Background: The human SAMHD1 protein has dNTP triphosphatase activity and is involved in HIV-1 restriction and autoimmune syndrome.
Results: Purified SAMHD1 exhibits nuclease activity against single-stranded DNA and RNA.
Conclusion: The nuclease activity of SAMHD1 is associated with its HD domain.
Significance: Identification of nuclease activity in SAMHD1 provides novel insight into the mechanisms of HIV-1 restriction and regulation of autoimmune response.
Keywords: Autoimmune Diseases, DNA Enzymes, DNase, HIV-1, Ribonuclease, RNA, Aicardi-Goutières Syndrome, HD Domain, SAMHD1, Nuclease Activity
Abstract
The human HD domain protein SAMHD1 is implicated in the Aicardi-Goutières autoimmune syndrome and in the restriction of HIV-1 replication in myeloid cells. Recently, this protein has been shown to possess dNTP triphosphatase activity, which is proposed to inhibit HIV-1 replication and the autoimmune response by hydrolyzing cellular dNTPs. Here, we show that the purified full-length human SAMHD1 protein also possesses metal-dependent 3′→5′ exonuclease activity against single-stranded DNAs and RNAs in vitro. In double-stranded substrates, this protein preferentially cleaved 3′-overhangs and RNA in blunt-ended DNA/RNA duplexes. Full-length SAMHD1 also exhibited strong DNA and RNA binding to substrates with complex secondary structures. Both nuclease and dNTP triphosphatase activities of SAMHD1 are associated with its HD domain, but the SAM domain is required for maximal activity and nucleic acid binding. The nuclease activity of SAMHD1 could represent an additional mechanism contributing to HIV-1 restriction and suppression of the autoimmune response through direct cleavage of viral and endogenous nucleic acids. In addition, we demonstrated the presence of dGTP triphosphohydrolase and nuclease activities in several microbial HD domain proteins, suggesting that these proteins might contribute to antiviral defense in prokaryotes.
Introduction
Both prokaryotes and eukaryotes have developed complex defense mechanisms to protect their cells from viral invasions. In addition to the conventional innate and adaptive immune responses, novel antiviral systems are emerging that are based on pre-existing, constitutively expressed, intracellular proteins (restriction factors) and that comprise an intrinsic immunity system (1). These factors can be considered as the front line of antiviral defense, as they act during the very first steps of virus-host interactions in an immunologically naïve host, and they are often counterattacked by viral protective proteins. Several known intrinsic immunity mechanisms include the APOBEC3 class of cytidine deaminases, components of nuclear domain structures (ND10), DNA repair proteins RNF8 and RNF168, and two groups of viral capsid inhibitors (Fv (Fv1, Fv4, and Lv1) and TRIM5α) (1–3).
Recently, the uncharacterized human protein SAMHD1 has been shown to be a novel restriction factor that inhibits replication of the HIV-1 genome in myeloid cells (4–6). However, when these cells are infected by the HIV-2 strain, the viral protein Vpx (which is absent in HIV-1) directs SAMHD1 to ubiquitination by the cellular cullin 4-based E3 ligase CRL4DCAF1 and subsequent proteasomal degradation (4, 5). In addition, SAMHD1 has been shown to be associated with the Aicardi-Goutières autoimmune syndrome (AGS)2 and suggested to function as a negative regulator of the innate immune response, potentially blocking the immune response to endogenous nucleic acids (7, 8). AGS is a genetically heterogeneous disease associated with mutations in the genes encoding the major 3′→5′ exonuclease TREX1 (AGS1), the heterotrimeric RNase H2 (AGS2, AGS3, and AGS4), and SAMHD1 (AGS5). SAMHD1 is up-regulated in response to viral infections in mice and to immunostimulatory DNA, and mutations in SAMHD1 and other AGS genes induce a phenotype that resembles the effects of a congenital viral infection (7–9).
SAMHD1 is a two-domain protein (626 amino acids (aa)) with the N-terminal SAM (sterile alpha motif) domain (aa 45–110) and the C-terminal HD domain (aa 164–319). SAM domains are known to function as protein interaction or RNA-binding modules, whereas several characterized HD domain proteins have been shown to possess phosphodiesterase, phosphatase, dGTP triphosphatase, or nuclease activities (10–17). Recently, the purified HD domains of human SAMHD1 and full-length mouse SAMHD1 have been shown to possess dGTP-stimulated dNTP triphosphohydrolase activity (18–20). The crystal structure of the human SAMHD1 HD domain revealed a mixed α/β-fold, and site-directed mutagenesis confirmed the significance of HD motif residues for dGTP hydrolysis (18). The dNTP triphosphohydrolase activity of SAMHD1 has been proposed to reduce the intracellular dNTP level, restricting HIV-1 replication and preventing activation of the immune system (18, 19). This hypothesis was supported by the direct demonstration of the effects of SAMHD1 knockdown and expression on the intracellular dNTP pool of myeloid cells (21).
Because two other AGS-associated proteins (TREX1 and RNase H2) are nucleases and the expression of SAMHD1 is up-regulated by immunostimulatory DNA, it has been hypothesized that SAMHD1 might act as a nuclease (7). In addition, recent works have demonstrated the presence of nuclease activity in five different HD domain proteins, most of which are associated with the microbial antiviral defense system CRISPR (clustered regularly interspaced short palindromic repeat) (12, 17, 22, 23). However, two previous publications have revealed no nuclease activity in the truncated human (HD domain only) and full-length monkey SAMHD1 proteins (18, 19). Here, using a sensitive radioactivity-based nuclease assay and the full-length human SAMHD1 protein, we demonstrate the presence of a nuclease activity against single-stranded (ss) DNAs and RNAs, as well as against RNA in DNA/RNA hybrids. Experiments with isolated SAMHD1 domains and site-directed mutagenesis have revealed that both nuclease and dGTP triphosphatase activities are associated with the HD domain, but the SAM domain is required for maximal activity. Our data suggest that the biochemical function of SAMHD1 might not be limited to dGTP hydrolysis and that its nuclease activity could also contribute to HIV-1 restriction and autoimmune response through a direct cleavage of viral and endogenous nucleic acids.
EXPERIMENTAL PROCEDURES
Protein Purification and Mutagenesis
Full-length SAMHD1 and its isolated domains (SAM (aa 1–118) and HD (aa 120–626)), as well as Aq_1910, TM1547, AF1432, PA1124, and Escherichia coli Dgt, were overexpressed in E. coli and purified as His6 tag fusions using affinity, size-exclusion, and anion-exchange chromatography as described previously (24). Site-directed mutagenesis of SAMHD1 and Aq_1910 was performed using a protocol based on the QuikChange site-directed mutagenesis kit (Stratagene).
