Background: BMP signaling can maintain self-renewal of mouse ESCs together with LIF.
Results: Cochlin is a target gene of BMP and supports self-renewal of mouse ESCs through suppressing neural differentiation.
Conclusion: Cochlin is a downstream target of BMP signaling to mediate BMP functions in mouse ESCs.
Significance: Identification of more mediators would provide a better understanding of BMP signaling in mouse ESC fate control.
Keywords: Bone Morphogenetic Protein (BMP), Cell Signaling, Differentiation, Embryonic Stem Cell, SMAD Transcription Factor, Transcription Regulation, Coch, Self-renewal
Abstract
BMP4 maintains self-renewal of mouse embryonic stem cells (ESCs) in collaboration with LIF. Here, we report the identification of a novel key BMP target gene, cochlin (Coch) in mouse ESCs. Coch can be significantly up-regulated by BMP4 specifically in ESCs but not in somatic differentiated cells, and this up-regulation is dependent on the BMP signaling mediators Smad1/5 and Smad4. Overexpression of Coch can partially substitute BMP4 to promote self-renewal of mouse ESCs together with LIF, whereas knockdown of Coch impairs self-renewal marker gene expression even in the presence of both BMP4 and LIF. Further studies showed that COCH could mimic BMP4 in repressing neural differentiation of mouse ESCs upon LIF withdrawal and the inhibitory effect of BMP4 on neural differentiation is compromised by Coch knockdown. Taken together, our data suggest that COCH is a part of the downstream target network of BMP signaling and serves as another important effector to fine-tune mouse ESC fates.
Introduction
Embryonic stem cells (ESCs)3 are pluripotent stem cells that can indefinitely self-renew in vitro while at the same time possess the ability to differentiate into all the adult tissues, including germ cells (1, 2). Because of two remarkable properties (self-renewal and differentiation), ESCs are valuable for a wide range of applications, such as an in vitro model to study early embryo development, and most importantly, as a promising cell source for transplantation in regenerative medicine (1–4). It has been recognized that mouse ESC fate determination is delicately regulated by multiple internal transcription factors and external signaling pathways. Internal core transcription factors, such as OCT4/POU5F1, NANOG, and SOX2, can form an autoregulatory feedback circuit to maintain pluripotency of ESCs. These factors physically interact with each other and co-occupy the promoters of target genes to establish ESC-specific transcriptome (2, 5). Several epigenetic factors controlling histone modification and chromatin structures also play important roles during the switch between self-renewal and differentiation of ESCs (6–9). Appropriate activity of extracellular signal-regulated kinase (ERK) within the cells also plays a central role in the fate choices of mouse ESCs (10–12). Besides these intrinsic factors, extracellular cytokines such as leukemia inhibitory factor (LIF), transforming growth factor β (TGF-β) superfamily, and Wnt signaling have also been shown to govern ESC fate (13–15). Among them, LIF signaling has long been established to support mouse ESC self-renewal on standard serum- and feeder-dependent culture conditions (16). LIF transduces its signal through binding to a heterodimeric receptor complex consisting of gp130 and the low-affinity LIF receptor and then activates STAT3 through phosphorylation (15, 16). However, LIF alone is not enough to maintain mouse ESC pluripotency, and the cooperation between BMP4 and LIF is found to be sufficient for long-term self-renewal maintenance in the absence of serum and feeder cells (11, 15).
BMP4 is a TGF-β superfamily member, which plays multiple critical roles during embryogenesis as well as in tissue homeostasis by regulating a series of cellular processes including cell proliferation, adhesion, migration, differentiation, and apoptosis (17–19). BMPs transduce their signal through binding to the transmembrane type I and type II serine/threonine kinase receptor complexes. The ligand-activated receptors then phosphorylate and activate R-Smads (Smad1, -5, and -8), leading to the formation of a complex consisting of R-Smad and Co-Smad (Smad4). Translocation of the Smad complex from cytoplasm to the nucleus enables direct transcriptional regulation of target gene expression (17, 20–22).
Considering the importance of BMP signaling in mouse ESC fate control, it is of great value to learn how BMP signaling exerts its functions. ID family proteins were first described as critical downstream targets of BMP4 to promote self-renewal by inhibiting neural differentiation (15). Our previous work extended the repertoire of potential direct target genes of BMP signaling in mouse ESCs from a genome-wide perspective (23) and further identified another important direct target gene, Dusp9, which mediates BMP signaling to balance ERK activity (10). In the present study, we report the identification of cochlin (coagulation factor C homolog) (Coch), as another important direct target gene of BMP signaling, which promotes mouse ESC self-renewal by inhibition of neural differentiation.
EXPERIMENTAL PROCEDURES
Cell Culture, Reagents, and Antibodies
Feeder-free E14 and R1 mouse ES cells were grown on gelatin-coated dishes and cultured in DMEM (Dulbecco's modified Eagle's medium) supplemented with 15% Knockout Serum Replacement (KSR) (Invitrogen), 2 mm glutamine, 10−4 m nonessential amino acids, 10−4 m β-mercaptoethanol, 100 units/ml penicillin, 100 μg/ml streptomycin, and 1,000 units/ml mouse LIF. To examine the effect of Coch on the self-renewal of mouse ESCs, R1 cells were cultured in N2B27 medium containing LIF or LIF plus BMP. For neural differentiation, R1 cells were cultured in N2B27 medium without any cytokines, as described previously (23). NMuMG, NIH3T3, C2C12, and HeLa cells were cultured in DMEM supplemented with 10% FBS (Hyclone).
BMP4 was purchased from R&D. The antibodies used in this study are listed as follows: rabbit anti-Smad4 antibody was generated from human Smad4-linker (aa144–316)-His; Smad1 (sc-7965X) was purchased from Santa Cruz Biotechnology; anti-nestin antibody (MAB353) was purchased from Chemicon International.
