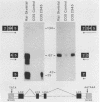
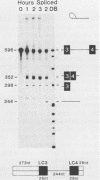
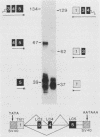
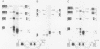Abstract
The mechanisms involved in the selective joining of appropriate 5' and 3' splice sites are still poorly understood in both constitutive and alternatively spliced genes. With two promoters associated with different exons, the myosin light-chain 1/3 gene generates two pre-mRNAs that also differ by the use of a pair of internal exons, 3 and 4, that are spliced in a mutually exclusive fashion. When the promoter upstream from exon 1 is used, only exon 4 is included. If the promoter upstream from exon 2 is used, only exon 3 is included. In an attempt to understand the molecular basis for the mutually exclusive behavior of these two exons and the basis of their specific selection, a number of minigene constructs containing exons 3 and 4 were tested in a variety of homologous or heterologous cis and trans environments. The results demonstrate that the mutually exclusive behavior of myosin light-chain exons 3 and 4 and selection between the two exons are cis regulated and are affected by the nature of the flanking sequences. Both exons competed for the common flanking 5' and 3' splice sites. Flanking exons were found that favored inclusion into mature mRNA of exon 3, exon 4, both, or neither, suggesting a specific cooperative interaction between certain 5' and 3' splice sites. Thus, alternative splicing of myosin light-chain 1/3 pre-mRNAs is regulated in cis by a hierarchy of compatibilities between pairs of 5' and 3' splice sites.
Full text
PDF

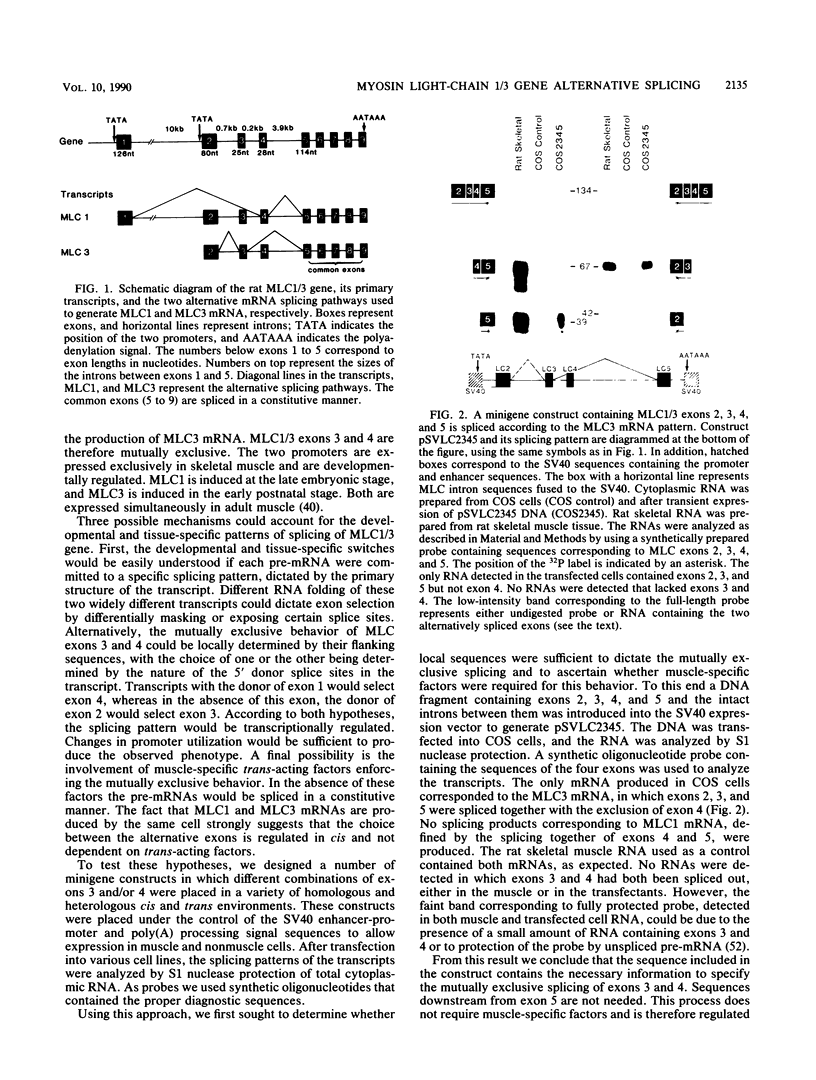
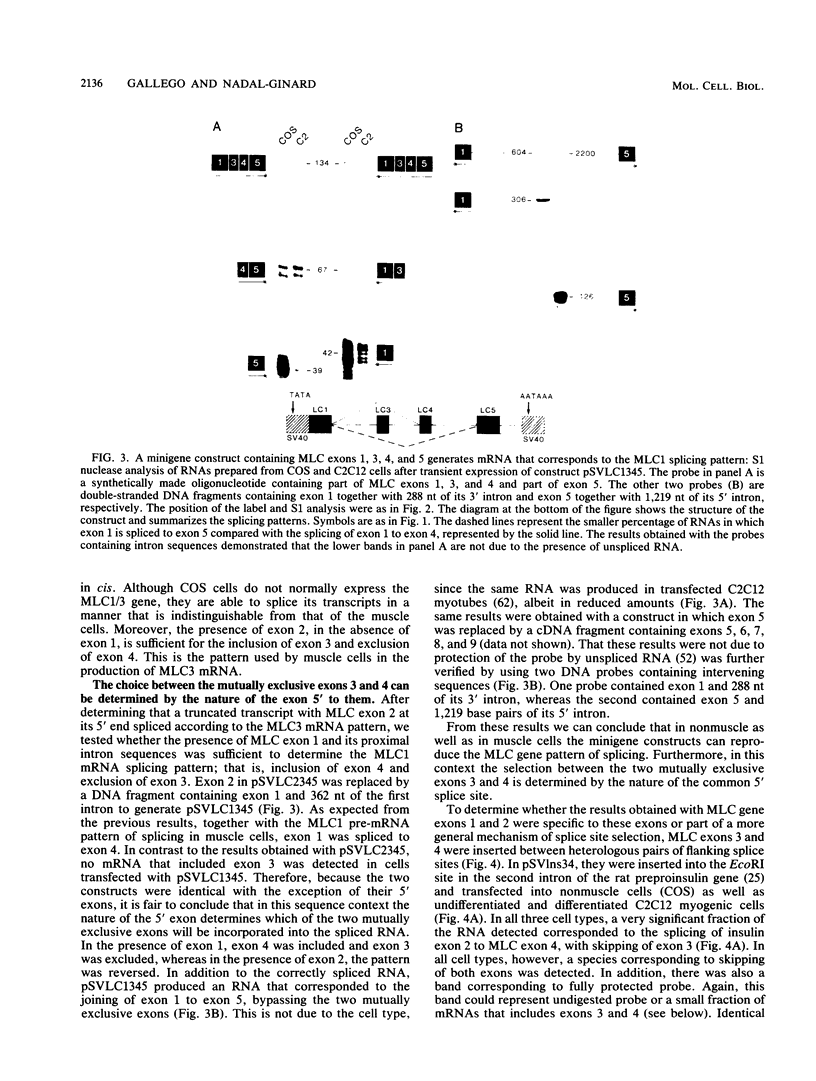
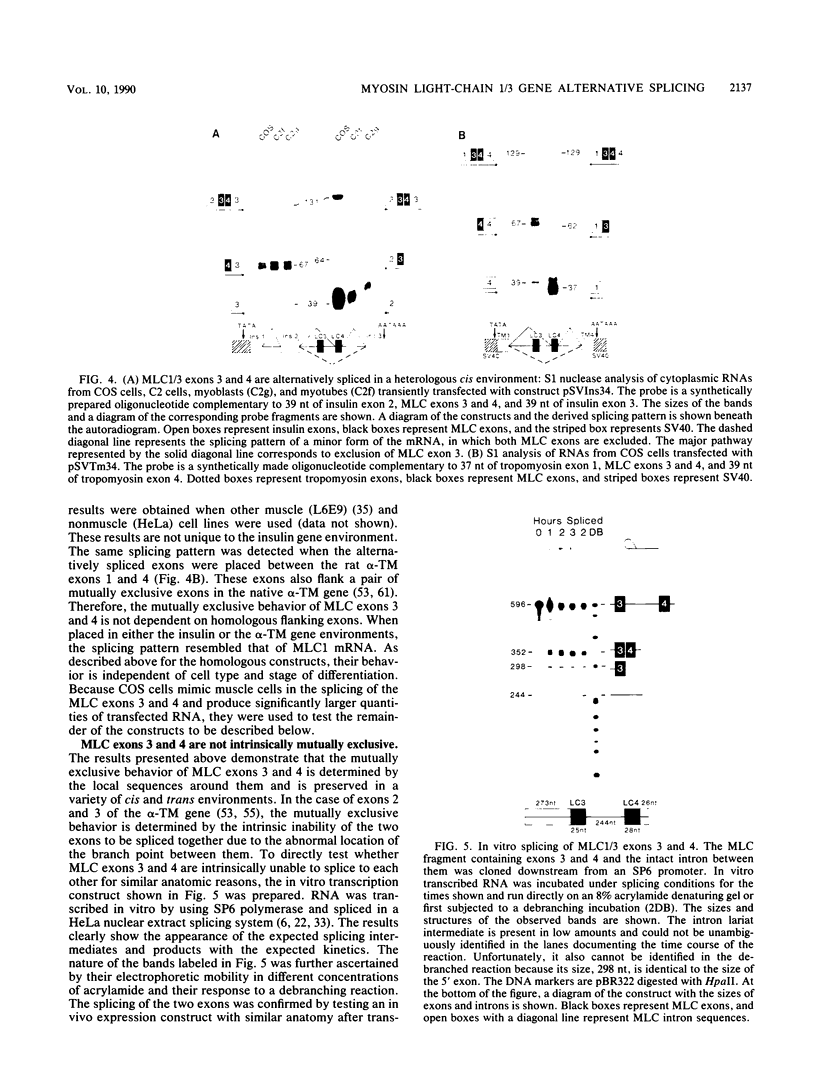
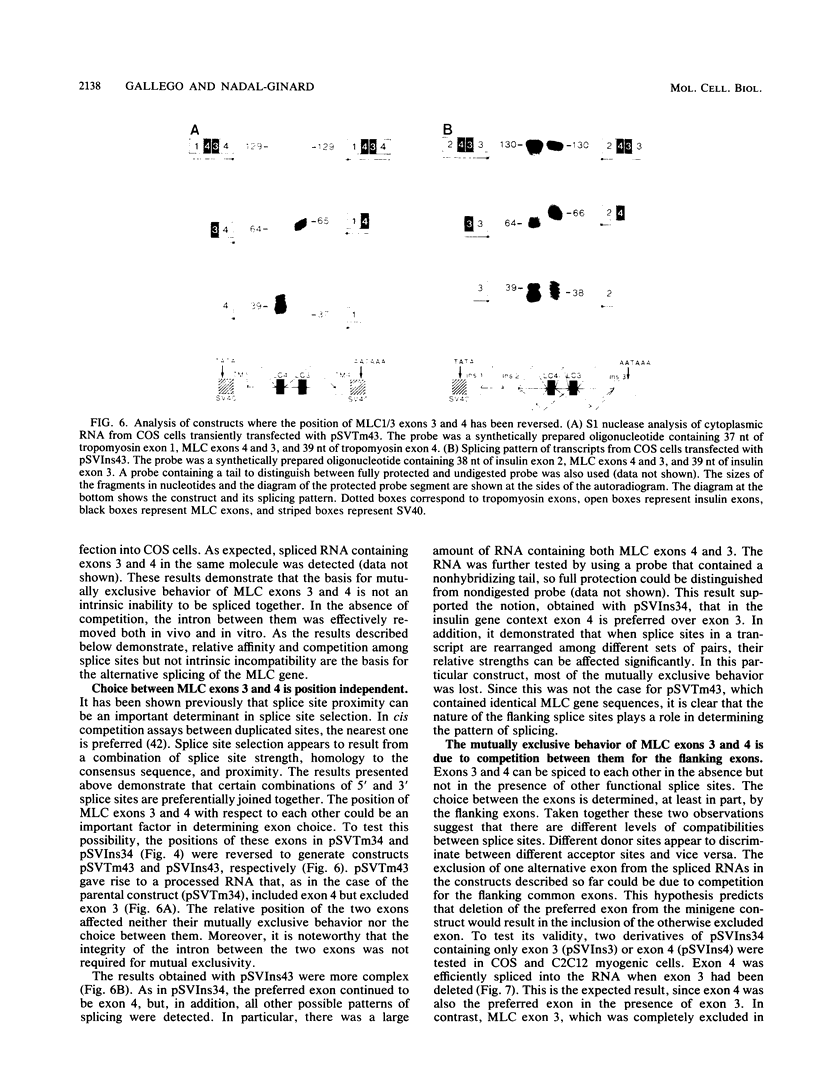




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aebi M., Hornig H., Weissmann C. 5' cleavage site in eukaryotic pre-mRNA splicing is determined by the overall 5' splice region, not by the conserved 5' GU. Cell. 1987 Jul 17;50(2):237–246. doi: 10.1016/0092-8674(87)90219-4. [DOI] [PubMed] [Google Scholar]
