Summary
Identifying the genetic mechanisms underlying phenotypic change is essential to understanding how gene regulatory networks and ultimately the genotype-to-phenotype map evolve. It is recognized that microRNAs (miRNAs) have the potential to facilitate evolutionary change [1–3]; however, there are no known examples of natural morphological variation caused by evolutionary changes in miRNA expression. Therefore, the contribution of miRNAs to evolutionary change remains unknown [1, 4]. Drosophila melanogaster subgroup species display a portion of trichome-free cuticle on the femur of the second leg called the “naked valley.” It was previously shown that Ultrabithorax (Ubx) is involved in naked valley variation between D. melanogaster and D. simulans [5, 6]. However, naked valley size also varies among populations of D. melanogaster, ranging from 1,000 up to 30,000 μm2. We investigated the genetic basis of intraspecific differences in the naked valley in D. melanogaster and found that neither Ubx nor shavenbaby (svb) [7, 8] contributes to this morphological difference. Instead, we show that changes in mir-92a expression underlie the evolution of naked valley size in D. melanogaster through repression of shavenoid (sha) [9]. Therefore, our results reveal a novel mechanism for morphological evolution and suggest that modulation of the expression of miRNAs potentially plays a prominent role in generating organismal diversity.
Highlights
► mir-92a represses shavenoid in the posterior femur to modulate naked valley size ► cis-regulatory changes in mir-92a cause the evolution of morphology in Drosophila ► Trichome pattern changes are caused by different regulatory factors ► Changes in miRNA expression might play a prominent role in phenotypic change
Results and Discussion
Intraspecific Variation in the Naked Valley
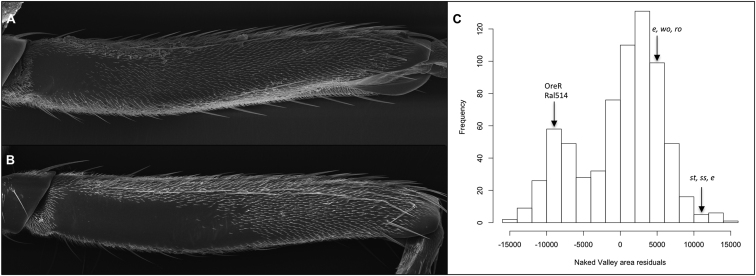
The naked valley exhibits considerable intraspecific variation in D. melanogaster, ranging from a trichome-free patch as small as 1,000 μm2 to a naked region of up to approximately 30,000 μm2 (Figure 1; see also Figure S1 available online). Moreover, small and large naked valley phenotypes segregate within natural D. melanogaster populations (Figures 1 and S1). In contrast, among D. simulans (Figure S1), D. mauritiana, and D. sechellia populations, as well as D. yakuba, we have only observed naked valley areas at the higher end of the size range (13,000 to 30,000 μm2). Therefore, small naked valleys (SNVs) appear to be a derived morphological feature within D. melanogaster, whereas larger naked valleys (LNVs) are likely to be ancestral with respect to the D. melanogaster species subgroup.
Figure 1.
Distribution of Naked Valley Sizes across D. melanogaster Populations
(A and B) Posterior femurs of the second legs of D. melanogaster strains Oregon-R (A) and e, wo, ro (B). Proximal is to the left and distal to the right in both panels.
(C) Bimodal frequency distribution of naked valley phenotypes (residuals of naked valley area regressed on femur length) of 679 male flies from isofemale lines of five populations sampled from Kenya, Turkey, Spain, and the USA. A minimum of three individuals were sampled for each isofemale line. Average values for strains Oregon-R, RAL514, e, wo, ro, and st, ss, e are indicated by arrows.
See also Figure S1.