Reporter Plasmids
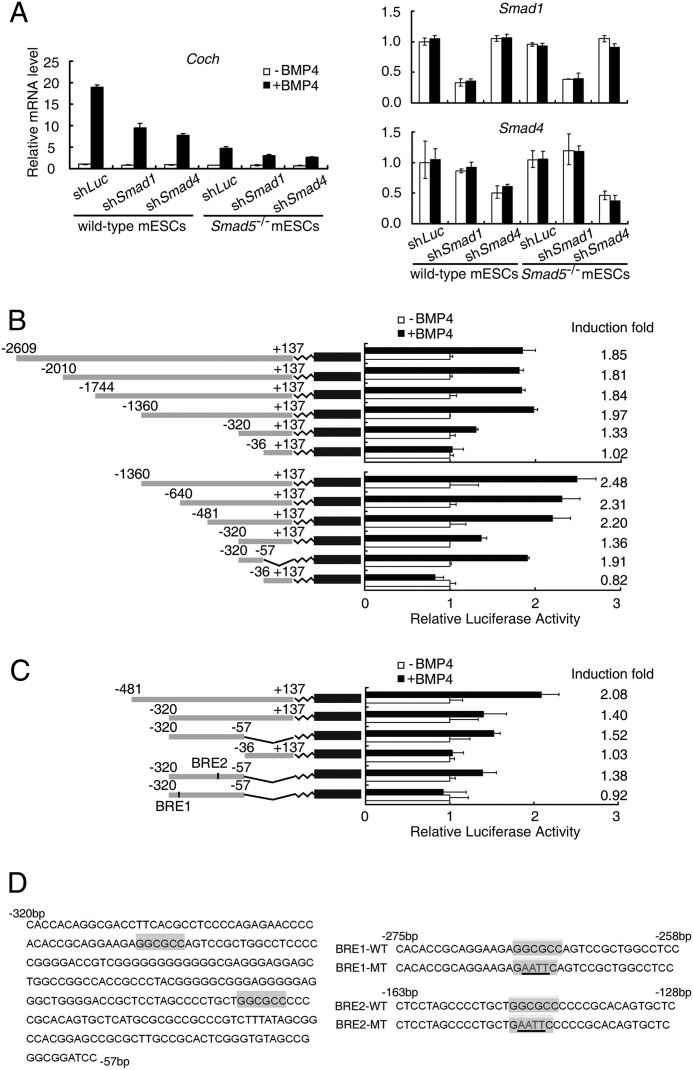
Mouse Coch (NC_000078) promoter constructs (−2609 bp ∼ +137 bp) were generated by PCR using mouse genomic DNA as a template and amplified fragments were cloned into the pGL3-basic reporter vector through MluI/BglII sites. A series of 5′ deletion constructs were generated using the longest Coch (−2609 bp ∼ +137 bp) fragment as the PCR template, and the PCR primers used are listed below: −2609 bp ∼ +137 bp construct: 5′-GTTACGCGTAGAACCAGCACCTTTCTG-3′ (sense); −2010 bp ∼ +137 bp construct: 5′-GCTACGCGTGACTCCTCCACCCTCCTT-3′ (sense); −1774 bp ∼ +137 bp construct: 5′-GTTACGCGTGACCATGCTGCCTGAGTT-3′ (sense); −1360 bp ∼ +137 bp construct: 5′-GTTACGCGTGCTGACGCACACTTCAGC-3′ (sense); −640 bp ∼ +137 bp construct: 5′-GTTACGCGTCAACAAATGACCCATCAG-3′ (sense); −481 bp ∼ +137 bp construct: 5′-GTTACGCGTTCTCAGAACCGGGTGGCT-3′ (sense); −320 bp ∼ +137 bp construct: 5′-GTTACGCGTCACCACAGGCGACCTTCA-3′ (sense); −36 bp ∼ +137 bp construct: 5′-GTTACGCGTGGAGGTGCATAGGGTCTG-3′ (sense). All of the above constructs were generated using the same antisense primer: 5′-GACAGATCTAGGCGTGAACGAGGTTCG-3′ (antisense). The −320 bp ∼ −57 bp construct was generated from −320 bp ∼ +137 bp construct by digesting the template with MluI and BamHI enzymes. Site-directed mutagenesis was performed using −320 bp ∼ −57 bp construct as a template. The wild-type GGCGCC sites were replaced by the mutated GaattC-mutation sequence (Fig. 2D).
FIGURE 2.
BMP4-induced Coch expression is dependent on Smad1/5 and Smad4. A, wild-type and Smad5 knockout (Smad5−/−) mouse ESCs were transfected with either firefly luciferase control (shLuc), Smad1 (shSmad1), or Smad4 (shSmad4) shRNA constructs, and then treated with 10 ng/ml BMP4 for 12 h before harvesting for qRT-PCR analysis of Coch mRNA levels. B and C, a series of luciferase reporter constructs with indicated Coch promoter regions were transfected into R1 ESCs followed by treatment with 10 ng/ml BMP4 for 24 h and then subjected to luciferase activity determination. The induction folds of the reporters responsive to BMP4 were shown at the right side of the figure. D, mutation of the BRE1 (GGCGCC) element to GAATTC abolished the response of the Coch promoter to BMP4 stimulation. Data were shown as mean ± S.E. (n = 3).
Construction of Lentiviral Plasmids
For overexpression of Coch, the coding sequence (CDS) of full-length mouse Coch with or without HA (hemagglutinin) tag (named COCH-1 and COCH-2, respectively) were cloned into pENTR1A backbone and then using LR clonase reactions (Invitrogen) to replace these CDS into the p2k7neo lentiviral vector under the control of EF1α promoter (24).
The short hairpin RNA (shRNA) targeting mouse Coch mRNA (NM_007728) or the control firefly luciferase gene was cloned into the pll3.7 lentiviral vector, which was modified with GFP region replaced by puromycin gene. The sequences of the shRNAs targeting mouse Coch are listed below: shCoch-1: 5′-CAGGGTTAGAACCAATTGT-3′; shCoch-2: 5′-GCCGATTTAATTTGCAGAA-3′. The shSmad1 and shSmad4 plasmids were generated previously (10, 23).
Lentivirus Production and Transduction
Lentivirus was produced by transfection of HEK293FT cells with lentiviral vectors, which contain the CDS or shRNA sequences of Coch using Lipofectamine 2000 (Invitrogen). After 3 days, the supernatant was collected and filtered through a 0.45 μm filter (Millipore). Virus-containing supernatants were added into plates of ES cells in the presence of 8 μg/ml polybrene. After incubation for 6 h at 37 °C, equal volume of complete mouse ESC culture medium (DMEM supplemented with 15% KSR, l-glutamine, nonessential amino acids, β-mercaptoethanol, and 1,000 units/ml LIF) was added and further incubated overnight. Then the supernatants were discarded, and cells were washed three or four times with PBS. Then the cells can be cultured with the complete ESC medium. In this study, Geneticin (250 μg/ml, Invitrogen) and puromycin (1000 μg/ml) were added into culture medium for 3 to 5 days to select cells with stable viral integration.