Preparation of DNA and RNA Substrates
The ssDNA or ssRNA oligonucleotide substrates (17–92 nucleotides (nt)) were purchased from Integrated DNA Technologies. The oligonucleotides were 5′-labeled using [γ-32P]ATP (6000 Ci/mmol; PerkinElmer Life Sciences) and T4 polynucleotide kinase (PNK; Fermentas) and purified using denaturing PAGE (8 m urea and 15% polyacrylamide). The labeled oligonucleotides were eluted from the gel, precipitated with 2% LiClO4 in acetone, washed with acetone, dried, and dissolved in diethylpyrocarbonate-treated Milli-Q water. The synthetic double-stranded (ds) DNAs and RNAs (supplemental Fig. S1) were prepared by annealing oligonucleotides DNA6+DNA9 and RNA2+RNA5, respectively; dsDNA and dsRNA substrates with 3′-overhangs with oligonucleotides DNA6+DNA7 and RNA2+DNA7, respectively; dsDNA and dsRNA substrates with 5′-overhangs with oligonucleotides DNA6+DNA8 and RNA2+DNA8, respectively; and dsDNA and dsRNA substrates with blunt ends with oligonucleotides DNA6+DNA9 and RNA2+DNA9, respectively. Uniformly labeled in vitro transcripts of HIV-1 gag and tat RNAs were synthesized using pKS-gag and pKS-tat constructs (25), [32P]UTP (3000 Ci/mmol; PerkinElmer Life Sciences), and a T7 RNA polymerase MAXIscript transcription kit (Ambion).
HPLC Assays of dNTP Hydrolysis
Hydrolysis of dGTP and other nucleotides by full-length SAMHD1, its isolated domains (SAM (aa 1–118) and HD (aa 120–626)), or Aq_1910 was assayed in reaction mixtures (75 μl) containing 50 mm Tris-HCl (pH 7.5), 10 mm MgCl2, 100 mm NaCl, 1 mm DTT, 5 mm dNTP, and SAMHD1 (22 μg; full-length or isolated domains) or other protein (1–5 μg). Reaction mixtures were incubated at 37 °C (SAMHD1) or at 60 °C (Aq_1910) and passed through Millipore Ultrafree-MC filtration devices to quench the reactions and remove the protein. Reaction products were analyzed by ion-pair reverse-phase HPLC using a Varian Pursuit C18 column (150 × 4.6 mm) and a Varian ProStar HPLC system. The mobile phase for separation of nucleotides consisted of two eluants: 0.1 m KH2PO4 (pH 6.0) with 8 mm tetrabutylammonium hydroxide and 0.1 m KH2PO4 (pH 6.0) with 8 mm tetrabutylammonium hydroxide and 30% methanol.
Nuclease Assays
The reaction mixture for DNase assays contained (in a final volume of 10 μl) 25 mm Tris-HCl (pH 8.0), 20 mm KCl, 10 mm MgCl2, 1 mm DTT, 0.1 μm 5′- or 3′-32P-labeled ssDNA, and the indicated protein. The solutions were incubated at 37 °C (for SAMHD1) or at 45 °C (for Aq_1910) for the indicated times and quenched by the addition of an equal volume of formamide loading buffer (12). The reaction products were separated by electrophoresis on 8 m urea and 8–15% polyacrylamide gels and visualized by autoradiography. The reaction mixture for RNase assays (10 μl) contained 25 mm Tris-HCl (pH 7.0), 100 mm KCl, 10 mm CaCl2, 1 mm DTT, 0.1 μm 5′-32P-labeled ssRNA, and 100 nm SAMHD1 (or other protein as indicated). The solutions were incubated at 37 °C (or 42 °C for Aq_1910) for 5–45 min and quenched by the addition of an equal volume of formamide loading buffer. The reaction products were resolved on 8 m urea and 6–15% polyacrylamide gels using Tris-borate/EDTA running buffer and visualized by autoradiography. For nucleotide size markers, an imidazole ladder produced by partial RNA cleavage by 2 m imidazole was used. Gel images (uncleaved substrates and reaction products) were quantified (as percent) using ImageQuant (GE Healthcare).
DNA/RNA Binding Assays
RNA binding was analyzed in reaction mixtures (10 μl) containing 25 mm Tris-HCl (pH 7.0), 1 mm MgCl2, 1 mm DTT, and 0.05 μm 32P-labeled RNA. Reaction mixtures for DNA-binding activity (10 μl) contained 25 mm Tris-HCl (pH 8.0), 1 mm MnCl2, 1 mm DTT, and 0.05 μm 32P-labeled DNA. Reaction mixtures were incubated at 25 °C for 5 min, analyzed on native 6–10% polyacrylamide gels, and visualized by phosphorimaging.