Mapping the Genetic Basis of Naked Valley Variation in D. melanogaster
It was previously shown that the Hox gene Ultrabithorax (Ubx) contributes to the difference in naked valley size between a D. melanogaster strain with a small naked valley and D. simulans [6]. Therefore, to determine whether Ubx is also responsible for intraspecific naked valley variation in D. melanogaster, we performed quantitative trait locus (QTL) mapping of naked valley size on chromosome 3 among backcross progeny from crosses between strains st, ss, e (LNV) and Oregon-R (SNV). We found a single QTL at 88.2 cM on chromosome 3 that explains up to 91% of the difference in naked valley size between the two parental strains (Figure S2A; Table S1), and, using a male F1 backcrossing strategy, we determined that the remaining effect (approximately 10%) is caused by chromosome 2 (p < 0.017, Bonferroni corrected pairwise comparison of means). Chromosomes X and 4 have no significant effect. Our mapping thus excludes both Ubx, which is at 58.8 cM on chromosome 3 (Figure 2), and the X-linked gene shavenbaby (svb), which is known to underlie variation in larval trichome patterns [10–12].
Figure 2.
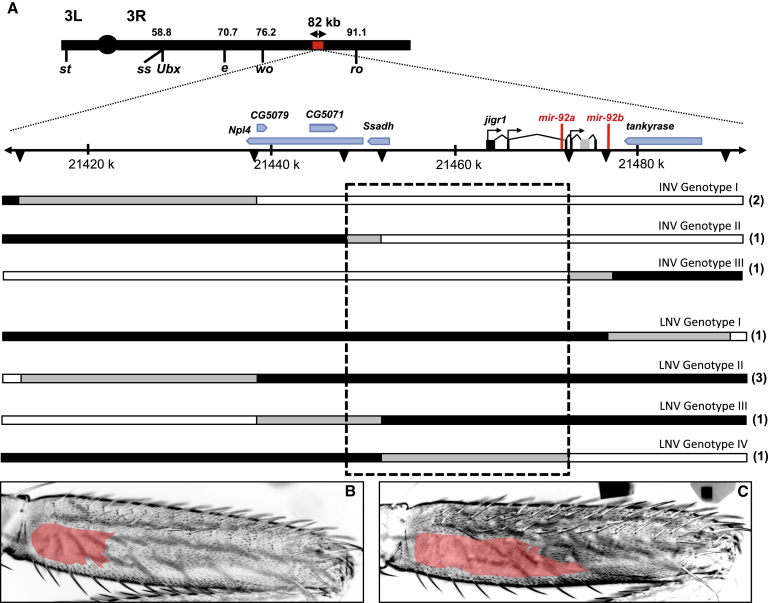
High-Resolution Mapping of the Causative Locus
(A) The topmost black bar represents chromosome 3, with the two arms (3L and 3R) indicated either side of the centromere (circle). The position of Ubx and selected QTL markers are shown below the bar, with their positions in cM indicated above. The section of chromosome 3R highlighted by the red bar represents the 82.2 kb evolved region identified by the first mapping experiment (Figure S2C), which is shown expanded below, and between the dotted diagonal lines, with the scale given in kb. The bars below the scale indicate the genotypes of selected recombinants with breakpoints in the 82.2 kb region (note that all flies also carried a nonrecombinant chromosome from strain e, wo, ro that is not shown). Positions of molecular markers (Table S2) are indicated by black triangles. The number of individual flies representing each of the selected recombinant genotypes illustrated is given in parentheses to the right. Chromosomal regions from strains e, wo, ro (large naked valley parental line) and Oregon-R (small naked valley parental line) are indicated in black and white, respectively. Chromosomal regions in gray indicate DNA where the parental strain identity was not determined. The dashed black box indicates the 25 kb region that underlies naked valley variation. INV and LNV indicate intermediate and large naked valley phenotypes, respectively.
(B and C) Representative examples of T2 posterior femurs from recombinant flies with either an INV (B) or LNV (C).