Luciferase Reporter Assay
Cells were seeded in 48-well plate and transfected with the reporter constructs when they reach to 30–50% confluent. pRenilla-TK vector was transfected as an internal control. Transfection was carried out with Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol. One day after transfection, 10 ng/ml BMP4 was added into the medium. Cells were lysed after BMP4 treatment for 24 h and then subjected to luciferase activity measurement. Each experiment was done in triplicate, and all the data are normalized to Renilla activity. The data were represented as the mean ± S.E. of three independent experiments.
RNA Isolation and Real-time PCR
Real-time PCR was performed as described previously (23). Briefly, the total RNA was isolated from the cells using TRIzol reagent (Invitrogen). Complementary DNA synthesis was primed with oligo dT (Takara) by reverse transcriptase (Toyobo). Real-time PCR was carried out using EvaGreen dye (Biotium) on the Mx3000P system. All the primers used are listed in supplemental Table S1.
Alkaline Phosphatase Staining, Immunostaining, and Western Blot
Alkaline phosphatase staining was performed using Alkaline Phosphatase Detection Kit (SCR004, Millipore) according to the manufacturer's instruction. Immunostaining and Western blot were carried out as described previously (10, 23).
Chromatin Immunoprecipitation (ChIP)
ChIP was carried out as described previously (10, 23). Briefly, cells were firstly cross-linked by 1% formaldehyde solution for 15 min. Then the cells were lysed with lysis buffer (1% SDS, 50 mm Tris pH 8.0, 5 mm EDTA, proteinase inhibitors) and sonicated to get DNA fragments ranging ∼300–500 bp on average. After centrifuging at 14,000 rpm for 10 min, the supernatant was collected and pre-absorbed by 50 μl of protein A beads (Zymed Laboratories Inc.). After incubation with 10 μg of antibodies (control IgG, anti-Smad1, and anti-Smad4) overnight at 4 °C, the immunocomplexes were collected with 50 μl of protein A beads by 3 h co-incubation and then washed sequentially with salt buffers. The bound immunocomplexes were eluted with elution buffer (25 mm Tris, pH 8.0, 10 mm EDTA, 0.5% SDS) by heating at 65 °C with occasional vortex for 15 min and crosslinking was reversed by overnight incubation at 65 °C. Immunoprecipitated DNA and input DNA were then purified by phenol:chloroform:isoamyl alcohol extraction and ethanol precipitation after treatment with RNase A and proteinase K. ChIP primers used in this study are listed in the supplemental Table S1.
RESULTS
BMP4 Up-regulates Coch Expression in ESCs
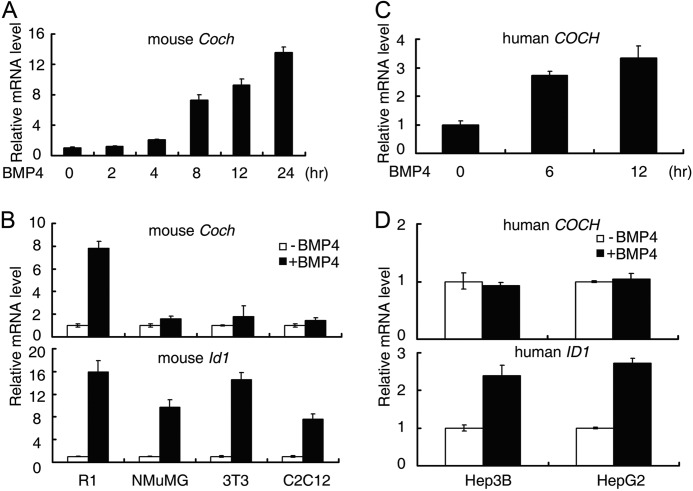
To identify direct downstream target genes of BMP signaling, we previously carried out Smad ChIP-chip and ChIP-seq combined with RNA expression microarray upon BMP4 stimulation in mouse ESCs (23). In addition to the previously characterized Id family genes (15), Dusp9 (10), Dypsl2, and Kdm6b (23), another gene named Coch exhibited significant up-regulation (2.1-fold change) in the RNA expression microarray data in response to BMP4 stimulation (10, 23). We confirmed this up-regulation by quantitative real-time PCR (qRT-PCR), and a dramatic up-regulation was observed (Fig. 1A). BMP4 treatment steadily induced Coch mRNA expression in E14 mouse ESCs, and the induction reached more than 12-fold after 24 h (Fig. 1A). Similar up-regulation could also be observed in R1 mouse ESCs (Fig. 1B), indicating Coch is a bona fide BMP target gene in mouse ESCs.
FIGURE 1.
BMP4 up-regulates Coch in ESCs. A and C, E14 mouse ESCs (A) or H1 human ESCs (C) were treated with 10 ng/ml BMP4 for the indicated time and then harvested for qRT-PCR analysis. B and D, mouse (B) or human (D) somatic cells were treated with 10 ng/ml BMP4 for 8 h before harvesting for qRT-PCR analysis. Data are shown as mean ± S.E. (n = 3).
To examine the cell type specificity of Coch up-regulation by BMP4, we treated differentiated mouse somatic cells NMuMG, NIH3T3, and C2C12 cells with 10 ng/ml BMP4 for 8 h. As shown in Fig. 1B, Coch did not exhibit any significant change while the classic BMP target gene, Id1, was consistently up-regulated in these cells. Thus, the up-regulation of Coch by BMP4 was specific to mouse ESCs.
In addition, we also found that human COCH is up-regulated by BMP4 in H1 human ESCs (Fig. 1C). Consistent with the results from mouse cells, human COCH did not respond to BMP4 in differentiated human somatic cells (Hep3B and HepG2), where ID1 was up-regulated by BMP4 (Fig. 1D). These data collectively indicate that BMP4 can up-regulate Coch expression, specifically in ESCs.
