Background: Targeted degradation by tripartite motif (TRIM) ligase-catalyzed polyubiquitin chain formation is critical for cell regulation and innate immunity.
Results: TRIM32-catalyzed chain formation requires oligomerization and uses a cooperative allosteric mechanism.
Conclusion: Kinetics suggest TRIM32 assembles polyubiquitin chains as E2-linked thioesters prior to en bloc target protein transfer.
Significance: A general mechanism for degradation signal assembly is revealed for the TRIM ligase superfamily.
Keywords: Enzyme Mechanisms, Kinetics, Ubiquitin, Ubiquitin-conjugating Enzyme (Ubc), Ubiquitin Ligase, TRIM25, TRIM32, Polyubiquitin Chain
Abstract
Ligation of polyubiquitin chains to proteins is a fundamental post-translational modification, often resulting in targeted degradation of conjugated proteins. Attachment of polyubiquitin chains requires the activities of an E1 activating enzyme, an E2 carrier protein, and an E3 ligase. The mechanism by which polyubiquitin chains are formed remains largely speculative, especially for RING-based ligases. The tripartite motif (TRIM) superfamily of ligases functions in many cellular processes including innate immunity, cellular localization, development and differentiation, signaling, and cancer progression. The present results show that TRIM ligases catalyze polyubiquitin chain formation in the absence of substrate, the rates of which can be used as a functional readout of enzyme function. Initial rate studies under biochemically defined conditions show that TRIM32 and TRIM25 are specific for the Ubc5 family of E2-conjugating proteins and, along with TRIM5α, exhibit cooperative kinetics with respect to Ubc5 concentration, with submicromolar [S]0.5 and Hill coefficients of 3–5, suggesting they possess multiple binding sites for their cognate E2-ubiquitin thioester. Mutation studies reveal a second, non-canonical binding site encompassing the C-terminal Ubc5α-helix. Polyubiquitin chain formation requires TRIM subunit oligomerization through the conserved coiled-coil domain, but can be partially replaced by fusing the catalytic domain to GST to promote dimerization. Other results suggest that TRIM32 assembles polyubiquitin chains as a Ubc5-linked thioester intermediate. These results represent the first detailed mechanistic study of TRIM ligase activity and provide a functional context for oligomerization observed in the superfamily.
Introduction
Post-translational conjugation of ubiquitin and ubiquitin-like proteins to polypeptide targets provides a diverse set of functional outcomes that regulate most aspects of cell homeostasis. The best understood of these functions is assembly of polyubiquitin chains on protein targets, resulting in degradation of the conjugated substrate by the 26S proteasome and disassembly of the polyubiquitin degradation signal (1). Through such targeted degradation, the ubiquitin system is a critical mechanism by which cells regulate steady-state protein concentrations and cellular homeostasis (2–4). Ligation of ubiquitin to target proteins functions through a conserved mechanism involving the sequential activities of three enzymes that catalyze new isopeptide bond formation between the C terminus of ubiquitin and primary amines present on the target (2, 5). The activating enzyme (E1)3 couples ATP hydrolysis to activation of the C terminus of ubiquitin to form a ternary complex comprising both E1-ubiquitin thioester and tightly bound ubiquitin adenylate moieties (6). The E1 ternary complex then binds any of a superfamily of cognate E2 proteins to catalyze transthiolation of the ubiquitin thioester between the conserved active site cysteines of E1 and E2 (7, 8). The second half-reaction of target protein ligation is then catalyzed by one of several hundred E3 ligases that couples new isopeptide bond formation to the aminolytic cleavage of the high energy E2-ubiquitin thioester (2).
Less understood is the mechanism by which polyubiquitin chains are synthesized (9), particularly among RING ligases that constitute a diverse group of conjugating enzymes sharing a common catalytic domain that coordinates two Zn2+ ions to stabilize specialized RING finger motifs (4). The RING domains constitute the functional cassette that is responsible for binding the cognate E2-ubiquitin thioester that functions as a co-substrate for the ligase and for catalysis of isopeptide bond formation (4). In the absence of substrate, RING ligases catalyze slow formation of high molecular weight free and autoconjugated polyubiquitin chains (10, 11). Observation of free polyubiquitin chain formation challenges the conventional view that chains are assembled on the target substrate sequentially by distal addition of ubiquitin moieties to a growing target-anchored ubiquitin chain (9). However, a mechanism proposed for substrate conjugation by the gp78 ligase in which polyubiquitin chains are first assembled on the cognate Ube2g2/Ubc7 ubiquitin carrier protein then transferred to the target protein en bloc anticipates free chain formation as the result of the side reaction of E2-chain thioester hydrolysis (12).
The tripartite motif (TRIM) superfamily constitutes the largest group of RING ligases with over 100 paralogs (13, 14). Members of the TRIM superfamily share a common tripartite RING-B box-coiled coil domain architecture and function in various aspects of cell regulation, development, innate immunity, and the etiology of certain cancers (13, 15). The B box and coiled coil domains are required for oligomerization of TRIM family members, although the role of oligomerization in the function of these enzymes is unclear (13). Additional domain(s) C-terminal to the coiled coil domain serve as targeting sites for recruiting protein substrates for ubiquitin conjugation (13). As with other RING ligases, the mechanism by which the TRIM paralogs catalyze formation of the polyubiquitin degradation signal is thought to proceed through sequential distal addition of ubiquitin to the growing chain appended to the target protein (2, 13), although this mechanism has not been empirically tested.
