Background: Iron-induced oligomerization of frataxin is still poorly understood.
Results: The molecular basis of iron-induced oligomerization of yeast and bacterial frataxin is revealed. Catalyzed ferroxidation is required for correct oligomerization of Yfh1.
Conclusion: Frataxin forms different oligomeric species at physiological conditions.
Significance: Iron availability controls frataxin oligomerization, which in turn may control the processes that require iron delivery by frataxin.
Keywords: Crystal Structure, Iron, Iron Metabolism, Metalloproteins, Protein Structure, Frataxin, Oligomerization, Protein-Protein Interactions, SAXS
Abstract
The role of the mitochondrial protein frataxin in iron storage and detoxification, iron delivery to iron-sulfur cluster biosynthesis, heme biosynthesis, and aconitase repair has been extensively studied during the last decade. However, still no general consensus exists on the details of the mechanism of frataxin function and oligomerization. Here, using small-angle x-ray scattering and x-ray crystallography, we describe the solution structure of the oligomers formed during the iron-dependent assembly of yeast (Yfh1) and Escherichia coli (CyaY) frataxin. At an iron-to-protein ratio of 2, the initially monomeric Yfh1 is converted to a trimeric form in solution. The trimer in turn serves as the assembly unit for higher order oligomers induced at higher iron-to-protein ratios. The x-ray crystallographic structure obtained from iron-soaked crystals demonstrates that iron binds at the trimer-trimer interaction sites, presumably contributing to oligomer stabilization. For the ferroxidation-deficient D79A/D82A variant of Yfh1, iron-dependent oligomerization may still take place, although >50% of the protein is found in the monomeric state at the highest iron-to-protein ratio used. This demonstrates that the ferroxidation reaction controls frataxin assembly and presumably the iron chaperone function of frataxin and its interactions with target proteins. For E. coli CyaY, the assembly unit of higher order oligomers is a tetramer, which could be an effect of the much shorter N-terminal region of this protein. The results show that understanding of the mechanistic features of frataxin function requires detailed knowledge of the interplay between the ferroxidation reaction, iron-induced oligomerization, and the structure of oligomers formed during assembly.
Introduction
Most organisms require iron-containing prosthetic groups such as heme and iron-sulfur clusters. However, the presence of iron in living cells and its potential toxicity under physiological conditions require iron chemistry to be tightly controlled. Iron toxicity mainly arises from the combination of ferrous iron with hydrogen peroxide in the Fenton reaction, which produces the highly reactive hydroxyl radical •OH. Additional toxic effects are related to the deposition of the insoluble Fe(OH)3 at physiological pH. Iron storage and detoxification proteins such as ferritin and the mitochondrial protein frataxin normally prevent these effects. They both detoxify Fe2+ by catalyzing the ferroxidation reaction that proceeds through two one-electron oxidations, with O2 or H2O2 as electron acceptor (1, 2). The oxidized iron is subsequently stored as a ferrihydrite biomineral within oligomeric species (3, 4). The toxicity of unligated iron may have implications in several neurodegenerative diseases (5). One of these is Friedreich ataxia, an autosomal recessive neurodegenerative disease of humans that affects 1 in 40,000 newborns (for review, see Ref. 6). Friedreich ataxia has been shown to be associated in most patients with reduced levels of frataxin, caused by expanded GAA repeats in the first intron of the gene encoding the protein (7). In addition, several mutations in the frataxin gene have been found to lead to Friedreich ataxia (7–12).
Frataxin has been directly implicated as an iron chaperone in both heme and iron-sulfur cluster biosynthesis and repair processes that take place inside the mitochondrial matrix (13–19). However, the details of its iron chaperone function, the mechanism of target recognition, and the mode of interactions with its targets are poorly understood. Several studies have demonstrated the propensity of human, yeast, and bacterial frataxin to assemble into oligomeric species (20–23). Thus, the frataxin homolog CyaY from Escherichia coli has been suggested to assemble into tetrameric species under anaerobic conditions (22) and larger oligomers under aerobic conditions (23). For the yeast frataxin homolog (Yfh1), iron-dependent formation of trimers, hexamers, and higher order oligomers, including 24- and 48-meric particles, has been suggested based on various biochemical studies under aerobic conditions (2, 15, 20, 24). Interestingly, the oligomeric species of Yfh1 can be easily dissociated into monomers, for example by the addition of reducing agents (15), suggesting that iron oxidation promotes oligomer formation and may be essential for oligomer stabilization.
Eukaryotic frataxins are initially synthesized in the cytoplasm as precursor polypeptides, which are imported to the mitochondrial matrix and processed to shorter forms. Whereas only one form of Yfh1 (residues 52–174) is normally detected in yeast mitochondria, human frataxin (FXN),5 which is expressed in the cytoplasm as a 210-residue polypeptide, has been observed to be present in mitochondria in different forms with variable length of the N-terminal part (21, 25–28). Thus, the variants with the longer N termini (FXN56–210 and FXN42–210) are often found assembled into larger structures in vivo and during expression in E. coli (21, 27). The larger oligomers can be disassembled irreversibly into stable monomers by the addition of SDS (29). The two other known shorter variants of FXN, FXN81–210 and FXN78–210, could not be demonstrated to adopt higher order oligomeric states in vitro (27, 30). The nature of the oligomers formed by human frataxin is not known, although its assembly has been shown to be essential for the detoxification of redox-active iron (29). These data suggest that frataxin from different organisms as well as the different isoforms with different N-terminal lengths may follow different oligomerization pathways and may even assemble into different types of oligomers. However, it should be noted that the research community is still divided about the ability of frataxin to oligomerize in vivo and the functional importance of the oligomers (for review, see Ref. 31).
The x-ray crystallographic structure of the trimeric form of the Y73A variant of yeast frataxin suggested that oligomer formation was essential for the formation of the ferroxidation site of the protein and revealed details of the interactions that result in trimer stabilization (32). The low resolution electron microscopic structures of iron-free and iron-loaded 24-meric particles of the same variant of the protein suggested that the trimer could serve as a building block even for other higher order oligomers (33). Using small angle x-ray scattering (SAXS), in the first systematic study of metal-induced oligomerization of wild-type Yfh1 it was also shown that, although at relatively high and non-physiological concentrations, Co2+ was able to induce the formation of oligomeric structures consisting of dimers, trimers, hexamers, and higher order oligomers (34). The work also suggested that the flexible N terminus of the protein might play an important role in the stabilization of the oligomers observed in this case.
Although frataxin from different organisms appears to behave differently with respect to oligomerization, the high degree of amino acid sequence and three-dimensional structure conservation within the family suggests the existence of common mechanistic features, which control frataxin propensity toward oligomer formation. Understanding this process at the three-dimensional structural level is of central importance for understanding the details of frataxin function and the interplay between the ferroxidation reaction, iron-induced oligomerization, and the architecture of the particles formed during oligomer assembly. In this study, using a combination of small-angle x-ray scattering, x-ray crystallography, and protein-protein docking, we have characterized the three-dimensional structure of yeast frataxin Yfh1 and E. coli CyaY oligomers formed during iron-dependent oligomerization of wild-type and a ferroxidation-deficient D79A/D82A variant of the protein (35).
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The mature form of wild-type Yfh1 and the D79A/D82A variant of Yfh1 were expressed and purified as described previously (36), except that an HR16/50 Superdex 75 column (GE Healthcare) was used for the final step of purification. CyaY was purified according to (37) with the difference that the second-step anion exchange using Macro-Prep High Q was not performed.
SAXS Data Collection and Analysis
For data collection, 3 mg/ml (∼0.22 mm) solution of wild-type Yfh1 and the D79A/D82A variant were incubated with ferrous ammonium sulfate at concentrations between 0.11 and 2.2 mm for 60 min at 30 °C according to previous protocols (15). The effect of the presence of magnesium ions on iron-induced oligomerization was assessed by repeating the incubation of wild-type Yfh1 at the highest iron concentration but in presence of 10 mm MgCl2. After incubation the samples were dialyzed using Slide-A-Lyzer MINI dialysis units (Thermo Fisher Scientific) against buffer containing 20 mm HEPES-NaOH and 100 mm NaCl. The higher protein concentrations (6 and 9 mg/ml protein) were used for measurements without added iron. Bacterial frataxin homolog CyaY was treated similarly.
SAXS data were collected at beamline I711 (38) at the MAX-lab synchrotron and at the EMBL X33 beamline at the DESY storage ring DORIS III (39, 40) as described previously (34). The data collected at X33 were reduced and processed initially using an automatic pipeline of scripts developed at EMBL Hamburg (41). Data were normalized to the intensity of the transmitted beam, and scattering of the buffer was subtracted. The processing was done using the ATSAS software package (42, 43). Forward scattering I(0) and the radius of gyration Rg were evaluated using the Guinier approximation (44). These parameters were also computed from the entire scattering patterns using the program GNOM (45), which provides the distance distribution functions P(r) and the maximum particle dimensions Dmax. Molecular weight estimates were made using lysozyme and bovine serum albumin as standards or from the excluded volume of the hydrated particle (the Porod volume Vp) (43, 46).
Flexibility Assessment and Modeling of Yfh1 D79A/D82A Variant and CyaY
To assess the flexibility of the D79A/D82A variant of Yfh1, we used ensemble optimization method (47). Two thousand monomers were created with the program RanCh from the ensemble optimization method program suite using one rigid body (residues 52–174, PDB entry 3OEQ). Subsequently, 2000 dimers were created by first generating low energy conformations of Yfh1 amino acid residues 52–174 using kinematic loop modeling (48) in the Rosetta software suite 3.2.1 (49). At the next stage these were combined uniquely in pairs of two (based on two monomers in the crystallographic trimer of PDB 3OEQ). Theoretical form-factors were calculated using CRYSOL (50). GAJOE (from the ensemble optimization method suite) was then used employing a genetic algorithm to select subsets of protein models to minimize the discrepancy between the experimental data and the average theoretical scattering for the subset. Multiple runs of GAJOE were performed using default parameters, except the number of curves per ensemble, which varied between 5 and 20.