Up-regulation of Coch by BMP4 Is Dependent on Smad1/5 and Smad4
BMP/Smad signaling regulates its downstream target genes through Smad1/5/8 and Smad4 (22). To examine whether induction of Coch by BMP4 was dependent on Smad proteins, we knocked down Smad1 as well as Smad4 in wild-type and Smad5−/− TC-1 mouse ESCs. As is shown in Fig. 2A, either Smad1 or Smad4 knockdown attenuated the up-regulation of Coch by BMP4, and the most dramatic attenuation of Coch induction was observed in Smad5 knockout ESCs with Smad4 knockdown, indicating that Smad1, Smad5, and Smad4 are responsible for BMP induction of Coch. The residual induction of Coch after shSmad1 in Smad5 knockout cells might result from incomplete deletion of Smad1. These data indicate that the induction of Coch by BMP4 is dependent on Smad1/5 and Smad4.
Smad1 and Smad4 Specifically Bind to a GGCGCC Motif in the Coch Promoter
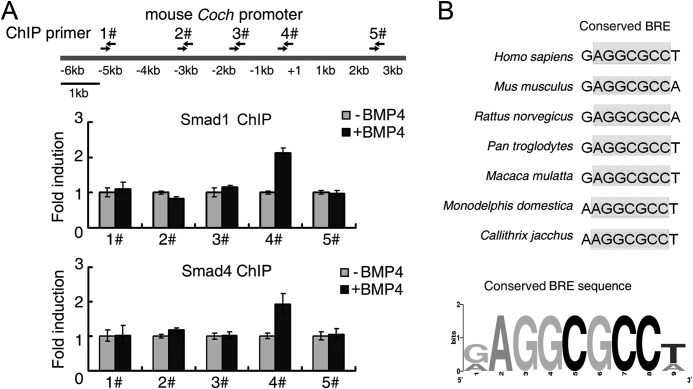
As Smad1 and Smad4 are transcription factors, they regulate the expression of downstream target genes by binding to specific DNA elements, such as CAGAC, GCCGnCGC, GGCGCC in gene promoters (25–29). Therefore, we attempted to identify the BMP responsive element (BRE) in the Coch promoter. To this end, we cloned putative mouse Coch promoter covering the −2609 base pair (bp) to +137 bp around the transcriptional start site (TSS) and inserted it into pGL3-basic luciferase reporter plasmid. Reporter assay showed that it responded to BMP4 stimulation by about 2-fold (Fig. 2B). Then, a series of deletion constructs were generated and tested for BMP4 responsiveness. As shown in Fig. 2B, the −1360 bp ∼ −36 bp region still retained the intact response to BMP. Further deletion within −1360 bp ∼ −36 bp region showed that two subregions (−481 bp ∼ −320 bp and −320 bp ∼ −57 bp) contained the minimal BREs for Coch up-regulation. The fragment of −320 bp ∼ −57 bp still retained partial response to BMP4 (Fig. 2, B and C). By scrutinizing the potential Smad binding elements within the −320 bp ∼ −57 bp region, we found two putative GC-rich BREs (BRE1 and BRE2), and mutation of key nucleotides within BRE1 but not in BRE2 totally abolished the transcription activation by BMP4 (Fig. 2, C and D), indicating that BRE1 is necessary for Coch up-regulation by BMP4. Further analysis of −480 bp to −320 bp region by either the region alone or mutation analysis of putative BREs in that region using reporter assay excluded the possibility that there are other functional BREs in that region (data not shown).
To examine whether Smad1/5 and Smad4 could physically bind to this region of Coch promoter upon BMP4 treatment, we conducted a ChIP assay using Smad1 and Smad4 antibodies. Five pairs of primers were designed to amplify the putative Smad binding region along the Coch promoter (supplemental Table S1). Only primer pair #4, whose amplicon covers the BRE1 region identified above, showed a binding signal for both Smad1 and Smad4 in response to BMP4 treatment (Fig. 3A). In addition, by comparing the putative Coch promoter region (3000 bp upstream of TSS and 500 bp downstream TSS) from several species, we found that the seven core nucleotides (AGGCGCC) of mouse BRE1 within the Coch promoter were conserved in these species (Fig. 3B). Thus, these data suggest that binding of Smad1 and Smad4 to the GGCGCC element of the Coch promoter is essential for BMP-induced Coch expression.
FIGURE 3.
Smad1/5 and Smad4 bind to the Coch promoter region. A, ChIP assay was performed with the antibodies against Smad1 or Smad4 in R1 cells treated with or without BMP4 for 6 h. The immunoprecipitated DNA was amplified by quantitative PCR with the primers detecting specific promoter regions denoted in the upper panel. Data are shown as mean ± S.E. (n = 3). B, GGCGCC element in the Coch promoter is conserved across species. The AGGCGCC consensus BRE motif was generated by WebLogo.
COCH Mediates BMP4-dependent Self-renewal in Mouse ESCs
BMP4 together with LIF can sustain mouse ESC self-renewal in serum- and feeder-free medium (15). Considering the ESC-specific regulation of Coch in mouse ESCs, we then asked whether Coch would act as a BMP target gene to promote self-renewal.