The current study investigates the mechanism of polyubiquitin chain formation catalyzed by TRIM32, the function of which is linked to tumorigenesis in squamous cell carcinoma and other cancers (16–18), the etiology of psoriasis (19), Alzheimer syndrome (20), and neuronal development/survival (21, 22). Defects in TRIM32 are additionally implicated in limb-girdle muscular dystrophy (23) and Bardet-Biedl syndrome (24). We have exploited the innate ability of TRIM32 to catalyze polyubiquitin chain formation in the absence of substrate as a functional readout of ligase activity (10). Selected experiments have been extended to two other members of the TRIM ligase superfamily to explore the generality of our observations. In this first mechanistic examination of TRIM ligase function, kinetic studies suggest that members of the TRIM superfamily function through a conserved allosteric cooperative mechanism requiring oligomerization. Additional rate data are consistent with an elongation mechanism in which polyubiquitin chains are first assembled as an E2-linked thioester prior to being appended en bloc to the target protein.
MATERIALS AND METHODS
Bovine ubiquitin, creatine phosphokinase, and apyrase were purchased from Sigma. Ubiquitin was further purified to apparent homogeneity (25) then radioiodinated by the chloramine-T procedure using carrier-free Na125I (PerkinElmer Life Sciences) to yield a specific radioactivity of 8000 cpm/pmol (26). Thrombin was purchased from GE Healthcare. Human erythrocyte Uba1 (UBE1) was purified to apparent homogeneity from outdated human blood (27). Active Uba1 was quantitated by the stoichiometric formation of 125I-ubiquitin thioester (27, 28). Recombinant human E2 proteins were those used previously (29), the activities of which were quantitated by the stoichiometric formation of E2-125I-ubiquitin thioester and compared with the expected activities based on protein content determined by their calculated 280 nm extinction coefficients (10, 28). Point mutants of Ubc5A were created using the QuikChange kit (Stratagene) (29, 30). Recombinant human IsoT (USP5) was expressed in Escherichia coli BL21(DE3) cultures harboring pRSIsoT (generous gift of Dr. Keith Wilkinson, Emory University School of Medicine) and purified to apparent homogeneity as described previously (31). The concentration of IsoT in the incubations was calculated from total protein determined spectrophotometrically using a theoretical ϵ280 nm extinction coefficient of 1.09 mg/ml−1.
Expression and Purification of Recombinant TRIM Proteins
Human TRIM32 cDNA (TRIM32; IMAGE clone BC003154) was used as a template for PCR-based subcloning of the full-length and truncated forms of the coding sequence into pPRO-EX-HTb and pGEX-4T1 vectors using the EcoRI/XhoI restriction sites. E. coli BL21(DE3) Arctic Express cells (Agilent Technologies) harboring pPRO-EX-HTb-TRIM32 were induced with 0.4 mm isopropyl 1-β-d-galactopyranoside in the presence of 0.1 mm ZnSO4 for 40 h at 15 °C. His6-TRIM32 was purified on a 1.5 × 3.0-cm HisTrap column (GE Healthcare) according to the manufacturer's instructions. The pPRO-EX-HTb-TRIM32ΔNHL plasmid was created by QuikChange mutation of the Ile352 codon to a STOP codon and was used to express the His6-TRIM32ΔNHL truncated protein, purification of which was performed as described for full-length TRIM32. The pGEX-4T1-TRIM32 construct was not suitable for production of full-length TRIM32 because thrombin treatment to remove GST also cleaved TRIM32 between the coiled coil and NHL domains. However, the construct was used to generate the GST-RING fusion by mutating the Leu98 codon to a STOP codon to yield pGEX-TRIM32ΔCL98. The pGEX-TRIM32ΔCL98 plasmid was used to express the GST-RING fusion protein as described for GST-E2 proteins and subsequent thrombin cleavage of GST-RING yielded the RING domain alone (29). The GST moiety was indispensable for RING domain purification and quantitation because the RING domain has neither tryptophan nor tyrosine residues and, thus, no appreciable absorbance at 280 nm. The GST-RING ruined thrombin construct was created by mutating the critical arginine within the thrombin cleavage site upstream of the multiple cloning site on the pGEX-4T1 plasmid to alanine. Quantitation of total protein for full-length and truncated TRIM32 species was accomplished by densitometry of Coomassie-stained bands and compared with a known amount of comparably stained GST protein following SDS-PAGE resolution. Recombinant TRIM proteins were flash frozen and stored in small aliquots at −80 °C. Aliquots were flash thawed and used only once.
