Background: How receptor binding by measles virus hemagglutinin (MV-H) triggers membrane fusion is unknown.
Results: Mutations in putative dimer-dimer interfaces of MV-H head domain tetramers inhibit fusion.
Conclusion: The proper dimer-dimer interactions of the MV-H head domain may play a role in fusion triggering.
Significance: One of the missing links between receptor binding and fusion triggering is revealed.
Keywords: Cell Surface Receptor, Membrane Fusion, Protein Conformation, Viral Protein, Virus Entry
Abstract
Measles virus (MV), an enveloped RNA virus belonging to the Paramyxoviridae family, enters the cell through membrane fusion mediated by two viral envelope proteins, an attachment protein hemagglutinin (H) and a fusion (F) protein. The crystal structure of the receptor-binding head domain of MV-H bound to its cellular receptor revealed that the MV-H head domain forms a tetrameric assembly (dimer of dimers), which occurs in two forms (forms I and II). In this study, we show that mutations in the putative dimer-dimer interface of the head domain in either form inhibit the ability of MV-H to support membrane fusion, without greatly affecting its cell surface expression, receptor binding, and interaction with the F protein. Notably, some anti-MV-H neutralizing monoclonal antibodies are directed to the region around the dimer-dimer interface in form I rather than receptor-binding sites. These observations suggest that the dimer-dimer interactions of the MV-H head domain, especially that in form I, contribute to triggering membrane fusion, and that conformational shift of head domain tetramers plays a role in the process. Furthermore, our results indicate that although the stalk and transmembrane regions may be mainly responsible for the tetramer formation of MV-H, the head domain alone can form tetramers, albeit at a low efficiency.
Introduction
To enter host cells, enveloped viruses must bind to cellular receptors and then fuse their envelope (a lipid bilayer membrane surrounding the viral nucleocapsid) with the plasma membrane or with the endosomal membrane after endocytosis. Paramyxoviruses are enveloped viruses containing a nonsegmented, negative strand RNA genome and include important human and animal viruses such as measles virus (MV),5 mumps virus, parainfluenza viruses, and Newcastle disease virus (NDV) (1). These viruses require the coordinated action of two envelope glycoproteins, an attachment protein, and a fusion (F) protein for their entry into cells (1). Binding of the attachment protein to its receptor is thought to trigger, by a poorly understood mechanism, a series of conformational changes of the adjacently located F protein leading to membrane fusion at the cell surface (1–7). Accordingly, the attachment protein, which is referred to as the hemagglutinin (H), hemagglutinin-neuraminidase (HN), or G protein depending on the virus, has three important functions in this process: receptor binding, interaction with the F protein, and triggering the F protein. Paramyxovirus H, HN, and G proteins consist of an N-terminal cytoplasmic tail, a transmembrane region, a membrane-proximal stalk region, and a C-terminal receptor-binding head domain, and the stalk region is likely involved in the interaction with the F protein (1, 2, 8–11).
MV remains a major cause of childhood morbidity and mortality worldwide despite the availability of highly effective vaccine (12). We previously reported the crystal structures of the MV attachment protein H (MV-H) in complex with its receptor, the signaling lymphocyte activation molecule (SLAM) (13). The complex forms a tetrameric assembly (dimer of dimers), which occurs in two forms (forms I and II) that differ by a shift of the two dimers relative to each other. From these findings, we proposed that a conformational shift of MV-H dimer of dimers acts as a mechanism of fusion triggering (13). Native-PAGE analysis, combined with bimolecular complementation, also revealed that the physiological MV-H forms tetramers or higher order oligomers (14). Furthermore, structural studies showed that soluble HN proteins (lacking the stalk region) of NDV and parainfluenza virus 5 (PIV5) exhibit tetramers (15, 16). On the other hand, the structure of the NDV HN ectodomain revealed that dimers of head domain dimers flank a four-helix bundle stalk (17). Only two of the four head domains interact with the tetrameric stalk, and no head domain dimer-dimer interface was observed in crystals. The stalk region of PIV5 was also found to form a four-helix bundle, like that of NDV (8).
To gain insight into the functional role of the tetramer formation of the MV-H head domain, we introduced various mutations at the dimer-dimer interfaces based on the crystal structures. The mutations inhibited the ability of MV-H to support membrane fusion, without affecting its cell surface expression, receptor binding, and interaction with the F protein. Thus, our results indicate that the dimer-dimer interactions of MV-H head domain tetramers contribute to triggering membrane fusion.
EXPERIMENTAL PROCEDURES
Construction of expression plasmids, protein preparation and surface plasmon resonance analysis were performed as previously described (18).
Flow Cytometry Analysis
HEK293T cells were transfected with expression plasmids encoding the full-length MV-H or its mutants. At 36 h post-transfection, the cells were incubated with human polyclonal antibody against MV (19), followed by Alexa Fluor 488-conjugated anti-human IgG (Molecular Probes, Inc.). The cells were analyzed by flow cytometry.