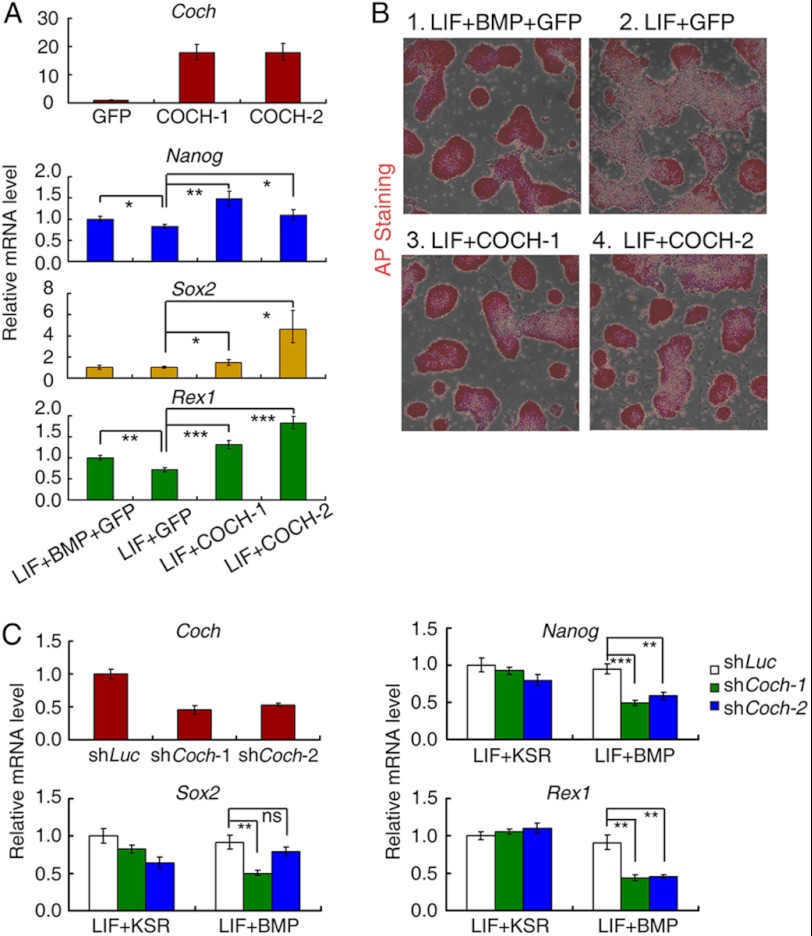
To examine whether overexpression of COCH could mimick BMP4 function to maintain self-renewal of mouse ESCs, we generated GFP- and COCH-overexpressing stable R1 ESCs by lentiviral infection (COCH-1 expressing untagged COCH, and COCH-2 expressing HA-tagged COCH at the C terminus), and cultured them either in LIF only or LIF+BMP4 supplemented N2B27 medium for 5 days. The self-renewal status was determined by the expression of pluripotency markers: Nanog, Rex1, and Sox2 as well as the alkaline phosphatase (AP) staining assay. The GFP-expressing stable cell line cultured in N2B27 medium with LIF plus BMP for 5 days was shown as positive control. As is shown in Fig. 4A, expression of Nanog, Rex1, and Sox2 deceased to differential extent in LIF only medium compared with LIF plus BMP condition in GFP overexpression cells; while overexpression of COCH recovered the self-renewal maker expression even in the presence of LIF only. This result was confirmed by the AP staining assay showing that COCH-overexpressed colonies cultured under the LIF-only condition have comparable positive staining intensity to those cultured under LIF-plus-BMP medium (Fig. 4B). These data suggest that overexpression of COCH can support mouse ESC self-renewal in the presence of LIF.
FIGURE 4.
COCH mediates BMP4 function to sustain self-renewal of mouse ESCs. A, R1 mouse ESCs stably expressing GFP (GFP) or COCH (COCH-1 and COCH-2) were cultured in N2B27 medium containing LIF only for 5 days, and the self-renewal marker expression was determined by qRT-PCR. R1 mouse ESCs stably expressing GFP (GFP) cells were cultured in N2B27 medium containing LIF + BMP and are shown as the positive control. B, COCH-overexpressing cells as well as GFP stably expressing cells were cultured in N2B27 medium containing LIF only for 5 days and then subjected to AP staining. GFP stably expressing R1 cells cultured in N2B27 medium containing LIF + BMP was used as the positive control. C, Coch knockdown (shCoch-1 and shCoch-2) and control (shLuc) R1 cells were cultured in KSR medium supplemented with LIF or N2B27 medium containing LIF+BMP or LIF only for 5 days, and the self-renewal marker expression was determined by qRT-PCR. Data are shown as mean ± S.E. (n = 3) (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
To determine whether Coch was essential for BMP4-dependent self-renewal, we stably knocked down Coch expression using two independent shRNA constructs and then evaluated the pluripotency marker gene expression in the LIF-plus-BMP4 condition. Compared with control luciferase knockdown cells, Coch knockdown significantly attenuated the expression of Nanog, Rex1, and Sox2 when cells were cultured under LIF-plus-BMP4 medium (Fig. 4C). In contrast, Coch knockdown had little effect on these markers when cells were cultured in standard knockout serum replacement (KSR) medium with little BMP4, indicating that the COCH function of self-renewal is dependent on the presence of BMP4 (Fig. 4C). Taken together, these results show that COCH can function downstream of BMP4 signaling to promote mouse ESC self-renewal.
COCH Prevents Neural Differentiation
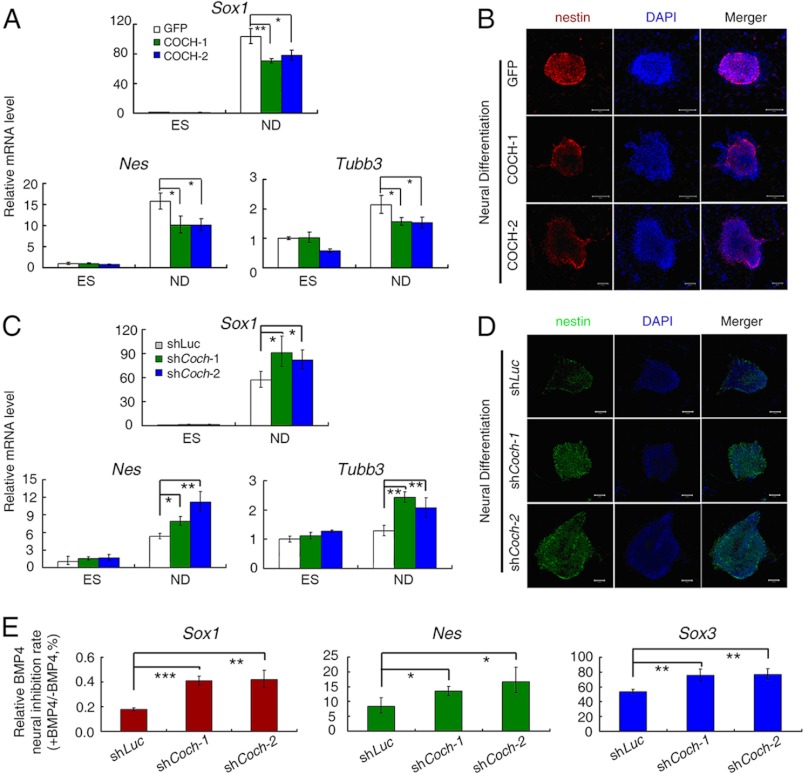
Without LIF and BMP4, mouse ESCs spontaneously undergo neural differentiation when cultured in N2 and B27 neurotrophic factor-supplemented medium (N2B27 medium), while addition of BMP4 can potently inhibit this process (15). Given the significant up-regulation of Coch by BMP4 in ESCs, we hypothesized that COCH might have functional roles during BMP4 inhibition of neural differentiation. To test this possibility, we stably overexpressed COCH in mouse ESCs with lentiviral infection and examined whether COCH overexpression could inhibit neural differentiation. As shown in Fig. 5A, compared with GFP overexpression control, overexpression of COCH by two independent COCH lentiviral constructs significantly inhibited neural differentiation, as evidenced by the decreased mRNA expression of neural markers: Sox1, Nes (nestin), and Tubb3 at day 5 of neural differentiation in N2B27 medium. The inhibition of neural differentiation by COCH overexpression was further confirmed by the anti-nestin immunostaining analysis at day 5 of neural differentiation (Fig. 5B). It was noteworthy that the inhibitory effect by COCH was not as potent as BMP4, consistent with the concept that BMP4 inhibition of neural differentiation can be orchestrated by multiple direct target genes downstream of BMP4 such as Id family genes, Dpysl2, Kdm6b (Jmjd3), and Dusp9 (10, 15, 23).