Human TRIM25 cDNA (TRIM25; IMAGE clone BC016924) was used as a template for PCR-based subcloning of the full-length coding sequence into the EcoRI/XhoI restriction sites of pGEX-4T1 to yield pGEX-4T1-TRIM25. Recombinant TRIM25 protein was expressed and purified identically to TRIM32 with the exception that the resulting GST-TRIM25 was processed with thrombin because the ligase was not cleaved during proteolytic processing of the GST moiety. Human TRIM5α (corresponding in sequence to NM_033034) was subcloned from pCDN3.1-Trim5α (generous gift of Dr. Ronald Luftig, Louisiana State University School of Medicine) into pGEX4T1 using the EcoRI/XhoI restriction sites to yield pGEX4T1-TRIM5α. Recombinant TRIM5α was expressed in the Rosetta strain of E. coli BL21 cells (Millipore) and purified identically to TRIM25.
In Vitro Polyubiquitin Chain Formation Assays
Formation of 125I-ubiquitin chains was measured at 37 °C in 25-μl reactions containing 50 mm Tris-HCl (pH 7.5), 2 mm ATP, 10 mm MgCl2, 10 mm creatine phosphate, 0.5 IU of creatine phosphokinase, 1 mm DTT, 0.1 mg/ml of BSA, 5 μm 125I-ubiquitin, and the indicated concentrations of Uba1, E2, and TRIM protein. After 10 min, reactions were quenched by addition of 25 μl of SDS sample buffer, then incubated at 100 °C for 3 min and resolved by 12% (w/v) SDS-PAGE (10, 28). The resulting 125I-polyubiquitin chains were visualized by autoradiography of the dried gels then, unless otherwise indicated, associated radioactivity in the stacker corresponding to free polyubiquitin chains was determined by excising the regions of conjugates and quantitating associated radioactivity by γ-counting (10, 32). Kinetic assays measured the initial rates of chain formation, for which less than 5% of total 125I-ubiquitin was conjugated, and under E3-limiting conditions, confirmed by the independence of rate on [Uba1]o (10). Sigmoidal kinetics for TRIM32-catalyzed chain formation were verified by the linearity of the resulting Hill plot; however, values for [S]0.5 and the Hill coefficient n were calculated by nonlinear regression analysis using Grafit 5.0 (Erithacus Software Ltd.).
IsoT-catalyzed Polyubiquitin Disassembly
In vitro chains were formed as described above and reactions were quenched with 1 IU of apyrase for 5 min at 37 °C quantitatively to deplete ATP before adding DTT to a final concentration of 10 mm and incubating an additional 5 min at 37 °C to cleave polyubiquitin chains from their E2 thioester linkage. To this reaction apparently homogeneous recombinant IsoT was added to a final concentration of 100 μm (zero time) after which aliquots were removed at the indicated times and quenched with an equal volume of SDS sample buffer. After 60 min, a second aliquot of IsoT was added and incubation was continued an additional 30 min. Samples were resolved by SDS-PAGE and autoradiographed.
Assembly of Polyubiquitin Chains on Ubc5AC85S-125I-Ubiquitin Oxyester
The Ubc5AC85S-125I-ubiquitin oxyester was synthesized and purified by Superdex 75 FPLC on a 1 × 30-cm column as described previously for Ubc2bC88S-125I-ubiquitin oxyester (33). Eighty nm Ubc5AC85S-125I-ubiquitin oxyester was incubated at 37 °C in a 30-μl volume containing 50 mm Tris-HCl (pH 7.5), 2 mm ATP, 10 mm MgCl2, 10 mm creatine phosphate, 0.5 IU of creatine phosphokinase, 1 mm DTT, 0.1 mg/ml of BSA, 80 nm Uba1, 100 nm Ubc5A, 10 μm TRIM32 protein, and 5 μm ubiquitin (as indicated) for 10 min. Reactions were quenched with SDS sample buffer and resolved by 12% SDS-PAGE. The gel was dried and bands were visualized by autoradiography, excised from the gel, and associated radioactivity was quantitated by γ-counting (10, 32).
Glutaraldehyde Cross-linking
Oligomerization of TRIM32 and GST-TRIM were demonstrated by glutaraldehyde cross-linking essentially as described by Peng et al. (34). Reactions of 20 μl final volume containing 50 mm Tris-HCl (pH 7.5), the indicated concentrations of glutaraldehyde, and either 2.9 μm TRIM32ΔNHL or 2.3 μm GST-RING were incubated 5 min at 22 °C before quenching by addition of 20 μl of SDS sample buffer. Reactions were then incubated an additional 30 min at 37 °C before resolving at 12% (w/v) SDS-PAGE. Proteins were electrophoretically transferred to PVDF membrane (Millipore) and detected with rabbit anti-human TRIM32 antibody directed against the RING domain (Abcam, ab50555). Bound antibody was visualized using horseradish peroxidase-conjugated goat anti-rabbit IgG antibody and Pierce ECL Western blot substrate (Thermo Scientific) according to the manufacturer's instructions.