To verify that variation in Ubx is not responsible for differences in the naked valley in D. melanogaster, we carried out two further experiments. First, we repeated our chromosome 3 mapping strategy with two different D. melanogaster strains, RAL514 and ebony (e), white ocelli (wo), rough (ro), which have SNVs and LNVs, respectively. QTL mapping using these three recessive markers confirmed the position of a single, large-effect QTL on chromosome 3 at 79.7 to 89.7 cM (2 LOD interval), between wo and ro (Figure S2A; Table S1). Second, we generated flies with recombinant third chromosomes: homozygous for the Ubx allele from a LNV background (UbxL) and homozygous for the QTL region from a SNV background (QTLS), and vice versa (UbxS and QTLL) (Figure S2B). The size of the naked valley of these flies was determined by the background from which the QTL region originated (Figure S2), and no significant effect could be attributed to Ubx: flies homozygous for UbxL and QTLS had a small naked valley, whereas flies homozygous for UbxS and QTLL had a large naked valley. Furthermore, the effect on naked valley area of homozygosity for QTLL or QTLS was consistent with the QTL mapping results (Figure S2). Our mapping results therefore showed that neither Ubx nor svb contributes to naked valley variation in D. melanogaster.
To fine map the causative locus or loci in the QTL region, we took advantage of the large effect of the QTL and employed the visible flanking markers wo and ro to screen for recombinants. We measured the naked valley area of these flies and scored them for microsatellite, nucleotide, and restriction fragment length polymorphism markers (Figures 2 and S2C). This strategy allowed us to map the causative locus to a region of 25 kb that contains only four genes: part of Npl4 ortholog, Succinic semialdehyde dehydrogenase (Ssadh), jing interacting gene regulatory 1 (jigr1), and mir-92a (Figures 2 and S2C). Npl4 ortholog is thought to be the homolog of the yeast nuclear pore protein [13]. Ssadh encodes a ubiquitously expressed metabolic enzyme [14], and jigr1 has been implicated in axonal guidance [15]. None of these protein-coding genes is known to be involved in trichome development. However, genome-wide analysis has shown that miR-92a and its seed relatives have the unique ability to induce trichome loss when ectopically expressed during wing development [9, 16] (Figure S3). Therefore, mir-92a represented a strong candidate for the evolution of the naked valley.
Functional Analysis of mir-92a in Naked Valley Development
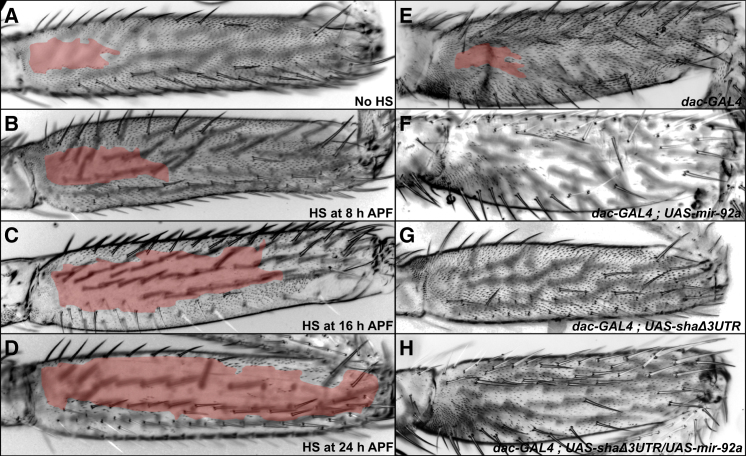
To investigate the role of miR-92a in naked valley development, we overexpressed UAS-mir-92a [16] using a heat-shock-GAL4 driver in pupal legs between 8 and 24 hr after puparium formation (APF), when the naked valley pattern is determined [6]. Whereas control flies displayed comparatively small naked valleys, overexpressing mir-92a by applying heat shock at 8, 16, or 24 hr APF resulted in flies with progressive loss of trichomes and therefore larger naked valleys (Figure 3). Indeed, the posterior T2 femurs of flies heat shocked at 24 hr APF displayed only a few trichomes (Figure 3D). Driving UAS-mir-92a with dac-GAL4 (which is expressed in the developing femur [17]) also resulted in loss of trichomes and an enlarged naked valley with respect to controls (Figures 3E and 3F). These experiments show that mir-92a can repress trichomes on the femur and that variation in mir-92a expression modulates the size of the naked valley.
Figure 3.
mir-92a Represses Trichome Development on the T2 Femur
Uniform expression of mir-92a represses trichome formation progressively depending on developmental timing of overexpression induced by heat shock. Tan shading indicates the extent of trichome-free cuticle. See also Figure S3.