- Andreadis A., Gallego M. E., Nadal-Ginard B. Generation of protein isoform diversity by alternative splicing: mechanistic and biological implications. Annu Rev Cell Biol. 1987;3:207–242. doi: 10.1146/annurev.cb.03.110187.001231. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Breitbart R. E., Andreadis A., Nadal-Ginard B. Alternative splicing: a ubiquitous mechanism for the generation of multiple protein isoforms from single genes. Annu Rev Biochem. 1987;56:467–495. doi: 10.1146/annurev.bi.56.070187.002343. [DOI] [PubMed] [Google Scholar]
- Brody E., Abelson J. The "spliceosome": yeast pre-messenger RNA associates with a 40S complex in a splicing-dependent reaction. Science. 1985 May 24;228(4702):963–967. doi: 10.1126/science.3890181. [DOI] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eperon L. P., Estibeiro J. P., Eperon I. C. The role of nucleotide sequences in splice site selection in eukaryotic pre-messenger RNA. Nature. 1986 Nov 20;324(6094):280–282. doi: 10.1038/324280a0. [DOI] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Frendewey D., Keller W. Stepwise assembly of a pre-mRNA splicing complex requires U-snRNPs and specific intron sequences. Cell. 1985 Aug;42(1):355–367. doi: 10.1016/s0092-8674(85)80131-8. [DOI] [PubMed] [Google Scholar]
- Fu X. Y., Ge H., Manley J. L. The role of the polypyrimidine stretch at the SV40 early pre-mRNA 3' splice site in alternative splicing. EMBO J. 1988 Mar;7(3):809–817. doi: 10.1002/j.1460-2075.1988.tb02879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furdon P. J., Kole R. Inhibition of splicing but not cleavage at the 5' splice site by truncating human beta-globin pre-mRNA. Proc Natl Acad Sci U S A. 1986 Feb;83(4):927–931. doi: 10.1073/pnas.83.4.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfinkel L. I., Davidson N. Developmentally regulated expression of a truncated myosin light-chain 1F/3F gene. Mol Cell Biol. 1987 Oct;7(10):3826–3829. doi: 10.1128/mcb.7.10.3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981 Jan;23(1):175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- Grabowski P. J., Seiler S. R., Sharp P. A. A multicomponent complex is involved in the splicing of messenger RNA precursors. Cell. 1985 Aug;42(1):345–353. doi: 10.1016/s0092-8674(85)80130-6. [DOI] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Green M. R. Pre-mRNA splicing. Annu Rev Genet. 1986;20:671–708. doi: 10.1146/annurev.ge.20.120186.003323. [DOI] [PubMed] [Google Scholar]