RESULTS
Conservation of E2 Specificity within the TRIM Ligase Family
An important step in the functional characterization of ubiquitin ligases requires identification of their cognate E2 for supporting the conjugation reaction because ligases typically function efficiently with members of only a single E2 family (10). However, such identification is not trivial because the E2 proteins share similar amino acid sequences and folds within their highly conserved ∼150 residue core catalytic domains (5, 35). These inherent structural similarities result in the potential for spurious binding interactions between E2 and E3 proteins at high concentrations of the interactors or long incubation times that frequently lead to ambiguities in the literature regarding correct E2-E3 cognate pairs, as has been discussed previously (5, 10). TRIM32 is reported to function with isozymes of the Ubc5 family of E2 proteins and the unrelated UbcH6 family (36, 37). To quantitatively re-examine the question of the cognate E2 for TRIM32, the biochemically defined functional E2 screen of Fig. 1 was performed under E3 limiting initial velocity conditions and equivalent 50 nm concentrations of active E2 protein, determined in activity assays by their stoichiometric formation of 125I-ubiquitin thioester (10, 28). The former conditions guarantee that the resulting autoradiographic intensity for conjugates formed in the reaction reflect a relative rate, whereas the latter functional assays obviate variations in E2 concentration originating from differences in specific activities of the E2 proteins when defined by total protein (10).
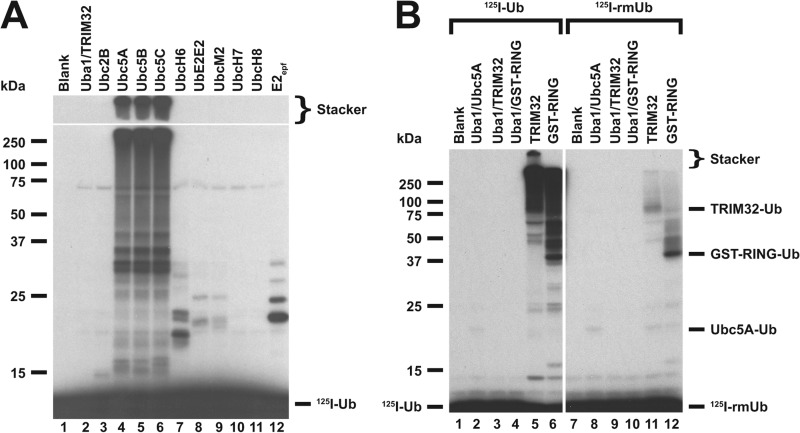
FIGURE 1.
E2 screen for TRIM32-catalyzed 125I-ubiquitin conjugation. A, autoradiogram of SDS-PAGE-resolved assays of TRIM32-catalyzed 125I-ubiquitin chain formation. Assays were conducted as described under “Materials and Methods” in the presence of 44 nm Uba1, 50 nm of the indicated E2, and 800 nm TRIM32. B, autoradiogram of SDS-PAGE resolved assays conducted identically to panel A but in the presence of 1.6 μm TRIM32 or 27 μm GST-RING protein and 460 nm Ubc5A as indicated. Incubations additionally contained 5 μm wild type (left subpanel) or reductively methylated (rmUB, right subpanel) 125I-ubiquitin. The left and right panels were exposed to normalize to account for the difference in specific radioactivity of the polypeptides. For panels A and B, migration positions for relative molecular weight standards are shown to the left. The position of the 5% stacker gel (Stacker), monoubiquitinated TRIM32 (Mwcalc = 80 kDa), monoubiquitinated GST-RING (Mwcalc = 44 kDa), monoubiquitinated Ubc5A (Mwcalc = 25 kDa), and free 125I-ubiquitin are shown to the right. Deviation of the apparent relative molecular weight of the monoubiquitinated species from their calculated molecular masses (Mwcalc) reflects non-ideality of ubiquitin due to its partial unfolding in SDS.
No conjugation was observed in the presence of Uba1 and TRIM32 alone (Fig. 1A, lane 2). Significant conjugation is observed only upon addition of the Ubc5 isozymes (Fig. 1A, lanes 4–6), indicating that these paralogs are the cognate E2 species for TRIM32. Conjugation supported by the Ubc5 isoforms is sufficiently robust that a significant fraction of the adducts appear at the top of the resolving gel or fail to migrate into the 5% (w/v) stacker (indicated). The minor 125I-ubiquitin adduct bands below 50 kDa observed with some E2 species result from E3-independent E2 autoubiquitination (10). The inability of an equivalent concentration of active UbcH6 to similarly support TRIM32 conjugation (lane 7) suggests that the E3-independent autoubiquitination noted earlier for this paralog (29) may have been mistaken for TRIM32-catalyzed conjugation or that the concentration of UbcH6, determined from the protein concentration (37), was sufficiently high to force an otherwise low affinity binding interaction not detected under the current biochemically defined conditions. The E3-dependent signal observed in Fig. 1A is due to formation of 125I-polyubiquitin chains because substitution of reductively methylated ubiquitin (125I-rmUb), in which all lysine residues have been stoichiometrically blocked with methyl groups to render them unable to form isopeptide bonds (38), significantly ablates overall conjugation (Fig. 1B, lanes 5 versus 11). The small amount of conjugates observed in the right panel of Fig. 1B represent monoubiquitinated TRIM32 (lane 11) or monoubiquitinated GST-RING (lane 12). Moreover, an observation that ladders of polyubiquitin chains extend upward from full-length TRIM32 (Fig. 1B, lane 5) is consistent with significant autoubiquitination of the ligase.