(A–D) Posterior of T2 femurs of F1 flies from the cross between HS-GAL4 and UAS-mir-92a.
(A) Posterior T2 femur of a control F1 fly that was not heat shocked.
(B–D) F1 flies heat shocked at 8 hr (B), 16 hr (C), and 24 hr (D) after puparium formation (APF).
(E) Posterior T2 femur of the dac-GAL4 control line.
(F) UAS-mir-92a expression driven by dac-GAL4 represses trichome formation throughout the posterior femur.
(G) UAS-shaΔ3UTR driven by dac-GAL4 results in the development of ectopic trichomes and removes the naked valley.
(H) Simultaneous overexpression of UAS-shaΔ3UTR and UAS-mir-92a using dac-GAL4 leads to rescue of trichome formation and removes the naked valley.
Comparison of the sequences of mir-92a between D. melanogaster strains with large and small naked valleys shows that the 22 nt sequence that constitutes the mature miRNA and the flanking 200 bp immediately upstream and downstream are identical in both strains (Figure S4A). This suggests that differences in one or more cis-regulatory regions (for example, enhancer sequences or splice sites) of mir-92a, rather than changes to the primary structure of this miRNA or differential arm usage, are responsible for naked valley evolution. To test this hypothesis, we carried out in situ hybridization against the primary miRNA transcripts to assess expression of mir-92a at 24 hr APF in the legs of D. melanogaster strains with LNVs and SNVs (e, wo, ro and Oregon-R). We found that pri-mir-92a expression in the posterior T2 femurs is expanded in e, wo, ro compared to Oregon-R (Figure 4D and 4E). This finding is consistent with the difference in the size of the naked valleys between these strains and therefore supports the notion that changes in the regulation of mir-92a expression underlie intraspecific variation in the naked valley.
Figure 4.
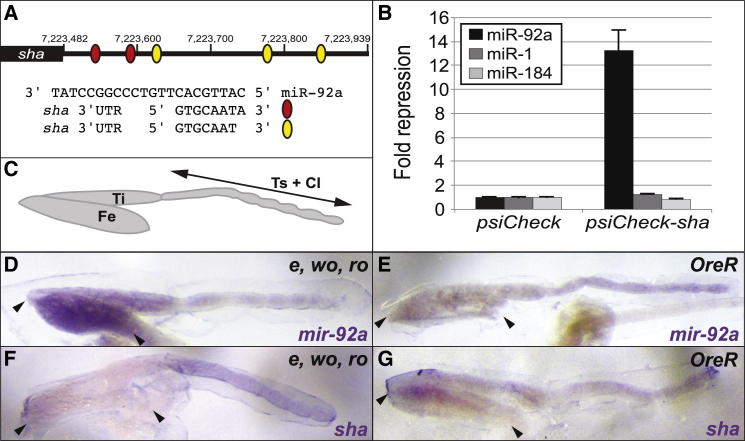
Differential Expression of mir-92a Underlies Naked Valley Variation through Repression of sha
(A) The sha 3′ UTR contains five highly conserved, canonical seed-match sites for miR-92a (see also Figure S4). The black rectangle represents the sha coding region and the black line the 3′ UTR. Numbering is with respect to the base-pair position on chromosome 2R. Red and yellow ovals represent predicted seed-match sites for miR-92a consensus sequences shown aligned with the mature miR-92a sequence.
(B) Luciferase sensor assays in S2 cells show that the sha 3′ UTR confers >13-fold repression in response to ectopic miR-92a but is unaffected by control miR-1 and miR-184. Error bars show the SD from four independent transfections.
(C) Representation of the T2 pupal leg showing the femur (Fe), tibia (Ti), tarsa (Ts), and claws (Cl).
(D–G) Expression of pri-mir-92a (D and E) and sha (F and G), in the pupal T2 legs of strains e, wo, ro and Oregon-R at 24 hr APF. Arrowheads indicate the femur in each picture.