We used the program OLIGOMER (42) for fitting of observed scattering curves of CyaY by weighted combinations of theoretical form-factors from the 20 models in PDB entry 1SOY (51). Low resolution ab initio models were generated with the programs DAMMIF and GASBOR (52, 53) and averaged using the DAMAVER suite (54).
Modeling of Iron-induced Oligomers
For fitting SAXS data, which were collected from Yfh1 samples incubated at six different Fe2+:Yfh1 ratios (1:2, 1:1, 2:1, 4:1, 7:1, and 10:1), we used an approach similar to that described previously (34). Briefly, we assembled a pool of models using NMR conformers from PDB entry 2GA5 (55), the asymmetric unit (a monomer), and the crystallographic trimer from PDB entry 3OEQ (34) as well as locally refined variants of the crystallographic trimer using Rosetta 3.2.1 symmetric docking (56). Hexamer, dodecamer, and 24-mer models were prepared as follows. Two crystallographic trimers were docked into the single-particle EM reconstruction of the iron-loaded 24-mer (33) and refined using the Rosetta 3.2.1 docking program (applying the “local refine” flag). Guided by the original EM reconstruction, the trimer-trimer interface of the Rosetta-refined hexamer model (Fig. 6) was used to generate a dodecamer and 24-mer complexes; these three models were subsequently used in SAXS fitting procedures, which also included the original model obtained in the EM reconstruction of the iron-loaded Yfh1 24-mer. In addition, by combining 18-, 21-, and 24-subunit oligomers, particles containing 42–81 monomers were created in an attempt to reconstruct the large rod-shaped particles observed earlier in EM micrographs (20, 21). CRYSOL (50) was used to calculate form-factors from Yfh1 models in the pool described above. For fitting of observed scattering curves to the weighted combinations of form-factors, we used OLIGOMER (42).
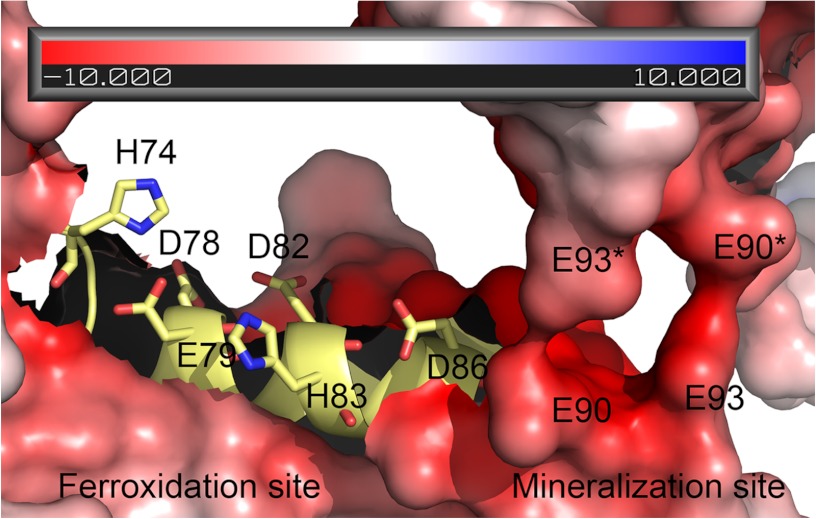
FIGURE 6.
The ferroxidation and mineralization sites of yeast frataxin in the context of the hexamer structure. The surface is colored according to electrostatic potential calculated using the APBS tools (75) plugin in PyMOL with standard settings. The increasing red represents increasing negative charge, white is neutral charge, and increasing blue represents positive charge. The asterisk denotes amino acids from the adjacent subunit on the other trimer. Amino acid side chains are shown as sticks.
Tetrameric CyaY was built based on the analysis of the interactions, which stabilize Yfh1 hexamers. It was noted that the core of the hexamer structure constituted a tetramer with extensive interactions between the monomers. Several conserved amino acids were found to be involved in these interactions. CyaY monomers were then superimposed on these Yfh1 monomers, and the resulting tetramer was used to generate a dimer. A pool of form-factors was then generated as described above, including also NMR conformers from PDB entry 1SOY.
X-ray Crystallography
Three-dimensional crystals were grown anaerobically under the conditions described by Söderberg et al. (34). Before flash-freezing in liquid nitrogen, the crystals were soaked for 2–5 s under anaerobic conditions in a cryoprotective solution containing 2 m (NH4)2SO4, 4% γ-butyrolactone, 20% glycerol, and 0.1 m Tris HCl, pH 8.5. In addition, 4 mm ferrous ammonium sulfate was added to the solution. X-ray data to 3.0 Å resolution were collected at the MAX-Lab synchrotron, beamline I911-3. XDS software (57) was used for indexing, integration, scaling, and merging the data. PHASER (58) software was used for molecular replacement using the previously determined crystallographic structure of the Y73A variant of Yfh1 (PDB entry 2FQL). Refinement was carried out using alternate runs of the program PHENIX (59). Each run was followed by inspection and manual rebuilding of the model using COOT 0.4.1 (60). For TLS refinement, the whole monomeric unit (residues 52–172) was considered as one TLS group. Splitting of the structure into two (52–60 and 61–172) or three groups (52–60, 61–75, and 76–172) did not result in any improvement in map quality or model geometry. The mean value for the B-factors was refined to 93 Å2. For localization of metal positions, anomalous Fourier maps were calculated using the Bijvoet differences (F+ − F−) as coefficients. The metal density peaks were clearly visible both in the Fo − Fc and anomalous difference density maps at levels of up to 8 and 2.5 σ, respectively. The occupancy of the metal was determined by assigning different values to the occupancy followed by inspection of the difference electron density. At the occupancy value of 1, no residual electron density was present, whereas the occupancy and the B-factor of the metal was refined to a value similar to that of the neighboring protein groups, which indicates the correctness of the chosen occupancy value. The coordinates have been deposited to the PDB (PDB entry 4EC2). A summary of data collection and refinement statistics is presented in Table 2.
TABLE 2.
X-ray data collection and refinement of Yfh1 Y73A variant soaked with Fe2+
r.m.s.d., root mean square deviation.
| Yfh1 soaked with Fe2+ | |
|---|---|
| Data collection | |
| Beamline | I911-2 |
| Space group | I213 |
| Wavelength (Å) | 1.038 |
| Cell dimensions, a = b = c (Å) | 121.57 |
| Resolution range (Å) | 30.0-3.0 (3.08-3.00) |
| Completeness (%) | 96.9 (93.3) |
| I/σ(I) | 11.6 (1.8) |
| No. of unique reflections | 11,254 (791) |
| Rmergea | 0.064 (0.63) |
| Refinement | |
| Rcrystb (Rfree) | 0.25 (0.31) |
| r.m.s.d.bond (Å) | 0.006 |
| r.m.s.d.angles (°) | 1.271 |
a Rmerge = Σ|Ii − 〈I〉|/ΣI, where Ii is an individual intensity measurement, and 〈I〉 is the average intensity for this reflection.
b Rcryst = Σ|Fobs − Fcalc|/ΣFobs, where Fobs and Fcalc are the observed and calculated structure factor amplitudes, respectively. Rfree is the same as Rcryst but was calculated on 5% of the data excluded from refinement. Values in the parentheses correspond to the highest resolution bin.
Sequence Analysis
For sequence analysis, in addition to sequences from organisms for which biochemical and biophysical data were available, we used sequences from randomly chosen organisms. Of the final 47 sequences (supplemental Fig. S4), 25 were of bacterial origin, and 22 were of eukaryotic origin. The ClustalW2 multiple alignment tool (61) at EBI (62) was used for initial alignment of the sequences. The alignment was then manually corrected in Jalview 2.3 (63) using the structures of the monomeric yeast frataxin, human frataxin, and the bacterial analog CyaY (55, 64, 65).
For analysis of the variability of the amino acid residues at different positions in the sequence, Shannon entropy is often used as a measure. Here we use the method of Fornasari et al. (66), which is based on the so-called reduced Shannon entropy, calculated for each alignment position i using the expression,
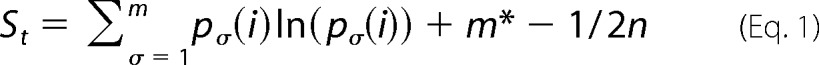 |
In the first part of the equation, which represents a standard Shannon entropy calculation, σ is a given class of amino acids according to Ptitsyn (67) ((1)-aromatics (Phe, Tyr, and Trp), (2)-bulky aliphatics (Lei, Ile, Val, and Met), (2)-small non-polar (Gly and Ala), (4)-acidic or amides (Glu, Asp, Gln, and Asn), (5)-basic (Lys, Arg, His), (6)-with hydroxyl (Ser and Thr), (7)-others (Pro and Cys)). This classification is introduced to account for conservative substitutions within the amino acid sequence; m is the number of classes (seven in this case); p(i) is the probability that the ith position in the sequence is occupied by a residue of class σ. The second term was introduced as a correction for a systematic bias in the estimation of Si. Here, m* is the number of amino acid classes for which p(i) ≠ 0, and n is the number of sequences in the alignment. Subsequently, the average entropy of the whole polypeptide chain calculated in this way was used to normalize Si, the value of which may depend on the choice of the aligned sequences, their number, and their diversity (68). Thus, Si < 1 implies a stronger conservation relative to the polypeptide as a whole. In addition, we calculated the average residual entropy for residues involved in monomer-monomer interactions at the trimer interface and the residues located at the interface between two trimers (Table 3).
TABLE 3.
