It is crucial for the continuation of life that, as a cell replicates and divides, the genetic information encoded in its DNA (its genome) is duplicated and passed on accurately to the two new daughter cells. All of the genome must be replicated, with nothing left unreplicated and nothing replicated more than once. This apparently simple problem is made complicated by the way in which eukaryotic cells (those with a defined nucleus) go about copying their large genomes. ‘Replication forks’ progress along double- stranded DNA, copying both strands at a rate of 10 to 100 base pairs per second. At this rate, a single replication fork would take years to duplicate a typical genome. So, to speed things up, eukaryotes initiate replication forks at thousands of different places — called replication ‘origins’ — scattered throughout the genome.
This raises a new problem, however: replication forks must never be initiated on DNA that has already replicated during that cell-division cycle. Although general principles have been proposed to explain how cells prevent such re-replication, the precise mechanism is unknown. Writing in this issue, Nishitani and colleagues1 and Maiorano and co-workers2 now suggest the importance of a protein known as Cdt1 in this process.
Previous experiments have shown that precise DNA duplication is achieved by separating the initiation of replication into two phases3. In the first phase, which occurs early in the cell cycle, replication origins are ‘licensed’ by becoming loaded with the RLF- M (for replication-licensing factor type ‘M’) complex of MCM proteins4 (also known as MCM/P1 proteins). In the second phase, an activity that promotes the DNA-replication (S) phase of the cell cycle — S-phase-promoting factor, or SPF — swings into action. SPF initiates a single pair of replication forks at each licensed origin. As replication forks proceed from the origin, MCM proteins are removed, thereby resetting the origin to the ‘unlicensed’ state. To ensure that initiation occurs no more than once at each origin, licensing activity must cease completely before SPF becomes active. Even a small amount of licensing activity present during S phase could lead to the re-replication of part of the genome. According to the new papers1,2, the Cdt1 protein acts in the tightly controlled pathway that promotes licensing only early in the cell cycle.
The cdt1 gene was initially identified in the fission yeast Schizosaccharomyces pombe as a gene that is essential for DNA replication. It is periodically expressed under the control of the transcription factor Cdc10 (ref. 5). These properties are shared by the cdc18 gene in S. pombe and its homologue, CDC6, in other eukaryotes. Cdc6/Cdc18 is essential for licensing replication origins6 (Fig. 1). Overexpression of Cdc6/Cdc18 in S. pombe causes DNA to replicate several times within a single cell cycle7,8, probably as a consequence of keeping the licensing system active late in the cycle, after DNA replication has started.
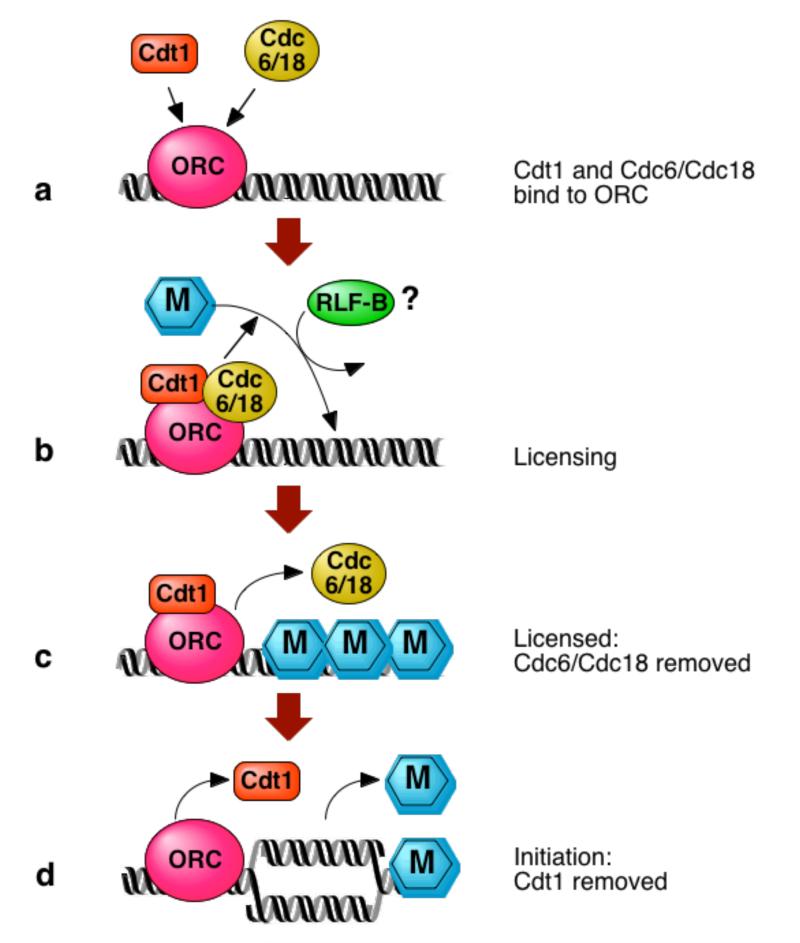
Figure 1.
