Background: DOCK1 is an atypical Rac activator.
Results: Activation of the PDGF receptor induces DOCK1 translocation to the dorsal ruffles through association with phosphatidic acid. Blocking of this interaction impairs dorsal, but not peripheral, ruffle formation.
Conclusion: Phosphatidic acid acts as a lipid anchor for DOCK1 during dorsal ruffle formation.
Significance: A novel regulatory mechanism for dorsal ruffle formation was identified.
Keywords: Cell Signaling, Gene Knock-out, Guanine Nucleotide Exchange Factor (GEF), Lipid-binding Protein, Phosphatidic Acid, Phospholipase D, Rac, DOCK Proteins, Localization, Membrane Ruffle
Abstract
Activation of receptor tyrosine kinases leads to the formation of two different types of plasma membrane structures: peripheral ruffles and dorsal ruffles. Although the formation of both ruffle types requires activation of the small GTPase Rac, the difference in kinetics suggests that a distinct regulatory mechanism operates for their ruffle formation. DOCK1 and DOCK5 are atypical Rac activators and are both expressed in mouse embryonic fibroblasts (MEFs). We found that although PDGF-induced Rac activation and peripheral ruffle formation were coordinately regulated by DOCK1 and DOCK5 in MEFs, DOCK1 deficiency alone impaired dorsal ruffle formation in MEFs. Unlike DOCK5, DOCK1 bound to phosphatidic acid (PA) through the C-terminal polybasic amino acid cluster and was localized to dorsal ruffles. When this interaction was blocked, PDGF-induced dorsal ruffle formation was severely impaired. In addition, we show that phospholipase D, an enzyme that catalyzes PA synthesis, is required for PDGF-induced dorsal, but not peripheral, ruffle formation. These results indicate that the phospholipase D-PA axis selectively controls dorsal ruffle formation by regulating DOCK1 localization.
Introduction
Activation of receptor tyrosine kinases (RTKs)3 by growth factors leads to the formation of actin-based structures known as membrane ruffles. Membrane ruffles are highly dynamic and F-actin-rich structures and are classified into two types: peripheral ruffles and dorsal ruffles, according to their morphology and localization (1). Peripheral ruffles are formed by the bending upward of the leading edge and are associated with cell motility (2). On the other hand, dorsal ruffles are the unique membrane structures characterized by a ring-shaped morphology and are thought to play important roles in macropinocytosis, internalization of cell surface receptors, and three-dimensional migration and invasion (3–5). The formation of both peripheral and dorsal ruffles requires activation of the small GTPase Rac (5–7). Notably, the kinetics of these structures are not identical with peripheral ruffles being formed continuously upon stimulation with growth factors, whereas dorsal ruffles typically do not form until minutes after stimulation, and their formation is more transient (1, 5, 8, 9). Therefore, the mechanism controlling activation and localization of Rac may be distinct between peripheral and dorsal ruffle formation.
Like other small GTPases, Rac functions as a molecular switch by cycling between GDP-bound inactive and GTP-bound active states, and stimulus-induced formation of the active Rac is mediated by guanine nucleotide exchange factors (GEFs) (10). There are two distinct families of Rac GEFs: Dbl homology domain-containing proteins and DOCK proteins (10, 11). Until recently, Dbl homology domain containing proteins have been considered to be the universal GEFs in eukaryotes. However, accumulating evidence indicates that the DOCK proteins act as major Rac GEFs in varied biological settings. For example, DOCK2, which is predominantly expressed in hematopoietic cells, plays key roles in migration and activation of leukocytes (12–17). On the other hand, DOCK1 (also known as DOCK180) and DOCK5 are widely expressed in various tissues and are known to regulate phagocytosis, cell migration, axon guidance, and/or myoblast fusion (18–22). Although these molecules do not contain Dbl homology domain, they mediate the GTP-GDP exchange reaction for Rac through the DOCK homology region 2 (DHR-2) (also know as CZH2 or Docker) domain (11, 23, 24).
Upon stimulation with growth factors, Rac is preferentially activated in peripheral and dorsal ruffles (2, 9), which is likely to be the result of localization of Rac GEFs. The Rac GEFs contain a variety of localization motifs such as pleckstrin homology (PH) domain and DHR-1 domain, both of which bind to phosphatidylinositol 3,4,5-trisphosphate (PIP3) (10, 25, 26), a lipid product of PI3Ks. PI3Ks have important roles in membrane ruffle formation probably by regulating local concentration of PIP3 (1, 2). However, PIP3 production alone cannot explain the difference in kinetics of peripheral and dorsal ruffle formation. On the other hand, phosphatidic acid (PA) is a negatively charged phospholipid that can function as a lipid anchor by binding directly to positively charged sites on effector proteins (27). In response to growth factors, PA is partly generated through hydrolysis of phosphatidylcholine (PC) by phospholipase D (PLD) (28, 29). Although the activity of PLD has been implicated in growth factor-induced macropinocytosis (30), the underlying mechanism remains largely unknown.
By analyzing mouse embryonic fibroblasts (MEFs) deficient in DOCK1 and/or DOCK5, we found that these GEFs have different roles in PDGF-induced peripheral and dorsal ruffle formation. In this study, we provide evidence that the PLD-PA axis selectively controls dorsal ruffle formation by regulating DOCK1 localization.
EXPERIMENTAL PROCEDURES
Mice
Mice selectively lacking DOCK1 SH3 domain (D1d/d) have been generated by crossing mice with a DOCK1 allele containing floxed exon 2 with EIIa-Cre transgenic mice, as previously reported (21). DOCK5-deficient (D5−/−) mice, PLD1-deficient (PLD1−/−) mice, and PLD2-deficient (PLD2−/−) mice have been described elsewhere (22, 31, 32). Mice homozygous for DOCK1 alleles containing floxed exon 1 (DOCK1flox/flox) will be described elsewhere. All animals were maintained in specific pathogen-free conditions in the animal facility of Kyushu University, and experiments were done in accordance with the guidelines of the Committee of Ethics of Animal Experiments of Kyushu University.
Reagents
l-α-PA was obtained from Avanti Polar Lipid. l-α-PC, l-α-phosphatidylethanolamine (PE), l-α-phosphatidylserine (PS), fibronectin, and 5-fluoro-2-indolyl des-clorohalopemide (FIPI) (33) were obtained from Sigma-Aldrich. PIP3- or phosphatidylinositol 3,4-bisphosphate (PI(3,4)P2)-coated beads were obtained from Echelon Biosciences. Alexa Fluor 488-conjugated anti-GFP antibody, and Alexa Fluor 546- or 647-conjugated phalloidin were obtained from Invitrogen. Anti-GST (B-14), anti-DOCK1 (C-19), and anti-β-actin (I-19) antibodies were obtained from Santa Cruz Biotechnology. Anti-Rac antibody (23A8) was obtained from Millipore. PDGF-BB was obtained from Peprotech. Polyclonal anti-DOCK5 antibody was produced by immunization of rabbits with peptides corresponding to the C-terminal portion (residue 1853–1868) of mouse DOCK5.