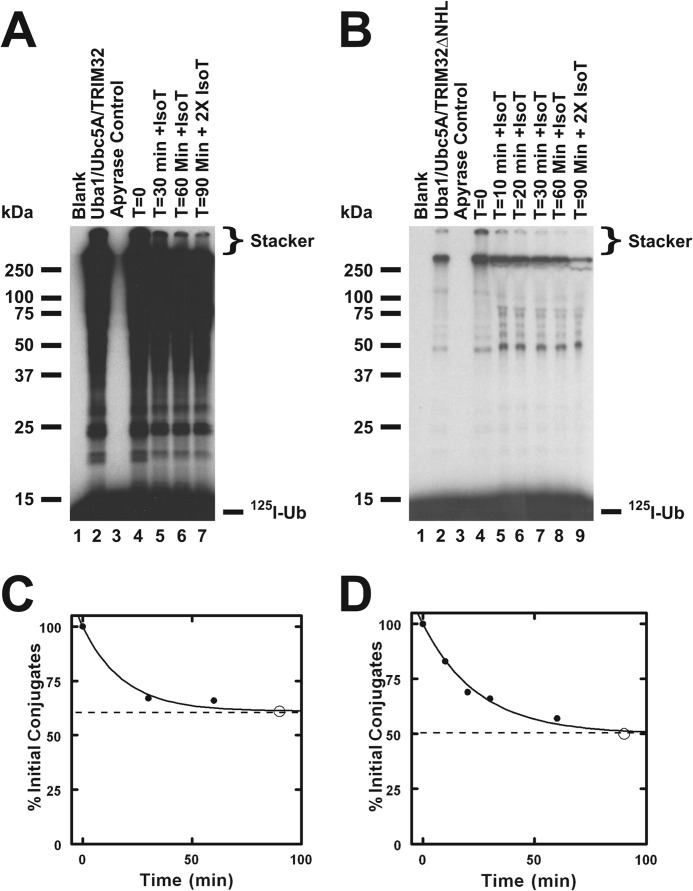
The ability of TRIM32 to form both free and anchored chains is demonstrated by the disappearance of a portion of the high molecular weight 125I-ubiquitin signal upon treatment with the ubiquitin-specific protease IsoT, which disassembles only free polyubiquitin chains (39) (Fig. 2, A and C). At the long exposure use in panel A of Fig. 2 to emphasize the labile signal in the stacker, one also observes loss of the lower molecular mass signal immediately above 25 kDa upon incubation with IsoT (lane 4 versus 5–7). That a fraction of the chains are refractory to disassembly by IsoT, even after further incubation with additional enzyme (Fig. 1C, open circle, and Fig. 2A, lane 7), demonstrates that ∼50% of the signal is due to anchored chains under these conditions, presumably representing autoubiquitination of TRIM32.
FIGURE 2.
TRIM32 and TRIM32ΔNHL form free and conjugated polyubiquitin chains. Incubations identical to those of Fig. 1 were conducted with Ubc5A and either TRIM32 (panel A) or TRIM32ΔNHL (panel B) for 10 min then ATP was removed by the addition of apyrase as described under “Materials and Methods.” Following addition of human recombinant IsoT to a final concentration of 100 μm, aliquots were taken at the indicated times. After 60 min an additional aliquot of IsoT was added and the reaction was incubated an additional 30 min. Migration positions for the relative molecular weight standards are shown to the left. Position of the 5% stacking gel and free 125I-ubiquitin are shown to the right. Panels C and D, total conjugated 125I-ubiquitin above 25 kDa, including the stacker gel, was quantified by associated radioactivity as described under “Materials and Methods” for the time points of panels A and B, respectively (closed circles). Open circles indicate conjugates remaining after addition of the second aliquot of IsoT. Data were fit to first order kinetics by nonlinear regression (solid line) to yield the end point (dashed line).
We also tested the specificity of TRIM25, which has been reported to function with a variety of E2 paralogs including UbcH8 (40, 41), Ubc5A (42, 43), Ubc5C (44), UbcH6 (41), and Ubc13 (43), all of which are members of the Ubc4/5-like clade (5). An E2 screen identical to that of Fig. 1A demonstrates that TRIM25-catalyzed polyubiquitin chain formation is supported exclusively by members of the Ubc5 family (not shown). Finally, a third E2 screen with recombinant TRIM5α also confirmed an earlier report that the ligase functions only with the three Ubc5 isozymes (45), not shown. Thus, of the three representative TRIM paralogs tested, all are supported exclusively by the Ubc5 family of ubiquitin carrier proteins, suggesting Ubc5 ortholog specificity may be conserved among the TRIM family members.