Normalized average Shannon entropy Si of residues involved in yeast frataxin trimer and hexamer interfaces
For eukaryotes, the area-weighted average Shannon entropy of the surface and interface is presented using the surface and trimer/hexamer interface packing calculated on the basis of yeast frataxin structure.
| Surface region | Eukaryotes | Bacteria |
|---|---|---|
| 〈Si〉 trimer interface | 0.76 | 0.91 |
| 〈Si〉 hexamer 1 interface | 0.69 | 0.93 |
| 〈Si〉 hexamer 2 interface | 0.60 | 0.84 |
| Ssurf | 1.01>a | |
| Sinterf trimer | 0.75>a | |
| Sinterf hexamer 1 | 0.83>a | |
| Sinterf hexamer 2 | 0.53>a |
a Based on yeast frataxin surface. Si < 1 means that position i is better conserved than the chain average, whereas Si > 1 means that it is more divergent. Hexamers 1 and 2 were obtained from the EM reconstruction of the 24-mer of the iron-free and iron-loaded Y73A variant of Yfh1, respectively (19, 20) after Rosetta refinement (see “Experimental Procedure”).
Using the PISA server (69), the accessible surface area for the yeast frataxin trimer (PDB entry 3OEQ) was calculated. In addition, we calculated the buried surface area for each amino acid residue contributing to the interface between two subunits in the crystallographic trimer (PDB entry 3OEQ) as well as between trimers in the hexamer model described above. Area-weighted average values Sinterf and Ssurf (Table 3) could then be calculated using the general equation from Dey et al. (68),
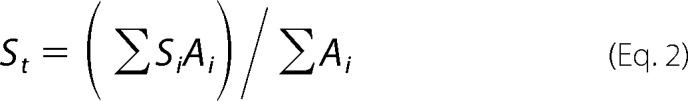 |
For Ssurf, Ai is the area that amino acid residue i contributes to the accessible surface area, and for Sinterf, Ai it is the area that amino acid residue i contributes to the buried surface area.
RESULTS
Iron-induced Oligomerization of Wild-type Yfh1 and D79A/D82A Yfh1 Variant
Earlier studies using SAXS showed that wild-type Yfh1 in solution has a highly flexible N terminus, a radius of gyration Rg of 1.80 nm, and an estimated maximum dimension of the molecule (Dmax) of 6.8 nm (34). Here we examined the solution structure of the D79A/D82A variant in buffer conditions similar to those used previously for wild-type Yfh1. Although deficient in ferroxidation activity, this yeast frataxin variant was still able to contribute to iron accumulation and formation of insoluble iron core (35). Supplemental Table S1 summarizes the structural parameters extracted from the SAXS experimental data, whereas supplemental Fig. S1a shows representative models of D79A/D82A Yfh1 monomer and dimer from the optimized ensemble, as selected by the program GAJOE for fitting the SAXS data. The overall parameters were essentially similar to those obtained earlier for the wild-type protein, although the D79A/D82A variant was prone to dimer formation (20% dimers and 80% monomers), much like wild-type Yfh1 in the presence of glycerol (34).
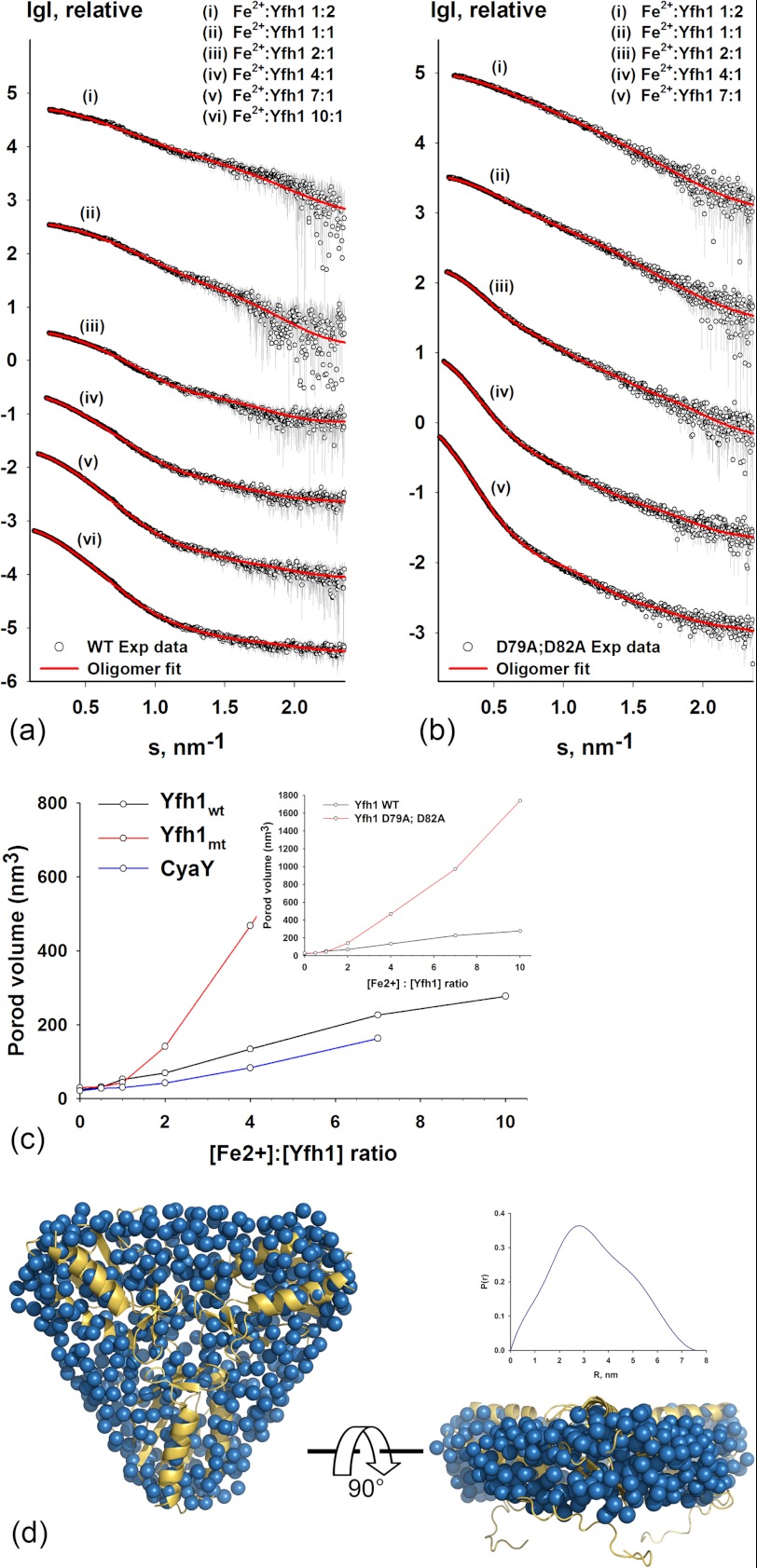
SAXS scattering data in the presence of iron and the corresponding fitting curves are shown for wild-type Yfh1 and the D79A/D82A variant in Figs. 1, a and b, respectively. Analysis of the Guinier region (supplemental Fig. S2, a and b) confirmed that the samples were not aggregated after the addition of Fe2+. The incubation of the proteins with Fe2+ at six different iron-to-protein ratios (1:2, 1:1, 2:1, 4:1, 7:1, and 10:1) resulted in clearly different scattering profiles for wild-type Yfh1 and the D79A/D82A variant. This may also be illustrated by the dependence of the excluded volume of the hydrated particle (Porod volume (46)) calculated from the SAXS data on iron concentration. As shown in Fig. 1c, the Porod volume increased much more rapidly for the D79A/D82A variant than for wild-type Yfh1 (Fig. 1c, red versus black curve), indicating that larger oligomers were formed in response to higher iron concentrations. A similar trend could be followed for Rg and Dmax values (supplemental Table S1).
FIGURE 1.
Metal-dependent oligomerization of frataxin. Shown are experimental SAXS profiles of wild-type (a) and D79A/D82A Yfh1 (b) after incubation at different iron-to-protein ratios. The curves were displaced appropriately along the logarithmic axis for better visualization and overlaid on the corresponding fits. The experimental data are shown as circles, and the corresponding fit to a pool of oligomers is shown as red lines (see Fig. 2 and Table 1 for details of the oligomeric content). The data are presented as the logarithm of the scattering intensity versus the momentum transfer s = 4πsin(θ)/λ (where 2θ is the scattering angle, and λ is the x-ray wavelength) and are shown to a maximal momentum transfer of s = 2.5 nm−1. c, shown is growth of the Porod volume (calculated from the SAXS data in panels a and b) plotted against increasing Fe2+-to-protein ratios for wild-type Yfh1 (black), D79A/D82A Yfh1 (red), and CyaY (blue). The volume axis was set to better visualize differences between Yfh1and CyaY. The inset shows the complete range for D79A/D82A Yfh1 variant for comparison with wild-type Yfh1. d, shown is a ribbon representation of the x-ray structure of the yeast frataxin trimer superimposed on an ab initio model generated from the SAXS data. The insets show the distance distribution function P(r) used for ab initio modeling.