Co-immunoprecipitation
CHO/vv5–4 cells (20) were infected with vTF7–3, a recombinant vaccinia virus encoding the T7-RNA polymerase (21), at a multiplicity of infection of 1.0, and then co-transfected with expression plasmids (with the T7 promoter) encoding a His-tagged H protein and a FLAG-tagged F protein. At 24 h after transfection, the cells were lysed in the immunoprecipitation buffer (10 mm HEPES, pH 7.4, 50 mm sodium pyrophosphate, 50 mm sodium fluoride, 50 mm sodium chloride, 5 mm EDTA, 5 mm EGTA, 1% Triton X-100, protease inhibitors), followed by centrifugation. The supernatants were precleared by the protein A-Sepharose (GE Healthcare), and one-tenth of each supernatant was collected as a cell lysate sample. The rest of the supernatant was incubated with anti-FLAG monoclonal antibody M2-treated (Sigma-Aldrich) protein A-Sepharose. The immunoprecipitated samples were collected by centrifugation, washed three times each with buffer A (100 mm Tris, pH 7.6, 500 mm lithium chloride, 0.1% Triton X-100) and then buffer B (20 mm HEPES, pH 7.2, 2 mm EGTA, 10 mm magnesium chloride, 0.1% Triton X-100), and analyzed by SDS-PAGE under a reducing condition and immunoblotting with anti-His tag rabbit polyclonal antibody (Medical and Biological Laboratories Co., Ltd.).
Quantitative Fusion Assay
HEK293T cells were co-transfected with expression plasmids encoding MV-H, MV F protein (MV-F), firefly luciferase, and Renilla luciferase. At 5 h post-transfection, the cells were mixed with Vero/hSLAM cells (22) expressing the T7 polymerase (Vero/hSLAM-T7). The Renilla luciferase gene is encoded downstream of the T7 promoter, and its transcription is activated by fusion between Vero/hSLAM-T7 and HEK293T cells. At 24 h post-transfection, luciferase activity in the cells was analyzed using the Dual luciferase reporter assay system (Promega), according to the manufacturer's instructions. Renilla luciferase activity was divided by firefly luciferase activity (directed by the herpes simplex virus thymidine kinase promoter) to correct transfection efficiency.
Blue Native-PAGE and Immunoblot Analysis
HEK293S GnTI(−) cells (23) were transfected with expression plasmids encoding the full-length MV-H or its mutants. At 48 h post-transfection, the cells were treated with the NativePAGETM sample buffer (Invitrogen) containing Coomassie Brilliant Blue G-250 and digitonin (0.5%) or n-dodecyl-β-d-maltoside (0.25%), and the lysates were separated on a 4–12% native polyacrylamide gel. Soluble forms of the H proteins were treated with the NativePAGETM sample buffer containing Coomassie Brilliant Blue G-250 and separated on the same gel. The polypeptides on the gels were transferred onto a polyvinylidene difluoride membrane, which was then incubated with a monoclonal antibody (C-1) against MV-H (24), followed by peroxidase-conjugated goat anti-mouse IgG (Bio-Rad). The membrane was treated with Chemi-Lumi One Super (Nacalai Tesque), and chemiluminescent signals were detected and visualized using a VersaDoc 3000 imager (Bio-Rad).
RESULTS
Mutations in the Putative Dimer-Dimer Interfaces of MV-H Head Domain Tetramers
Because crystal structures and biochemical analysis of MV-H showed tetrameric formation (dimer of dimers) (13, 14), we reasoned that the dimer-dimer interaction of the MV-H head domain may be involved in triggering membrane fusion. To test this idea, we introduced amino acid substitutions that likely disrupt dimer-dimer interfaces in the MV-H head domain (Fig. 1, A and B). MV-H consists of an N-terminal cytoplasmic tail, a transmembrane region, a stalk region, and a C-terminal head domain (Fig. 1A) (1, 12). The stalk region is thought to interact with MV-F (25–27), like other paramyxoviruses (2, 8–11). In the present study, we used the H protein of the MV Edmonston strain (Ed-H), the soluble form of which had been utilized for our previous structural studies (13, 18).
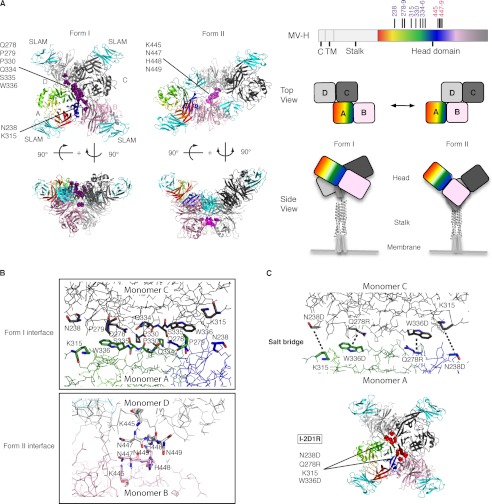
FIGURE 1.