It was previously shown that Ubx is involved in trichome development and the evolution of the naked valley between species, although the precise causative changes in Ubx have not yet been identified [5, 6]. The involvement of Ubx in interspecific differences was demonstrated in part via interspecific complementation tests, where flies carrying a single functional copy of Ubx from D. simulans had a larger naked valley that those with a single functional copy of Ubx from D. melanogaster in an otherwise identical genetic background [6]. However, these experiments also showed that flies with D. melanogaster chromosomes had consistently smaller naked valleys than those with D. simulans chromosomes irrespective of whether the D. melanogaster chromosome carried a nonfunctional Ubx, which Stern concluded was caused by the involvement of at least one other gene [6]. Because Stern’s findings can be interpreted as showing that flies with mir-92a from D. melanogaster have smaller naked valleys than those with this factor from D. simulans, it is possible that the evolution of mir-92a may also contribute to the difference between species, consistent with Stern’s evidence for the involvement of another gene [6].
miR-92a Regulates Naked Valley Size through Repression of shavenoid
Searches for relevant miR-92a targets showed that the shavenoid (sha) 3′ UTR contains five highly conserved, canonical seed-match sites (Figures 4A and S4B). sha is required for trichome development [18], and its predicted degree of targeting by an individual miRNA exceeds nearly all other genes in Drosophila [19]. Therefore, sha is well positioned to mediate changes in trichome patterning through altered miR-92a activity. We used luciferase sensor assays to show that the sha 3′ UTR is highly and specifically repressed by miR-92a, relative to several other miRNAs that had no effect (Figure 4B). Given that miRNAs often only fine tune their targets by 20% to 30%, even in ectopic tests, the 13-fold regulation we observed indicates a potent regulatory interaction between miR-92a and sha.
To test whether mir-92a regulates the size of the naked valley via sha, we coexpressed UAS-mir-92a with a sha construct lacking its 3′ UTR [18]. This suppressed the naked valley, and trichomes were found across the posterior T2 femur (Figures 3G and 3H). These results are consistent with the interpretation that miR-92a represses trichomes via downregulation of sha. In support of this, we found that sha was expressed in a smaller domain in the developing posterior T2 femur of the pupal legs of flies with a LNV compared to those with a SNV (Figures 4F and 4G). Therefore, our in situ results suggest that mRNA degradation is the possible mechanism of repression, but translational blocking could also be involved [9]. Furthermore, it remains possible that miR-92a also regulates other genes involved in trichome formation on the femur.
Although the exact mechanism of sha repression via mir-92a remains to be resolved, we and others [9, 11] provide compelling evidence that the sha 3′ UTR can be regulated by miR-92a, leading to phenotypic effects consistent with the function of this target gene. Interestingly, sha is a known target of the transcription factor Svb, which is thought to act as an input/output integrator to determine where trichomes will develop [10–12] and is a known hotspot for the evolution of larval trichomes [7, 8, 20, 21]. Although presumably Ubx acts upstream of svb during trichome development, our results show that the modulation of the expression of a downstream gene involved in cytoskeletal organization by a miRNA can also facilitate the evolution of trichome patterns.
Conclusions
We report here the first example of natural variation in the expression of a miRNA causing morphological change. Because the main role of miRNAs is to subtly modulate gene expression levels, variation in the expression and function of such factors appears to be an obvious mechanism to facilitate phenotypic evolution [2, 3]. Although the appearance of new miRNAs and evolutionary changes in the seed sequences of these factors or the 3′ UTRs of their targets have been described [22, 23], the phenotypic consequences of these genetic changes are not known. Therefore, our work represents the first experimental evidence that changes in the cis-regulatory sequences of miRNAs contribute to phenotypic evolution.
Experimental Procedures
Morphological Measurements
Dissected T2 legs were mounted in Hoyer’s medium and imaged under dark-field or differential interference contrast microscopy using a Leica DM5500 compound microscope and a DFC300 camera. The area of naked valley (μm2) was measured as the extent of the naked cuticle (without trichomes) starting at the base of the femur (red polygon in Figures 2B and 2C). Femur length (μm) was measured from the proximal end of the femur to the distalmost bristle along the ventral margin.