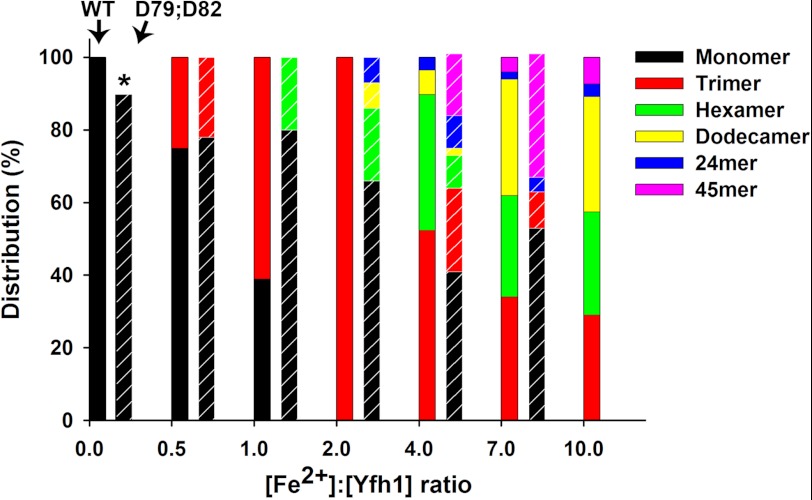
The program OLIGOMER was used to fit a set of form-factors from different oligomeric models (see “Experimental Procedure”) to the experimental SAXS data (Fig. 1a). It can be seen from the figure that despite the low χ2 value (0.6), for example at the metal to iron ratio of 1:2, at higher scattering angles (s values between 1.5–1.7) the fit to the experimental data is not perfect (Fig. 1a). This may indicate some rearrangements in the secondary structure elements of the monomers during oligomerization. Such rearrangement, which may be of functional interest, could be difficult to account for at the present resolution of the data, also taking into account the inhomogeneous nature of the solution with different oligomeric states. Analysis of the volume distributions of the oligomeric states showed that at an iron-to-protein ratio of 1:2, 25% of wild-type Yfh1 was in a trimeric form, whereas at a ratio of 2:1 this percentage increased to 100% (Table 1). Fig. 1d shows the fit of the x-ray structure of the yeast frataxin trimer to an ab initio model generated from the SAXS data. Further increase in the iron-to-protein ratio resulted in the formation of hexamers and higher-order oligomers, whereas the percentage of trimers decreased (Fig. 2 and Table 1). This suggests that in solution trimers serve as building blocks for higher order oligomers, consistent with the EM reconstruction of 24-meric particles of the Y73A variant of Yfh1 (33). At an iron-to-protein ratio of 4:1, the percentage of trimers decreased to about 49%, whereas a large fraction of hexamers, dodecamers, and 24-mers was formed (38, 7, and 4%, respectively; Fig. 2, supplemental Fig. S3, and Table 1). Interestingly, at the highest iron-to-protein ratio used for the wild-type protein (10:1), an approximately equimolar equilibrium between trimers, hexamers, and dodecamers was apparently established, with essentially no monomers remaining in solution. Notably, the fraction of higher order oligomers, such as the 24-meric spherical particles, remained rather low, suggesting that the experimental conditions used did not promote the stability of this particular complex.
TABLE 1.
The combinations of the models with different oligomeric states used for fitting to the experimental data (Fig. 1, a and b) are shown together with respective distributions in the mixture and the corresponding value of discrepancy (χ2)
| Fe2+:Yfh1 | Wild-type Yfh1 |
D79A/D82A Yfh1 |
||||
|---|---|---|---|---|---|---|
| Oligomers (subunits) | Distribution | χ2 | Oligomers | Distribution | χ2 | |
| % | % | |||||
| Apo | 1 | 100 | 1.04 | 1/2 | 81/19 | 0.99 |
| 1:2 | 1/3 | 75/25 | 0.56 | 1/3 | 78/22 | 1.03 |
| 1:1 | 1/3 | 39/61 | 0.47 | 1/6 | 80/20 | 1.08 |
| 2:1 | 3 | 100 | 0.49 | 1/6/12/24 | 66/20/7/7 | 1.02 |
| 4:1 | 3/6/12/24 | 52/38/7/4 | 0.52 | 1/3/6/12/24/45 | 41/23/9/2/9/17 | 1.1 |
| 7:1 | 3/6/12/24/45 | 34/28/32/2/4/ | 0.52 | 1/3/24/45/66/87 | 53/10/4/10/17/7 | 1.38 |
| 10:1 | 3/6/12/24/45 | 29/29/32/4/7 | 0.62 | |||
FIGURE 2.
Iron-induced oligomerization of yeast frataxin. A bar diagram shows the percentage of various types of oligomers of wild-type Yfh1 (solid fill) and D79A/D82A Yfh1 variant (lined fill) at different Fe2+-to-Yfh1 ratios (see Table 1 for exact numbers and for χ2 discrepancy of the fitting).
To understand the role of the ferroxidation reaction in iron-dependent oligomerization, we analyzed the iron-dependent oligomerization of the ferroxidation-deficient D79A/D82A variant. The results demonstrated a distinctly different oligomerization pattern compared with wild-type Yfh1 (Fig. 2 and Table 1). At an iron-to-protein ratio of 1:2, the fraction of trimers in solution was ∼20%, a value similar to that of the wild-type protein. However, at a ratio of 1:1 there were 20% hexamers and essentially no free trimers. A further increase in the iron-to-protein ratio (2:1) required hexamers, dodecamers (20 and 7%, respectively), and even higher order oligomers such as 24-meric particles to be included in the modeling. In contrast to wild-type Yfh1, which was present in the trimeric form at this ratio (2:1), 66% of the D79A/D82A variant protein still remained in the monomeric form. At the iron-to-protein ratio of 4:1 and 7:1, the highest ratios tested, a large fraction of frataxin oligomers was bigger than 24-mers. It has been shown that such oligomers have amyloid-type appearance and are highly heterogeneous in length (see EM images from Ref. 21). This makes the estimation of oligomer size and monomer composition at higher iron-to-protein ratios highly inaccurate. In addition, uncontrolled iron oxidation in solution in conditions of a deficient Fe2+ binding site and absence of ferroxidation activity as well as iron accumulation within the larger particles (35) will reduce the concentration of freely available iron in solution. This may in turn affect the proportion of smaller oligomers like trimers and hexamers, which may remain in solution. Therefore, the presented fitting and oligomer distribution at these concentrations should be considered as a qualitative indication of the behavior to be compared with wild-type protein oligomerization rather than a quantitative description of the actual equilibrium between different oligomeric states at a certain iron concentration. At an iron-to-protein ratio of 10, which was the highest ratio at which wild-type Yfh1 was analyzed, the D79A/D82A variant protein could not be analyzed due to the presence of large aggregates in the sample.
Iron Binding to Yfh1 and the Role of the Metal in Oligomerization
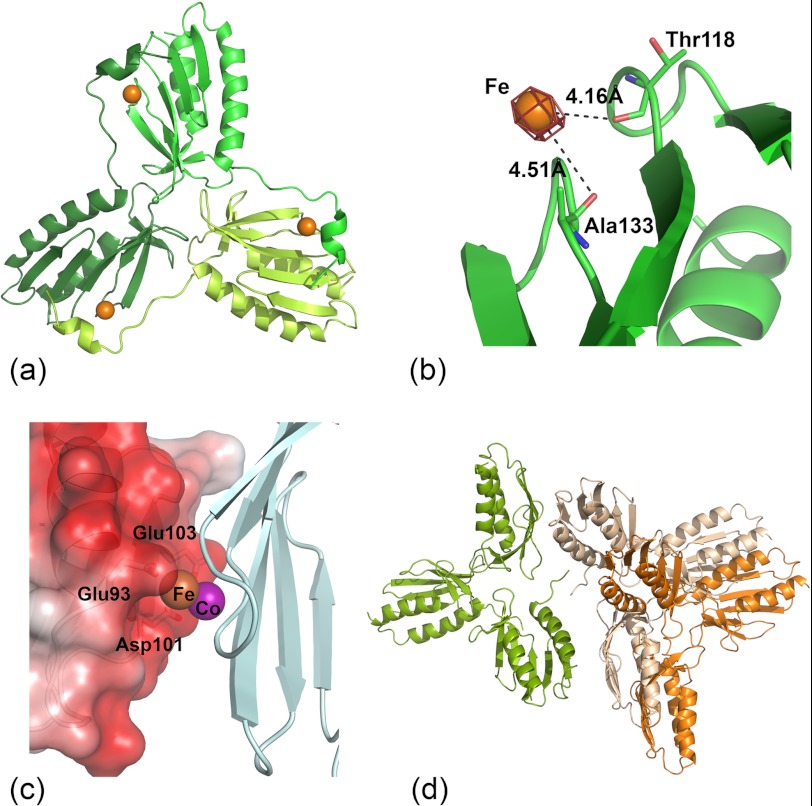
To obtain a complex of Yfh1 with iron, crystals of the Yfh1 Y73A variant were soaked anaerobically for a few seconds with ferrous ammonium sulfate. Longer soaking times resulted in cracking of the crystals, presumably as a result of a conformational change triggered by metal binding. X-ray data were collected to 3.0 Å resolution (Table 2), and the previously determined structure of the Yfh1 Y73A variant (PDB entry 2FQL) was used to obtain phase information. Our previous crystallographic work showed that the three-dimensional structure of frataxin trimers preloaded with iron contained a metal atom bound in the channel at the 3-fold axis of the trimer (32). With the current data, quick soaking with ferrous iron showed a peak both in the Fo − Fc and anomalous difference density maps at levels of up to 8 and 2.5 σ, respectively. When assigned occupancy of 1, no residual electron density in the Fo − Fc map could be observed, and the B-factor of the metal could be refined to a value similar to that of the surrounding protein groups. Fig. 3a shows the position of the iron atom in the context of the trimer structure, whereas Fig. 3b shows a close-up with details of the binding site. The metal binds on top of the β-sheet, between β-strands 3 and 4, and is covered by N-terminal residues Val-52—Lys-72 from a neighboring frataxin monomer. At this position bound iron could presumably contribute to the stabilization of the N terminus in solution and thus promote trimer stability.
FIGURE 3.