- Hampson R. K., La Follette L., Rottman F. M. Alternative processing of bovine growth hormone mRNA is influenced by downstream exon sequences. Mol Cell Biol. 1989 Apr;9(4):1604–1610. doi: 10.1128/mcb.9.4.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfman D. M., Ricci W. M. Branch point selection in alternative splicing of tropomyosin pre-mRNAs. Nucleic Acids Res. 1989 Jul 25;17(14):5633–5650. doi: 10.1093/nar/17.14.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedes D. H., Steitz J. A. Accurate 5' splice-site selection in mouse kappa immunoglobulin light chain premessenger RNAs is not cell-type-specific. Proc Natl Acad Sci U S A. 1987 Nov;84(22):7928–7932. doi: 10.1073/pnas.84.22.7928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedes D. H., Steitz J. A. Correct in vivo splicing of the mouse immunoglobulin kappa light-chain pre-mRNA is dependent on 5' splice-site position even in the absence of transcription. Genes Dev. 1988 Nov;2(11):1448–1459. doi: 10.1101/gad.2.11.1448. [DOI] [PubMed] [Google Scholar]
- Krainer A. R., Maniatis T., Ruskin B., Green M. R. Normal and mutant human beta-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. 1984 Apr;36(4):993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- Lamond A. I., Konarska M. M., Sharp P. A. A mutational analysis of spliceosome assembly: evidence for splice site collaboration during spliceosome formation. Genes Dev. 1987 Aug;1(6):532–543. doi: 10.1101/gad.1.6.532. [DOI] [PubMed] [Google Scholar]
- Libri D., Marie J., Brody E., Fiszman M. Y. A subfragment of the beta tropomyosin gene is alternatively spliced when transfected into differentiating muscle cells. Nucleic Acids Res. 1989 Aug 25;17(16):6449–6462. doi: 10.1093/nar/17.16.6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomedico P. T. Use of recombinant DNA technology to program eukaryotic cells to synthesize rat proinsulin: a rapid expression assay for cloned genes. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5798–5802. doi: 10.1073/pnas.79.19.5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery D. E., Van Ness B. G. Comparison of in vitro and in vivo splice site selection in kappa-immunoglobulin precursor mRNA. Mol Cell Biol. 1988 Jun;8(6):2610–2619. doi: 10.1128/mcb.8.6.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery D. E., Van Ness B. G. In vitro splicing of kappa immunoglobulin precursor mRNA. Mol Cell Biol. 1987 Apr;7(4):1346–1351. doi: 10.1128/mcb.7.4.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Reed R. The role of small nuclear ribonucleoprotein particles in pre-mRNA splicing. Nature. 1987 Feb 19;325(6106):673–678. doi: 10.1038/325673a0. [DOI] [PubMed] [Google Scholar]
- Mardon H. J., Sebastio G., Baralle F. E. A role for exon sequences in alternative splicing of the human fibronectin gene. Nucleic Acids Res. 1987 Oct 12;15(19):7725–7733. doi: 10.1093/nar/15.19.7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj I. W. A binding consensus: RNA-protein interactions in splicing, snRNPs, and sex. Cell. 1989 Apr 7;57(1):1–3. doi: 10.1016/0092-8674(89)90164-5. [DOI] [PubMed] [Google Scholar]
- Mayeda A., Ohshima Y. Short donor site sequences inserted within the intron of beta-globin pre-mRNA serve for splicing in vitro. Mol Cell Biol. 1988 Oct;8(10):4484–4491. doi: 10.1128/mcb.8.10.4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadal-Ginard B. Commitment, fusion and biochemical differentiation of a myogenic cell line in the absence of DNA synthesis. Cell. 1978 Nov;15(3):855–864. doi: 10.1016/0092-8674(78)90270-2. [DOI] [PubMed] [Google Scholar]
- Nelson K. K., Green M. R. Splice site selection and ribonucleoprotein complex assembly during in vitro pre-mRNA splicing. Genes Dev. 1988 Mar;2(3):319–329. doi: 10.1101/gad.2.3.319. [DOI] [PubMed] [Google Scholar]
- Ohshima Y., Gotoh Y. Signals for the selection of a splice site in pre-mRNA. Computer analysis of splice junction sequences and like sequences. J Mol Biol. 1987 May 20;195(2):247–259. doi: 10.1016/0022-2836(87)90647-4. [DOI] [PubMed] [Google Scholar]
