylococcus aureus grown in nutrient, dextrose and brain heart bovine broths measured using a proton transfer reaction mass spectrometer. J Breath Res. 2009;3:8. doi: 10.1088/1752-7155/3/2/027001. [DOI] [PubMed] [Google Scholar]
- Palka-Santini M, Cleven BE, Eichinger L, Krönke M, Krut O. Large scale multiplex PCR improves pathogen detection by DNA microarrays. BMC Microbiol. 2009;9:1. doi: 10.1186/1471-2180-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlou A, Turner AP, Magan N. Recognition of anaerobic bacterial isolates in vitro using electronic nose technology. Lett Appl Microbiol. 2002;35:366–369. doi: 10.1046/j.1472-765X.2002.01197.x. [DOI] [PubMed] [Google Scholar]
- Perl T, Bödeker B, Jünger M, Nolte N, Vautz W. Alignment of retention time obtained from multicapillary column gas chromatography used for VOC analysis with ion mobility spectrometry. Anal Bioanal Chem. 2010;397:2385–2394. doi: 10.1007/s00216-010-3798-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perl T, Jünger M, Vautz W, Nolte J, Kuhns M, Borg-von Zepelin M, Quintel M. Detection of characteristic metabolites of Aspergillus fumigatus and Candida species using ion mobility spectrometry—metabolic profiling by volatile organic compounds. Mycoses. 2011;54:e828–e837. doi: 10.1111/j.1439-0507.2011.02037.x. [DOI] [PubMed] [Google Scholar]
- Phillips M, Altorki N, Austin JH, Cameron RB, Cataneo RN, Kloss R, Maxfield RA, Munawar MI, Pass HI, Rashid A, Rom WN, Schmitt P, Wai J. Detection of lung cancer using weighted digital analysis of breath biomarkers. Clin Chim Acta. 2008;393:76–84. doi: 10.1016/j.cca.2008.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rello J, Gallego M, Mariscal D, Sonora R, Valles J. The value of routine microbial investigation in ventilator-associated pneumonia. Am J Respir Crit Care Med. 1997;156:196–200. doi: 10.1164/ajrccm.156.1.9607030. [DOI] [PubMed] [Google Scholar]
- Roehl RE. Environmental and process applications for ion mobility spectrometry. Appl Spectrosc Rev. 1991;26:1–57. doi: 10.1080/05704929108053459. [DOI] [Google Scholar]
- Ruzsanyi V, Sielemann S, Baumbach JI. Determination of microbial volatile organic compounds (MVOC) using IMS with different ionization sources. Int J Ion Mobil Spectrom. 2002;5:138–142. [Google Scholar]
- Schöller CE, Gürtler H, Pedersen R, Molin S, Wilkins K. Volatile metabolites from Actinomycetes. J Agric Food Chem. 2002;50:2615–2621. doi: 10.1021/jf0116754. [DOI] [PubMed] [Google Scholar]
- Schulz S, Dickschat JS. Bacterial volatiles: the smell of small organisms. Nat Prod Rep. 2007;24:814–842. doi: 10.1039/b507392h. [DOI] [PubMed] [Google Scholar]
- Seng P, Drancourt M, Gouriet F, La Scola B, Fournier PE, Rolain JM, Raoult D. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009;49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- Shnayderman M, Mansfield B, Yip P, Clark HA, Krebs MD, Cohen SJ, Zeskind JE, Ryan ET, Dorkin HL, Callahan MV, Stair TO, Gelfand JA, Gill CJ, Hitt B, Davis CE. Species-specific bacteria identification using differential mobility spectrometry and bioinformatics pattern recognition. Anal Chem. 2005;77:5930–5937. doi: 10.1021/ac050348i. [DOI] [PubMed] [Google Scholar]
- Smith GB, Eiceman GA, Walsh MK, Critz SA, Andazola E, Ortega E, Cadena F. Detection of Salmonella typhimurium by hand-held ion mobility spectrometer: a quantitative assessment of response characteristics. Field Anal Chem Technol. 1997;1:213–226. doi: 10.1002/(SICI)1520-6521(1997)1:4<213::AID-FACT4>3.0.CO;2-W. [DOI] [Google Scholar]
- Snyder AP, Shoff DB, Eiceman GA, Blyth DA, Parsons JA. Detection of bacteria by ion mobility spectrometry. Anal Chem. 1991;63:526–529. doi: 10.1021/ac00005a028. [DOI] [PubMed] [Google Scholar]
- Stotzky G, Schenck S. Volatile organic compounds and microorganisms. CRC Crit Rev Microbiol. 1976;4:333–382. doi: 10.3109/10408417609102303. [DOI] [PubMed] [Google Scholar]
- Vautz W, Baumbach JI. Exemplar application of multi-capillary column ion mobility spectrometry for biological and medical purpose. Int J Ion Mobil Spectrom. 2008;11:35–41. doi: 10.1007/s12127-008-0007-4. [DOI] [Google Scholar]
- Vautz W, Bödeker B, Baumbach JI, Bader S, Westhoff M, Perl T. An implementable approach to obtain reproducible reduced ion mobility. Int J Ion Mobil Spectrom. 2009;12:47–57. doi: 10.1007/s12127-009-0018-9. [DOI] [Google Scholar]
- Vautz W, Nolte J, Bufe A, Baumbach JI, Peters M. Analyses of mouse breath with ion mobility spectrometry: a feasibility study. J Appl Physiol. 2010;108:697–704. doi: 10.1152/japplphysiol.00658.2009. [DOI] [PubMed] [Google Scholar]
- Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J Bacteriol. 2003;185:2061–2065. doi: 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhoff M, Litterst P, Freitag L, Baumbach JI. Ion mobility spectrometry in the diagnosis of sarcoidosis: results of a feasibility study. J Physiol Pharmacol. 2007;58(Suppl 5):739–751. [PubMed] [Google Scholar]
- Yoo SM, Choi JH, Lee SY, Yoo NC. Applications of DNA microarray in disease diagnostics. J Microbiol Biotechnol. 2009;19:635–646. [PubMed] [Google Scholar]
- Zoller HF, Clark WM. The production of volatile fatty acids by bacteria of the dysentery group. J Gen Physiol. 1921;3:325–330. doi: 10.1085/jgp.3.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]