The x-ray structure of the Y73A variant of Yfh1 in complex with Fe2+. a, shown is a ribbon representation of the three-dimensional structure of the Y73A variant of yeast frataxin in complex with Fe2+. The metal is shown as an orange sphere. b, a close-up shows Fe2+ superimposed on the Fo − Fc difference electron density map (mesh representation) contoured at 5.0 σ. Protein amino acid groups are shown as sticks. c, the Fe2+-binding site is superimposed on the hexamer model used for fitting of SAXS data (Fig. 1). The Co2+ ion (magenta) bound to the crystallographic structure of E. coli CyaY (PDB entry 2EFF (70)) is also shown for comparison of metal positions. The surface representation is colored according to electrostatic potential. d, a ribbon representation shows a hexamer structure obtained from the Rosetta refinement and used for fitting of the SAXS data. Orange and white represent hexamer conformation before and after Rosetta refinement, respectively.
There is no direct coordination of the metal to any amino acid side chain, because the closest protein polar groups, the carboxyls of residues Thr-118 and Ala-133, are located ∼4 Å from the metal (Fig. 3b). However, because the metal is most probably hydrated, it may interact with protein groups via solvent molecules, which are not visible at the present resolution of the data. A comparison with cobalt binding to CyaY shows that the position of one of the cobalt atoms (PDB entry 2EFF (70)) is very similar to iron position in the current structure (Fig. 3c). As shown in Fig. 3c, both iron and cobalt, when analyzed in the context of the hexamer used to fit the SAXS data, are bound at the trimer-trimer interface. A monomer from the second trimer contributes an acidic surface, which includes the highly conserved Glu-93 and Asp-101. In fact, as shown in the sequence analysis below, both surfaces involved in hexamer-hexamer contacts contain highly conserved residues.
Solution Structure and Iron-induced Oligomerization of Bacterial Frataxin CyaY
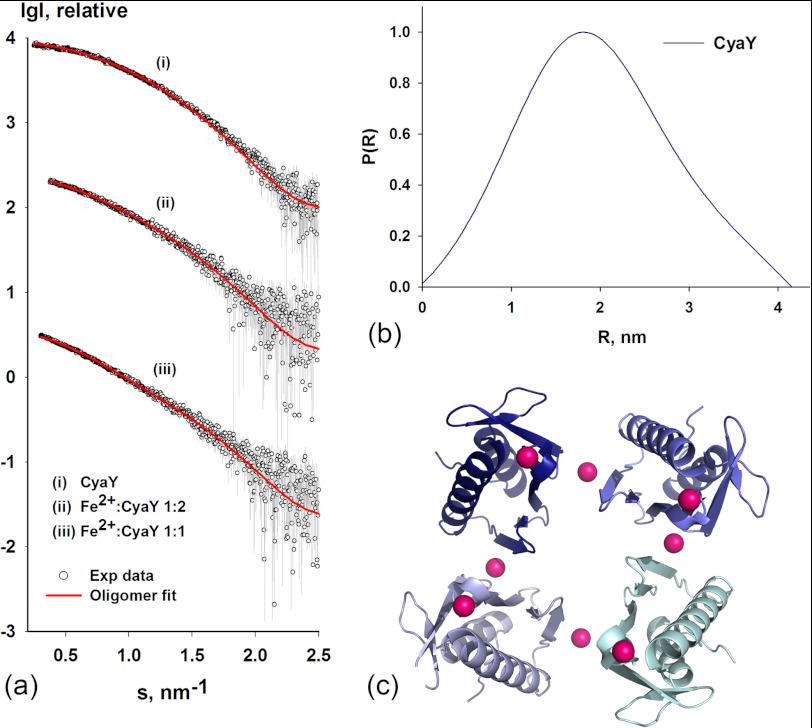
As mentioned above, earlier studies showed the importance of the N terminus of frataxin in the formation of oligomeric species (30, 32). Although bacterial and eukaryotic frataxin show a high degree of conservation of the core structure, both at the level of the amino acid sequence and at the level of the three-dimensional structure, the amino acid content and length of the N-terminal part of the sequences are highly variable. To study the consequences of the shorter N-terminal amino acid sequence in bacterial frataxin, we collected SAXS data on E. coli CyaY. As with Yfh1, no concentration effects were observed in this case. Rg and Dmax were found to be 1.38 ± 0.05 and 4.2 ± 0.1 nm, respectively (supplemental Table S2). Thus, Dmax as well as Rg were smaller than the corresponding values for Yfh1. When we used OLIGOMER to estimate the volume fraction of the different CyaY NMR conformers (PDB entry 1SOY (51)), which could be used to fit the SAXS data, conformer number 6 showed the best fit (Fig. 4a). To verify the model, we also performed ab initio modeling using the program GASBOR (53). Twenty GASBOR models had NSD (normalized spatial discrepancy) in the narrow range of 0.754–0.779, which is in the range of the values normally accepted for model quality. The GASBOR models also agreed with the globular fold of the selected conformer. Superposition of the best ab initio model on conformer 6 from PDB entry 1SOY is shown in supplemental Fig. S3b. The good fit of the NMR model to the SAXS data reflects the high homogeneity of the CyaY structure in solution. In contrast, for wild-type Yfh1 two different NMR models contributing volume fractions of 32 and 68%, respectively, had to be used to attain good fit to the experimental SAXS data mainly due to the higher flexibility of the N-terminal part of the structure (34).
FIGURE 4.
SAXS measurement of E. coli frataxin CyaY. a, shown are experimental SAXS profiles of E. coli CyaY before and after incubation with Fe2+ at two different Fe2+-to-protein ratios. The curves were displaced appropriately along the logarithmic axis for better visualization. The experimental data are shown as circles, and the fit to a pool of oligomers is shown as red lines (see supplemental Table 3S for details of the oligomeric content). The data are presented as the logarithm of the scattering intensity versus the momentum transfer s = 4πsin(θ)/λ (where 2θ is the scattering angle, and λ is the x-ray wavelength) and shown to a maximal momentum transfer of s = 2.5 nm−1. b, the distance distribution function P(r) used for ab initio modeling. c, shown is a ribbon representation of the CyaY tetramer used to fit experimental SAXS profiles of CyaY after incubation with Fe2+. The Co2+ ion (magenta) bound to the crystallographic structure of E. coli CyaY (PDB entry 2EFF (70)) is also shown.
To test the ability of CyaY to assemble into large oligomers in the presence of iron, we carried out SAXS measurements in the same fashion as for Yfh1. The molecular mass of the particles in solution was estimated as described above using the Porod volume (43). As shown in Fig. 1c, the increase in iron concentration resulted in an increase in the Porod volume, essentially in the same manner as for wild-type Yfh1. This demonstrates that CyaY may also form oligomeric species in the presence of iron despite the short N-terminal part.
As noted above, it has been suggested that CyaY tetramers may be formed at anaerobic conditions in the presence of Fe2+ (22). However, the way by which the monomers were arranged into tetrameric species was not known. As described under “Experimental Procedures,” we used Yfh1 hexamers to construct the initial CyaY tetramers (Fig. 4c). The figure also shows the cobalt atoms, observed earlier to bind to CyaY (70). One of these positions was close to bound cobalt in cobalt-soaked crystals of the Y73A variant of Yfh1 (34), whereas the second is similar to the position of bound iron observed in the current work (Fig. 3c). This suggested that the constructed tetramer model was largely correct. Fitting to the SAXS data showed that at the lowest iron-to-protein ratio (1:2), around 9% of CyaY had formed tetramers. When we doubled the relative amount of iron (1:1 ratio) 20% of the protein was in a tetrameric state (supplemental Table S3). At higher iron-to-protein ratios the SAXS data could not be fit using our limited pool of oligomeric particles, suggesting that higher state assemblies were formed. Using this tetramer arrangement it would be possible to construct a 24-meric particle. However, modeling of these structures at this stage is difficult because no guiding structural data from other methods, for example, electron microscopy, exist.
Conservation of Oligomer Interfaces in the Frataxin Family
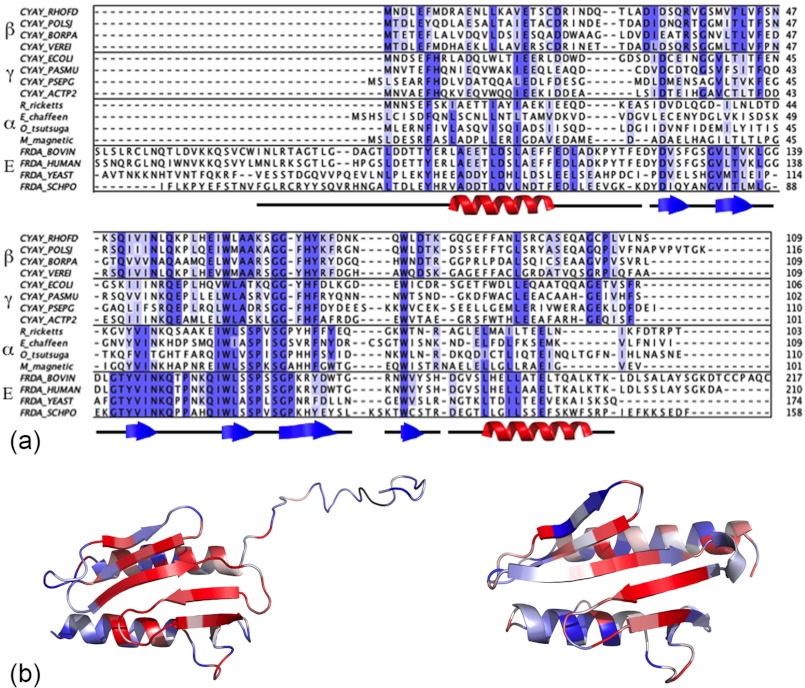
To obtain an insight into the amino acid determinants of frataxin oligomerization, we compared the sequences from different species (Fig. 5a). For a quantitative description of variability at various positions within the sequences, Shannon entropy was calculated as described under “Experimental Procedure.” After normalizing the entropy at each position of the alignment to the average entropy, the interaction interfaces in the trimer and hexamer (trimer-trimer contacts) structures were analyzed.