RESULTS
Nuclease and dGTP Hydrolase Activities of SAMHD1 and Its Isolated Domains
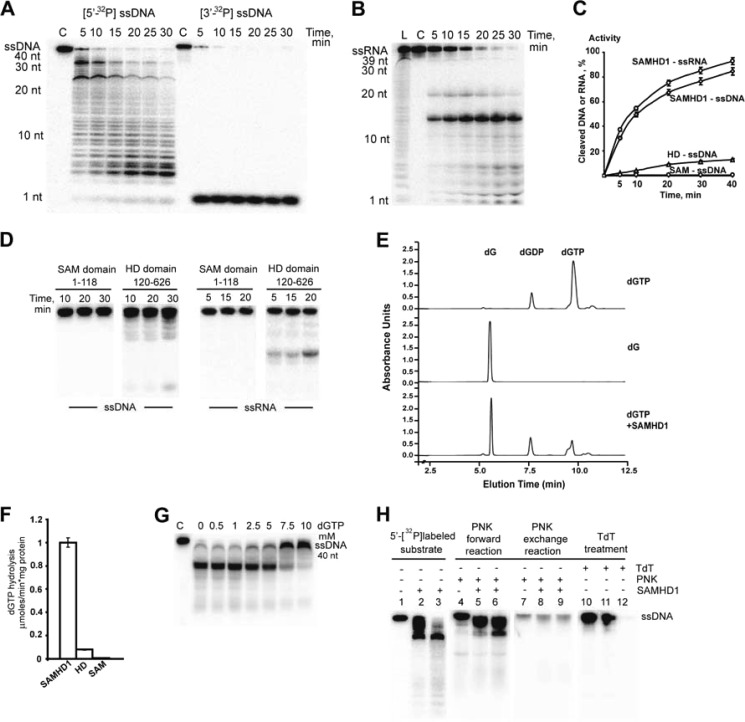
Full-length human SAMHD1 was overexpressed in E. coli and analyzed for the presence of nuclease activity using 5′-32P-labeled ssDNAs and ssRNAs as substrates. These assays revealed an exonuclease-like cleavage of both substrates with an optimal pH of 7.0–8.0 (Fig. 1, A and B). Without the addition of divalent metal cations, the protein showed low cleavage of both ssRNA and ssDNA that was completely suppressed by EDTA (supplemental Fig. S2), suggesting that it was likely supported by a protein-associated metal ion. The cleavage of ssDNA was stimulated by the addition of Mg2+ (optimal concentration of 7.5–10.0 mm), Ca2+, and Mn2+, whereas Ca2+ addition stimulated the cleavage of ssRNA (data not shown). These three metal ions also supported hydrolysis of dGTP by SAMHD1 (supplemental Fig. S2). Under optimal reaction conditions, SAMHD1 hydrolyzed ssRNA with a slightly higher rate than ssDNA (Fig. 1C). The elution profile of SAMHD1 from the size-exclusion and anion-exchange columns correlated with the peaks of DNase and dGTP triphosphatase activities (supplemental Fig. S2), confirming the association of both activities with this protein. With the 3′-labeled ssDNA as substrate, SAMHD1 produced one short (1 nt long) product, whereas the 5′-labeled substrate was cleaved with the formation of a series of products of different lengths, with the shortest product containing 1 nt (Fig. 1A). These results indicate that SAMHD1 is a 3′→5′ exonuclease that cleaves 1 nt at a time from the 3′-end of the substrate.
FIGURE 1.
Nuclease activity of the full-length human SAMHD1 and its domains. A and B, time points of ssDNA and ssRNA cleavage, respectively, by full-length SAMHD1. The 5′- or 3′-32P-labeled ssDNA or ssRNA substrate was incubated without protein (C lane) or with 100 nm SAMHD1 at 37 °C and analyzed by denaturing gel electrophoresis. L, ladder. C, time point of cleavage of 32P-labeled ssDNA (40 nt) or ssRNA (39 nt) by full-length SAMHD1 (100 nm), the SAM domain (100 nm), or the HD domain (100 nm). To calculate nuclease activity (as percent of the maximal activity of full-length SAMHD1), the gel images were quantified using ImageQuant TL software (GE Healthcare). D, time points of ssDNA (left panels) and ssRNA (right panels) cleavage by isolated SAM and HD domains. The 5′-32P-labeled ssDNA or ssRNA substrate was incubated with 0.3 μg of SAM or HD domain (as indicated) at 37 °C. E, HPLC analysis of the reaction products generated by SAMHD1 during hydrolysis of dNTPs. Shown are elution profiles of the reaction products separated by ion-pair reverse-phase HPLC (monitored at 254 nm). The upper two profiles show control samples incubated without enzyme (dGTP and deoxyguanosine (dG) standards; 5 mm each), whereas the lower profile shows the sample incubated with the enzyme for 15 min. F, dGTP triphosphatase activity of full-length SAMHD1 and its isolated SAM and HD domains. G, cleavage of ssDNA by SAMHD1 in the presence of dGTP. H, cleavage products of SAMHD1. Shown is the PNK or TdT labeling of the DNA cleavage products. Lanes 1–3, 5′-32P-labeled ssDNA6 (40 nt, 0.1 μm) was incubated at 37 °C in the absence (lane 1) or presence of 250 nm (lane 2) of 400 nm (lane 3) SAMHD1 for 25 min without the addition of PNK or TdT. Lanes 4–9, PNK-treated reactions. Unlabeled ssDNA (0.1 μm) was incubated at 37 °C in the absence (lanes 4–7) or presence of 250 nm (lanes 5 and 8) or 400 nm (lanes 6 and 9) SAMHD1 for 20 min, followed by treatment with PNK (30 min) in the 5′-end labeling (forward; lanes 4–6) or phosphate exchange (lanes 7–9) reactions. Lanes 10–12, TdT treatment reactions. Unlabeled ssDNA (0.1 μm) was incubated at 37 °C in the absence (lane 10) or presence of 250 nm (lane 11) or 500 nm (lane 12) SAMHD1 for 20 min, followed by treatment with TdT (30 min) in the 3′-end labeling reaction.
The purified individual SAM domain (aa 1–118) of SAMHD1 showed no cleavage of ssDNA or ssRNA, whereas the isolated HD domain (aa 120–626) exhibited nuclease activity that was approximately five times lower than that of the full-length protein (Fig. 1D). Similarly, the dGTP triphosphatase activity of SAMHD1 was also associated with its HD domain, which showed several times lower activity than the full-length protein (Fig. 1, E and F), suggesting that the SAM domain is required for the maximal nuclease and dGTP hydrolase activities of SAMHD1. The addition of dGTP (2.5–10 mm) inhibited the nuclease activity of SAMHD1 (Fig. 1G), implying that both activities of this protein are associated with the same active site. The labeling of ssDNA cleavage products using PNK (both 5′-phosphorylation and exchange reactions) or terminal deoxynucleotidyl transferase (TdT; nucleotide addition at the 3′-OH end) showed that SAMHD1 produced products containing 5′-OH and 3′-phosphates (Fig. 1H), which is in line with the formation of a 5′-OH-containing product (guanosine) during hydrolysis of dGTP. The metal-dependent Cas3 HD domain nuclease MJ0384 from Methanocaldococcus jannaschii also cleaves ssDNA with the formation of 5′-OH and 3′-phosphates (12), suggesting that the HD domain nucleases use the same catalytic mechanism.