FIGURE 5.
COCH suppresses neural differentiation of mouse ESCs. A, R1 ESCs stably expressing GFP or COCH were cultured in normal KSR medium (ES) or N2B27 medium to allow neural differentiation for 5 days (ND). mRNA levels of the neural differentiation markers Sox1, Nes, and Tubb3 were determined by qRT-PCR. B, R1 ESCs stably expressing GFP or COCH were cultured in N2B27 medium for 5 days and were then subjected to anti-nestin immunostaining analysis. Scale bars: 50 μm. C, Coch knockdown R1 mouse ES cells (shCoch-1 and shCoch-2), and the control luciferase knockdown cells (shLuc) were cultured in normal KSR medium (ES) or N2B27 medium (ND) for 5 days, and then harvested for qRT-PCR analysis. D, Coch knockdown R1 mouse ESCs or control cells (shLuc) were cultured in N2B27 medium for 5 days, and then subjected to anti-nestin immunostaining analysis. Scale bars: 50 μm. E, Coch knockdown R1 mouse ESCs (shCoch-1 and shCoch-2) and control (shLuc) cells were cultured in N2B27 medium with or without BMP4 for 5 days and then harvested for qRT-PCR. BMP4 neural inhibition rate (+BMP4/−BMP4) was presented by the ratio of the corresponding mRNA levels of +BMP4 to −BMP4. Quantitation data are shown as mean ± S.E. (n = 3) (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
Next, we knocked down Coch to examine its effects on neural differentiation. As expected, Coch knockdown enhanced neural differentiation as evidenced by the increased expression of Sox1, Nes, and Tubb3 mRNAs (Fig. 5C) and nestin protein (Fig. 5D). Furthermore, an increased expression of the neural markers Sox1, Nes, and Sox3 caused by Coch knockdown was still observed in the presence of BMP4 (Fig. 5E), indicating that the inhibition of neural differentiation by BMP4 could be partially impaired by Coch knockdown. Collectively, these results suggest that Coch contributes to the repressive function of BMP4 in neural differentiation.
COCH Attenuates ERK Activation
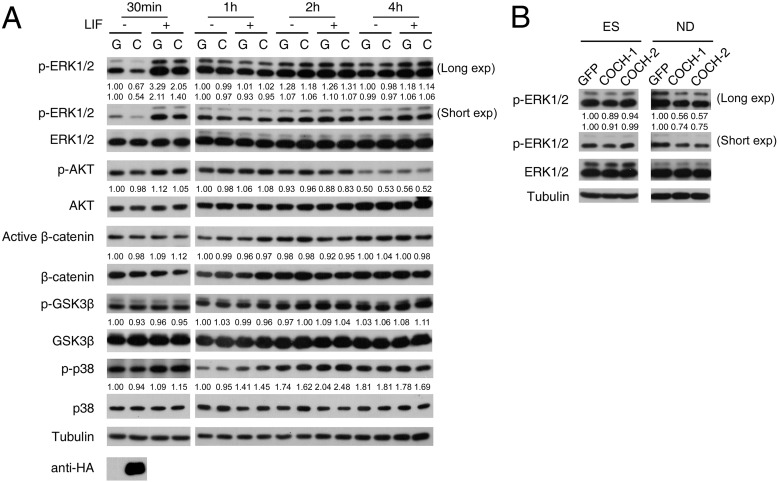
To address the molecular mechanism underlying the COCH effect, we examined the effect of COCH on the major signaling pathways involved in ESC fate control. The conditioned media were collected from HEK293T cells transfected with either GFP or COCH plasmids for 36 h. After treatment with these conditioned media, R1 ESCs were harvested for immunoblotting to examine the activation of various signaling molecules. As shown in Fig. 6A, among the molecules we have examined, COCH decreased ERK phosphorylation, regardless of the presence of LIF, at 30 min, but this inhibition was not observed at the later time points. The inhibition of ERK by COCH was also confirmed in COCH-overexpressed stable R1 cells during neural differentiation (Fig. 6B).
FIGURE 6.
COCH has an inhibitory effect on ERK activity. A, R1 ESCs were treated with the conditioned medium, which was collected from HEK293T cells transfected with GFP control (G) or HA- tagged COCH (C) plasmids, for the indicated times in the presence or absence of LIF. The cell lysates were then harvested for immunoblotting with indicated antibodies. Anti-HA immunoblotting shows COCH expression in the conditioned medium. B, R1 ESCs stably expressing GFP or COCH were cultured in normal KSR medium (ES) or N2B27 medium to allow neural differentiation (ND) for 5 days. ERK phosphorylation was then examined. Numbers indicate the relative band intensity of the phosphorylated proteins after normalization with their corresponding total protein levels.
DISCUSSION
In this study, we report that Coch is a BMP target and is involved in BMP-mediated pluripotency maintenance of mouse embryonic stem cells. BMP-induced Coch expression depends on Smad1/5 and Smad4, which bind to the Coch promoter. We have further identified in the promoter region a GC-rich element, which is required for BMP/Smad signal activation of Coch expression. Functionally, Coch promotes stemness and prevents neural differentiation of mouse ESCs.