FIGURE 5.
Multiple sequence alignment of frataxin amino acid sequences. a, shown are four frataxin sequences each from eukaryotes and β-, γ-, and α-proteobacteria from a large multiple sequence alignment shown in supplemental Fig. S3. Secondary structure elements according to yeast frataxin crystal structure (PDB entry 3OEQ) are shown along the alignment. b, shown is a ribbon representation of yeast frataxin (left) and CyaY (right) monomers colored according to normalized average Shannon entropy. Red represents low entropy (high conservation), and blue represents high entropy (low conservation).
Table 3 shows that in comparison to the polypeptide sequence as a whole, for eukaryotic frataxin the entropy is clearly lower for interfaces that are involved in the stabilization of trimers and trimer-trimer contacts. On the other hand, for the bacterial frataxin homolog only the positions that correspond to the trimer-trimer interface had significantly lower entropy than the polypeptide sequence as a whole, which supports our interpretation of the experimental SAXS data.
DISCUSSION
The present work is the first detailed analysis of iron-induced oligomerization of yeast and bacterial frataxin. The results reveal the solution structure of Yfh1 and CyaY oligomers formed during assembly and suggest their stabilization mechanisms. We show that in solution at aerobic conditions and at iron-to-protein ratio of up to 2:1 Yfh1 forms trimers, which in turn at higher iron content build up iron-stabilized hexamers and higher order oligomers. In contrast, in the case of CyaY, iron-induced formation of tetrameric complexes appears to be the initial step in oligomerization.
The 2:1 iron-to-protein ratio required for Yfh1 trimer stabilization suggests that there should be two metal-binding sites per monomer and that the occupation of these sites is required for the stabilization of the trimeric structure, whereas the formation of higher order oligomers requires additional metal ions. The 2-iron/monomer ratio agrees with previous studies, which showed the presence of two iron binding sites/monomer in yeast frataxin by the use of nuclear magnetic resonance (NMR) and x-ray absorption spectroscopy (XAS) (55, 71). The dissociation constants for these sites were shown to be in the micro-molar range (Kd within the range of 2–3 μm), showing a relatively low affinity metal binding sites. Because the primary aim of this work was the study of Fe2+ binding to frataxin, strictly anaerobic conditions were employed, in contrast to our experiments, which were carried out aerobically, mimicking the physiological conditions. It should be noted that the yeast frataxin-catalyzed ferroxidation reaction is rather slow and requires ∼30 min for completion (2), suggesting that iron was in the ferric form in our experiments. The earlier studied crystallographic structure of the Y73A variant of Yfh1, which was aerobically preloaded with iron at conditions similar to those used in the current study, showed one metal ion bound in the channel at the 3-fold axis of the trimer. Although the resolution of the structure was only 3.5 Å, the metal appeared to be coordinated to three water molecules, which in turn were coordinated to the side chains of three aspartates (the invariant Asp-143) within the channel. No other metal ions could be detected in the crystal structure, presumably due to the combination of factors like the low resolution of the x-ray data and low occupancy of the metal binding sites due to the above-mentioned low metal binding affinity.
In the NMR experiments by Cook et al. (71), metal binding to Yfh1 was found to affect two regions of the protein. The first region included several residues from helix 1 (Ala-77—His-95), Lys-128, and Trp-131 from strand 4 as well as some residues closer to the end of helix 2. All these amino acids were found to undergo amide chemical shift perturbations, which as noted by the authors, can be attributed to local changes in the chemical environment of the residues involved. The second set, apart from the ferroxidation site residues Asp-78, Asp -82, Asp -83, and Asp -86 in helix 1 (Fig. 6), included Glu-103, Glu-112, and Thr-118 from β-strands 2 and 3, respectively. These residues had undergone significant amide resonance line broadening, which suggests that they are located in close proximity to a paramagnetic metal ion. In the crystal structure presented in this work the iron is bound between β-strands 3 and 4, approximately at 6 Å from the side chain of Thr-118 and 9.8 Å from Glu-112. Both these distances are within the range for amide resonance line broadening (10 Å). However, in our data we did not find a convincing electron density, which could be attributed to a bound metal at the ferroxidation site, presumably a result of the short soaking time used. It should be noted that in their interpretation the authors excluded the possibility of oligomerization of frataxin under the conditions of the experiment. However, among the residues listed above to have undergone amide chemical shift perturbations, the authors identified Lys-128, which is located at the 3-fold axis of the trimer, and Trp-131, which is located at the trimer interaction surface in the hexamers used here to fit the SAXS data. In addition, Glu-90 and Glu-93, shown on Fig. 6 to be within the mineralization site formed by 2 trimers, were also among the residues that were found to undergo amide chemical shift perturbations. These results suggest that at the anaerobic conditions used, contrary to the assumption of the authors that frataxin was in a monomeric state (71), oligomerization may still have taken place. At the conditions of the experiment (iron-to-protein ratio of 2) the oligomers formed are presumably hexamers, similarly to the observed hexamer formation in our experiments with the ferroxidation-deficient D79A/D82A variant of Yhf1 (Fig. 2). It should also be noted that apart from the absence of Mg2+-ions, the buffer solutions used in our SAXS experiments were essentially identical to those used by the authors (71). To assess the effect of magnesium on the oligomerization state of the protein, we performed SAXS experiments in the presence of 10 mm MgCl2. We found that in these conditions and at an iron-to-protein ratio of 10:1, the average particle volume notably increased from 277 to 999 nm3 (data not shown), clearly indicating that additional oligomerization or aggregation had taken place. Contrary to the suggested monomer-stabilizing effect at anaerobic conditions (71), magnesium ions at aerobic conditions appear to facilitate the interactions between oligomeric particles once they are formed.
The presented data clearly show that ferroxidation activity and presumably iron binding to the mineralization site are two critical factors both for correct oligomerization and for the stabilization of higher order oligomers of frataxin. As shown on Fig. 6, the oligomerization process described in the present work places the ferroxidation and mineralization sites in close proximity to each other, suggesting a mechanism according to which oxidation of iron immediately results in its displacement from the ferroxidation site to the mineralization site. This explains why metal binding at the ferroxidation site could not be observed in the crystal structure of aerobically iron-loaded trimer (32). The preference of the ferroxidation-deficient D79A/D82A variant of Yfh1 to form higher order oligomers, which require ferric iron for stabilization, suggests that frataxin may bind ferric iron from solution, thus contributing to additional iron detoxification (15). However, the high percentage of the remaining monomers in solution (Fig. 2) shows that this process is less efficient and probably proceeds at a slower rate as compared with iron detoxification by wild-type protein. The importance of the ferroxidation reaction and ferric iron in oligomer stabilization was also confirmed by earlier data, which showed that oligomeric species of yeast frataxin could be dissolved into monomers by the addition of reducing agents (15).
The slow rate of the ferroxidation reaction observed earlier (2) suggests that even in the presence of oxygen frataxin may serve as an iron chaperone, delivering iron to processes like heme and iron-cluster biosynthesis. It has even been suggested that the conditions inside mitochondria are reminiscent of the anaerobic conditions of the NMR experiments used to study iron binding to frataxin (71). Based on this it was concluded that frataxin primarily exists as a monomeric protein in mitochondria. However, the mitochondrial matrix is not a strictly anaerobic environment, with oxygen concentrations ranging between 3 and 30 μm in tissues, whereas cells directly exposed to atmospheric oxygen, such as pulmonary alveoli cells or yeast cells grown aerobically contain even higher amounts of oxygen (for review, see Ref. 72). In addition to the results presented in this work, earlier biochemical studies have demonstrated that in vitro under aerobic conditions in the presence of Fe2+, Yfh1 monomers may assemble into trimers and higher order oligomers that bind iron and progressively convert it to a ferric mineral (2, 4, 15, 73). It has also been suggested that this process could take place in yeast, where increments in mitochondrial iron content were found to be associated with Yfh1 oligomerization (24). On the other hand, in a different study oligomerization of Yfh1 in yeast was not associated with iron accumulation, which led to the conclusion that iron was not stored in Yfh1 oligomers (74). However, in this study cells were cultured in the absence of iron, which was added at a later stage of their growth. Although these conditions could affect iron uptake by frataxin, the iron-storage function of Yfh1 in vivo, which is presumably dependent on the amount of available iron, still needs to be conclusively demonstrated (for review, see Ref. 31).
The functional role of frataxin oligomers was also studied in the work by Li et al. (18), which showed that iron-sulfur cluster synthesis on the iron-sulfur cluster scaffold protein Isu1 is faster for wild-type Yfh1, incubated at a metal-to-Yfh1 ratio of 2:1, than for the Yfh1 Y73A variant, which assembles into high order oligomers independently of the presence of iron (18, 32). Presumably, “iron-free oligomers” by competing with the “proper,” iron-containing, oligomers may build unproductive complexes with the proteins of the iron-sulfur cluster synthesis machinery. The authors also showed that under anaerobic conditions, where oligomerization of Yfh1 is reduced, the rate of iron-sulfur cluster synthesis was only slightly higher in the presence of Yfh1. In contrast, the synthesis rate under aerobic conditions was significantly higher, again demonstrating that the ferroxidation reaction and the formation of proper oligomeric species are essential for frataxin interactions with its target proteins.
The results of this work also suggest that the formation of CyaY oligomers is initiated by tetramer formation. This also confirms earlier data, which based on analytical centrifugation, suggested that CyaY in the presence of iron could form tetrameric complexes (22). This clearly shows the role of the shorter N-terminal sequence in CyaY, compared with Yfh1 and the FXN42–210 and FXN56–210, forms of human frataxin. This also agrees with the analysis of the conservation pattern of the amino acid sequences of frataxin from different organisms, which suggests that in contrast to yeast frataxin, the basic oligomer-building structure in bacterial frataxin oligomers does not need to be a trimer. It also suggests that different forms of human frataxin, depending on the length of the N-terminal sequence, may form different oligomeric species. In other words, the length of the N-terminal part could be used in the regulation of frataxin function.