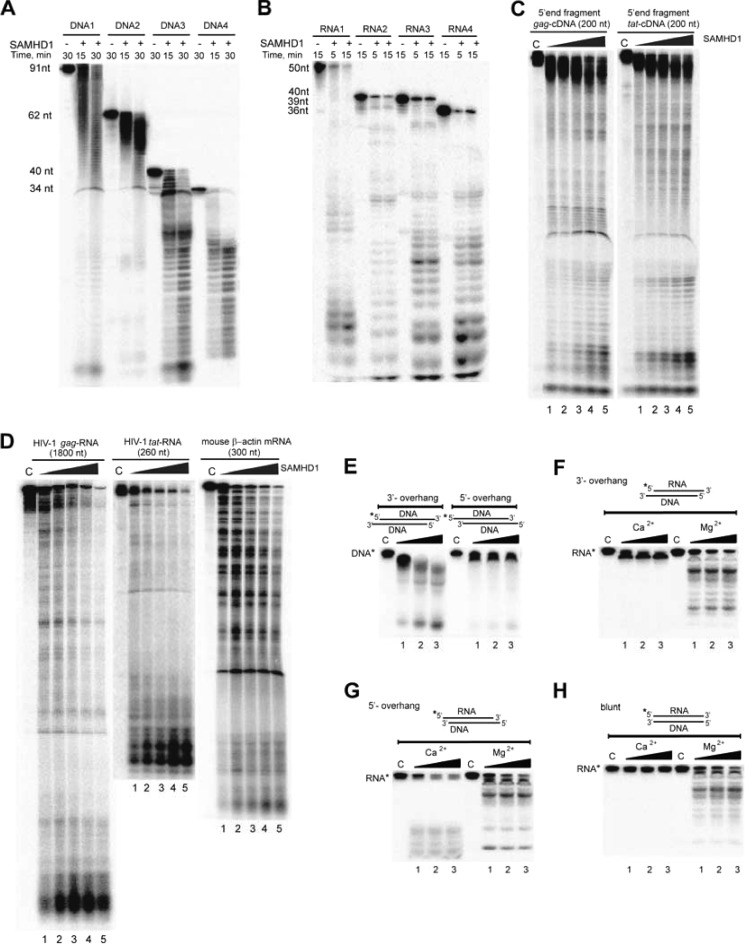
Full-length SAMHD1 exhibited no apparent nucleic acid sequence or substrate length preference and showed an exonuclease-like cleavage pattern with both linear ssDNAs and ssRNAs, whereas no cleavage was observed using the circular ssDNA of the M13mp18 phage as substrate (Fig. 1, A and B, and Fig. 2, A and B; data not shown for M13mp18 cleavage). The full-length protein also degraded long ssRNAs and ssDNAs, including the HIV-1 gag and tat RNAs produced in vitro by the T7 RNA polymerase (Fig. 2, C and D), suggesting that SAMHD1 can potentially degrade viral nucleic acids in vivo. The optimal reaction conditions and cleavage pattern of the SAMHD1 nuclease are similar to that of the Cas3 HD domain nuclease MJ0384, which is an effector protein of the microbial CRISPR antiviral defense system (12).
FIGURE 2.
A and B, cleavage of the 32P-labeled ssDNA or ssRNA with different lengths and sequences by SAMHD1 (500 nm). C and D, cleavage of the uniformly 32P-labeled in vitro transcripts of HIV-1 gag (1800 nt) and tat (260 nt) RNAs and the 5′-end fragments (200 nt) of HIV-1 gag and tat DNAs (as indicated). The substrates (0.1 μm) were incubated with 0.13, 0.25, 0.5, 0.7, or 0.8 μm SAMHD1 (lanes 1–5) at 37 °C for 45 min. C lane, control. E–H, cleavage of complex nucleic acid substrates by SAMHD1. Shown is the cleavage of dsDNA (E) and DNA-RNA complexes with a 3′-overhang (F), 5′-overhang (G), or blunt ends (H) by 0.4, 0.6, or 0.9 μm SAMHD1 (lanes 1–3) in the presence of Mg2+ or Ca2+. Asterisks indicate the labeled strand in the complex substrate.
In the presence of Mg2+, SAMHD1 did not cleave DNA in dsDNA or DNA-RNA complexes with blunt ends, but it cleaved the double-stranded substrates containing 3′-overhangs and, more slowly, substrates with 5′-overhangs (Fig. 2, E–H). Together with the data presented in Fig. 2 (E–H), the cleavage pattern of SAMHD1 with double-stranded substrates suggests that SAMHD1 is primarily a 3′→5′ exonuclease with low endonuclease or 5′→3′ exonuclease activity. In the presence of Ca2+ and with double-stranded substrates containing the 5′-32P-labeled RNA strand, this protein showed no activity against dsRNA with blunt ends or DNA-RNA complexes with 3′-overhangs and low activity against DNA-RNA complexes with 5′-overhangs (Fig. 2, F–H). However, in the presence of Mg2+, SAMHD1 showed increased cleavage of the labeled RNA strand in DNA-RNA complexes or dsRNA (Fig. 2, F–H), indicating that like RNase H2 (26), SAMHD1 has a preference for RNA in DNA-RNA complexes.
Nucleic Acid-binding Activity of SAMHD1 and Its Domains
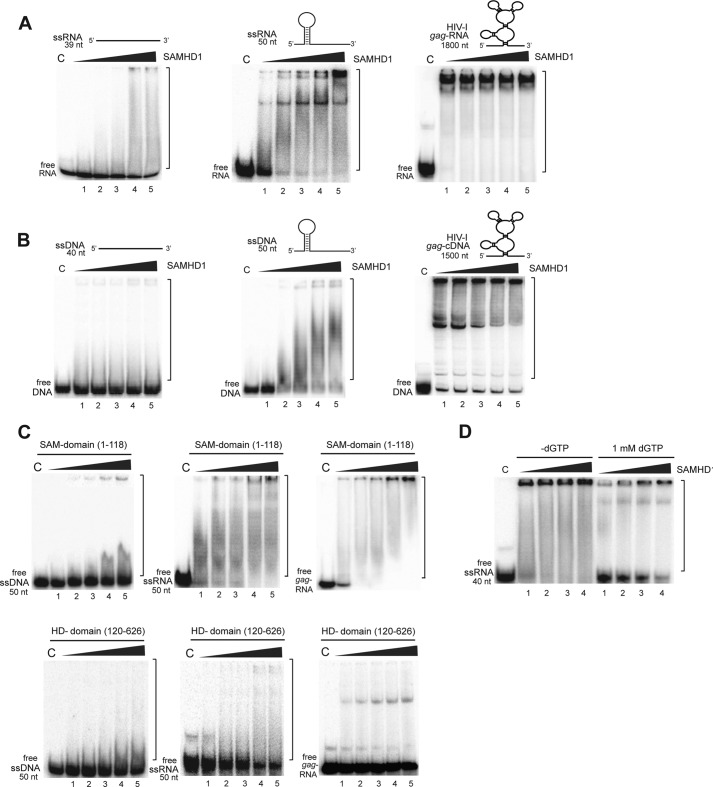
Purified full-length human SAMHD1 showed low binding to linear ssRNA and ssDNA, whereas no binding was observed to double-stranded substrates (Fig. 3, A and B). The binding was stronger to ssDNA and ssRNA substrates containing a stem-loop and was further increased with long substrates containing complex secondary structures such as the HIV-1 gag RNA or gag cDNA (Fig. 3, A and B). This suggests that, in vivo, SAMHD1 is likely to have a high affinity for the HIV-1 genomic RNA and DNA, which are known to contain numerous secondary structures (27). The purified SAM domain (aa 1–118) also exhibited substantial binding to both ssDNA and ssRNA, whereas the isolated HD domain (aa 120–626) showed a lower binding activity (Fig. 3C). This is consistent with the observation of two patches of positively charged residues located near the active site of the SAMHD1 HD domain (Protein Data Bank code 3U1N) and its structural homolog Aq_1910 from Aquifex aeolicus (code 2HEK) (supplemental Fig. S2). Similar to nuclease activity, the addition of dGTP inhibited binding of SAMHD1 to nucleic acids (Fig. 3D). Thus, both SAMHD1 domains contribute to nucleic acid binding, and the full-length protein exhibits a preference for nucleic acids with complex secondary structures, such as in viral ssRNA or ssDNA.