Plasmids
For expression of N-terminally tagged GFP fusion proteins and mCherry in mammalian cells, pCI-GFP and pCI-mCherry vectors were created by subcloning a cDNA encoding EGFP or mCherry (Clontech) into pCI (Promega), respectively. The cDNA encoding the WT or mutant DOCK1 or DOCK5 or the PH domain of SOS (residues 423–551) was subcloned into pCI-GFP. The pCI vector encoding DOCK1 with the HA tag at its C terminus has been created by PCR. The pcDNA vector encoding V5-tagged ELMO1 has been described previously (34). The retroviral vector expressing Cre recombinase was created using pMxs-IRES-GFP (35). To expresses the DOCK1 chimera molecule encoding the C-terminal region of DOCK5 (designated D1/5C), the Dock1 gene corresponding to the C-terminal region (residues 1553–1865) was replaced with the Dock5 gene encoding amino acid residues 1578–1868 in pCI-GFP. Site-directed mutagenesis was performed using the method of inverse PCR.
To bacterially express recombinant proteins encoding GST at the N terminus, the cDNA encoding the C-terminal region of DOCK1 (residues 1610–1865), DOCK5 (residues 1634–1868), lactadherin-C2 (residues 277–463), or Akt-PH (residues 1–167) was subcloned into pGEX-6P vector (GE Healthcare). Recombinant proteins were expressed in ArcticExpress bacterial strain (Agilent) and then purified with glutathione-Sepharose 4B (GE Healthcare) according to the manufacturer's instructions.
Cell Culture and Transfection
For preparation of MEFs, embryonic day 13.5 embryos were trypsinized and cultured in DMEM supplemented with nonessential amino acids, sodium pyruvate, penicillin-streptomycin, and 10% FCS (complete DMEM). MEFs were transfected with WT or mutant DOCK1 or DOCK5 constructs by using Nucleofector kit V (Lonza), except for pCI-GFP vector encoding the C-terminal fragment of DOCK1 (residues 1612–1865). For transfection of this plasmid and pCI-GFP SOS PH into MEFs, Lipofectamine 2000 (Invitrogen) was used. Recombinant retrovirus was prepared as previously described (17) and used to infect DOCK1flox/flox MEFs. Twenty-four hours after the last infection, cells were washed with complete DMEM and cultured for 96 h for PDGF stimulation. Transient transfection into HEK293T cells was performed with polyethylenimine.
Pulldown Assay and Immunoblotting
For Rac activation assay, aliquots of the cell extracts were kept for total lysate controls, and the remaining extracts were incubated with GST fusion Rac-binding domain of PAK1 at 4 °C for 60 min. The bound proteins and the same amounts of total lysates were analyzed by SDS-PAGE. Immunoblotting was performed with relevant antibodies.
Immunofluorescence Microscopy
MEFs prepared from various knock-out mouse lines were seeded on fibronectin-coated glass-bottomed dish (Matsunami Glass). After serum starvation overnight, the cells were stimulated with PDGF (30 ng/ml) for the specified times. The cells were then fixed with 4% paraformaldehyde for 10 min, permeabilized with 0.2% Triton X-100-PBS for 5 min, blocked with 1% BSA-PBS for 1 h, and stained with Alexa Fluor 546-conjugated phalloidin. In some experiments, the cells were preincubated with FIPI (750 nm) (33) or dimethyl sulfoxide (DMSO) for 45 min before stimulation with PDGF (30 ng/ml).
For ratiometric analyses, the GFP fusion WT or mutant DOCK1 or DOCK5 constructs (3 μg) were coexpressed with pCI-mCherry (1 μg) in MEFs, and the cells were cultured on fibronectin-coated glass-bottomed dish in the presence of sodium butyrate (1.5 mm) for 48 h. After stimulation with PDGF (50 ng/ml) for 7 min, the cells were stained with Alexa Fluor 488-conjugated anti-GFP antibody and Alexa Fluor 647-conjugated phalloidin. The ratio images of GFP/mCherry were created with MetaMorph software (Molecular Devices) and were used to represent the efficiency of the ratio. For rescue experiments, the GFP fusion WT or mutant DOCK1 constructs (3 μg) were expressed in D1d/d MEFs before stimulation with PDGF (50 ng/ml). All of the samples were analyzed with a laser scanning confocal microscope (LSM510 META; Zeiss).
Lipid Binding Assay
For preparation of liposomes, a mixture of PC, PE, and specified concentration of PA or PS was dissolved in chloroform and dried in SpeedVac concentrator (Savant Instruments). The lipid powder was resuspended in TBS (20 mm Tris-HCl, 0.15 m NaCl, pH 7.5) and incubated at 37 °C for 30 min followed by vigorous vortexing for 10 min. Before use, liposomes were precipitated at 20,000 × g for 5 min and washed twice with TBS.
The GST-tagged recombinant proteins were mixed with liposomes (400 μg) or lipid-coated beads in TBS supplemented with Nonidet P-40 (final 0.005% for liposomes and 0.25% for lipid-coated beads) to make 1 ml of solution. The mixture was incubated for 1.5 h at room temperature and washed twice (for liposomes) or four times (for lipid-coated beads) with TBS containing 0.005% or 0.25% of Nonidet P-40 followed by centrifugation at 20,000 × g (for liposomes) or 300 × g (for lipid-coated beads) at 4 °C. The binding proteins were immunoblotted using anti-GST antibody.
To assay proteins expressed in HEK293T cells for lipid binding, cells transfected with the specified plasmid DNAs were suspended in 0.6 ml of TBS supplemented with 1 mm EDTA, 1 mm PMSF, and 0.2% Nonidet P-40, and subjected to a single freeze and thaw cycle in liquid nitrogen and a 37 °C water bath. Insoluble debris was removed by centrifugation at 20,000 × g for 15 min at 4 °C. The supernatants were diluted 40-fold with TBS containing 1 mm EDTA and 1 mm PMSF and then subjected to liposome binding assays at 4 °C as described above.
Measurement for PA Production
MEFs from WT and PLD1/2 double deficient (PLD1/2−/−) mice were labeled with 32Pi and stimulated with PDGF (30 ng/ml). There was no difference in 32Pi incorporation into phospholipids between them (data not shown). Cellular PA levels at the specified times were measured by thin layer chromatography as previously described (36).
Statistical Analyses
Unless stated otherwise, statistical analyses were performed by using two-tailed Student's t test.