Further studies should shed more light on the possible role of the different oligomeric forms of frataxin in vivo, the role of the conserved surface areas in interaction with other proteins in the mitochondria, and the role of the N terminus of the protein in oligomer stability.
Acknowledgments
The bacterial frataxin CyaY expression plasmid was kindly provided by Dr. Tibor Bedekovics (Mayo Clinic, College of Medicine, Rochester, MN). We also thank the EMBL-Hamburg and Lund university for synchrotron beam time allocation at DESY (Hamburg, Germany) and Max-Lab, respectively. Clement Blanchet and Tomás Plivelic are acknowledged for technical support at beam line X33 (DESY) and I711 (MAX-Lab), respectively, and Dr. I André for assistance with the Rosetta program.
This work was supported, in whole or in part, by National Institutes of Health Grant AG15709 (NIA; to G. I.). This work was also supported by the Swedish Research Council (Vetenskapsrådet), a MAX IV/ESS grant from the Natural Science Faculty of Lund University, the Crafoord Foundation, and the Carl Trygger Foundation (to S. A. K.).

This article contains supplemental Tables S1–S3 and Figs. S1–S4.
- FXN
- frataxin
- SAXS
- small angle x-ray scattering.
REFERENCES
- 1. Chasteen N. D., Harrison P. M. (1999) Mineralization in ferritin. an efficient means of iron storage. J. Struct. Biol. 126, 182–194 [DOI] [PubMed] [Google Scholar]
- 2. Park S., Gakh O., Mooney S. M., Isaya G. (2002) The ferroxidase activity of yeast frataxin. J. Biol. Chem. 277, 38589–38595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lewin A., Moore G. R., Le Brun N. E. (2005) Formation of protein-coated iron minerals. Dalton Trans. 22, 3597–3610 [DOI] [PubMed] [Google Scholar]
- 4. Nichol H., Gakh O., O'Neill H. A., Pickering I. J., Isaya G., George G. N. (2003) Structure of frataxin iron cores. An X-ray absorption spectroscopic study. Biochemistry 42, 5971–5976 [DOI] [PubMed] [Google Scholar]
- 5. Kell D. B. (2009) Iron behaving badly. Inappropriate iron chelation as a major contributor to the aetiology of vascular and other progressive inflammatory and degenerative diseases. BMC Med. Genomics 2, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Koeppen A. H. (2011) Friedreich's ataxia. Pathology, pathogenesis, and molecular genetics. J. Neurol. Sci. 303, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Campuzano V., Montermini L., Moltò M. D., Pianese L., Cossée M., Cavalcanti F., Monros E., Rodius F., Duclos F., Monticelli A., Zara F., Cañizares J., Koutnikova H., Bidichandani S. I., Gellera C., Brice A., Trouillas P., De Michele G., Filla A., De Frutos R., Palau F., Patel P. I., Di Donato S., Mandel J. L., Cocozza S., Koenig M., Pandolfo M. (1996) Friedreich's ataxia. Autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 271, 1423–1427 [DOI] [PubMed] [Google Scholar]
- 8. Bidichandani S. I., Ashizawa T., Patel P. I. (1997) Atypical Friedreich ataxia caused by compound heterozygosity for a novel missense mutation and the GAA triplet-repeat expansion. Am. J. Hum. Genet. 60, 1251–1256 [PMC free article] [PubMed] [Google Scholar]
- 9. Bartolo C., Mendell J. R., Prior T. W. (1998) Identification of a missense mutation in a Friedreich's ataxia patient. Implications for diagnosis and carrier studies. Am. J. Med. Genet. 79, 396–399 [PubMed] [Google Scholar]
- 10. Forrest S. M., Knight M., Delatycki M. B., Paris D., Williamson R., King J., Yeung L., Nassif N., Nicholson G. A. (1998) The correlation of clinical phenotype in Friedreich ataxia with the site of point mutations in the FRDA gene. Neurogenetics 1, 253–257 [DOI] [PubMed] [Google Scholar]
- 11. Cossée M., Dürr A., Schmitt M., Dahl N., Trouillas P., Allinson P., Kostrzewa M., Nivelon-Chevallier A., Gustavson K. H., Kohlschütter A., Müller U., Mandel J. L., Brice A., Koenig M., Cavalcanti F., Tammaro A., De Michele G., Filla A., Cocozza S., Labuda M., Montermini L., Poirier J., Pandolfo M. (1999) Friedreich's ataxia. Point mutations and clinical presentation of compound heterozygotes. Ann. Neurol. 45, 200–206 [DOI] [PubMed] [Google Scholar]
- 12. De Castro M., García-Planells J., Monrós E., Cañizares J., Vázquez-Manrique R., Vílchez J. J., Urtasun M., Lucas M., Navarro G., Izquierdo G., Moltó M. D., Palau F. (2000) Genotype and phenotype analysis of Friedreich's ataxia compound heterozygous patients. Hum. Genet. 106, 86–92 [DOI] [PubMed] [Google Scholar]
- 13. Lesuisse E., Santos R., Matzanke B. F., Knight S. A., Camadro J. M., Dancis A. (2003) Iron use for haeme synthesis is under control of the yeast frataxin homologue (Yfh1). Hum. Mol. Genet. 12, 879–889 [DOI] [PubMed] [Google Scholar]
- 14. Yoon T., Cowan J. A. (2004) Frataxin-mediated iron delivery to ferrochelatase in the final step of heme biosynthesis. J. Biol. Chem. 279, 25943–25946 [DOI] [PubMed] [Google Scholar]
- 15. Park S., Gakh O., O'Neill H. A., Mangravita A., Nichol H., Ferreira G. C., Isaya G. (2003) Yeast frataxin sequentially chaperones and stores iron by coupling protein assembly with iron oxidation. J. Biol. Chem. 278, 31340–31351 [DOI] [PubMed] [Google Scholar]
- 16. Yoon T., Cowan J. A. (2003) Iron-sulfur cluster biosynthesis. Characterization of frataxin as an iron donor for assembly of [2Fe-2S] clusters in ISU-type proteins. J. Am. Chem. Soc. 125, 6078–6084 [DOI] [PubMed] [Google Scholar]
- 17. Lill R. (2009) Function and biogenesis of iron-sulphur proteins. Nature 460, 831–838 [DOI] [PubMed] [Google Scholar]
- 18. Li H., Gakh O., Smith D. Y., 4th, Isaya G. (2009) Oligomeric yeast frataxin drives assembly of core machinery for mitochondrial iron-sulfur cluster synthesis. J. Biol. Chem. 284, 21971–21980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bulteau A. L., O'Neill H. A., Kennedy M. C., Ikeda-Saito M., Isaya G., Szweda L. I. (2004) Frataxin acts as an iron chaperone protein to modulate mitochondrial aconitase activity. Science 305, 242–245 [DOI] [PubMed] [Google Scholar]
- 20. Adamec J., Rusnak F., Owen W. G., Naylor S., Benson L. M., Gacy A. M., Isaya G. (2000) Iron-dependent self assembly of recombinant yeast frataxin. Implications for Friedreich ataxia. Am. J. Hum. Genet. 67, 549–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cavadini P., O'Neill H. A., Benada O., Isaya G. (2002) Assembly and iron-binding properties of human frataxin, the protein deficient in Friedreich ataxia. Hum. Mol. Genet. 11, 217–227 [DOI] [PubMed] [Google Scholar]
- 22. Bou-Abdallah F., Adinolfi S., Pastore A., Laue T. M., Dennis Chasteen N. (2004) Iron binding and oxidation kinetics in frataxin CyaY of Escherichia coli. J. Mol. Biol. 341, 605–615 [DOI] [PubMed] [Google Scholar]
- 23. Layer G., Ollagnier-de Choudens S., Sanakis Y., Fontecave M. (2006) Iron-sulfur cluster biosynthesis. Characterization of Escherichia coli CyaY as an iron donor for the assembly of [2Fe-2S] clusters in the scaffold IscU. J. Biol. Chem. 281, 16256–16263 [DOI] [PubMed] [Google Scholar]
- 24. Gakh O., Smith D. Y., 4th, Isaya G. (2008) Assembly of the iron-binding protein frataxin in Saccharomyces cerevisiae responds to dynamic changes in mitochondrial iron influx and stress level. J. Biol. Chem. 283, 31500–31510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Koutnikova H., Campuzano V., Koenig M. (1998) Maturation of wild-type and mutated frataxin by the mitochondrial processing peptidase. Hum. Mol. Genet. 7, 1485–1489 [DOI] [PubMed] [Google Scholar]
- 26. Schmucker S., Argentini M., Carelle-Calmels N., Martelli A., Puccio H. (2008) The in vivo mitochondrial two-step maturation of human frataxin. Hum. Mol. Genet. 17, 3521–3531 [DOI] [PubMed] [Google Scholar]
- 27. Gakh O., Bedekovics T., Duncan S. F., Smith D. Y., 4th, Berkholz D. S., Isaya G. (2010) Normal and Friedreich ataxia cells express different isoforms of frataxin with complementary roles in iron-sulfur cluster assembly. J. Biol. Chem. 285, 38486–38501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Condò I., Ventura N., Malisan F., Rufini A., Tomassini B., Testi R. (2007) In vivo maturation of human frataxin. Hum. Mol. Genet. 16, 1534–1540 [DOI] [PubMed] [Google Scholar]
- 29. O'Neill H. A., Gakh O., Park S., Cui J., Mooney S. M., Sampson M., Ferreira G. C., Isaya G. (2005) Assembly of human frataxin is a mechanism for detoxifying redox-active iron. Biochemistry 44, 537–545 [DOI] [PubMed] [Google Scholar]
- 30. O'Neill H. A., Gakh O., Isaya G. (2005) Supramolecular assemblies of human frataxin are formed via subunit-subunit interactions mediated by a non-conserved amino-terminal region. J. Mol. Biol. 345, 433–439 [DOI] [PubMed] [Google Scholar]