MV-H head domain tetramer (dimer of dimers). A, left panel, top and side views of the MV-H-SLAM complex. The residues replaced in mutant MV-H proteins are indicated in purple (form I) and pink (form II). SLAM is indicated in cyan. Right panel, schematics of MV-H. The cytoplasmic tail (C), transmembrane region (TM), stalk region, and head domain are indicated with mutated positions. Monomers A (rainbow colors) and B (pink) form one dimer, whereas monomers C (dark gray) and D (light gray) form another. The colors of the respective MV-H monomers are the same in schematics and crystal structures. Upon receptor binding, conformational changes involving the dimer-dimer interfaces of the head domain may occur, which would induce structural rearrangements of the stalk region. The modules within the central region of the stalk have been shown to be involved in contact with the F protein and its triggering (7, 8, 17, 30, 31). B, the dimer-dimer interfaces of MV-H tetramers. Form I and II dimer-dimer interfaces are shown with replaced amino acids. C, the expected salt bridge formation in the mutant I-2D1R (upper panel). Mutated amino acid residues in I-2D1R are indicated by red circles (lower panel).
Based on MV-H crystal structures (13), we selected amino acid residues that are located at the dimer-dimer interfaces (Fig. 1B) and substituted alanine or aspartic acid for them. The latter residue was employed to disrupt the interfaces more strongly through its charged side chain. The mutant MV-H proteins were named according to the dimer-dimer interface affected (form I or II), the number of residues replaced, and the residue(s) substituted (A or D). Mutants that have the putative dimer-dimer interface in form I mutated are I-6A (Q278A, P279A, P330A, Q334A, S335A, and W336A) and I-2D2A (P330D, Q334D, S335A, and W336A), whereas those that have the putative interface in form II mutated are II-1D (K445D) and II-3D (N447D, H448D, and N449D). Lys-445 may not be directly involved in the dimer-dimer interface, but the replacement of this charged residue was expected to affect the dimer-dimer interaction in form II. The mutant II-4D (K445D, N447D, H448D, and N449D) has the combined substitutions of these two mutants II-1D and II-3D. We also generated another mutant I-2D1R (N238D, Q278R, and W336D), in which the substitutions were expected to generate salt bridges at the interface in form I and immobilize its dimer-dimer interaction (Fig. 1C). Together, we produced six types of MV-H mutants in which the putative dimer-dimer interaction was altered in form I or II.
Cell Surface Expression, Interaction with the F Protein, and Receptor Binding of MV-H Mutants
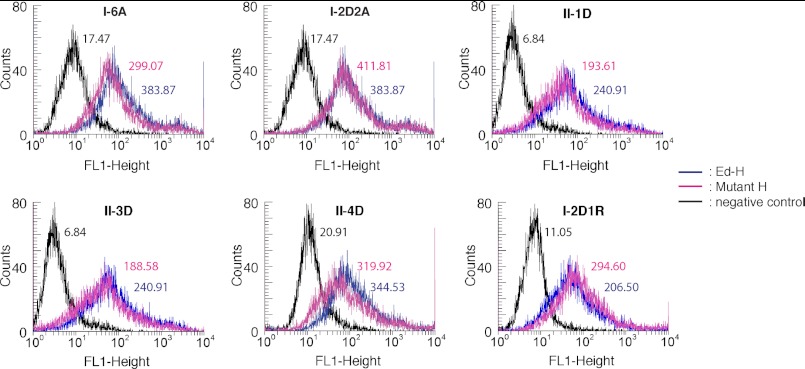
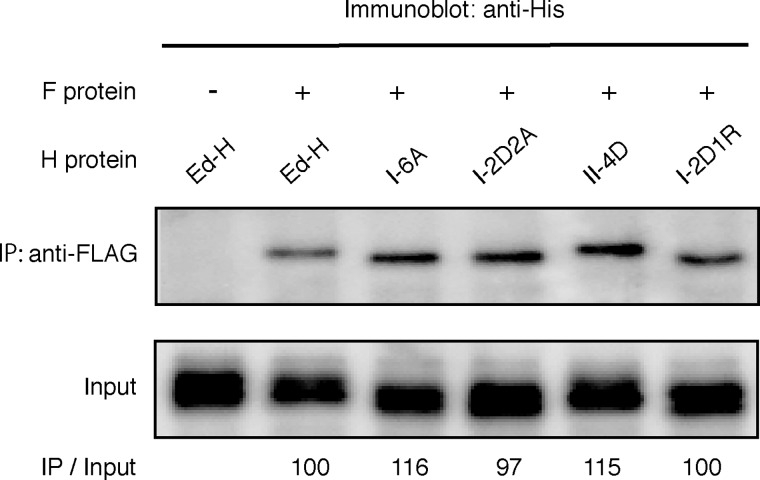
To ascertain that these mutant H proteins are produced normally (except fusion triggering ability) in cells, the parental Ed-H and mutant proteins were transiently expressed in HEK293T cells. Flow cytometry revealed that all mutant proteins were expressed on the cell surface at levels comparable with or slightly lower than that of Ed-H (Fig. 2). Furthermore, mutant H proteins (only the combined mutant II-4D was examined among form II mutants) co-precipitated with the F protein as comparably as the parental Ed-H protein (Fig. 3), verifying that these substitutions at the dimer-dimer interfaces of the MV-H head domain do not affect the interaction between the stalk region and the F protein. We also assessed the abilities of these mutants to bind to cellular receptors SLAM and CD46 by surface plasmon resonance analysis. For this analysis, MV-H proteins were purified as a soluble form (residues 149–617) that contains the head domain together with part of the stalk region containing a disulfide bond-forming cysteine residue at position 154. The receptors were also prepared as soluble molecules. Surface plasmon resonance analysis indicated that Ed-H and mutants bind to SLAM with similar affinities (Table 1). I-2D2A and II-4D also bound to CD46 as efficiently as Ed-H, whereas I-6A and I-2D1R exhibited four to six times higher Kd values as compared with Ed-H. Although some alterations were observed, none of the mutations introduced at the dimer-dimer interfaces of the head domain greatly affected the cell surface expression, receptor binding, and interaction with the F protein of MV-H.