RESULTS
Differential Role of DOCK1 and DOCK5 in Peripheral and Dorsal Ruffle Formation
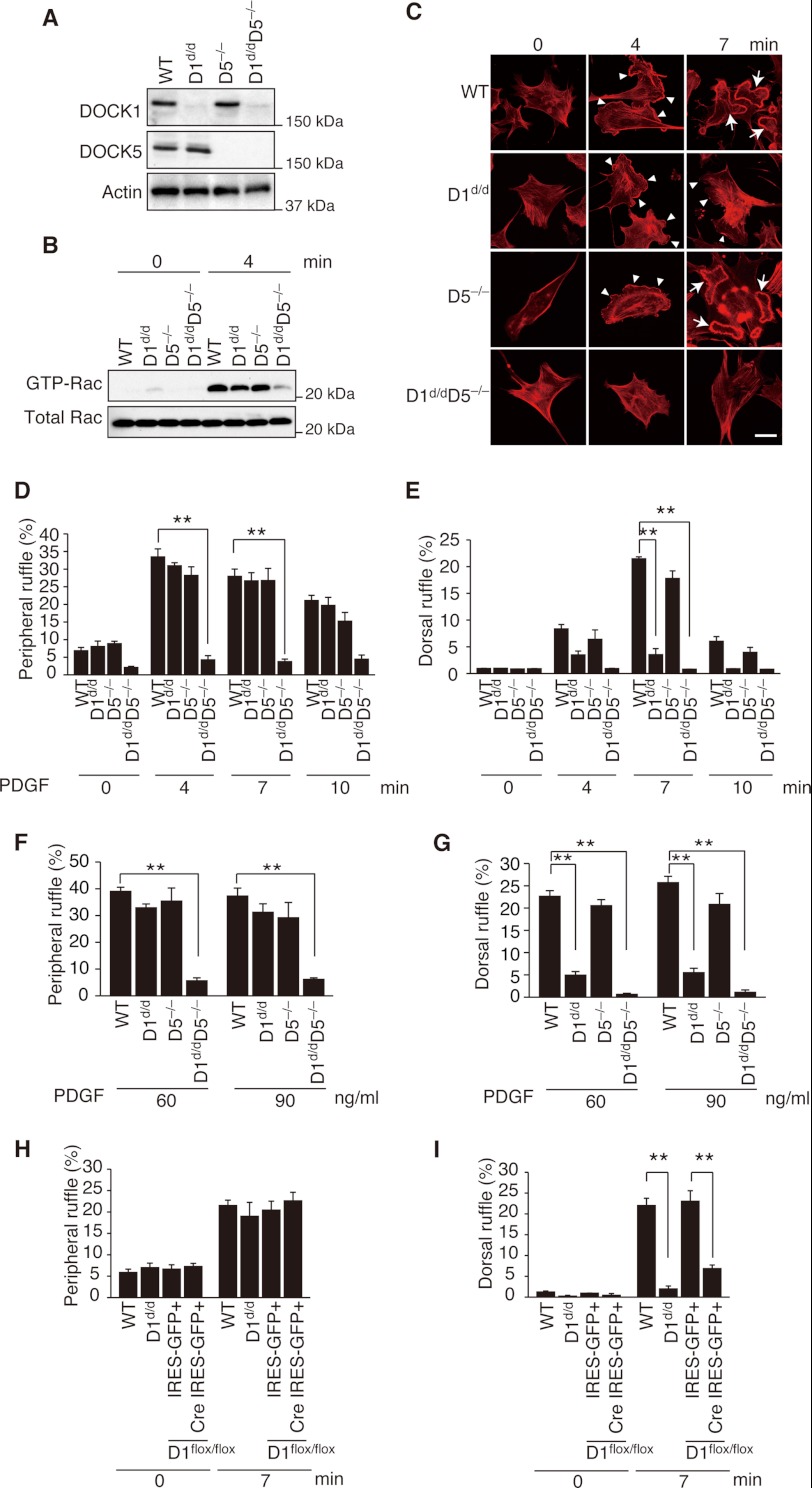
DOCK1 and DOCK5 are members of the atypical Rac GEFs (11), both of which are expressed in MEFs (Fig. 1A). To examine the role of DOCK1 and DOCK5 in membrane ruffle formation under physiological conditions, we prepared MEFs from mice lacking DOCK1 SH3 domain (D1d/d) and DOCK5-deficient (D5−/−) mice (21, 22). DOCK1 interacts with ELMO through the N-terminal region containing SH3 domain and a putative α-helical region (18, 37). Because this interaction is required to inhibit ubiquitination and degradation of DOCK1 (21), DOCK1 expression was markedly reduced in D1d/d MEFs when analyzed by Western blotting (Fig. 1A).
FIGURE 1.
Differential role of DOCK1 and DOCK5 in RTK-mediated peripheral and dorsal ruffle formation. A, immunoblot analysis for DOCK1 and DOCK5 expression in MEFs prepared from WT and mutant mice. Actin was used as a loading control. B, activation of Rac was analyzed for MEFs from WT and mutant mice before and after stimulation with PDGF. The data are representative of two independent experiments. C, after stimulation with PDGF (30 ng/ml) for the indicated times, WT and mutant MEFs were stained with phalloidin. Arrowheads and arrows indicate peripheral ruffles and dorsal ruffles, respectively. Scale bar, 50 μm. D and E, summaries for PDGF-induced peripheral (D) and dorsal (E) ruffle formation in C. The data are expressed as percentages of cells with ruffle formation (means ± S.E.) and are from three separate experiments. In each experiment, more than 100 cells were analyzed. Statistical analysis was performed by using analysis of variance followed by Bonferroni correction. **, p < 0.01. F and G, after stimulation with PDGF at 60 or 90 ng/ml for 4 (F) or 7 (G) minutes, WT and mutant MEFs were stained with phalloidin. Summaries for PDGF-induced peripheral (F) and dorsal (G) ruffle formation are shown. The data were analyzed as described in D and E. **, p < 0.01. H and I, following retroviral transduction of Cre IRES-GFP or IRES-GFP alone, DOCK1flox/flox MEFs were stimulated with PDGF (30 ng/ml) for 7 min and stained with phalloidin. WT and D1d/d MEFs without transfection were used as controls. Summaries for PDGF-induced peripheral (H) and dorsal (I) ruffle formation are shown. **, p < 0.01.
We first examined the role of DOCK1 or DOCK5 in RTK-mediated Rac activation. In response to PDGF stimulation, Rac was activated in D1d/d and D5−/− MEFs, as well as in WT MEFs (Fig. 1B). In contrast, PDGF-induced Rac activation was severely impaired in MEFs lacking the expression of both DOCK1 and DOCK5 (D1d/dD5−/−; Fig. 1B). Consistent with this finding, PDGF-induced peripheral ruffle formation occurred normally in D1d/d MEFs and D5−/− MEFs but was almost completely lost in D1d/dD5−/− MEFs (Fig. 1, C, D, and F). These results indicate that in MEFs, DOCK1 and DOCK5 are major Rac GEFs acting downstream of PDGF RTK and regulate coordinately peripheral ruffle formation. Surprisingly, however, PDGF-induced dorsal ruffle formation was severely impaired in D1d/d MEFs at any concentrations tested (Fig. 1, C, E, and G), despite the normal expression of DOCK5 (Fig. 1A). To further confirm this finding, we used MEFs from DOCK1flox/flox mice. A similar defect was observed in DOCK1flox/flox MEFs when Dock1 gene expression was abrogated by retrovirally expressing Cre recombinase (Fig. 1, H and I).