BMP and LIF are two extracellular signals that cooperate to maintain mouse ESC self-renewal (15). We have previously employed genome-wide approaches to identify genes whose expression change upon BMP4 treatment and mapped the Smad binding sites across the genome (23). A couple of target genes of BMP signaling have been identified, and one of them, Dusp9, has been demonstrated to be a critical crosstalk node between extrinsic BMP signal and intrinsic ERK signal, thus contributing to robust mouse ESC self-renewal (10). Coch was among the genes whose expression altered upon BMP4 treatment, and our ChIP assay also revealed Smad binding in its promoter region. Consistently, up-regulation of Coch was also reported upon BMP2 treatment in mouse E14 ES cells (30), or it also could be a target gene of TGF-β2 (31).
We demonstrated that Coch was up-regulated by BMP4 at the mRNA level, and the up-regulation was dependent on both Smad1/5 and Smad4. We showed that the Coch promoter could respond to BMP4 stimulation, and Smad1 and Smad4 directly bound to a GGCGCC element in the Coch promoter. The observation that the GGCGCC element is evolutionarily conserved across multiple mammalian species further implicated the importance of BMP regulation of Coch. Interestingly, like Dusp9, Coch is specifically up-regulated by BMP in both mouse and human ESCs, but not in differentiated somatic cells. The underlying mechanism is currently unclear.
BMP4 can maintain self-renewal of mouse ESCs in collaboration with LIF through its downstream target genes, such as Ids (15) and Dusp9 (10). We found that Coch, as a BMP/Smad target, could also contribute to BMP functions in maintenance of mouse ESC pluripotency. When overexpressed, COCH partially substituted BMP4 to maintain the pluripotency of mouse ESCs, while knockdown of Coch impaired the self-renewal. BMP signaling supports mouse ESC self-renewal mainly by inhibiting differentiation to the neural lineages (10, 15, 23). Consistent with this, overexpression of COCH mimicking BMP4 inhibited neural differentiation, and knockdown of Coch attenuated BMP-mediated neural inhibition. Thus, like previously identified BMP target genes Ids and Dusp9, Coch supports mouse ESC self-renewal via inhibition of neural lineage commitment.
How COCH, as an extracellular matrix component, inhibits neural differentiation is still an open question. We tested whether COCH influenced the key signaling pathways involved in ESC fate determination. Among the signaling molecules we have examined, only ERK activity was significantly decreased at 30 min with COCH-conditioned medium treatment. ERK activity is tightly controlled in embryonic stem cells (summarized in Li et al. (10)). Based on our data, we speculate that COCH may exert its function by interfering with ERK activity. It has been shown that transient interference of ERK activity could generate significant but distinct biological effects compared with sustained ERK perturbation (32–34). Therefore, the transient inhibition of ERK by Coch could partially explain the functions of Coch in mediating BMP4 effects.
Moreover, as shown in Fig. 6B, we tested ERK activity of COCH-overexpressing cells under both self-renewal (ES) and neural differentiation (ND) conditions. Overexpression of COCH could significantly inhibit ERK activity in the neural differentiation. Thus, the transient inhibitory effect by COCH-conditioned medium in normal ESCs and inhibition in neural differentiation by COCH overexpression collectively argue that COCH may function through repressing ERK activity. However, we still could not exclude other possibilities by which Coch may exert its function. The detailed mechanism requires further investigation.
The Coch gene encodes the COCH protein, which is highly conserved in chicken, mouse, and human. Several mutations in human COCH have been reported to be highly associated with DFNA9 (deafness, autosomal-dominant 9), an autosomal-dominant non-syndromic hearing loss disorder. COCH is highly expressed in human and mouse inner ear, but its DFNA9-causing mechanisms remain elusive. Thus, we for the first time observed that Coch can be highly induced by BMP4 in mouse ESCs, suggesting that Coch may have important roles in stem cell fate determination and in early development, in addition to the previously known functions in the inner ear. Although Coch knockout mice were viable (35), it is interesting to note that Coch is highly expressed in the placenta during postimplantation development and can be specifically up-regulated by LIF in luminal epithelial of uterus during embryo implantation (35).
Acknowledgments
We thank Bing Zhao, Yi Ding, and Benyu Ma for technical assistance.
This work was supported by grants from the National Key Basic Research Program of China (973 Program) (2011CB943803) and the National Natural Science Foundation of China (30930050, 30921004) (to Y. G. C.).

This article contains supplemental Table S1.
- ESC
- embryonic stem cell
- BMP
- bone morphogenic protein
- BRE
- BMP responsive element
- ChIP
- chromatin immunoprecipitation
- Coch
- cochlin
- GFP
- green fluorescence protein
- LIF
- leukemia inhibitory factor
- shRNA
- short hairpin RNA
- AP
- alkaline phosphatase.