FIGURE 2.
Flow cytometry analysis of mutant MV-H proteins. HEK293T cells were transfected with an empty vector (black) or expression plasmids encoding Ed-H (blue) or respective mutant H proteins (pink) and examined for the cell surface expression of MV-H. The numbers next to the peaks indicate the mean fluorescence intensities of respective samples.
FIGURE 3.
Co-immunoprecipitation of Ed-H or mutant MV-H proteins with the MV-F protein in transiently transfected CHO/vv5–4 cells. The F protein is FLAG-tagged, and Ed-H and all mutant H proteins are His-tagged. CHO/vv5–4 cell lysates were immunoprecipitated (IP) with anti-FLAG M2 monoclonal antibody and immunoblotted with an anti-His tag rabbit polyclonal antibody as described under “Experimental Procedures.” The signal intensities of IP samples were quantified by using a VersaDoc 3000 imager, divided by those of the input samples, and indicated below each band.
TABLE 1.
Surface plasmon resonance analysis of interactions of mutant MV-H proteins with SLAM or CD46
Analyte, protein injected in solution; ligand, protein immobilized on CM5 chip.
| Ligand | Analyte | Kd |
|---|---|---|
| μm | ||
| SLAM | Ed-H | 0.17 |
| I-6A | 0.14 | |
| I-2D2A | 0.19 | |
| II-4D | 0.23 | |
| I-2D1R | 0.18 | |
| CD46 | Ed-H | 1.28 |
| I-6A | 7.73 | |
| I-2D2A | 2.24 | |
| II-4D | 1.15 | |
| I-2D1R | 5.04 |
Fusion Support Activity of MV-H Mutants
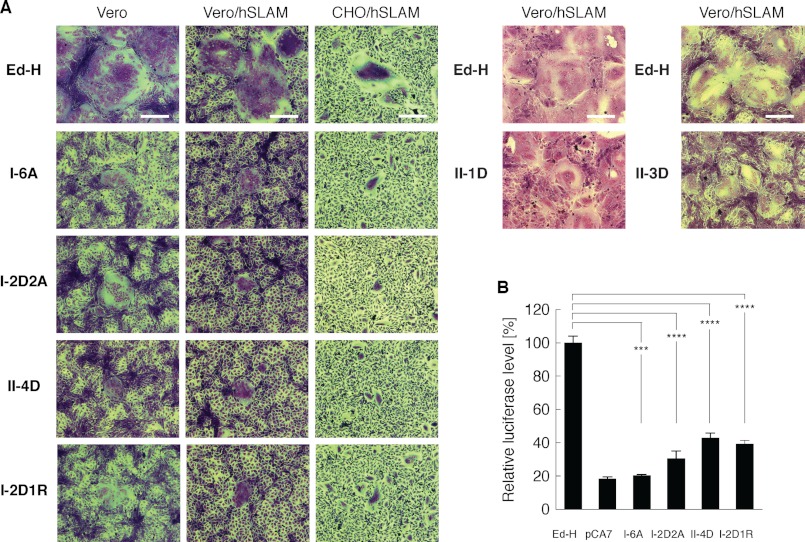
We then examined whether mutations in the putative dimer-dimer interfaces of the head domain affect fusion support activity of MV-H. The mutant H proteins were transiently expressed, together with the F protein, in Vero (CD46+), Vero/hSLAM (CD46+, SLAM+), and CHO/hSLAM (SLAM+) cells (22). II-1D and II-3D were examined only in Vero/hSLAM cells. The cells transfected with any of the mutant H proteins produced smaller sizes of syncytia, compared with those transfected with Ed-H (Fig. 4A). II-1D and II-3D still induced relatively large syncytia, but the combined mutant II-4D induced highly reduced sizes of syncytia like other form I mutants. A quantitative fusion assay also revealed that all of I-6A, I-2D2A, II-4D, and I-2D1R exhibit reduced fusion support activity (Fig. 4B). The results indicate that MV-H exhibits decreased fusion support activity, when its putative dimer-dimer interface of the head domain in form I or II is altered. Reduction in CD46 binding (Table 1) may explain why I-6A and I-2D1R did not efficiently support fusion in Vero cells, but it does not explain their inability to support efficient fusion in Vero/hSLAM and CHO/hSLAM cells. Thus, the decrease in receptor binding cannot be the main reason for the reduced ability of these H protein mutants to support membrane fusion.