DOCK1, but Not DOCK5, Accumulates at the Dorsal Ruffle Membrane Depending on the C-terminal Region
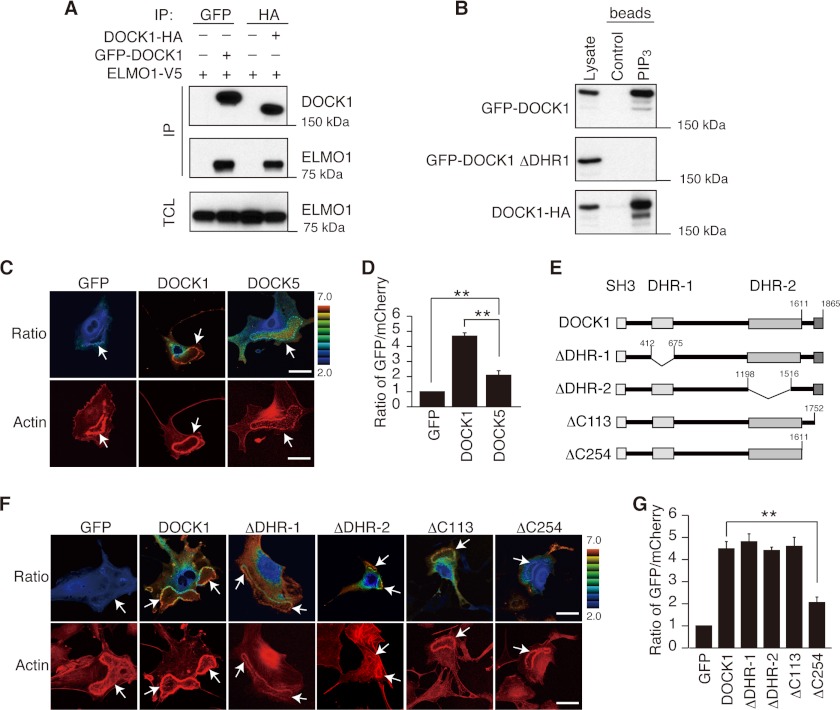
Having found that DOCK1, but not DOCK5, is required for PDGF-induced dorsal ruffle formation in MEFs, we next compared localization of these Rac GEFs by expressing as the N-terminal GFP fusion proteins. We confirmed that the presence of an N-terminal GFP does not interfere with binding to ELMO and PIP3 (Fig. 2, A and B). When GFP fusion DOCK1 (DOCK1-GFP) was expressed in WT MEFs, it readily accumulated at the dorsal ruffle membrane in response to PDGF stimulation (Fig. 2, C and D). However, such localization was scarcely found with DOCK5-GFP (Fig. 2, C and D), raising the possibility that DOCK1 may contain a special localization motif that targets it to dorsal ruffle membrane.
FIGURE 2.
DOCK1, but not DOCK5, accumulates at the dorsal ruffle membrane in response to PDGF stimulation. A, following expression of the N-terminally GFP-tagged DOCK1 and the C-terminally HA-tagged DOCK1 in HEK293T cells in combination with V5-tagged ELMO1, cell extracts were immunoprecipitated (IP) with anti-GFP antibody or anti-HA antibody. Immunoblotting was carried out with anti-DOCK1 antibody or anti-V5 antibody. TCL, total cell lysate. B, following expression of the N-terminally GFP-tagged WT or mutant DOCK1 or the C-terminally HA-tagged DOCK1 in HEK293T cells, cell extracts were pulled down with PIP3-coated beads. Immunoblotting was carried out with anti-GFP antibody or anti-HA antibody. C, following expression of GFP, DOCK1-GFP, or DOCK5-GFP with mCherry in WT MEFs, cells were stimulated with PDGF for 7 min. Dorsal ruffles (arrows) were identified by staining cells with phalloidin. Scale bar, 50 μm. D, quantification of data from C. The data are expressed as the ratios of GFP to mCherry (means ± S.E.) after normalization of the GFP value to an arbitrary value of 1. More than 20 cells were analyzed for each category. Statistical analysis was performed by using analysis of variance followed by Bonferroni correction. **, p < 0.01. E, schematic representation of DOCK1 mutants used in this experiment. F, following expression of WT or mutant DOCK1-GFP (ΔDHR-1, ΔDHR-2, ΔC113, or ΔC254) with mCherry in WT MEFs, cells were stimulated with PDGF for 7 min. Dorsal ruffles (arrows) were identified by staining cells with phalloidin. Scale bar, 50 μm. G, quantification of data from F. The data are expressed as the ratios of GFP to mCherry (means ± S.E.) after normalization of the GFP value to an arbitrary value of 1. More than 20 cells were analyzed for each category. **, p < 0.01.
To determine the region of DOCK1 required for localization to the dorsal ruffle membrane, we expressed several deletion mutants of DOCK1-GFP in WT MEFs. DOCK1 binds to PIP3 through its DHR-1 domain and mediates the GTP-GDP exchange reaction for Rac via its DHR-2 domain (23–26). We found that DOCK1 was localized normally to the dorsal ruffle membrane even when these domains are deleted (designated ΔDHR-1 and ΔDHR-2; Fig. 2, E–G). Likewise, deletion of the C-terminal 113 amino acid residues (residues 1753–1865) (ΔC113; Fig. 2E), which contains a proline-rich sequence that is known to bind to the SH3 domain of Crk or Nck (38, 39), did not affect DOCK1 localization (Fig. 2, F and G). However, accumulation of DOCK1 at the dorsal ruffle membrane was markedly suppressed by deleting the C-terminal 254 amino acid residues (residues 1612–1865) (ΔC254; Fig. 2, E–G), suggesting that the amino acid residues from 1612 to 1752 contain an important localization motif.
DOCK1 Binds to PA through the C-terminal Polybasic Amino Acid Cluster
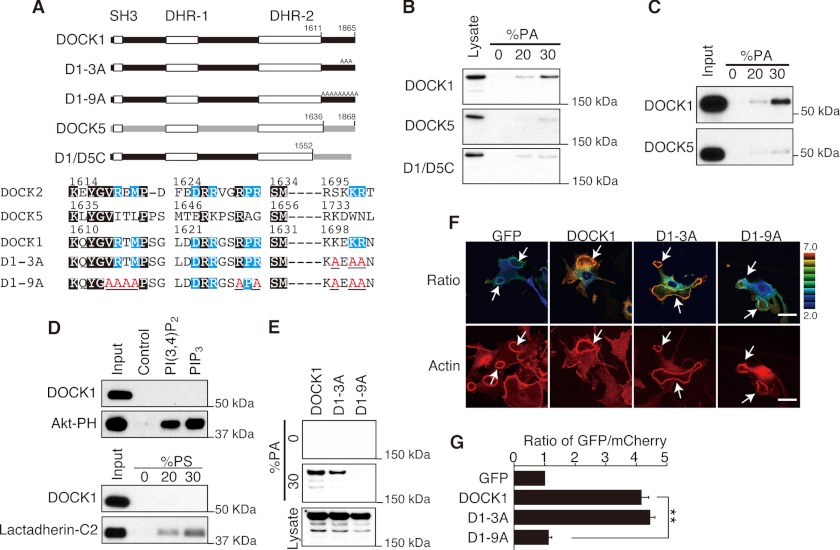
PA functions as a lipid anchor by binding directly to positively charged sites on effector proteins (27). The C-terminal region of DOCK1 contains several basic amino acid clusters. This amino acid sequence has a similarity to that of DOCK2, which is known to bind to PA (40, 41), but is distinct from that of DOCK5 (Fig. 3A). Therefore, we next examined whether DOCK1 binds to PA. Although no binding was found when HEK293T cell lysates containing DOCK5 were incubated with lipid vesicles containing PA, DOCK1 showed definite binding to PA-containing vesicles (Fig. 3B). This PA binding was abolished when the C-terminal region of DOCK1 was replaced with that of DOCK5 (D1/D5C; Fig. 3B), indicating that this region is important for PA binding. Consistent with this, the C-terminal fragment of DOCK1 binds effectively to lipid vesicles containing PA in vitro (Fig. 3C), but not to other acidic phospholipids such as PIP3, PI(3,4)P2, and PS (Fig. 3D). These results indicate that the C-terminal region of DOCK1 interacts directly and selectively with PA.