- 31. Vaubel R. A., Isaya G. (2012) Iron-sulfur cluster synthesis, iron homeostasis, and oxidative stress in Friedreich ataxia. Mol. Cell. Neurosci., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Karlberg T., Schagerlöf U., Gakh O., Park S., Ryde U., Lindahl M., Leath K., Garman E., Isaya G., Al-Karadaghi S. (2006) The structures of frataxin oligomers reveal the mechanism for the delivery and detoxification of iron. Structure 14, 1535–1546 [DOI] [PubMed] [Google Scholar]
- 33. Schagerlöf U., Elmlund H., Gakh O., Nordlund G., Hebert H., Lindahl M., Isaya G., Al-Karadaghi S. (2008) Structural basis of the iron storage function of frataxin from single-particle reconstruction of the iron-loaded oligomer. Biochemistry 47, 4948–4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Söderberg C. A., Shkumatov A. V., Rajan S., Gakh O., Svergun D. I., Isaya G., Al-Karadaghi S. (2011) Oligomerization propensity and flexibility of yeast frataxin studied by X-ray crystallography and small-angle X-ray scattering. J. Mol. Biol. 414, 783–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gakh O., Park S., Liu G., Macomber L., Imlay J. A., Ferreira G. C., Isaya G. (2006) Mitochondrial iron detoxification is a primary function of frataxin that limits oxidative damage and preserves cell longevity. Hum. Mol. Genet. 15, 467–479 [DOI] [PubMed] [Google Scholar]
- 36. Gakh O., Adamec J., Gacy A. M., Twesten R. D., Owen W. G., Isaya G. (2002) Physical evidence that yeast frataxin is an iron storage protein. Biochemistry 41, 6798–6804 [DOI] [PubMed] [Google Scholar]
- 37. Bedekovics T., Gajdos G. B., Kispal G., Isaya G. (2007) Partial conservation of functions between eukaryotic frataxin and the Escherichia coli frataxin homolog CyaY. Fems. Yeast. Res. 7, 1276–1284 [DOI] [PubMed] [Google Scholar]
- 38. Knaapila M., Svensson C., Barauskas J., Zackrisson M., Nielsen S. S., Toft K. N., Vestergaard B., Arleth L., Olsson U., Pedersen J. S., Cerenius Y. (2009) A new small-angle x-ray scattering set-up on the crystallography beamline I711 at MAX-lab. J. Synchrotron. Radiat. 16, 498–504 [DOI] [PubMed] [Google Scholar]
- 39. Blanchet C. E., Zozulya A. V., Kikhney A. G., Franke D., Konarev P. V., Shang W. F., Klaering R., Robrahn B., Hermes C., Cipriani F., Svergun D. I., Roessle M. (2012) Instrumental setup for high-throughput small- and wide-angle solution scattering at the X33 beamline of EMBL Hamburg. J. Appl. Crystallogr. 45, 489–495 [Google Scholar]
- 40. Roessle M. W., Klaering R., Ristau U., Robrahn B., Jahn D., Gehrmann T., Konarev P., Round A., Fiedler S., Hermes C., Svergun D. (2007) Upgrade of the small-angle X-ray scattering beamline X33 at the European Molecular Biology Laboratory, Hamburg. J. Appl. Crystallogr. 40, S190–S194 [Google Scholar]
- 41. Franke D., Kikhney A. G., Svergun D. I. (2012) Automated acquisition and analysis of small angle X-ray scattering data. Nucl. Instrum. Methods Phys. Res. A 689, 52–59 [Google Scholar]
- 42. Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H. J., Svergun D. I. (2003) PRIMUS. A Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282 [Google Scholar]
- 43. Petoukhov M., Franke D., Shkumatov A., Tria G., Kikhney A., Gajda M., Gorba C., Mertens H., Konarev P., Svergun D. (2012) New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 45, 342–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Guinier A. (1939) La diffraction des rayons X aux très petits angles; application a l'étude de phénomènes ultramicroscopiques. Ann. Phys. (Paris) 12, 161–237 [Google Scholar]
- 45. Svergun D. I. (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 [Google Scholar]
- 46. Porod G. (1982) in Small Angle X-Ray Scattering (Glatter O., K. O., ed) pp 17–51, Academic Press, London [Google Scholar]
- 47. Bernadó P., Mylonas E., Petoukhov M. V., Blackledge M., Svergun D. I. (2007) Structural characterization of flexible proteins using small-angle X-ray scattering. J. Am. Chem. Soc. 129, 5656–5664 [DOI] [PubMed] [Google Scholar]
- 48. Mandell D. J., Coutsias E. A., Kortemme T. (2009) Sub-angstrom accuracy in protein loop reconstruction by robotics-inspired conformational sampling. Nat. Methods 6, 551–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Leaver-Fay A., Tyka M., Lewis S. M., Lange O. F., Thompson J., Jacak R., Kaufman K., Renfrew P. D., Smith C. A., Sheffler W., Davis I. W., Cooper S., Treuille A., Mandell D. J., Richter F., Ban Y. E., Fleishman S. J., Corn J. E., Kim D. E., Lyskov S., Berrondo M., Mentzer S., Popović Z., Havranek J. J., Karanicolas J., Das R., Meiler J., Kortemme T., Gray J. J., Kuhlman B., Baker D., Bradley P. (2011) ROSETTA3. An object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 487, 545–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Svergun D., Barberato C., Koch M. H. J. (1995) CRYSOL. A program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28, 768–773 [Google Scholar]
- 51. Nair M., Adinolfi S., Pastore C., Kelly G., Temussi P., Pastore A. (2004) Solution structure of the bacterial frataxin ortholog, CyaY. Mapping the iron binding sites. Structure 12, 2037–2048 [DOI] [PubMed] [Google Scholar]
- 52. Franke D., Svergun D. I. (2009) DAMMIF, a program for rapid ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 42, 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Svergun D. I., Petoukhov M. V., Koch M. H. (2001) Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 80, 2946–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Volkov V. V., Svergun D. I. (2003) Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 36, 860–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. He Y., Alam S. L., Proteasa S. V., Zhang Y., Lesuisse E., Dancis A., Stemmler T. L. (2004) Yeast frataxin solution structure, iron binding, and ferrochelatase interaction. Biochemistry 43, 16254–16262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. André I., Bradley P., Wang C., Baker D. (2007) Prediction of the structure of symmetrical protein assemblies. Proc. Natl. Acad. Sci. U.S.A. 104, 17656–17661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX. A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) ClustalW and ClustalX Version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 62. Goujon M., McWilliam H., Li W., Valentin F., Squizzato S., Paern J., Lopez R. (2010) A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38, W695–W699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Clamp M., Cuff J., Searle S. M., Barton G. J. (2004) The Jalview Java alignment editor. Bioinformatics 20, 426–427 [DOI] [PubMed] [Google Scholar]
- 64. Dhe-Paganon S., Shigeta R., Chi Y. I., Ristow M., Shoelson S. E. (2000) Crystal structure of human frataxin. J. Biol. Chem. 275, 30753–30756 [DOI] [PubMed] [Google Scholar]
- 65. Cho S. J., Lee M. G., Yang J. K., Lee J. Y., Song H. K., Suh S. W. (2000) Crystal structure of Escherichia coli CyaY protein reveals a previously unidentified fold for the evolutionarily conserved frataxin family. Proc. Natl. Acad. Sci. U.S.A. 97, 8932–8937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Fornasari M. S., Laplagne D. A., Frankel N., Cauerhff A. A., Goldbaum F. A., Echave J. (2004) Sequence determinants of quaternary structure in lumazine synthase. Mol. Biol. Evol. 21, 97–107 [DOI] [PubMed] [Google Scholar]
- 67. Ptitsyn O. B. (1998) Protein folding and protein evolution. Common folding nucleus in different subfamilies of c-type cytochromes? J. Mol. Biol. 278, 655–666 [DOI] [PubMed] [Google Scholar]
- 68. Dey S., Pal A., Chakrabarti P., Janin J. (2010) The subunit interfaces of weakly associated homodimeric proteins. J. Mol. Biol. 398, 146–160 [DOI] [PubMed] [Google Scholar]
- 69. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 70. Pastore C., Franzese M., Sica F., Temussi P., Pastore A. (2007) Understanding the binding properties of an unusual metal-binding protein. A study of bacterial frataxin. FEBS J. 274, 4199–4210 [DOI] [PubMed] [Google Scholar]
- 71. Cook J. D., Bencze K. Z., Jankovic A. D., Crater A. K., Busch C. N., Bradley P. B., Stemmler A. J., Spaller M. R., Stemmler T. L. (2006) Monomeric yeast frataxin is an iron-binding protein. Biochemistry 45, 7767–7777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Turrens J. F. (2003) Mitochondrial formation of reactive oxygen species. J. Physiol. 552, 335–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wang T., Craig E. A. (2008) Binding of yeast frataxin to the scaffold for Fe-S cluster biogenesis, Isu. J. Biol. Chem. 283, 12674–12679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Seguin A., Bayot A., Dancis A., Rogowska-Wrzesinska A., Auchère F., Camadro J. M., Bulteau A. L., Lesuisse E. (2009) Overexpression of the yeast frataxin homolog (Yfh1). Contrasting effects on iron-sulfur cluster assembly, heme synthesis, and resistance to oxidative stress. Mitochondrion 9, 130–138 [DOI] [PubMed] [Google Scholar]
- 75. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Electrostatics of nanosystems. Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]