Polyubiquitin Chain Formation Requires Oligomerization of the RING Domain
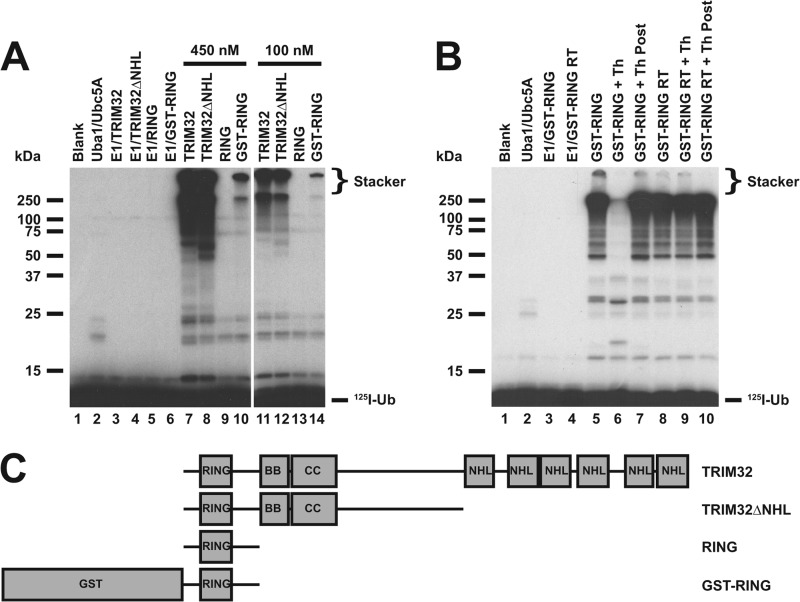
The RING domain is conserved among nearly all of the TRIM family members (13, 46) as well as several hundred other ligases possessing distinct domain architectures (4). A panel of truncated TRIM32 proteins was generated to investigate the minimum portion of TRIM32 necessary to form polyubiquitin chains, schematically summarized in Fig. 3C. Conjugation assays similar to those of Fig. 1A were conducted under E3-limiting initial velocity conditions so that the autoradiographic intensity of the resulting autoradiogram would be proportional to rate (10). Additionally, assays were performed at different E3 concentrations to ensure the conjugation observed was E3 limiting, as shown by the proportionality of rate to the TRIM construct concentration. Fig. 3A demonstrates that truncation of the NHL domains has no effect on polyubiquitin chain formation because equimolar TRIM32 or TRIM32ΔNHL qualitatively exhibits nearly identical rates of 125I-ubiquitin chain formation (Fig. 3A, lanes 7, 8, 11, and 12) compared with controls with Uba1 and Ubc5A alone (lane 2) or Uba1 and TRIM32 alone (lanes 3–6). However, additional truncation of the B box and coiled coil domains to yield the RING domain alone abrogates chain formation (lanes 9 and 13). The ability to form chains is partially rescued by fusion of the free RING domain to GST (lanes 10 and 14), presumably due to the propensity of GST to dimerize (47, 48) and partially substitute for B box- and coiled coil domain-mediated oligomerization (46). Apparent loss of chain formation following in situ processing of GST from the RING (Fig. 3B, lane 5 versus 6) is not due to artifactual processing the polyubiquitin chains by thrombin because the protease has no effect on chains when added after the chain formation reaction has been quenched (Fig. 3B, lane 7). Nor is loss of chain formation due to direct inhibition by thrombin, because the GST-RING ruined thrombin fusion protein harboring a mutated thrombin cleavage site is not similarly inhibited by thrombin treatment (lane 9). Although the processed RING domain alone is incapable of catalyzing polyubiquitin chain formation, it is able to undergo automonoubiquitination to yield a unique band of about 18 kDa (Fig. 3B, lane 6), formation of which can serve as a functional readout of RING domain activity.
FIGURE 3.
Polyubiquitin chain formation requires multimerization. A, incubations similar to those of Fig. 1 were conducted with TRIM32 (lanes 7 and 11), TRIM32ΔNHL (lanes 8 and 12), RING domain alone formed by processing with 90 IU/ml of thrombin for 30 min (lanes 9 and 13), or a GST-RING fusion (lanes 10 and 14). Reactions contained 48 nm Uba1 and either 450 (left subpanel) or 100 nm (right subpanel) of the indicated E3 protein in the absence (lanes 3–6) or presence (lanes 7–14) of 280 nm Ubc5A. B, thrombin-dependent processing of GST from the RING moiety is responsible for abrogation of chain formation. Conjugation reactions contained 34 nm Uba1 and 18 μm GST-RING protein in the absence (lanes 3 and 4) or presence of 600 nm Ubc5A (lanes 4–10). The GST-RING or GST-RING ruined thrombin in which the thrombin cleavage site had been mutated to obviate processing were preincubated in the absence (lanes 5 and 8) or presence (lanes 6 and 9) of 90 IU/ml of thrombin for 30 min before addition to the conjugation assay. Alternatively, thrombin treatment was conducted after quenching the conjugation reaction (lanes 7 and 10). For both panels, migration positions for relative molecular weight standards are shown to the left. The position of the 5% stacking gel (Stacker), free 125I-ubiquitin, and mono-125I-ubiquitinated RING are shown to the right. C, schematic representation of constructs used in panels A and B.