Fly Lines and Crosses
The stocks st, ss, e (Drosophila Genetic Resource Center 101760), Oregon-R, e, wo, ro (BL496), and RAL514 [24] were used for mapping experiments. The stocks st, ss, e and Oregon-R were also used to generate reciprocal homozygous recombinant lines for chromosome III (Figure S2B). Transgenic fly stocks used for functional analysis included w; dacGAL4/Cyo (referred to as dac-GAL4 [17]), w; P(w(+mc)==GAL4-HSP70PB) (HS-GAL4; a gift from Clive Wilson, University of Oxford), UAS-DsRed-mir-92a (referred to as UAS-mir-92a) [16], w[∗]; P(w(+mC)=UAS-shaGFP)3 (referred to as UAS-shaΔ3UTR) [18], bx-GAL4, ptc-GAL4, and sd-GAL4. All flies and crosses were maintained under standard fly culture conditions. Heat-shock experiments were conducted as described previously [25]. Naked valley sizes were also surveyed from populations of D. simulans and D. melanogaster [24, 26].
QTL Mapping
Two independent backcross mapping populations were generated for QTL mapping. First, we backcrossed F1 virgin female progeny (from the cross of st, ss, e to Oregon-R) to male st, ss, e flies. For the second mapping population, we backcrossed F1 virgin female progeny (from the cross of e, wo, ro to RAL514) to male e, wo, ro. Resultant backcross progeny were phenotyped for naked valley area and T2 femur length and genotyped on chromosome 3 (see Table S2 for genetic markers used). QTL analysis was performed using standard interval mapping with extended Haley-Knott regression [27] with the R package [28]. Additive allelic effects were estimated by fitting linear models for the significant QTL. All analyses were performed with and without femur length as a covariate.
The contribution of chromosomes X, 2, and 4 to variation in naked valley area was assayed by backcrossing male F1 progeny from a cross between Oregon-R and st, ss, e to st, ss, e females and comparing naked valley area between backcross progeny homozygous or heterozygous for each of these three chromosomes in a homozygous st, ss, e chromosome 3 background.
Fine-Scale Mapping
To fine-scale map the causative locus responsible for naked valley variation in the QTL region on 3R, we generated and screened approximately 1,000 recombinants between markers e and msb and between markers wo and ro in backcross progeny from crosses between D. melanogaster strains st, ss, e and Oregon-R and between strains e, wo, ro and Oregon-R, respectively. We then measured the naked valley area and femur length of recombinants and mapped the recombination breakpoints using 20 microsatellite, nucleotide, and restriction-site polymorphism markers.
Luciferase Assays
The shavenoid 3′ UTR and downstream genomic sequence were cloned into psiCHECK2 (Promega) using a Cold Fusion cloning kit (System Biosciences) and the following primers: Cf_sha3utr_fwd, CCACCTGTTCCTGTAGCGGCCGCATTAGGCTATGCTTAAGTGC; Cf_sha3utr_rev, CCTTCACAAAGATCCCTCGAGTGAACGCAAAAGTAGCGC. Luciferase assays were carried out as described previously [29]. Briefly, cells were seeded in a 96-well plate at ∼1.2 million cells per ml, 100 μl per well. Each well was transfected with 12.5 ng Ub-Gal4 plasmid, 25 ng UAS-mir plasmid, and 25 ng pSicheck-derived plasmid using Effectene transfection reagent (QIAGEN). After 3 days, results were read using a Dual-Glo luciferase assay (Promega) and a luminometer (Turner).
In Situ Hybridizations
In situ hybridization was carried out using a standard protocol with DIG-labeled antisense RNA probes. Pupae were fixed at 24 hr APF for 1 hr in 4% formaldehyde after pupal cases were removed. In situ hybridizations were performed with the same concentration of probe for each strain, and the nitro blue tetrazolium/5-bromo-4-chloro-3′-indolyphosphate reaction was stopped at the same time. pri-mir-92a and shavenoid sequences were cloned into a TOPO PCR4 vector (Invitrogen) using GCAAAATGATGTGAGGCGTA and TCATAAGCAAAATACGAGACAT and AGGAGGATATGGGCATTGTG and TGAACATGGGTGAACTGGAA primer pairs, respectively, following the manufacturer’s protocol. M13 forward and reverse primers were used to linearize the DNA. T3 RNA polymerase was used to generate the DIG-labeled riboprobes.