FIGURE 3.
Binding of ssRNA or ssDNA by SAMHD1 and its domains. SAMHD1 at 0.13, 0.25, 0.5, 0.6, or 0.8 μm (lanes 1–5) was incubated with ssRNA (A) or ssDNA (B), and complex formation was analyzed on native polyacrylamide gel. The predicted structures of the substrates used are shown schematically on the top of gel images. C, ssDNA or ssRNA binding by isolated SAM and HD domains at 0.13, 0.25, 0.5, 0.6, or 0.8 μm (lanes 1–5). D, ssRNA binding by SAMHD1 in the presence of dGTP. C lane, control.
Nuclease and dGTP Triphosphatase Activities of Other HD Domain Proteins
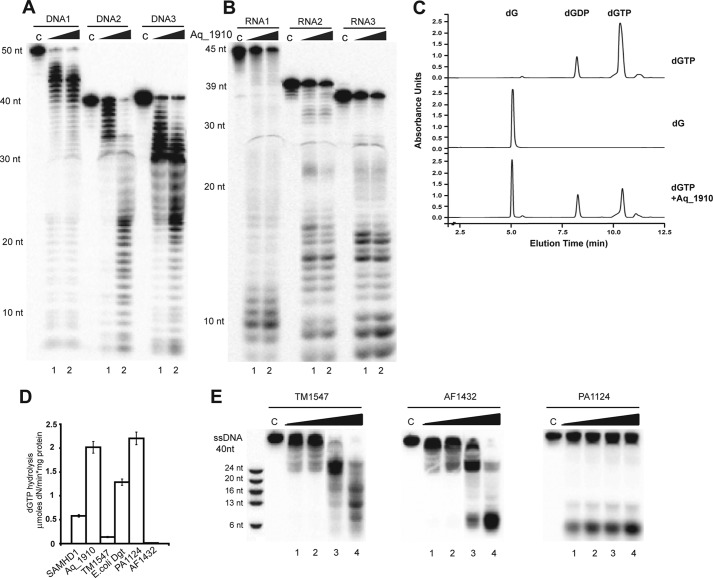
The HD domain of human SAMHD1 has low sequence similarity (25.4% sequence identity) to the HD domain protein Aq_1910 from the thermophilic bacterium A. aeolicus (supplemental Fig. S2), whose crystal structure has been determined by a structural genomics project (28). A Dali search for structural homologs of SAMHD1 identified the Aq_1910 structure as the top structural homolog (Z-score of 32.1 and root mean square deviation of 3.3). The structure of Aq_1910 also revealed the presence of a phosphate molecule and metal ion (Zn2+) bound near the catalytic HD motif (28), but it has a higher resolution (2.0 Å) than the structure of the SAMHD1 HD domain (3.1 Å) (18). We found that purified Aq_1910 also exhibited both dGTP triphosphatase and nuclease activities against ssDNAs and ssRNAs (Fig. 4, A–C). The reaction requirements and substrate cleavage pattern of the Aq_1910 nuclease were similar to those of human SAMHD1 (data not shown). In addition, we identified the presence of both ssDNase and dGTP triphosphatase activities in the purified microbial HD domain proteins TM1547 from Thermotoga maritima and PA1124 from Pseudomonas aeruginosa, which also show low sequence similarity to SAMHD1 (<25% sequence identity) (Fig. 4, D and E, and supplemental Fig. S2). In contrast, AF1432 from Archaeoglobus fulgidus and MJ0384 possessed only ssDNase activity (no dGTPase activity), whereas the E. coli dGTPase Dgt showed no nuclease activity (Fig. 4, D and E). Recently, the HD domain protein EF1143 from Enterococcus faecalis was found to exhibit both dGTP triphosphatase and nuclease activities (22). Thus, HD domain proteins can have either nuclease or dGTPase activity or both activities simultaneously.
FIGURE 4.
Nuclease and dGTP triphosphatase activities of other HD domain proteins. A and B, hydrolysis of different ssDNA substrates (39–50 nt; A) or ssRNA substrates (36–45 nt; B) by Aq_1910. C, HPLC analysis of the reaction products generated by Aq_1910 during hydrolysis of dGTP. Elution profiles are shown (254 nm). D, hydrolysis of dGTP (5 mm) at 37 °C by 4 μm SAMHD1 (15 min at 37 °C), 0.75 μm E. coli Dgt (10 min at 37 °C), 0.9 μm PA1124 (10 min at 37 °C), 1 μm Aq_1910 (30 min at 60 °C), and 8 μm TM1547 (30 min at 60 °C). E, cleavage of ssDNA by TM1547, AF1432, and PA1124. C lane, control.