FIGURE 3.
DOCK1 binds to PA through the C-terminal polybasic amino acid cluster. A, schematic representation of DOCK1 mutants used in this experiment. Amino acid residues conserved between DOCK1 and DOCK2 but different from those of DOCK5 are marked in blue, and the mutated amino acid residues are labeled in red. B, extracts from HEK293T cells expressing GFP-tagged DOCK1, DOCK5, or D1/D5C were pulled down with PA-containing lipid vesicles. C, GST fusion protein encoding the C-terminal fragment of DOCK1 or DOCK5 was pulled down with lipid vesicles containing PA. D, GST fusion protein encoding the C-terminal fragment of DOCK1, Akt-PH, or lactadherin-C2 was pulled down with PIP3- or PI(3,4)P2-coated beads or PS-containing lipid vesicles. GST fusion Akt-PH or lactadherin-C2 was used as a positive control for PIP3/PI(3,4)P2 binding or PS binding, respectively. E, extracts from HEK293T cells expressing DOCK1-GFP or its mutants (D1–3A or D1–9A) were pulled down with PA-containing lipid vesicles. F, following expression of WT or mutant DOCK1-GFP (D1–3A or D1–9A) with mCherry in WT MEFs, cells were stimulated with PDGF for 7 min. Dorsal ruffles (arrows) were identified by staining cells with phalloidin. Scale bar, 50 μm. G, quantification of data from F. The data are expressed as the ratio of GFP to mCherry (means ± S.E.) after normalization of the GFP value to an arbitrary value of 1. More than 20 cells were analyzed for each category. **, p < 0.01.
To determine the amino acid residues of DOCK1 critical for PA binding, we mutated several basic amino acid residues to alanine. For example, DOCK1 has the amino acid sequence KEKR at positions 1699–1702, which corresponds to SKKR in DOCK2 and KDWN in DOCK5 (Fig. 3A). When the DOCK1-GFP mutant (designated D1–3A) encoding alanine instead of the three basic residues in this sequence was expressed in HEK293T cells, PA binding was diminished, but only to a modest extent (Fig. 3E). However, by mutating six additional residues to alanine (designated D1–9A), PA binding of DOCK1 was severely impaired (Fig. 3E). Having found that D1–3A and D1–9A have different binding capacity to PA, we compared the localization of these DOCK1-GFP mutants during dorsal ruffle formation. When D1–3A was expressed in WT MEFs, D1–3A readily accumulated at the dorsal ruffle membrane in response to PDGF stimulation (Fig. 3, F and G). However, such localization was scarcely found with D1–9A (Fig. 3, F and G). Collectively, these results suggest that the C-terminal polybasic amino acid cluster of DOCK1 is important for binding to PA and targeting to the dorsal ruffle membrane.
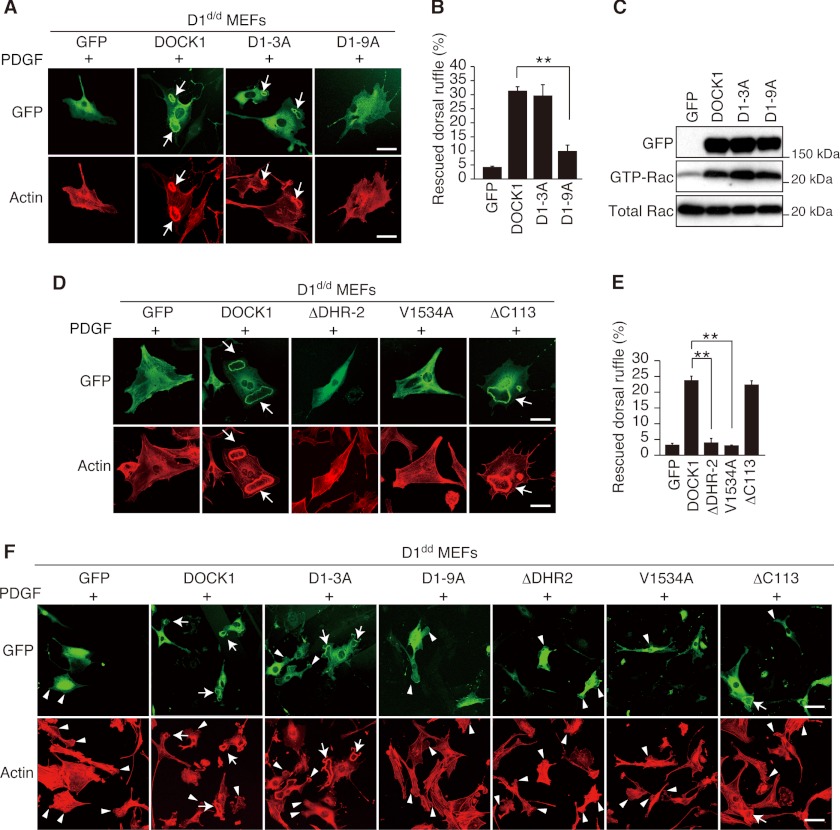
The Association with PA Is Required for DOCK1-mediated Dorsal Ruffle Formation
To examine whether PA binding of DOCK1 is required for dorsal ruffle formation, we expressed several DOCK1-GFP mutants in D1d/d MEFs. When WT DOCK1 and D1–3A were expressed in D1d/d MEFs, dorsal ruffle formation was induced as in WT MEFs (Fig. 4, A, B, and F). However, the expression of D1–9A failed to restore dorsal ruffle formation (Fig. 4, A, B, and F), despite the fact that this mutant retains full Rac GEF activity (Fig. 4C). It is clear that DOCK1 regulates dorsal ruffle formation by acting as a Rac GEF, because dorsal ruffle formation was not restored by expressing the ΔDHR-2 or V1534A mutant encoding alanine instead of valine at position 1534 that is known to function as a nucleotide sensor (42) (Fig. 4, D–F). Thus, these results indicate that the association with PA is required for DOCK1-mediated dorsal ruffle formation.
FIGURE 4.
The association of DOCK1 with PA is required for dorsal ruffle formation. A, following expression of WT or mutant DOCK1-GFP (D1–3A or D1–9A) in D1d/d MEFs, cells were stimulated with PDGF for 7 min. Formation of dorsal ruffles was defined by staining cells with phalloidin. Scale bar, 50 μm. B, quantification of data from A. The data are expressed as percentages of cells with dorsal ruffles (means ± S.E.) and are from three separate experiments. For each category, more than 200 cells were analyzed. **, p < 0.01. C, following expression of WT or mutant DOCK1-GFP (D1–3A or D1–9A) in HEK293T cells, cell extracts were subjected to Rac activation assay. D, following expression of WT or mutant DOCK1-GFP (ΔDHR-2, V1534A, or ΔC113) in D1d/d MEFs, cells were stimulated with PDGF for 7 min. Formation of dorsal ruffles was defined by staining cells with phalloidin. Scale bar, 50 μm. E, quantification of data from D. The data are expressed as percentages of cells with dorsal ruffles (means ± S.E.) and are from four separate experiments. For each category, more than 200 cells were analyzed. Statistical analysis was performed by using analysis of variance followed by Bonferroni correction. **, p < 0.01. F, low magnification images of D1d/d MEFs stimulated with PDGF after expression of WT or mutant DOCK1. Formation of peripheral (arrowheads) and dorsal ruffles (arrows) was defined by staining cells with phalloidin. Scale bar, 100 μm.