To confirm that fusion of GST to the RING domain rescues the functional phenotype of chain formation observed for full-length TRIM32, the activity of the GST-RING fusion was tested in the presence of 125I-rmUb. Substitution of 125I-rmUb for wild type 125I-ubiquitin in the GST-RING-catalyzed chain formation assay results in significant loss in conjugation, indicating that the GST-RING fusion also forms polyubiquitin chains (Fig. 1B, lanes 6 versus 12). Furthermore, the chains of ubiquitin formed by the GST-RING fusion also partition between free and conjugated forms because the former are susceptible to disassembly by IsoT (not shown), as observed for full-length TRIM32 and TRIM32ΔNHL (Fig. 2).
The TRIM Proteins Exhibit Cooperative Kinetics with Respect to [Ubc5]o
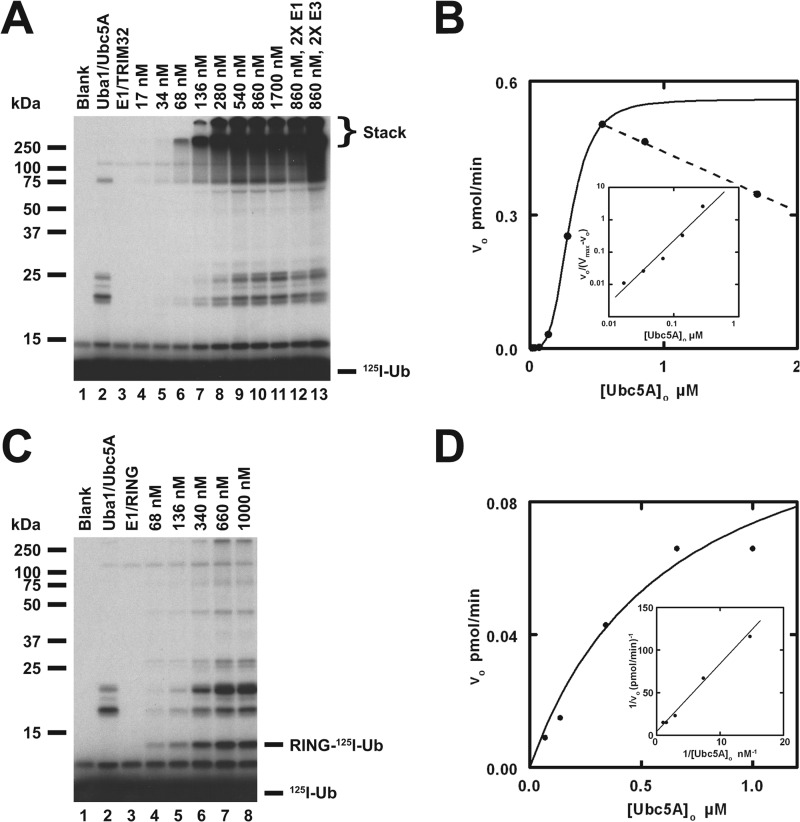
Prior work from our laboratory has demonstrated that rates of polyubiquitin chain formation can serve as a facile reporter function for kinetically characterizing E3 activity (10, 33, 49). We applied this approach to expand on the semiquantitative data from Figs. 1–3. Under TRIM32-limiting conditions, 125I-ubiquitin chain formation exhibits a marked dependence on Ubc5A concentration (Fig. 4A). Because the assays were conducted under initial velocity conditions, autoradiographic intensity is proportional to the initial rate. That the reactions are E3 limiting and, therefore, reflect the intrinsic rate of TRIM32 is demonstrated by the increase in conjugation when TRIM32 (lane 13) but not Uba1 (lane 12) is doubled (Fig. 4A). The latter control additionally confirms that the assays contain sufficient Uba1 to maintain all Ubc5A charged as the corresponding Ubc5A-125I-ubiquitin thioester, the actual substrate for TRIM32 (33, 49). Excision of the lanes from the gel and quantitation of the associated radioactivity by γ-counting allows one to calculate the corresponding rate of polyubiquitin chain formation based on the specific radioactivity of the 125I-ubiquitin (10, 32, 49). The dependence of the initial rate for formation of free chains present in the stacker gel on the concentration of Ubc5A-125I-ubiquitin thioester, measured as total active Ubc5A, is sigmoidal below 600 nm but exhibits pronounced substrate inhibition at higher E2 concentrations (Fig. 4B, dashed line). The sigmoidal dependence indicates TRIM32 shows allosteric cooperativity with respect to its charged E2 substrate, which is the first example of such behavior by a ubiquitin ligase. The sigmoidal dependence on [Ubc5A]o in Fig. 4B is confirmed by the linearity of the corresponding Hill plot (inset) and the excellent nonlinear regression fit of the data to the Hill equation (solid line). Cooperativity requires multiple conformationally linked binding sites for the Ubc5A-125I-ubiquitin thioester substrate, the lower limit of which is defined by n, whereas substrate inhibition at higher concentrations is consistent with ordered binding of Ubc5A-125I-ubiquitin thioester to these multiple sites during the catalytic cycle of chain formation. In these kinetic measurements we chose to focus on rates of free chain formation because identification of such homogeneous products are unambiguous and well resolved in the stacker from the lower molecular weight anchored chains. However, analysis of the rates of anchored chain formation shows results similar to those of free chains, consistent with partitioning of a common intermediate between free and anchored chains.