- Padgett R. A., Grabowski P. J., Konarska M. M., Seiler S., Sharp P. A. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- Parker B. A., Stark G. R. Regulation of simian virus 40 transcription: sensitive analysis of the RNA species present early in infections by virus or viral DNA. J Virol. 1979 Aug;31(2):360–369. doi: 10.1128/jvi.31.2.360-369.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periasamy M., Strehler E. E., Garfinkel L. I., Gubits R. M., Ruiz-Opazo N., Nadal-Ginard B. Fast skeletal muscle myosin light chains 1 and 3 are produced from a single gene by a combined process of differential RNA transcription and splicing. J Biol Chem. 1984 Nov 10;259(21):13595–13604. [PubMed] [Google Scholar]
- Reed R., Maniatis T. A role for exon sequences and splice-site proximity in splice-site selection. Cell. 1986 Aug 29;46(5):681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- Reed R., Maniatis T. Intron sequences involved in lariat formation during pre-mRNA splicing. Cell. 1985 May;41(1):95–105. doi: 10.1016/0092-8674(85)90064-9. [DOI] [PubMed] [Google Scholar]
- Reed R., Maniatis T. The role of the mammalian branchpoint sequence in pre-mRNA splicing. Genes Dev. 1988 Oct;2(10):1268–1276. doi: 10.1101/gad.2.10.1268. [DOI] [PubMed] [Google Scholar]
- Ruby S. W., Abelson J. An early hierarchic role of U1 small nuclear ribonucleoprotein in spliceosome assembly. Science. 1988 Nov 18;242(4881):1028–1035. doi: 10.1126/science.2973660. [DOI] [PubMed] [Google Scholar]
- Ruiz-Opazo N., Nadal-Ginard B. Alpha-tropomyosin gene organization. Alternative splicing of duplicated isotype-specific exons accounts for the production of smooth and striated muscle isoforms. J Biol Chem. 1987 Apr 5;262(10):4755–4765. [PubMed] [Google Scholar]
- Ruskin B., Green M. R. An RNA processing activity that debranches RNA lariats. Science. 1985 Jul 12;229(4709):135–140. doi: 10.1126/science.2990042. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Krainer A. R., Maniatis T., Green M. R. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell. 1984 Aug;38(1):317–331. doi: 10.1016/0092-8674(84)90553-1. [DOI] [PubMed] [Google Scholar]