FIGURE 4.
Fusion support activity of mutant MV-H proteins. A, Vero, Vero/hSLAM, and CHO/hSLAM cells were transfected with expression plasmids encoding full-length Ed-H or its mutant H proteins (2 μg) plus that encoding MV-F (2 μg). At 16 (CHO/hSLAM) or 36 h (Vero and Vero/hSLAM) after transfection, the cells were subjected to Giemsa staining and observed under a light microscope. Scale bars, 200 μm. B, quantitative fusion assay of mutant MV-H proteins. The levels of cell-cell fusion were determined as described under “Experimental Procedures.” The fusion level of the cells transfected with the expression plasmid encoding Ed-H was set to 100%. pCA7, an empty vector used as a negative control. The data represent the means ± S.D. of triplicate samples. Compared with cells expressing Ed-H, those expressing I-6A, I-2D2A, II-4D, and I-2D1R showed significantly reduced cell-cell fusion (t test, p = 0.00088, 0.000041, 0.000038, and 0.000093, respectively; ****, p < 0.0001; ***, p < 0.001. The alpha level for all tests was defined as 0.05).
Blue Native-PAGE Analysis of MV-H Structure
We also assessed MV-H oligomerization using blue native-PAGE analysis. The full-length MV-H has been shown to exhibit a tetramer formation, when expressed in cells (13, 14). However, it is unknown whether the MV-H head domain alone assumes a tetrameric structure. Therefore, we first examined whether the MV-H head domain can oligomerize upon expression in cells. For this analysis, we used the soluble form of MV-H (residues 149–617), the ectodomain that we had utilized for our crystallization of MV-H bound or unbound to SLAM (13, 18). The soluble form of Ed-H exhibited three discrete bands migrating at ∼400, 200, and 100 kDa on blue native-PAGE (Fig. 5A). It is known that there is a discrepancy between the calculated molecular weight of a protein and its mobility on blue native-PAGE (14), and therefore we assumed that the three bands on blue native-PAGE represent tetramers, dimers, and monomers of the soluble MV-H, respectively. Among the three forms, dimers were predominant, consistent with the crystal structure of the NDV HN ectodomain, in which the head domain dimers were shown not to associate with each other to form tetramers (17). Importantly, tetramers were also detected albeit at a lower level, indicating that tetramers of the MV-H-SLAM complex were not artifacts in crystallization. All of the MV-H mutants also exhibited the three forms, with the dimers most abundant. Notably, I-2D2A exhibited a reduced level of tetramers, but the other three mutants still formed almost comparable levels of tetramers to Ed-H.
FIGURE 5.
Blue native-PAGE and immunoblot analysis of mutant MV-H proteins. A, blue native-PAGE analysis of soluble forms of Ed-H and its mutant H proteins. B, blue native-PAGE analysis of full-length MV-H proteins. Lysates from HEK293S GnTI(−) cells that were transiently transfected with plasmids encoding the parental Ed-H or its mutant H proteins and then treated with digitonin (Dig) or n-dodecyl-β-d-maltoside (DDM) were subjected to blue native-PAGE and immunoblot analysis with anti-MV-H antibody. The numbers (kDa) indicate the migration pattern of a native protein standard (A and B).
On the other hand, the full-length Ed-H transiently expressed in cells predominantly produced tetramers after solubilization with digitonin (Fig. 5B). When n-dodecyl-β-d-maltoside, a harsher detergent, was used for solubilization, the dimer form of MV-H emerged. These results are consistent with those reported previously (14). All of the full-length MV-H mutants, including I-2D2A whose soluble form hardly formed tetramers, also produced tetramers. The results indicate that the stalk and transmembrane regions are sufficient for tetramer formation, in agreement with previous reports on tetramer formation of paramyxovirus attachment proteins (17, 28, 29).