Acknowledgments
Work in A.P.M.’s group was funded by the Austrian Science Fund (FWF): M1059-B09, and Oxford Brookes University. Work in E.C.L.’s group was supported by the National Institute of General Medical Sciences of the National Institutes of Health (R01-GM083300). We thank Virginie Orgogozo for fruitful discussions about the project and comments on the manuscript.
Published: February 28, 2013
Footnotes
Supplemental Information includes four figures and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.cub.2013.02.018.
Accession Numbers
Mapping data have been deposited at Dryad (http://datadryad.org/) with the DOI http://dx.doi.org/10.5061/dryad.qd88b.
Supplemental Information
References
- 1.Li J., Zhang Z. miRNA regulatory variation in human evolution. Trends Genet. 2013;29:116–124. doi: 10.1016/j.tig.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 2.Niwa R., Slack F.J. The evolution of animal microRNA function. Curr. Opin. Genet. Dev. 2007;17:145–150. doi: 10.1016/j.gde.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 3.Ronshaugen M., Biemar F., Piel J., Levine M., Lai E.C. The Drosophila microRNA iab-4 causes a dominant homeotic transformation of halteres to wings. Genes Dev. 2005;19:2947–2952. doi: 10.1101/gad.1372505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alonso C.R., Wilkins A.S. The molecular elements that underlie developmental evolution. Nat. Rev. Genet. 2005;6:709–715. doi: 10.1038/nrg1676. [DOI] [PubMed] [Google Scholar]
- 5.Davis G.K., Srinivasan D.G., Wittkopp P.J., Stern D.L. The function and regulation of Ultrabithorax in the legs of Drosophila melanogaster. Dev. Biol. 2007;308:621–631. doi: 10.1016/j.ydbio.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stern D.L. A role of Ultrabithorax in morphological differences between Drosophila species. Nature. 1998;396:463–466. doi: 10.1038/24863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sucena E., Delon I., Jones I., Payre F., Stern D.L. Regulatory evolution of shavenbaby/ovo underlies multiple cases of morphological parallelism. Nature. 2003;424:935–938. doi: 10.1038/nature01768. [DOI] [PubMed] [Google Scholar]
- 8.Sucena E., Stern D.L. Divergence of larval morphology between Drosophila sechellia and its sibling species caused by cis-regulatory evolution of ovo/shaven-baby. Proc. Natl. Acad. Sci. USA. 2000;97:4530–4534. doi: 10.1073/pnas.97.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schertel C., Rutishauser T., Förstemann K., Basler K. Functional characterization of Drosophila microRNAs by a novel in vivo library. Genetics. 2012;192:1543–1552. doi: 10.1534/genetics.112.145383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chanut-Delalande H., Fernandes I., Roch F., Payre F., Plaza S. Shavenbaby couples patterning to epidermal cell shape control. PLoS Biol. 2006;4:e290. doi: 10.1371/journal.pbio.0040290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delon I., Chanut-Delalande H., Payre F. The Ovo/Shavenbaby transcription factor specifies actin remodelling during epidermal differentiation in Drosophila. Mech. Dev. 2003;120:747–758. doi: 10.1016/s0925-4773(03)00081-9. [DOI] [PubMed] [Google Scholar]
- 12.Stern D.L., Orgogozo V. The loci of evolution: how predictable is genetic evolution? Evolution. 2008;62:2155–2177. doi: 10.1111/j.1558-5646.2008.00450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeHoratius C., Silver P.A. Nuclear transport defects and nuclear envelope alterations are associated with mutation of the Saccharomyces cerevisiae NPL4 gene. Mol. Biol. Cell. 1996;7:1835–1855. doi: 10.1091/mbc.7.11.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rothacker B., Ilg T. Functional characterization of a Drosophila melanogaster succinic semialdehyde dehydrogenase and a non-specific aldehyde dehydrogenase. Insect Biochem. Mol. Biol. 2008;38:354–366. doi: 10.1016/j.ibmb.