- SCHERER W. F., SYVERTON J. T., GEY G. O. Studies on the propagation in vitro of poliomyelitis viruses. IV. Viral multiplication in a stable strain of human malignant epithelial cells (strain HeLa) derived from an epidermoid carcinoma of the cervix. J Exp Med. 1953 May;97(5):695–710. doi: 10.1084/jem.97.5.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seraphin B., Rosbash M. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell. 1989 Oct 20;59(2):349–358. doi: 10.1016/0092-8674(89)90296-1. [DOI] [PubMed] [Google Scholar]
- Shapiro M. B., Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987 Sep 11;15(17):7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A. Splicing of messenger RNA precursors. Science. 1987 Feb 13;235(4790):766–771. doi: 10.1126/science.3544217. [DOI] [PubMed] [Google Scholar]
- Sisodia S. S., Cleveland D. W., Sollner-Webb B. A combination of RNase H and S1 nuclease circumvents an artefact inherent to conventional S1 analysis of RNA splicing. Nucleic Acids Res. 1987 Mar 11;15(5):1995–2011. doi: 10.1093/nar/15.5.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. W., Nadal-Ginard B. Mutually exclusive splicing of alpha-tropomyosin exons enforced by an unusual lariat branch point location: implications for constitutive splicing. Cell. 1989 Mar 10;56(5):749–758. doi: 10.1016/0092-8674(89)90678-8. [DOI] [PubMed] [Google Scholar]
- Smith C. W., Patton J. G., Nadal-Ginard B. Alternative splicing in the control of gene expression. Annu Rev Genet. 1989;23:527–577. doi: 10.1146/annurev.ge.23.120189.002523. [DOI] [PubMed] [Google Scholar]
- Smith C. W., Porro E. B., Patton J. G., Nadal-Ginard B. Scanning from an independently specified branch point defines the 3' splice site of mammalian introns. Nature. 1989 Nov 16;342(6247):243–247. doi: 10.1038/342243a0. [DOI] [PubMed] [Google Scholar]
- Solnick D. Alternative splicing caused by RNA secondary structure. Cell. 1985 Dec;43(3 Pt 2):667–676. doi: 10.1016/0092-8674(85)90239-9. [DOI] [PubMed] [Google Scholar]