DISCUSSION
How receptor binding by a paramyxovirus attachment protein induces conformational changes of the associated F protein to trigger membrane fusion is not well understood. In this study, we generated MV-H mutants, in which mutations were introduced in the putative dimer-dimer interfaces of head domain tetramers. They were all expressed on the cell surface, bound to cellular receptors (SLAM and CD46), and associated with the F protein, almost like the parental H protein, yet they failed to support membrane fusion efficiently. These results indicate that the dimer-dimer interactions of the MV-H head domain play a role in triggering membrane fusion.
X-ray crystallography has revealed the MV-H head domain tetramer (dimer of dimers) that occurs in two forms. The finding that mutations in the putative dimer-dimer interface in either form inhibits the ability of MV-H to support membrane fusion indicates that both tetramer forms are involved in fusion triggering. Although more information, including the structure of the entire MV-H ectodomain, is required, one interpretation of our results is that upon receptor binding, conformational changes of the MV-H head domain occur involving both forms I and II, which in turn trigger the activation of the F protein. Because structural rearrangements of the H stalk of MV and the closely related canine distemper virus are shown to trigger the activation of the F protein (7, 30, 31), the conformational changes of MV-H head domain tetramers must be transmitted to the stalk to trigger membrane fusion (Fig. 1A).
On the other hand, the recently reported crystal structure of the NDV HN ectodomain did not reveal the contact between two head domain dimers (17). Our blue native-PAGE analysis also showed that the soluble MV-H (the head domain plus part of the stalk) predominantly forms dimers. However, tetramers were also present albeit at a lower level, indicating that the head domain alone can form tetramers. It should be noted that the soluble MV-H head domain alone exhibited homodimer in crystal structures, as well as in gel filtration chromatography (18), whereas the MV-H head domain in complex with its receptor SLAM formed tetramer (13). Therefore, it is possible that receptor binding facilitates and/or stabilizes tetramer formation of the MV-H head domain. Tetramers thus formed may undergo further conformational changes involving both forms I and II, as described above.
Furthermore, there are structural and functional differences between MV-H and HN proteins of other paramyxoviruses using sialic acid-bearing receptors (including NDV and PIV5). First, the arrangement of the MV-H head domain is somehow different from that of the NDV HN protein, including relative positions of β-sheets and orientation of monomers forming a dimer (18). Second, MV-H binds proteinaceous receptors at the lateral surface of the β-propeller fold, whereas HN proteins bind sialic acid at the top (13, 18, 32, 33). Third, receptor binding by MV-H is thought to release the F protein from the preassembled H-F oligomers, thereby allowing spontaneous conformational changes of the F protein (2–5), whereas HN proteins of sialic acid receptor-using paramyxoviruses, upon receptor binding, appear to cause the conformational change of the F protein by actively acting on it (3, 6). Thus, it is possible that the fusion-triggering mechanism is also different between MV and sialic acid receptor-using paramyxoviruses. This contention may be consistent with a recent report that the receptor-binding head domain of the PIV5 HN protein is entirely dispensable for the activation of the F protein (34).
Although the cell surface expression, receptor binding, and interaction with the F protein were not greatly affected, the mutations introduced in the putative dimer-dimer interfaces of the H protein head domain may have disrupted membrane fusion for unknown reasons other than their effects on the ability of the H protein to trigger membrane fusion. For example, these mutations may have affected the structure/function of the H protein such that our assays could not detect its defects in expression, receptor binding, and/or interaction with the F protein. However, the importance of the dimer-dimer interactions of MV-H head domain tetramers in fusion triggering is also supported by other observations. First, Brindley et al. (31) demonstrated using transcomplementation experiments that receptor binding to only one dimer of the MV-H head domain dimer of dimers can induce F protein triggering mediated by the stalks of the other dimer. The results suggest that receptor binding and F protein triggering could be communicated across two MV-H dimers, either at the head domain or at the stalk region. Second, anti-MV-H neutralizing monoclonal antibodies I-29 and BH38 were found to be directed to the region around the dimer-dimer interface in form I rather than receptor-binding sites (13, 18, 35, 36). Escape mutants from I-29 possessed the substitutions at position 313 or 314 of MV-H (36), whereas those from BH38 had substitutions at position 296 or 310 (35). It is likely that these antibodies exert their neutralizing activity by affecting the dimer-dimer interaction of MV-H. Third, an asparagine at position 53 of SLAM, an N-linked glycosylation site, is located at the interface between SLAM and MV-H monomer only in form II, and an asparagine to glutamine substitution at this position greatly affects MV entry and syncytium formation (13). This substitution may facilitate stable formation of form II by removing carbohydrates between SLAM and MV-H in form II, thereby facilitating fusion triggering.
Our present results with MV-H mutants, together with these previous observations by us and others, strongly indicate that the dimer-dimer interactions of MV-H head domain play an essential role in triggering membrane fusion. Presumably, the tetramer formation and subsequent conformational shift (involving the dimer-dimer interfaces) of the MV-H head domain that may occur upon receptor binding would induce structural rearrangements of the stalk region, which in turn cause conformational changes of the F protein.