Site-directed Mutagenesis of SAMHD1 and Aq_1910
Alanine replacement mutagenesis of the four HD motif residues of SAMHD1 (His-167, His-206, Asp-207, and Asp-311) and Aq_1910 (His-54, His-84, Asp-85, and Asp-161) resulted in almost complete loss of their nuclease and dGTP hydrolase activities (Fig. 5). In SAMHD1, these mutations also greatly reduced binding to ssDNA and ssRNA (Fig. 5, B and C). Alanine replacement of several conserved or semiconserved residues located in the active sites of Aq_1910 (His-88, His-93, and Thr-155) and SAMHD1 (His-210, Lys-304, Arg-305, Lys-312, Tyr-315, and Tyr-374) also produced strong negative effects on the nuclease and dGTP hydrolase activities of these proteins (Fig. 5, A, D, and E). In contrast, alanine substitution of non-conserved residues Lys-288, Ser-302, and Lys-312 of SAMHD1 produced mutant proteins with significant nuclease activity (Fig. 5A). Most known SAMHD1 mutations associated with AGS involve amino acid replacements of highly conserved residues located in the HD domain, but the HD motif residues are not affected in the reported mutant proteins (7). We purified four SAMHD1 proteins containing the AGS-related amino acid substitutions (R143H, Q149A, M385V, and Q549A) and found that these proteins showed reduced nuclease activity (Fig. 5A). The SAMHD1-associated SAM domains contain no absolutely conserved residues (supplemental Fig. S3), and alanine substitution of three semiconserved Arg residues (Arg-69, Arg-97, and Arg-106) located on the surface of the SAM domain (Protein Data Bank code 2E8O) resulted in proteins with significant nuclease activity and ssDNA binding (Fig. 5, A and B). These data indicate that both nuclease and dNTP triphosphatase activities are associated with the HD motif residues, which also contribute to nucleic acid binding.
FIGURE 5.
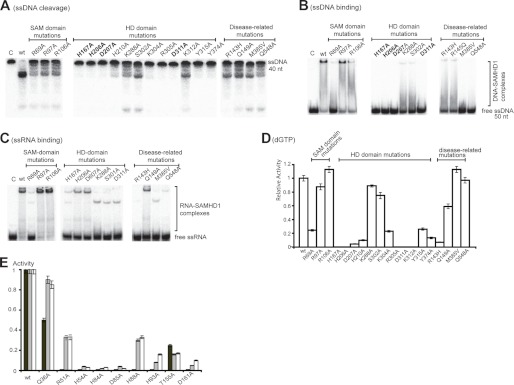
Alanine replacement mutagenesis of SAMHD1 and Aq_1910. A–D, cleavage of ssDNA (A), binding of ssDNA (B) and ssRNA (C), and dGTP triphosphatase activity (D) by the wild-type and mutant SAMHD1 proteins. C lane, control. E, cleavage of dGTP (black bars), ssDNA (gray bars), and ssRNA (white bars) substrates by purified wild-type and mutant proteins.
DISCUSSION
Most of the characterized HD domain proteins show metal-dependent phosphohydrolase activity against various nucleotides and nucleic acids (12, 14, 22, 29). Our results show that, in addition to dNTP hydrolysis, the full-length human SAMHD1 protein possesses nuclease activity toward ssDNAs, ssRNAs, and RNA in DNA/RNA hybrids. Like the characterized CRISPR-associated HD domain nucleases (Cas3), SAMHD1 cleaves nucleic acid substrates mainly as a 3′→5′ exonuclease and shows no obvious cleavage sequence preference. Both the dGTP triphosphatase and nuclease activities of SAMHD1 exhibit similar reaction requirements and are associated with the HD motif residues of the HD domain, suggesting a common active site and catalytic mechanism. Although the SAM domain of human SAMHD1 is not critical for its enzymatic activities, the presence of this domain significantly increases both activities probably through the contribution to substrate binding.
Thus, like two other human proteins associated with AGS and HIV restriction (TREX1 and RNase H2), SAMHD1 has nuclease activity, and its role in HIV-1 restriction and autoimmune response might not be limited to the degradation of DNA precursors (dNTPs). TREX1 is active against ssDNAs and dsDNAs, with the highest activity against dsDNA with 3′-overhangs, whereas RNase H2 degrades RNA in DNA/RNA hybrids (26, 30). It has been suggested that these nucleases are involved in clearing the cellular nucleic acid debris, the accumulation of which results in triggering the innate immune response that is normally induced by viral nucleic acids (31–34). Similar to these nucleases, SAMHD1 cleaves ssDNA and RNA in DNA/RNA hybrids and is also active against ssRNAs and dNTPs. Like TREX1, the expression of the SAMHD1 gene has been shown to be stimulated by immunostimulatory DNA as part of the interferon-stimulatory DNA response, a cytosolic Toll-like receptor-independent antiviral pathway that detects DNA and triggers immune activation (7). It has been proposed that the absence of TREX1, RNase H2, or SAMHD1 activity results in the accumulation of endogenous nucleic acids, which are sensed as “non-self,” inducing an interferon-mediated immune response (8). The demonstration of nuclease activity in SAMHD1 provides further support for this model. This is also in line with the observation that the Vpx-mediated enhancement of HIV infectivity in myeloid cells correlates with the increased accumulation of viral cDNAs (35). Interestingly, human TREX1 has been shown to degrade the excess of cytosolic HIV-1 DNA generated during the reverse transcription step, helping the integrated HIV virus to avoid detection by nucleic acid sensors (36). Because SAMHD1 is localized in the nucleus (7), it has been proposed that it might act on components of the pre-integration complex after nuclear entry, or it might be exported to the cytoplasm during the early phases of infection (6).
Recent progress in our understanding of antiviral mechanisms in different organisms has revealed interesting similarities between these systems in eukaryotes and prokaryotes. For example, the E. coli dGTPase Dgt (an HD domain protein) has been shown to bind to ssDNA with high affinity, and its increased expression in E. coli cells resulted in a 5-fold decrease in the intracellular concentration of dGTP (37, 38). Surprisingly, Dgt overexpression also suppressed the replication of bacteriophage T7 with a mutated gene 1.2, probably because Dgt depletes the dGTP pool, and new phage DNA cannot be produced (39). In vitro, the purified gene 1.2 protein has been shown to bind specifically to Dgt, resulting in inhibition of its dGTPase activity and binding to ssDNA (38). SAMHD1 also restricts HIV-1 replication by depleting the intracellular dNTP pool and is counteracted by the HIV-2 accessory protein Vpx (4, 5, 21). The T7 gene 1.2 protein and lentiviral Vpx share no sequence homology, and they represent a remarkable example of functional similarity of the anti-restriction strategy of prokaryotic and eukaryotic viruses. Another case is the microbial antiviral defense system CRISPR, which employs the HD domain-containing proteins (Cas3) as effector nucleases degrading viral nucleic acids (12, 17). Thus, HD domain proteins play an important role in antiviral defense and represent a universal component of the antiviral armamentarium in both prokaryotes and eukaryotes. Sequenced genomes contain multiple genes encoding unknown HD domain proteins, and their experimental characterization will likely reveal additional antiviral factors.