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 15.Sun X., Morozova T., Sonnenfeld M. Glial and neuronal functions of the Drosophila homolog of the human SWI/SNF gene ATR-X (DATR-X) and the jing zinc-finger gene specify the lateral positioning of longitudinal glia and axons. Genetics. 2006;173:1397–1415. doi: 10.1534/genetics.106.057893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bejarano F., Bortolamiol-Becet D., Dai Q., Sun K., Saj A., Chou Y.T., Raleigh D.R., Kim K., Ni J.Q., Duan H. A genome-wide transgenic resource for conditional expression of Drosophila microRNAs. Development. 2012;139:2821–2831. doi: 10.1242/dev.079939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heanue T.A., Reshef R., Davis R.J., Mardon G., Oliver G., Tomarev S., Lassar A.B., Tabin C.J. Synergistic regulation of vertebrate muscle development by Dach2, Eya2, and Six1, homologs of genes required for Drosophila eye formation. Genes Dev. 1999;13:3231–3243. doi: 10.1101/gad.13.24.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren N., He B., Stone D., Kirakodu S., Adler P.N. The shavenoid gene of Drosophila encodes a novel actin cytoskeleton interacting protein that promotes wing hair morphogenesis. Genetics. 2006;172:1643–1653. doi: 10.1534/genetics.105.051433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruby J.G., Stark A., Johnston W.K., Kellis M., Bartel D.P., Lai E.C. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frankel N., Erezyilmaz D.F., McGregor A.P., Wang S., Payre F., Stern D.L. Morphological evolution caused by many subtle-effect substitutions in regulatory DNA. Nature. 2011;474:598–603. doi: 10.1038/nature10200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGregor A.P., Orgogozo V., Delon I., Zanet J., Srinivasan D.G., Payre F., Stern D.L. Morphological evolution through multiple cis-regulatory mutations at a single gene. Nature. 2007;448:587–590. doi: 10.1038/nature05988. [DOI] [PubMed] [Google Scholar]
- 22.Zorc M., Skok D.J., Godnic I., Calin G.A., Horvat S., Jiang Z., Dovc P., Kunej T. Catalog of microRNA seed polymorphisms in vertebrates. PLoS ONE. 2012;7:e30737. doi: 10.1371/journal.pone.0030737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen K., Rajewsky N. Natural selection on human microRNA binding sites inferred from SNP data. Nat. Genet. 2006;38:1452–1456. doi: 10.1038/ng1910. [DOI] [PubMed] [Google Scholar]
- 24.Mackay T.F., Richards S., Stone E.A., Barbadilla A., Ayroles J.F., Zhu D., Casillas S., Han Y., Magwire M.M., Cridland J.M. The Drosophila melanogaster Genetic Reference Panel. Nature. 2012;482:173–178. doi: 10.1038/nature10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stern D.L. The Hox gene Ultrabithorax modulates the shape and size of the third leg of Drosophila by influencing diverse mechanisms. Dev. Biol. 2003;256:355–366. doi: 10.1016/s0012-1606(03)00035-6. [DOI] [PubMed] [Google Scholar]
- 26.Pool J.E., Corbett-Detig R.B., Sugino R.P., Stevens K.A., Cardeno C.M., Crepeau M.W., Duchen P., Emerson J.J., Saelao P., Begun D.J., Langley C.H. Population genomics of Sub-Saharan Drosophila melanogaster: African diversity and non-African admixture. PLoS Genet. 2012;8:e1003080. doi: 10.1371/journal.pgen.1003080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haley C.S., Knott S.A. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity (Edinb.) 1992;69:315–324. doi: 10.1038/hdy.1992.131. [DOI] [PubMed] [Google Scholar]
- 28.Broman K.W., Wu H., Sen S., Churchill G.A. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- 29.Okamura K., Hagen J.W., Duan H., Tyler D.M., Lai E.C. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.