Abstract
Controlled protein degradation terminates a range of biochemical activities in living cells. New results show that degradation of a component of the licensing system is needed to prevent the re-replication of chromosomes during the cell-division cycle.
How would you ensure that a biochemical reaction is irreversible? This is a key problem in living organisms, because many of their control systems must progress unidirectionally through a series of different states. During the cell-division cycle, for instance, chromosomal DNA must first be duplicated (in the so-called S phase), and then the two copies must be segregated to daughter cells (during mitosis). Organisms usually solve the problem of irreversibility by inactivating the proteins that catalyse each biochemical reaction after they have carried out their task. There are several ways of achieving this, but the most direct is simply to destroy the proteins. For example, it is known that the exit from mitosis is made irreversible by the degradation of mitotic proteins. Elsewhere in this issue (page xxx), Zhong and colleagues1 show that the ability to precisely replicate chromosomal DNA also depends on protein degradation. Zhong et al. identify the protein CUL-4 as being essential for the timely degradation of another protein, Cdt1 — a component of the cellular system that ‘licenses’ DNA replication. In the absence of CUL-4, cells continuously replicate their DNA without progressing into mitosis.
Proteolysis — the degradation of proteins into their constituent amino acids — is typically carried out by a cellular machine known as the 26S proteasome. Proteins are targeted to this machine by being covalently tagged with a short peptide called ubiquitin. So, the regulation of proteolysis depends on the activity of the ubiquitin ligase enzymes that attach ubiquitin to doomed proteins. One such enzyme is the anaphase-promoting complex, which is activated at the end of mitosis to enforce mitotic exit by ubiquitinating key proteins such as the cyclins (Fig. 1). CUL-4, a member of the cullin family of proteins, forms an essential part of another ubiquitin ligase2. Zhong et al.1 have now found that, during larval development of the nematode worm Caenorhabditis elegans, the CUL-4 gene is expressed to a high level in proliferating cells. This hints that CUL-4 is involved in regulating cell-cycle progression — so what, if anything, is its role?
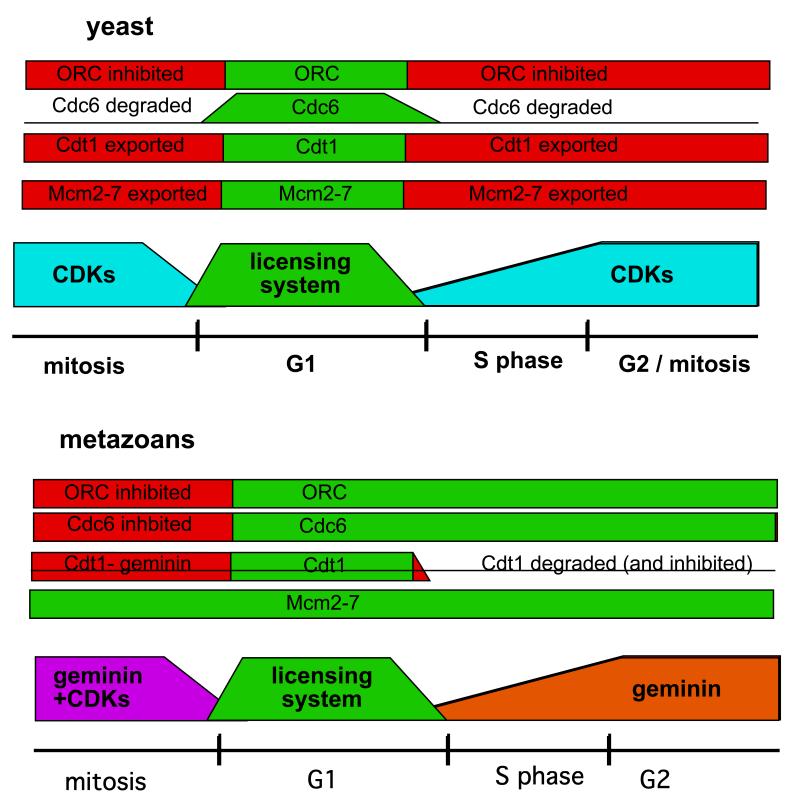
Figure 1.
Protein degradation and the cell cycle. Along the bottom are shown the four phases of the cell-division cycle: G1, S (when chromosomal DNA is precisely duplicated), G2 and mitosis (when the duplicated DNA is partitioned into two daughter cells). Cyclin-dependent kinases (CDKs) are activated during late G1 and are inactivated at the end of mitosis. The replication licensing system is only active in late mitosis and G1. The lines above show the relative abundances of cyclin proteins (required for CDK activity) and Cdt1 (essential for replication licensing). Cyclins are destroyed at the end of mitosis (along with the Cdt1 inhibitor geminin) by means of the anaphase-promoting complex (APC/C), a ubiquitin ligase; activation of the APC/C is dependent on CDKs. Zhong et al.1 have now shown that Cdt1 is destroyed at the end of G1, in a manner that is dependent on the activity of the CUL-4 ubiquitin ligase. We speculate that Cdt1 becomes a CUL-4 substrate following phosphorylation by CDKs.