Acknowledgments
We thank K. Maenaka, S. Watanabe, M. Takeda, and S. Ohno for discussion; T. Saitoh and D. Kohda for technical advice; and M. B. A. Oldstone for reagents.
This study was supported by grants from the Ministry of Health, Labor and Welfare of Japan and by Grants-in-Aid for Scientific Research 21249032 and 24115005 from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
- MV
- measles virus
- NDV
- Newcastle disease virus
- HN
- hemagglutinin-neuraminidase
- H protein
- hemagglutinin protein
- F protein
- fusion protein
- SLAM
- signaling lymphocyte activation molecule
- PIV
- parainfluenza virus
- Ed-H
- H protein of MV Edmonston strain.
REFERENCES
- 1. Lamb R. A., Parks G. D. (2007) Paramyxoviridae: The viruses and their replication, in Fields Virology (Knipe D. M., Howley P. M., Griffin D. E., Lamb R. A., Martin M. A., Roizman B., Straus S. E., eds) 5th Ed., pp. 1449–1496, Lippincott Williams & Wilkins, Philadelphia [Google Scholar]
- 2. Iorio R. M., Mahon P. J. (2008) Paramyxoviruses. Different receptors-different mechanisms of fusion. Trends. Microbiol. 16, 135–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Smith E. C., Popa A., Chang A., Masante C., Dutch R. E. (2009) Viral entry mechanisms. The increasing diversity of paramyxovirus entry. FEBS J. 276, 7217–7227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Plemper R. K., Brindley M. A., Iorio R. M. (2011) Structural and mechanistic studies of measles virus illuminate paramyxovirus entry. PLoS Pathog. 7, e1002058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee B., Ataman Z. A. (2011) Modes of paramyxovirus fusion. A Henipavirus perspective. Trends. Microbiol. 19, 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Connolly S. A., Leser G. P., Jardetzky T. S., Lamb R. A. (2009) Bimolecular complementation of paramyxovirus fusion and hemagglutinin-neuraminidase proteins enhances fusion. Implications for the mechanism of fusion triggering. J. Virol. 83, 10857–10868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Navaratnarajah C. K., Negi S., Braun W., Cattaneo R. (2012) Membrane fusion triggering. Three modules with different structure and function in the upper half of the measles virus attachment protein stalk. J. Biol. Chem. 287, 38543–38551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bose S., Welch B. D., Kors C. A., Yuan P., Jardetzky T. S., Lamb R. A. (2011) Structure and mutagenesis of the parainfluenza virus 5 hemagglutinin-neuraminidase stalk domain reveals a four-helix bundle and the role of the stalk in fusion promotion. J. Virol. 85, 12855–12866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Melanson V. R., Iorio R. M. (2006) Addition of N-glycans in the stalk of the Newcastle disease virus HN protein blocks its interaction with the F protein and prevents fusion. J. Virol. 80, 623–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tanabayashi K., Compans R. W. (1996) Functional interaction of paramyxovirus glycoproteins. Identification of a domain in Sendai virus HN which promotes cell fusion. J. Virol. 70, 6112–6118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang Z., Mirza A. M., Li J., Mahon P. J., Iorio R. M. (2004) An oligosaccharide at the C-terminus of the F-specific domain in the stalk of the human parainfluenza virus 3 hemagglutinin-neuraminidase modulates fusion. Virus. Res. 99, 177–185 [DOI] [PubMed] [Google Scholar]
- 12. Griffin D. E. (2007) Measles virus, in Fields Virology (Knipe D. M., Howley P. M., Griffin D. E., Lamb R. A., Martin M. A., Roizman B., Straus S. E., eds), 5th Ed., pp. 1551–1585, Lippincott Williams & Wilkins, Philadelphia [Google Scholar]
- 13. Hashiguchi T., Ose T., Kubota M., Maita N., Kamishikiryo J., Maenaka K., Yanagi Y. (2011) Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM. Nat. Struct. Mol. Biol. 18, 135–141 [DOI] [PubMed] [Google Scholar]
- 14. Brindley M. A., Plemper R. K. (2010) Blue native PAGE and biomolecular complementation reveal a tetrameric or higher-order oligomer organization of the physiological measles virus attachment protein H. J. Virol. 84, 12174–12184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yuan P., Thompson T. B., Wurzburg B. A., Paterson R. G., Lamb R. A., Jardetzky T. S. (2005) Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure 13, 803–815 [DOI] [PubMed] [Google Scholar]