- Solnick D., Lee S. I. Amount of RNA secondary structure required to induce an alternative splice. Mol Cell Biol. 1987 Sep;7(9):3194–3198. doi: 10.1128/mcb.7.9.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strehler E. E., Periasamy M., Strehler-Page M. A., Nadal-Ginard B. Myosin light-chain 1 and 3 gene has two structurally distinct and differentially regulated promoters evolving at different rates. Mol Cell Biol. 1985 Nov;5(11):3168–3182. doi: 10.1128/mcb.5.11.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streuli M., Saito H. Regulation of tissue-specific alternative splicing: exon-specific cis-elements govern the splicing of leukocyte common antigen pre-mRNA. EMBO J. 1989 Mar;8(3):787–796. doi: 10.1002/j.1460-2075.1989.tb03439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulfendahl P. J., Kreivi J. P., Akusjärvi G. Role of the branch site/3'-splice site region in adenovirus-2 E1A pre-mRNA alternative splicing: evidence for 5'- and 3'-splice site co-operation. Nucleic Acids Res. 1989 Feb 11;17(3):925–938. doi: 10.1093/nar/17.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek D. F., Smith C. W., Nadal-Ginard B. The rat alpha-tropomyosin gene generates a minimum of six different mRNAs coding for striated, smooth, and nonmuscle isoforms by alternative splicing. Mol Cell Biol. 1988 Feb;8(2):679–694. doi: 10.1128/mcb.8.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe D., Saxel O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature. 1977 Dec 22;270(5639):725–727. doi: 10.1038/270725a0. [DOI] [PubMed] [Google Scholar]
- Zhuang Y. A., Goldstein A. M., Weiner A. M. UACUAAC is the preferred branch site for mammalian mRNA splicing. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2752–2756. doi: 10.1073/pnas.86.8.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y., Leung H., Weiner A. M. The natural 5' splice site of simian virus 40 large T antigen can be improved by increasing the base complementarity to U1 RNA. Mol Cell Biol. 1987 Aug;7(8):3018–3020. doi: 10.1128/mcb.7.8.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in U1 snRNA suppresses a 5' splice site mutation. Cell. 1986 Sep 12;46(6):827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- Zillmann M., Rose S. D., Berget S. M. U1 small nuclear ribonucleoproteins are required early during spliceosome assembly. Mol Cell Biol. 1987 Aug;7(8):2877–2883. doi: 10.1128/mcb.7.8.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]