Acknowledgments
We thank Rosalind Kim (University of California, Berkeley, CA) for the Aq_1910 expression plasmid, Elena Dobrovetsky (Structural Genomics Consortium, Toronto, Canada) for providing the SAMHD1 clone from the Mammalian Gene Collection, and Andrew Mouland (McGill University, Montreal, Canada) for providing the HIV-1 gag and tat plasmids for RNA synthesis. We thank all members of the Structural Proteomics in Toronto (SPiT) Centre for help in conducting the experiments.
This work was supported by the Government of Canada through Genome Canada and Ontario Genomics Institute Grant 2009-OGI-ABC-1405, Ontario Research Fund Grant ORF-GL2–01-004, and a Natural Sciences and Engineering Research Council (NSERC) Discovery team grant (to A. F. Y.).

This article contains supplemental Figs. S1–S3.
- AGS
- Aicardi-Goutières autoimmune syndrome
- aa
- amino acids
- ss
- single-stranded
- nt
- nucleotides
- PNK
- T4 polynucleotide kinase
- ds
- double-stranded
- TdT
- terminal deoxynucleotidyl transferase.
REFERENCES
- 1. Bieniasz P. D. (2004) Intrinsic immunity: a front-line defense against viral attack. Nat. Immunol. 5, 1109–1115 [DOI] [PubMed] [Google Scholar]
- 2. Lilley C. E., Chaurushiya M. S., Boutell C., Everett R. D., Weitzman M. D. (2011) The intrinsic antiviral defense to incoming HSV-1 genomes includes specific DNA repair proteins and is counteracted by the viral protein ICP0. PLoS Pathog. 7, e1002084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Weitzman M. D., Lilley C. E., Chaurushiya M. S. (2010) Genomes in conflict: maintaining genome integrity during virus infection. Annu. Rev. Microbiol. 64, 61–81 [DOI] [PubMed] [Google Scholar]
- 4. Laguette N., Sobhian B., Casartelli N., Ringeard M., Chable-Bessia C., Ségéral E., Yatim A., Emiliani S., Schwartz O., Benkirane M. (2011) SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hrecka K., Hao C., Gierszewska M., Swanson S. K., Kesik-Brodacka M., Srivastava S., Florens L., Washburn M. P., Skowronski J. (2011) Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Berger A., Sommer A. F., Zwarg J., Hamdorf M., Welzel K., Esly N., Panitz S., Reuter A., Ramos I., Jatiani A., Mulder L. C., Fernandez-Sesma A., Rutsch F., Simon V., König R., Flory E. (2011) SAMHD1-deficient CD14+ cells from individuals with Aicardi-Goutières syndrome are highly susceptible to HIV-1 infection. PLoS Pathog. 7, e1002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rice G. I., Bond J., Asipu A., Brunette R. L., Manfield I. W., Carr I. M., Fuller J. C., Jackson R. M., Lamb T., Briggs T. A., Ali M., Gornall H., Couthard L. R., Aeby A., Attard-Montalto S. P., Bertini E., Bodemer C., Brockmann K., Brueton L. A., Corry P. C., Desguerre I., Fazzi E., Cazorla A. G., Gener B., Hamel B. C., Heiberg A., Hunter M., van der Knaap M. S., Kumar R., Lagae L., Landrieu P. G., Lourenco C. M., Marom D., McDermott M. F., van der Merwe W., Orcesi S., Prendiville J. S., Rasmussen M., Shalev S. A., Soler D. M., Shinawi M., Spiegel R., Tan T. Y., Vanderver A., Wakeling E. L., Wassmer E., Whittaker E., Lebon P., Stetson D. B., Bonthron D. T., Crow Y. J. (2009) Mutations involved in Aicardi-Goutières syndrome implicate SAMHD1 as regulator of the innate immune response. Nat. Genet. 41, 829–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Crow Y. J., Rehwinkel J. (2009) Aicardi-Goutières syndrome and related phenotypes: linking nucleic acid metabolism with autoimmunity. Hum. Mol. Genet. 18, R130–R136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hartman Z. C., Kiang A., Everett R. S., Serra D., Yang X. Y., Clay T. M., Amalfitano A. (2007) Adenovirus infection triggers a rapid, MyD88-regulated transcriptome response critical to acute-phase and adaptive immune responses in vivo. J. Virol. 81, 1796–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schultz J., Ponting C. P., Hofmann K., Bork P. (1997) SAM as a protein interaction domain involved in developmental regulation. Protein Sci. 6, 249–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Aravind L., Koonin E. V. (1998) The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem. Sci. 23, 469–472 [DOI] [PubMed] [Google Scholar]
- 12. Beloglazova N., Petit P., Flick R., Brown G., Savchenko A., Yakunin A. F. (2011) Structure and activity of the Cas3 HD nuclease MJ0384, an effector enzyme of the CRISPR interference. EMBO J. 30, 4616–4627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oussenko I. A., Sanchez R., Bechhofer D. H. (2002) Bacillus subtilis YhaM, a member of a new family of 3′→5′ exonucleases in Gram-positive bacteria. J. Bacteriol. 184, 6250–6259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zimmerman M. D., Proudfoot M., Yakunin A., Minor W. (2008) Structural insight into the mechanism of substrate specificity and catalytic activity of an HD domain phosphohydrolase: the 5′-deoxyribonucleotidase YfbR from Escherichia coli. J. Mol. Biol. 378, 215–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kornberg S. R., Lehman I. R., Bessman M. J., Simms E. S., Kornberg A. (1958) Enzymatic cleavage of deoxyguanosine triphosphate to deoxyguanosine and tripolyphosphate. J. Biol. Chem. 233, 159–162 [PubMed] [Google Scholar]
- 16. Kim C. A., Bowie J. U. (2003) SAM domains: uniform structure, diversity of function. Trends Biochem. Sci. 28, 625–628 [DOI] [PubMed] [Google Scholar]
- 17. Sinkunas T., Gasiunas G., Fremaux C., Barrangou R., Horvath P., Siksnys V. (2011) Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 30, 1335–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Goldstone D. C., Ennis-Adeniran V., Hedden J. J., Groom H. C., Rice G. I., Christodoulou E., Walker P. A., Kelly G., Haire L. F., Yap M. W., de Carvalho L. P., Stoye J. P., Crow Y. J., Taylor I. A, Webb M. (2011) HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480, 379–382 [DOI] [PubMed] [Google Scholar]