REFERENCES
- 1. Wobus A. M., Boheler K. R. (2005) Embryonic stem cells: prospects for developmental biology and cell therapy. Physiol. Rev. 85, 635–678 [DOI] [PubMed] [Google Scholar]
- 2. Young R. A. (2011) Control of the embryonic stem cell state. Cell 144, 940–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Evans M. J., Kaufman M. H. (1981) Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156 [DOI] [PubMed] [Google Scholar]
- 4. Martin G. R. (1981) Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. U.S.A. 78, 7634–7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cole M. F., Young R. A. (2008) Mapping key features of transcriptional regulatory circuitry in embryonic stem cells. Cold Spring Harb Symp Quant Biol. 73, 183–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bibikova M., Laurent L. C., Ren B., Loring J. F., Fan J. B. (2008) Unraveling epigenetic regulation in embryonic stem cells. Cell Stem Cell 2, 123–134 [DOI] [PubMed] [Google Scholar]
- 7. Boyer L. A., Plath K., Zeitlinger J., Brambrink T., Medeiros L. A., Lee T. I., Levine S. S., Wernig M., Tajonar A., Ray M. K., Bell G. W., Otte A. P., Vidal M., Gifford D. K., Young R. A., Jaenisch R. (2006) Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441, 349–353 [DOI] [PubMed] [Google Scholar]
- 8. Lee T. I., Jenner R. G., Boyer L. A., Guenther M. G., Levine S. S., Kumar R. M., Chevalier B., Johnstone S. E., Cole M. F., Isono K., Koseki H., Fuchikami T., Abe K., Murray H. L., Zucker J. P., Yuan B., Bell G. W., Herbolsheimer E., Hannett N. M., Sun K., Odom D. T., Otte A. P., Volkert T. L., Bartel D. P., Melton D. A., Gifford D. K., Jaenisch R., Young R. A. (2006) Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125, 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Christophersen N. S., Helin K. (2010) Epigenetic control of embryonic stem cell fate. J. Exp. Med. 207, 2287–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li Z., Fei T., Zhang J., Zhu G., Wang L., Lu D., Chi X., Teng Y., Hou N., Yang X., Zhang H., Han J. D., Chen Y. G. (2012) BMP4 Signaling Acts via dual-specificity phosphatase 9 to control ERK activity in mouse embryonic stem cells. Cell Stem Cell 10, 171–182 [DOI] [PubMed] [Google Scholar]
- 11. Ying Q. L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. (2008) The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li Z., Chen Y. G. (2012) Fine-tune of intrinsic ERK activity by extrinsic BMP signaling in mouse embryonic stem cells. Protein Cell 3, 401–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sato N., Meijer L., Skaltsounis L., Greengard P., Brivanlou A. H. (2004) Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat. Med. 10, 55–63 [DOI] [PubMed] [Google Scholar]
- 14. Williams R. L., Hilton D. J., Pease S., Willson T. A., Stewart C. L., Gearing D. P., Wagner E. F., Metcalf D., Nicola N. A., Gough N. M. (1988) Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature 336, 684–687 [DOI] [PubMed] [Google Scholar]
- 15. Ying Q. L., Nichols J., Chambers I., Smith A. (2003) BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 115, 281–292 [DOI] [PubMed] [Google Scholar]
- 16. Niwa H., Burdon T., Chambers I., Smith A. (1998) Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 12, 2048–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Massagué J., Chen Y. G. (2000) Controlling TGF-β signaling. Genes Dev. 14, 627–644 [PubMed] [Google Scholar]
- 18. Shi Y., Massagué J. (2003) Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 113, 685–700 [DOI] [PubMed] [Google Scholar]
- 19. Fei T., Chen Y. G. (2010) Regulation of embryonic stem cell self-renewal and differentiation by TGF-β family signaling. Sci. China Life Sci. 53, 497–503 [DOI] [PubMed] [Google Scholar]
- 20. Massagué J. (1998) TGF-β signal transduction. Annu. Rev. Biochem. 67, 753–791 [DOI] [PubMed] [Google Scholar]
- 21. Feng X. H., Derynck R. (2005) Specificity and versatility in TGF-β signaling through Smads. Annu. Rev. Cell Dev. Biol. 21, 659–693 [DOI] [PubMed] [Google Scholar]
- 22. Massagué J., Seoane J., Wotton D. (2005) Smad transcription factors. Genes Dev. 19, 2783–2810 [DOI] [PubMed] [Google Scholar]
- 23. Fei T., Xia K., Li Z., Zhou B., Zhu S., Chen H., Zhang J., Chen Z., Xiao H., Han J. D., Chen Y. G. (2010) Genome-wide mapping of SMAD target genes reveals the role of BMP signaling in embryonic stem cell fate determination. Genome Res. 20, 36–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kee K., Angeles V. T., Flores M., Nguyen H. N., Reijo Pera R. A. (2009) Human DAZL, DAZ, and BOULE genes modulate primordial germ-cell and haploid gamete formation. Nature 462, 222–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dennler S., Itoh S., Vivien D., ten Dijke P., Huet S., Gauthier J. M. (1998) Direct binding of Smad3 and Smad4 to critical TGFβ-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 17, 3091–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jonk L. J., Itoh S., Heldin C. H., ten Dijke P., Kruijer W. (1998) Identification and functional characterization of a Smad binding element (SBE) in the JunB promoter that acts as a transforming growth factor-β, activin, and bone morphogenetic protein-inducible enhancer. J. Biol. Chem. 273, 21145–21152 [DOI] [PubMed] [Google Scholar]
- 27. Kusanagi K., Inoue H., Ishidou Y., Mishima H. K., Kawabata M., Miyazono K. (2000) Characterization of a bone morphogenetic protein-responsive Smad-binding element. Mol. Biol. Cell 11, 555–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ishida W., Hamamoto T., Kusanagi K., Yagi K., Kawabata M., Takehara K., Sampath T. K., Kato M., Miyazono K. (2000) Smad6 is a Smad1/5-induced smad inhibitor. Characterization of bone morphogenetic protein-responsive element in the mouse Smad6 promoter. J. Biol. Chem. 275, 6075–6079 [DOI] [PubMed] [Google Scholar]
- 29. Korchynskyi O., ten Dijke P. (2002) Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J. Biol. Chem. 277, 4883–4891 [DOI] [PubMed] [Google Scholar]
- 30. Alarcón C., Zaromytidou A. I., Xi Q., Gao S., Yu J., Fujisawa S., Barlas A., Miller A. N., Manova-Todorova K., Macias M. J., Sapkota G., Pan D., Massagué J. (2009) Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-β pathways. Cell 139, 757–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bhattacharya S. K., Gabelt B. T., Ruiz J., Picciani R., Kaufman P. L. (2009) Cochlin expression in anterior segment organ culture models after TGFβ2 treatment. Invest Ophthalmol Vis. Sci. 50, 551–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Murphy L. O., Smith S., Chen R. H., Fingar D. C., Blenis J. (2002) Molecular interpretation of ERK signal duration by immediate early gene products. Nat. Cell Biol. 4, 556–564 [DOI] [PubMed] [Google Scholar]
- 33. Sasagawa S., Ozaki Y., Fujita K., Kuroda S. (2005) Prediction and validation of the distinct dynamics of transient and sustained ERK activation. Nat. Cell Biol. 7, 365–373 [DOI] [PubMed] [Google Scholar]
- 34. Murphy L. O., Blenis J. (2006) MAPK signal specificity: the right place at the right time. Trends Biochem. Sci. 31, 268–275 [DOI] [PubMed] [Google Scholar]
- 35. Rodriguez C. I., Cheng J. G., Liu L., Stewart C. L. (2004) Cochlin, a secreted von Willebrand factor type a domain-containing factor, is regulated by leukemia inhibitory factor in the uterus at the time of embryo implantation. Endocrinology 145, 1410–1418 [DOI] [PubMed] [Google Scholar]