Steps in the licensing of a DNA replication origin. a, Cdt1 (as shown by Nishitani et al.1 and Maiorano et al.2) and Cdc6/Cdc18 bind to the origin-recognition complex (ORC) at an origin of DNA replication. b, Several MCM protein complexes, represented by ‘M’, are then assembled onto the origin to ‘license’ it for replication. The activity of replication-licensing factor B (RLF-B) might be involved at this point. c, As a consequence of replication licensing, Cdc6/Cdc18 is displaced from the origin. d, As DNA replication starts, all of the Cdt1 is removed from the origins. MCM proteins are removed as the DNA replicates.
Nishitani et al.1 now show that Cdt1 has a similar function to Cdc6/Cdc18, and is required for origin licensing. They show that, in cells in which Cdc6/Cdc18 is only slightly overexpressed (at levels that alone do not induce significant re-replication of DNA), coexpression of Cdt1 causes high levels of re-replication. The inability of over-expressed Cdt1 to induce over-replication by itself indicates that it might need to be less tightly repressed later in the cell cycle than is Cdc6/Cdc18. In a parallel series of experiments, Maiorano et al.2 cloned the Xenopus laevis (frog) CDT1 gene, and investigated its function in cell-free extracts that support cell-cycle progression in vitro. The results of Maiorano et al., showing that the Xenopus Cdt1 protein is required to license DNA by loading the MCM proteins onto it, are consistent with those obtained in S. pombe.
These papers also show that, in both fission yeast and frog, Cdt1 itself associates with DNA. This association is dependent on the presence of the origin-recognition complex (ORC)2, as is the association of Cdc6/Cdc18 with DNA6. In fission yeast extracts, Cdt1 and Cdc6/Cdc18 can be precipitated together by an antibody against just one of the proteins, suggesting that they might form a complex. However, they can also associate with DNA independently of one another. These results indicate that, early in the G1 phase of the cell cycle (the stage preceding S phase), Cdt1 and Cdc6/Cdc18 may bind together to ORCs on DNA (Fig. 1a), where they are required for licensing to occur (Fig. 1b).
A further essential licensing activity, termed RLF-B, is probably also required at this stage9. The protein underlying RLF-B activity is unknown, and could theoretically correspond to Cdt1. However, the behaviour of RLF-B differs from that of Cdt1: unlike Cdt1, RLF-B activity does not associate with unlicensed DNA9. Cdt1 also seems to behave differently from Cdc6/Cdc18, as, in the frog system, Cdt1 remains associated with DNA right up until the start of S phase (Fig. 1d), whereas Cdc6/Cdc18 is displaced from DNA immediately after origins are licensed10 (Fig. 1c). Further studies will be needed before we fully understand the roles of these proteins in origin licensing.
The existence of this complex series of events in origin licensing probably reflects the essential function of this system in maintaining genomic integrity. And it is probably even more important to prevent the re-licensing of replicated origins (and hence the re-initiation of DNA replication) in multi-cellular organisms than it is in single-celled ones. The occasional re-replication event would have little effect on the proliferative capacity of a strain of single-celled organisms such as yeast. But, by generating a cancer-causing mutation, it might be fatal to a multicellular organism. Re-assembling the Cdt1 protein — which is regulated in different ways (transcriptionally or post-translationally) in yeast1 and frogs2 — at replication origins represents yet another step that is necessary for re-licensing to occur. We can expect the regulation of the replication licensing system to become more complex still.
References
- 1.Nishitani H, Lygerou Z, Nishimoto T, Nurse P. Nature. 2000;404:625–628. doi: 10.1038/35007110. [DOI] [PubMed] [Google Scholar]
- 2.Maiorano D, Moreau J, Méchali M. Nature. 2000;404:622–625. doi: 10.1038/35007104. [DOI] [PubMed] [Google Scholar]
- 3.Blow JJ, Laskey RA. Nature. 1988;332:546–548. doi: 10.1038/332546a0. [DOI] [PubMed] [Google Scholar]
- 4.Tye BK. Annu. Rev. Biochem. 1999;68:649–686. doi: 10.1146/annurev.biochem.68.1.649. [DOI] [PubMed] [Google Scholar]
- 5.Hofmann JF, Beach D. EMBO J. 1994;13:425–434. doi: 10.1002/j.1460-2075.1994.tb06277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coleman TR, Carpenter PB, Dunphy WG. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- 7.Kelly TJ, et al. Cell. 1993;74:371–382. doi: 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]
- 8.Nishitani H, Nurse P. Cell. 1995;83:397–405. doi: 10.1016/0092-8674(95)90117-5. [DOI] [PubMed] [Google Scholar]
- 9.Tada S, Chong JPJ, Mahbubani HM, Blow JJ. Curr. Biol. 1999;9:211–214. doi: 10.1016/s0960-9822(99)80092-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rowles A, Tada S, Blow JJ. J. Cell Sci. 1999;112:2011–2018. doi: 10.1242/jcs.112.12.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]