PLD-PA Axis Controls Dorsal Ruffle Formation
The PH domain of SOS is known to bind to PA (43). To visualize PA production during dorsal ruffle formation, we expressed the GFP fusion SOS-PH and the C-terminal fragment of DOCK1 (DOCK1 C254) in WT MEFs. As a result, we found that these biosensors strongly accumulated at the dorsal ruffle membrane upon stimulation with PDGF (Fig. 5A). These results suggest that PA can be produced at the site of dorsal ruffle in response to PDGF stimulation.
FIGURE 5.
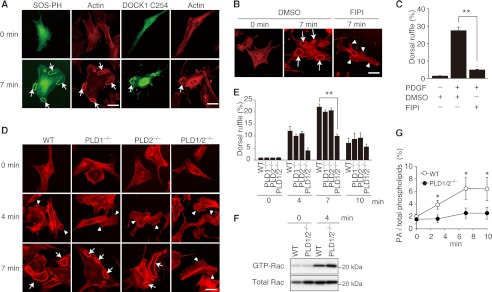
PLD-PA axis selectively controls dorsal ruffle formation. A, following expression of GFP fusion SOS-PH or DOCK1 C254 in WT MEFs, cells were stimulated with PDGF for 7 min. Dorsal ruffles (arrows) were identified by staining cells with phalloidin. Scale bar, 50 μm. B, WT MEFs were treated with FIPI or DMSO vehicle before stimulation with PDGF. Dorsal ruffles (arrows) and peripheral ruffles (arrowheads) were identified by staining cells with phalloidin. Scale bar, 50 μm. C, quantification of data from B. The data are expressed as percentages of cells with dorsal ruffles (means ± S.E.) and are from three separate experiments. In each experiment, more than 100 cells were analyzed. **, p < 0.01. D, upon stimulation with PDGF for the indicated times, WT and mutant MEFs were stained with phalloidin. Arrowheads or arrows indicate peripheral ruffles or dorsal ruffles, respectively. Scale bar, 50 μm. E, summary for PDGF-induced dorsal ruffle formation in D. The data are expressed as percentages of cells with dorsal ruffle formation (means ± S.E.) and are from three separate experiments. In each experiment, more than 100 cells were analyzed. **, p < 0.01. F, WT and PLD1/2−/− MEFs were stimulated with PDGF for 4 min, and Rac activation was analyzed biochemically. G, PDGF-induced PA production was compared between WT and PLD1/2−/− MEFs. The data are expressed as percentages of PA to total phospholipids (mean ± S.D.) of three independent experiments. *, p < 0.05.
This finding led us to examine whether PLD activity is required for dorsal ruffle formation. For this purpose, we treated WT MEFs with FIPI, a PLD-specific inhibitor (33). Compared with the control treated with DMSO, treatment with FIPI markedly suppressed PDGF-induced dorsal ruffle formation (Fig. 5, B and C). PLD family is composed of six members, all of which contain the invariant charged motif, HXXXXKXD (HKD), where the amino acids are histidine, any amino acid, lysine, and aspartic acid (28, 29). PLD1 and PLD2 are classical PLDs that produce PA through hydrolysis of PC (28, 29). On the other hand, PLD6 acts on cardiolipin to generate PA (44), yet the enzymatic activity and the substrate specificity of other PLD members remain unclear.
To examine whether PLD1 and PLD2 play roles in dorsal ruffle formation, we prepared MEFs from PLD1−/−, PLD2−/−, and PLD1/2−/− mice and stimulated them with PDGF. Single deficiency of PLD1 or PLD2 did not affect peripheral and dorsal ruffle formation (Fig. 5, D and E). However, whereas peripheral ruffle formation occurred normally even in PLD1/2−/− MEFs, double deficiency of PLD1 and PLD2 in MEFs significantly impaired PDGF-induced dorsal ruffle formation (Fig. 5, D and E). We found that although double deficiency of PLD1 and PLD2 did not affect global activation of Rac (Fig. 5F), PDGF-induced PA production was much reduced in PLD1/2−/− MEFs, as compared with that in WT MEFs (Fig. 5G). These results indicate that PLD1/2-mediated PA production is necessary to form dorsal ruffles effectively.
DISCUSSION
In this study, by analyzing MEFs deficient in DOCK1 and/or DOCK5, we have examined the role of these atypical Rac GEFs in PDGF-induced membrane ruffle formation. We found that although peripheral ruffle formation is coordinately regulated by DOCK1 and DOCK5, DOCK1 deficiency alone impairs dorsal ruffle formation. These results indicate that DOCK1 and DOCK5 have different roles in PDGF-induced peripheral and dorsal ruffle formation.
Reversible interactions between cytosolic proteins and phospholipids in the plasma membrane are important for a wide range of cellular functions. We found that, unlike DOCK5, DOCK1 binds to PA through the C-terminal polybasic amino acid cluster and accumulates effectively at the dorsal ruffle membrane in response to PDGF stimulation. When this interaction is blocked, PDGF-induced dorsal ruffle formation is severely impaired. Thus, the association with PA plays a key role in DOCK1-mediated dorsal ruffle formation. Although DOCK5 does not bind to PA, accumulation of DOCK5-GFP at dorsal ruffles was statistically significant in WT MEFs, as compared with GFP control. Because DOCK5 forms an heterodimer with DOCK1 (45), such heterodimerization may influence DOCK5 localization in this experiment.
In response to PDGF stimulation, signaling pools of PA can be formed through hydrolysis of PC by PLD (46). We found that deletion of PLD1 and PLD2 leads to impairment of dorsal, but not peripheral, ruffle formation. These results indicate that the PLD-PA pathway is crucial for PDGF-induced dorsal ruffle formation. It is currently unknown whether generation of PIP3 and PA is functionally linked during dorsal ruffle formation. However, several lines of evidence indicate that PIP3 activates PLD directly or indirectly (28, 29, 47, 48). Therefore, generation of PIP3 and PA may be sequentially regulated during dorsal ruffle formation.
In summary, we have shown that PLD-PA axis selectively controls dorsal ruffle formation by regulating DOCK1 localization. Our results thus define the novel regulatory mechanism for RTK-mediated dorsal ruffle formation.
Acknowledgments
We thank Ayumi Inayoshi and Meiko Sanematsu for technical assistance.
This work was supported by the Core Research for Evolutional Science and Technology program of Japan Science and Technology Agency (to Y. F.), the Project for Development of Innovative Research for Cancer Therapeutics from the Ministry of Education, Culture, Sports, Science and Technology (to Y. F.), Canadian Cancer Society Grant 019104 (to J.-F. C.), and Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (to Y. F. and F. S.).