- 16. Zaitsev V., von Itzstein M., Groves D., Kiefel M., Takimoto T., Portner A., Taylor G. (2004) Second sialic acid binding site in Newcastle disease virus hemagglutinin-neuraminidase. Implications for fusion. J. Virol. 78, 3733–3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yuan P., Swanson K. A., Leser G. P., Paterson R. G., Lamb R. A., Jardetzky T. S. (2011) Structure of the Newcastle disease virus hemagglutinin-neuraminidase (HN) ectodomain reveals a four-helix bundle stalk. Proc. Natl. Acad. Sci. U.S.A. 108, 14920–14925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hashiguchi T., Kajikawa M., Maita N., Takeda M., Kuroki K., Sasaki K., Kohda D., Yanagi Y., Maenaka K. (2007) Crystal structure of measles virus hemagglutinin provides insight into effective vaccines. Proc. Natl. Acad. Sci. U.S.A. 104, 19535–19540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yanagi Y., Cubitt B. A., Oldstone M. B. (1992) Measles virus inhibits mitogen-induced T cell proliferation but does not directly perturb the T cell activation process inside the cell. Virology 187, 280–289 [DOI] [PubMed] [Google Scholar]
- 20. Bair C. H., Chung C. S., Vasilevskaya I. A., Chang W. (1996) Isolation and characterization of a Chinese hamster ovary mutant cell line with altered sensitivity to vaccinia virus killing. J. Virol. 70, 4655–4666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fuerst T. R., Niles E. G., Studier F. W., Moss B. (1986) Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 83, 8122–8126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ono N., Tatsuo H., Hidaka Y., Aoki T., Minagawa H., Yanagi Y. (2001) Measles viruses on throat swabs from measles patients use signaling lymphocytic activation molecule (CDw150) but not CD46 as a cellular receptor. J. Virol. 75, 4399–4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reeves P. J., Callewaert N., Contreras R., Khorana H. G. (2002) Structure and function in rhodopsin. High-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc. Natl. Acad. Sci. U.S.A. 99, 13419–13424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McFarlin D. E., Bellini W. J., Mingioli E. S., Behar T. N., Trudgett A. (1980) Monospecific antibody to the haemagglutinin of measles virus. J. Gen. Virol. 48, 425–429 [DOI] [PubMed] [Google Scholar]
- 25. Lee J. K., Prussia A., Paal T., White L. K., Snyder J. P., Plemper R. K. (2008) Functional interaction between paramyxovirus fusion and attachment proteins. J. Biol. Chem. 283, 16561–16572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Paal T., Brindley M. A., St Clair C., Prussia A., Gaus D., Krumm S. A., Snyder J. P., Plemper R. K. (2009) Probing the spatial organization of measles virus fusion complexes. J. Virol. 83, 10480–10493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ennis M. K., Hu C., Naik S. K., Hallak L. K., Peng K. W., Russell S. J., Dingli D. (2010) Mutations in the stalk region of the measles virus hemagglutinin inhibit syncytium formation but not virus entry. J. Virol. 84, 10913–10917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McGinnes L., Sergel T., Morrison T. (1993) Mutations in the transmembrane domain of the HN protein of Newcastle disease virus affect the structure and activity of the protein. Virology 196, 101–110 [DOI] [PubMed] [Google Scholar]
- 29. Yuan P., Leser G. P., Demeler B., Lamb R. A., Jardetzky T. S. (2008) Domain architecture and oligomerization properties of the paramyxovirus PIV 5 hemagglutinin-neuraminidase (HN) protein. Virology 378, 282–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ader N., Brindley M. A., Avila M., Origgi F. C., Langedijk J. P., Örvell C., Vandevelde M., Zurbriggen A., Plemper R. K., Plattet P. (2012) Structural rearrangements of the central region of the morbillivirus attachment protein stalk domain trigger F protein refolding for membrane fusion. J. Biol. Chem. 287, 16324–16334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brindley M. A., Takeda M., Plattet P., Plemper R. K. (2012) Triggering the measles virus membrane fusion machinery. Proc. Natl. Acad. Sci. U.S.A. 109, E3018–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Santiago C., Celma M. L., Stehle T., Casasnovas J. M. (2010) Structure of the measles virus hemagglutinin bound to the CD46 receptor. Nat. Struct. Mol. Biol. 17, 124–129 [DOI] [PubMed] [Google Scholar]
- 33. Crennell S., Takimoto T., Portner A, Taylor G. (2000) Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat. Struct. Biol. 7, 1068–1074 [DOI] [PubMed] [Google Scholar]
- 34. Bose S., Zokarkar A., Welch B. D., Leser G. P., Jardetzky T. S., Lamb R. A. (2012) Fusion activation by a headless parainfluenza virus 5 hemagglutinin-neuraminidase stalk suggests a modular mechanism for triggering. Proc. Natl. Acad. Sci. U.S.A. 109, E2625–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bouche F. B., Ertl O. T., Muller C. P. (2002) Neutralizing B cell response in measles. Viral. Immunol. 15, 451–471 [DOI] [PubMed] [Google Scholar]
- 36. Hu A., Sheshberadaran H., Norrby E., Kövamees J. (1993) Molecular characterization of epitopes on the measles virus hemagglutinin protein. Virology 192, 351–354 [DOI] [PubMed] [Google Scholar]