To find out, Zhong et al. used RNA interference (RNAi) technology to reduce CUL-4 expression in C. elegans larvae. This caused several cellular abnormalities, the most dramatic of which was massive re-replication of DNA, generating cells with more than 30 times the normal DNA content. Cells normally use a strict control system to ensure that DNA replicates only once in each cell cycle; this involves replication being divided into two non-overlapping phases3. Prior to S phase, replication origins (the places on DNA where replication starts) are ‘licensed’ by being loaded with the Mcm2-7 proteins. Licensing also requires several other proteins, including Cdt1. During S phase, the replication machinery is assembled only at these licensed replication origins. As the DNA is replicated, the Mcm2-7 proteins are displaced from it, thereby ensuring that replicated DNA is unlicensed and cannot be replicated again.
In order for this to work properly, it is essential that the licensing system is inactivated once S phase has started. Consistent with results in other organisms4–6, Zhong et al. show that C. elegans cells contain Cdt1 only in late mitosis and G1 phase (the phase between mitosis and S phase; Fig. 1). Moreover, the authors find that reducing CUL-4 expression stabilizes Cdt1 in S phase — showing that CUL-4-mediated proteolysis is essential for the normal downregulation of Cdt1.
Is this stabilization of Cdt1 enough to explain why cells lacking CUL-4 over-replicate their DNA? Zhong et al. first provide several lines of evidence in support of the idea that the over-replication is due to ‘origin re-firing’ — the continuing re-licensing and re-activation of replication origins during S phase3. They go on to show that in strains heterozygous for Cdt1 (ie strains that have just one copy of the CDT1 gene rather than the normal two, and which therefore would be expected to have lowered Cdt1 levels), the re-replication induced by the reduction in CUL-4 levels is decreased five-fold. In contrast, strains with just one copy of the gene encoding another licensing protein, CDC6, show no major reduction in re-replication when CUL-4 levels are decreased.
These results suggest that CUL-4-mediated degradation of Cdt1 is essential for the inactivation of the licensing system in C. elegans. Further evidence that levels of Cdt1 are critical for preventing DNA re-replication was provided by a recent study of human cancer cells, where artificial overexpression of this protein was enough to cause significant re-replication7. It is currently unknown, however, exactly how these levels are achieved — that is, how the activity of CUL-4 is regulated. For other cullin-based ubiquitin ligases, the target protein must be phosphorylated before it can be ubiquitinated. Given the known role of cyclin-dependent kinases (CDKs) in preventing re-replication of DNA3, a plausible hypothesis is that as CDKs are activated at the onset of S phase, they phosphorylate Cdt1, thus directing it to CUL-4 for ubiquitination.
Previous studies in other organisms have identified several different ways by which the licensing system can be downregulated in S phase. These include inhibitory phosphorylation of licensing components by CDKs, and the inhibition of Cdt1 by a protein called geminin, as well as proteolysis3. In budding yeast, for example, three different regulatory mechanisms must be inactivated to promote modest re-replication of DNA8. One striking feature of Zhong and colleagues’ work is the apparent dependence of C. elegans cells exclusively on CUL-4-mediated degradation of Cdt1 — in the absence of which massive re-replication occurs.
Most cancer cells display genomic instability, and many also have dysregulated ubiquitin ligases, leading to the inappropriate ‘survival’ of their protein targets9. Significantly, CDT1 has been reported to have the potential to cause cells to become cancerous10, and in light of the new work1,7 we speculate that this comes about by aberrantly expressed CDT1 driving over-replication, thereby causing genomic instability. The extensive re-replication that occurs as a consequence of losing CUL-4 function in C. elegans suggests that, in some cases, the barriers to genomic instability — which in multicellular organisms can potentially lead to death from cancer — are less robust than might be imagined.
References
- 1.Zhong W, Feng H, Santiago FE, Kipreos ET. Nature. 2003;423:xxx–xxx. doi: 10.1038/nature01747. [DOI] [PubMed] [Google Scholar]
- 2.Zheng N, et al. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 3.Blow JJ, Hodgson B. Trends Cell Biol. 2002;12:72–78. doi: 10.1016/s0962-8924(01)02203-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishitani H, Lygerou Z, Nishimoto T, Nurse P. Nature. 2000;404:625–628. doi: 10.1038/35007110. [DOI] [PubMed] [Google Scholar]
- 5.Wohlschlegel JA, et al. Science. 2000;290:2309–2312. doi: 10.1126/science.290.5500.2309. [DOI] [PubMed] [Google Scholar]
- 6.Nishitani H, Taraviras S, Lygerou Z, Nishimoto T. J. Biol. Chem. 2001;276:44905–44911. doi: 10.1074/jbc.M105406200. [DOI] [PubMed] [Google Scholar]
- 7.Vaziri C, et al. Mol. Cell. 2003;11:997–1008. doi: 10.1016/s1097-2765(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 8.Nguyen VQ, Co C, Li JJ. Nature. 2001;411:1068–1073. doi: 10.1038/35082600. [DOI] [PubMed] [Google Scholar]
- 9.Spruck CH, Strohmaier HM. Cell Cycle. 2002;1:250–254. [PubMed] [Google Scholar]
- 10.Arentson E, et al. Oncogene. 2002;21:1150–1158. doi: 10.1038/sj.onc.1205175. [DOI] [PubMed] [Google Scholar]