- RTK
- receptor tyrosine kinase
- DHR
- DOCK homology region
- FIPI
- 5-fluoro-2-indolyl des-clorohalopemide
- GEF
- guanine nucleotide exchange factor
- MEF
- mouse embryonic fibroblast
- PA
- phosphatidic acid
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PH
- pleckstrin homology
- PS
- phosphatidylserine
- PIP3
- phosphatidylinositol 3,4,5-trisphosphate
- PI(3,4)P2
- phosphatidylinositol 3,4-bisphosphate
- PLD
- phospholipase D.
REFERENCES
- 1. Buccione R., Orth J. D., McNiven M. A. (2004) Foot and mouth. Podosomes, invadopodia and circular dorsal ruffles. Nat. Rev. Mol. Cell Biol. 5, 647–657 [DOI] [PubMed] [Google Scholar]
- 2. Ridley A. J., Schwartz M. A., Burridge K., Firtel R. A., Ginsberg M. H., Borisy G., Parsons J. T., Horwitz A. R. (2003) Cell migration. Integrating signals from front to back. Science 302, 1704–1709 [DOI] [PubMed] [Google Scholar]
- 3. Swanson J. A., Watts C. (1995) Macropinocytosis. Trends Cell Biol. 5, 424–428 [DOI] [PubMed] [Google Scholar]
- 4. Orth J. D., McNiven M. A. (2006) Get off my back! Rapid receptor internalization through circular dorsal ruffles. Cancer Res. 66, 11094–11096 [DOI] [PubMed] [Google Scholar]
- 5. Suetsugu S., Yamazaki D., Kurisu S., Takenawa T. (2003) Differential roles of WAVE1 and WAVE2 in dorsal and peripheral ruffle formation for fibroblast cell migration. Dev. Cell 5, 595–609 [DOI] [PubMed] [Google Scholar]
- 6. Lanzetti L., Palamidessi A., Areces L., Scita G., Di Fiore P. P. (2004) Rab5 is a signalling GTPase involved in actin remodelling by receptor tyrosine kinases. Nature 429, 309–314 [DOI] [PubMed] [Google Scholar]
- 7. Vidali L., Chen F., Cicchetti G., Ohta Y., Kwiatkowski D. J. (2006) Rac1-null mouse embryonic fibroblasts are motile and respond to platelet-derived growth factor. Mol. Biol. Cell 17, 2377–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Krueger E. W., Orth J. D., Cao H., McNiven M. A. (2003) A dynamin-cortactin-Arp2/3 complex mediates actin reorganization in growth factor-stimulated cells. Mol. Biol. Cell 14, 1085–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yoshida S., Hoppe A. D., Araki N., Swanson J. A. (2009) Sequential signaling in plasma-membrane domains during macropinosome formation in macrophages. J. Cell Sci. 122, 3250–3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schmidt A., Hall A. (2002) Guanine nucleotide exchange factors for Rho GTPases. Turning on the switch. Genes Dev. 16, 1587–1609 [DOI] [PubMed] [Google Scholar]
- 11. Côté J. F., Vuori K. (2002) Identification of an evolutionarily conserved superfamily of DOCK180-related proteins with guanine nucleotide exchange activity. J. Cell Sci. 115, 4901–4913 [DOI] [PubMed] [Google Scholar]
- 12. Fukui Y., Hashimoto O., Sanui T., Oono T., Koga H., Abe M., Inayoshi A., Noda M., Oike M., Shirai T., Sasazuki T. (2001) Haematopoietic cell-specific CDM family protein DOCK2 is essential for lymphocyte migration. Nature 412, 826–831 [DOI] [PubMed] [Google Scholar]
- 13. Sanui T., Inayoshi A., Noda M., Iwata E., Oike M., Sasazuki T., Fukui Y. (2003) DOCK2 is essential for antigen-induced translocation of TCR and lipid rafts, but not PKC-θ and LFA-1, in T cells. Immunity 19, 119–129 [DOI] [PubMed] [Google Scholar]
- 14. Kunisaki Y., Nishikimi A., Tanaka Y., Takii R., Noda M., Inayoshi A., Watanabe K., Sanematsu F., Sasazuki T., Sasaki T., Fukui Y. (2006) DOCK2 is a Rac activator that regulates motility and polarity during neutrophil chemotaxis. J. Cell Biol. 174, 647–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tanaka Y., Hamano S., Gotoh K., Murata Y., Kunisaki Y., Nishikimi A., Takii R., Kawaguchi M., Inayoshi A., Masuko S., Himeno K., Sasazuki T., Fukui Y. (2007) T helper type 2 differentiation and intracellular trafficking of the interleukin 4 receptor-α subunit controlled by the Rac activator Dock2. Nat. Immunol. 8, 1067–1075 [DOI] [PubMed] [Google Scholar]
- 16. Gotoh K., Tanaka Y., Nishikimi A., Inayoshi A., Enjoji M., Takayanagi R., Sasazuki T., Fukui Y. (2008) Differential requirement for DOCK2 in migration of plasmacytoid dendritic cells versus myeloid dendritic cells. Blood 111, 2973–2976 [DOI] [PubMed] [Google Scholar]
- 17. Gotoh K., Tanaka Y., Nishikimi A., Nakamura R., Yamada H., Maeda N., Ishikawa T., Hoshino K., Uruno T., Cao Q., Higashi S., Kawaguchi Y., Enjoji M., Takayanagi R., Kaisho T., Yoshikai Y., Fukui Y. (2010) Selective control of type I IFN induction by the Rac activator DOCK2 during TLR-mediated plasmacytoid dendritic cell activation. J. Exp. Med. 207, 721–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gumienny T. L., Brugnera E., Tosello-Trampont A. C., Kinchen J. M., Haney L. B., Nishiwaki K., Walk S. F., Nemergut M. E., Macara I. G., Francis R., Schedl T., Qin Y., Van Aelst L., Hengartner M. O., Ravichandran K. S. (2001) CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell 107, 27–41 [DOI] [PubMed] [Google Scholar]
- 19. Grimsley C. M., Kinchen J. M., Tosello-Trampont A. C., Brugnera E., Haney L. B., Lu M., Chen Q., Klingele D., Hengartner M. O., Ravichandran K. S. (2004) Dock180 and ELMO1 proteins cooperate to promote evolutionarily conserved Rac-dependent cell migration. J. Biol. Chem. 279, 6087–6097 [DOI] [PubMed] [Google Scholar]
- 20. Li X., Gao X., Liu G., Xiong W., Wu J., Rao Y. (2008) Netrin signal transduction and the guanine nucleotide exchange factor DOCK180 in attractive signaling. Nat. Neurosci. 11, 28–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sanematsu F., Hirashima M., Laurin M., Takii R., Nishikimi A., Kitajima K., Ding G., Noda M., Murata Y., Tanaka Y., Masuko S., Suda T., Meno C., Côté J. F., Nagasawa T., Fukui Y. (2010) DOCK180 is a Rac activator that regulates cardiovascular development by acting downstream of CXCR4. Circ. Res. 107, 1102–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Laurin M., Fradet N., Blangy A., Hall A., Vuori K., Côté J. F. (2008) The atypical Rac activator Dock180 (Dock1) regulates myoblast fusion in vivo. Proc. Natl. Acad. Sci. U.S.A. 105, 15446–15451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brugnera E., Haney L., Grimsley C., Lu M., Walk S. F., Tosello-Trampont A. C., Macara I. G., Madhani H., Fink G. R., Ravichandran K. S. (2002) Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat. Cell Biol. 4, 574–582 [DOI] [PubMed] [Google Scholar]