- 19. Powell R. D., Holland P. J., Hollis T., Perrino F. W. (2011) Aicardi-Goutières syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. J. Biol. Chem. 286, 43596–43600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goncalves A., Karayel E., Rice G. I., Bennett K. L., Crow Y. J., Superti-Furga G., Bürckstümmer T. (2012) SAMHD1 is a nucleic-acid binding protein that is mislocalized due to Aicardi-Goutières syndrome-associated mutations. Hum. Mutat. 33, 1116–1122 [DOI] [PubMed] [Google Scholar]
- 21. Lahouassa H., Daddacha W., Hofmann H., Ayinde D., Logue E. C., Dragin L., Bloch N., Maudet C., Bertrand M., Gramberg T., Pancino G., Priet S., Canard B., Laguette N., Benkirane M., Transy C., Landau N. R., Kim B., Margottin-Goguet F. (2012) SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol. 13, 223–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vorontsov I. I., Minasov G., Kiryukhina O., Brunzelle J. S., Shuvalova L., Anderson W. F. (2011) Characterization of the deoxynucleotide triphosphate triphosphohydrolase (dNTPase) activity of the EF1143 protein from Enterococcus faecalis and crystal structure of the activator-substrate complex. J. Biol. Chem. 286, 33158–33166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mulepati S., Bailey S. (2011) Structural and biochemical analysis of nuclease domain of clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein 3 (Cas3). J. Biol. Chem. 286, 31896–31903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Beloglazova N., Brown G., Zimmerman M. D., Proudfoot M., Makarova K. S., Kudritska M., Kochinyan S., Wang S., Chruszcz M., Minor W., Koonin E. V., Edwards A. M., Savchenko A., Yakunin A. F. (2008) A novel family of sequence-specific endoribonucleases associated with the clustered regularly interspaced short palindromic repeats. J. Biol. Chem. 283, 20361–20371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mouland A. J., Xu H., Cui H., Krueger W., Munro T. P., Prasol M., Mercier J., Rekosh D., Smith R., Barbarese E., Cohen E. A., Carson J. H. (2001) RNA trafficking signals in human immunodeficiency virus type 1. Mol. Cell. Biol. 21, 2133–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cerritelli S. M., Crouch R. J. (2009) Ribonuclease H: the enzymes in eukaryotes. FEBS J. 276, 1494–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Watts J. M., Dang K. K., Gorelick R. J., Leonard C. W., Bess J. W., Jr., Swanstrom R., Burch C. L., Weeks K. M. (2009) Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 460, 711–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Oganesyan V., Adams P. D., Jancarik J., Kim R., Kim S. H. (2007) Structure of O67745_AQUAE, a hypothetical protein from Aquifex aeolicus. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 63, 369–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yakunin A. F., Proudfoot M., Kuznetsova E., Savchenko A., Brown G., Arrowsmith C. H., Edwards A. M. (2004) The HD domain of the Escherichia coli tRNA nucleotidyltransferase has 2′,3′-cyclic phosphodiesterase, 2′-nucleotidase, and phosphatase activities. J. Biol. Chem. 279, 36819–36827 [DOI] [PubMed] [Google Scholar]
- 30. Mazur D. J., Perrino F. W. (1999) Identification and expression of the TREX1 and TREX2 cDNA sequences encoding mammalian 3′→5′ exonucleases. J. Biol. Chem. 274, 19655–19660 [DOI] [PubMed] [Google Scholar]
- 31. Crow Y. J., Leitch A., Hayward B. E., Garner A., Parmar R., Griffith E., Ali M., Semple C., Aicardi J., Babul-Hirji R., Baumann C., Baxter P., Bertini E., Chandler K. E., Chitayat D., Cau D., Déry C., Fazzi E., Goizet C., King M. D., Klepper J., Lacombe D., Lanzi G., Lyall H., Martínez-Frías M. L., Mathieu M., McKeown C., Monier A., Oade Y., Quarrell O. W., Rittey C. D., Rogers R. C., Sanchis A., Stephenson J. B., Tacke U., Till M., Tolmie J. L., Tomlin P., Voit T., Weschke B., Woods C. G., Lebon P., Bonthron D. T., Ponting C. P., Jackson A. P. (2006) Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutières syndrome and mimic congenital viral brain infection. Nat. Genet. 38, 910–916 [DOI] [PubMed] [Google Scholar]
- 32. Alarcón-Riquelme M.-E. (2006) Nucleic acid by-products and chronic inflammation. Nat. Genet. 38, 866–867 [DOI] [PubMed] [Google Scholar]
- 33. Yang Y. G., Lindahl T., Barnes D. E. (2007) Trex1 exonuclease degrades ssDNA to prevent chronic checkpoint activation and autoimmune disease. Cell 131, 873–886 [DOI] [PubMed] [Google Scholar]
- 34. Stetson D. B., Ko J. S., Heidmann T., Medzhitov R. (2008) Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 134, 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Goujon C., Rivière L., Jarrosson-Wuilleme L., Bernaud J., Rigal D., Darlix J. L., Cimarelli A. (2007) SIVSM/HIV-2 Vpx proteins promote retroviral escape from a proteasome-dependent restriction pathway present in human dendritic cells. Retrovirology 4, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yan N., Regalado-Magdos A. D., Stiggelbout B., Lee-Kirsch M. A., Lieberman J. (2010) The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat. Immunol. 11, 1005–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Myers J. A., Beauchamp B. B., Richardson C. C. (1987) Gene 1.2 protein of bacteriophage T7. Effect on deoxyribonucleotide pools. J. Biol. Chem. 262, 5288–5292 [PubMed] [Google Scholar]
- 38. Wurgler S. M., Richardson C. C. (1993) DNA binding properties of the deoxyguanosine triphosphate triphosphohydrolase of Escherichia coli. J. Biol. Chem. 268, 20046–20054 [PubMed] [Google Scholar]
- 39. Huber H. E., Beauchamp B. B., Richardson C. C. (1988) Escherichia coli dGTP triphosphohydrolase is inhibited by gene 1.2 protein of bacteriophage T7. J. Biol. Chem. 263, 13549–13556 [PubMed] [Google Scholar]