- 24. Meller N., Irani-Tehrani M., Kiosses W. B., Del Pozo M. A., Schwartz M. A. (2002) Zizimin1, a novel Cdc42 activator, reveals a new GEF domain for Rho proteins. Nat. Cell Biol. 4, 639–647 [DOI] [PubMed] [Google Scholar]
- 25. Côté J. F., Motoyama A. B., Bush J. A., Vuori K. (2005) A novel and evolutionarily conserved PtdIns(3,4,5)P3-binding domain is necessary for DOCK180 signalling. Nat. Cell Biol. 7, 797–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Premkumar L., Bobkov A. A., Patel M., Jaroszewski L., Bankston L. A., Stec B., Vuori K., Côté J. F., Liddington R. C. (2010) Structural basis of membrane targeting by the Dock180 family of Rho family guanine exchange factors (Rho-GEFs). J. Biol. Chem. 285, 13211–13222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stace C. L., Ktistakis N. T. (2006) Phosphatidic acid- and phosphatidylserine-binding proteins. Biochim. Biophys. Acta 1761, 913–926 [DOI] [PubMed] [Google Scholar]
- 28. McDermott M., Wakelam M. J., Morris A. J. (2004) Phospholipase D. Biochem. Cell Biol. 82, 225–253 [DOI] [PubMed] [Google Scholar]
- 29. Jenkins G. M., Frohman M. A. (2005) Phospholipase D. A lipid centric review. Cell Mol. Life Sci. 62, 2305–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Haga Y., Miwa N., Jahangeer S., Okada T., Nakamura S. (2009) CtBP1/BARS is an activator of phospholipase D1 necessary for agonistinduced macropinocytosis. EMBO J. 28, 1197–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen Q., Hongu T., Sato T., Zhang Y., Ali W., Cavallo J.-A., van der Velden A., Tian H., Di Paolo G., Nieswandt B., Kanaho Y., Frohman M. A. (2012) Key roles for the lipid signaling enzyme phospholipase D1 in the tumor microenvironment during tumor angiogenesis and metastasis. Sci. Signal. 5, ra79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sato T., Hongu T., Sakamoto M., Funakoshi Y., Kanaho Y. (2013) Molecular mechanism of N-formyl-methionyl-leucyl-phenylalanine-induced superoxide generation and degranulation in mouse neutrophils. Phospholipase D is dispensable. Mol. Cell Biol. 33, 136–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Su W., Yeku O., Olepu S., Genna A., Park J. S., Ren H., Du G., Gelb M. H., Morris A. J., Frohman M. A. (2009) 5-Fluoro-2-indolyl des-chlorohalopemide (FIPI), a phospholipase D pharmacological inhibitor that alters cell spreading and inhibits chemotaxis. Mol. Pharmacol. 75, 437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sanui T., Inayoshi A., Noda M., Iwata E., Stein J. V., Sasazuki T., Fukui Y. (2003) DOCK2 regulates Rac activation and cytoskeletal reorganization through interaction with ELMO1. Blood 102, 2948–2950 [DOI] [PubMed] [Google Scholar]
- 35. Nosaka T., Kawashima T., Misawa K., Ikuta K., Mui A. L., Kitamura T. (1999) STAT5 as a molecular regulator of proliferation, differentiation and apoptosis in hematopoietic cells. EMBO J. 18, 4754–4765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Okada T., Sakuma L., Fukui Y., Hazeki O., Ui M. (1994) Blockage of chemotactic peptide-induced stimulation of neutrophils by wortmannin as a result of selective inhibition of phosphatidylinositol 3-kinases. J. Biol. Chem. 269, 3563–3567 [PubMed] [Google Scholar]
- 37. Komander D., Patel M., Laurin M., Fradet N., Pelletier A., Barford D., Côté J. F. (2008) An α-helical extension of the ELMO1 pleckstrin homology domain mediates direct interaction to DOCK180 and is critical in Rac signaling. Mol. Biol. Cell 19, 4837–4851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hasegawa H., Kiyokawa E., Tanaka S., Nagashima K., Gotoh N., Shibuya M., Kurata T., Matsuda M. (1996) DOCK180, a major CRK-binding protein, alters cell morphology upon translocation to the cell membrane. Mol. Cell Biol. 16, 1770–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tu Y., Kucik D. F., Wu C. (2001) Identification and kinetic analysis of the interaction between Nck-2 and DOCK180. FEBS Lett. 491, 193–199 [DOI] [PubMed] [Google Scholar]
- 40. Nishikimi A., Fukuhara H., Su W., Hongu T., Takasuga S., Mihara H., Cao Q., Sanematsu F., Kanai M., Hasegawa H., Tanaka Y., Shibasaki M., Kanaho Y., Sasaki T., Frohman M. A., Fukui Y. (2009) Sequential regulation of DOCK2 dynamics by two phospholipids during neutrophil chemotaxis. Science 324, 384–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nishioka T., Frohman M. A., Matsuda M., Kiyokawa E. (2010) Heterogeneity of phosphatidic acid levels and distribution at the plasma membrane in living cells as visualized by a Föster resonance energy transfer (FRET) biosensor. J. Biol. Chem. 285, 35979–35987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kulkarni K., Yang J., Zhang Z., Barford D. (2011) Multiple factors confer specific Cdc42 and Rac protein activation by dedicator of cytokinesis (DOCK) nucleotide exchange factors. J. Biol. Chem. 286, 25341–25351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhao C., Du G., Skowronek K., Frohman M. A., Bar-Sagi D. (2007) Phospholipase D2-generated phosphatidic acid couples EGFR stimulation to Ras activation by Sos. Nat. Cell Biol. 9, 706–712 [DOI] [PubMed] [Google Scholar]
- 44. Choi S. Y., Huang P., Jenkins G. M., Chan D. C., Schiller J., Frohman M. A. (2006) A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis. Nat. Cell Biol. 8, 1255–1262 [DOI] [PubMed] [Google Scholar]
- 45. Patel M., Pelletier A., Côté J. F. (2011) Opening up on ELMO regulation. New insights into the control of Rac signaling by the DOCK180/ELMO complex. Small GTPases 2, 268–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fukami K., Takenawa T. (1992) Phosphatidic acid that accumulates in platelet-derived growth factor-stimulated Balb/c 3T3 cells is a potential mitogenic signal. J. Biol. Chem. 267, 10988–10993 [PubMed] [Google Scholar]
- 47. Hammond S. M., Jenco J. M., Nakashima S., Cadwallader K., Gu Q., Cook S., Nozawa Y., Prestwich G. D., Frohman M. A., Morris A. J. (1997) Characterization of two alternately spliced forms of phospholipase D1. J. Biol. Chem. 272, 3860–3868 [DOI] [PubMed] [Google Scholar]
- 48. Czech M. P. (2000) PIP2 and PIP3. Complex roles at the cell surface. Cell 100, 603–606 [DOI] [PubMed] [Google Scholar]