FIGURE 4.
TRIM32 exhibits cooperativity with respect to [Ubc5A]o. A, autoradiogram of the dependence of TRIM32-catalyzed 125I-ubiquitin chain formation on [Ubc5A]o (lanes 4–11). To verify that reactions were E3 limiting, E1 and E3 concentrations were doubled in lanes 12 and 13, respectively, whereas holding the E2 concentration at 860 nm. Reactions contained 44 nm Uba1, the indicated concentrations of Ubc5A, and 80 nm TRIM32 protein, as indicated. The position of the 5% stacking gel and free 125I-ubiquitin are shown to the right. B, plot of the initial velocity of chain formation (the band in the stacking gel from panel A) versus [Ubc5A]o. The sigmoidal curve is a nonlinear least squares fit to the Hill equation of the data from lanes 4–9, prior to the observed substrate inhibition in lanes 10 and 11. The sigmoidicity of the plot is verified by the linearity of the corresponding Hill plot (inset). The substrate inhibition observed above 540 nm is indicated by the dashed line. C, autoradiogram of the dependence of RING domain-catalyzed auto-125I-monoubiquitination on [Ubc5A]o (lanes 4–8). Reactions contained 72 nm Uba1, the indicated concentrations of active Ubc5A, and 9.4 μm RING domain protein, as indicated. The positions of the 5% stacking gel, RING-125I-ubiquitin, and free 125I-ubiquitin are shown on the right. The RING domain was generated prior to the assays by thrombin cleavage (20 IU of thrombin in 230 μl containing 118 μm GST-RING fusion protein for 30 min at room temperature). D, plot of the initial velocity of RING auto-125I-monoubiquitination versus [Ubc5A]o. The hyperbolic shape of the curve was verified by the linearity of the double-reciprocal plot in the inset. The hyperbolic curve was derived by nonlinear least squares fit to the data in lanes 4–8, using GraFit (Erithacus Software Ltd.).
Nonlinear regression fit of the data in Fig. 4B to the Hill equation allows calculation of Vmax (0.54 ± 0.01 pmol/min), the corresponding kcat as Vmax/[Trim32]o (4.5 ± 0.8 × 10−3 s−1) with [TRIM32]o expressed as total protein, the concentration of substrate yielding half-maximal velocity [S]0.5 (410 ± 10 nm), and the Hill coefficient n (5.1 ± 0.3) (Table 1). Parallel experiments similar to that of Fig. 4 revealed that TRIM25 and TRIM5α also exhibit cooperativity with respect to [Ubc5A]o and substrate inhibition at higher concentrations of the E2 (not shown). The resulting kinetic constants derived from nonlinear regression analysis of the respective data also corresponded well to that obtained with TRIM32 (Table 1). This suggests that cooperativity and the mechanism of polyubiquitin chain formation are conserved catalytic features of the TRIM superfamily.
TABLE 1.
Summary of kinetic constants
| kcata | [S]0.5 | n | |
|---|---|---|---|
| s−1 | nm | ||
| TRIM32 | 4.5 ± 0.8 × 10−3 | 410 ± 10 | 5.1 ± 0.3 |
| TRIM25 | 5.2 ± 0.1 × 10−2 | 460 ± 20 | 3.6 ± 0.5 |
| TRIM5α | 0.28 ± 0.05 × 10−3 | 280 ± 43 | 2.9 ± 0.6 |
| TRIM32ΔNHL | 5.6 ± 0.3 × 10−3 | 340 ± 12 | 5.1 ± 0.7 |
| GST-RING | 26 ± 8 × 10−3 | 5000 ± 840 | 2.0 ± 0.1 |
a Values of kcat determined as Vmax/[E3]o, where [E3]o is calculated from total TRIM protein.
Allosteric cooperativity was observed with TRIM32ΔNHL (not shown), consistent with the ability of the TRIM enzymes to oligomerize through their conserved B box-coiled coil domains in the absence of the NHL targeting domains and yielded kinetics similar to wild type TRIM32 when normalized to enzyme concentration (Table 1). To confirm that subunit association through the B box-coiled coil region is required for the cooperative allosteric response, we also examined the kinetics of a GST-RING construct derived from the catalytic domain of TRIM32 (not shown) and the corresponding free RING domain following in situ processing with thrombin (Fig. 4, C and D). The GST-RING fusion protein exhibited attenuated allosteric cooperativity (n = 2 ± 0.1) with a significantly reduced apparent affinity for Ubc5A ([S]0.5 = 5.0 ± 0.8 μm), suggesting a diminished coupling between binding sites (Table 1). These observations indicate that the NHL domains are not essential for either chain formation or cooperativity. Although the B box and coiled coil domains are required for oligomerization and chain formation catalyzed by full-length TRIM32, truncation of these oligomerization domains does not completely abolish the potential for cooperativity when an alternative mechanism for tethering the RING domains is substituted by fusion to GST, which is known to dimerize with micromolar affinity (34, 50). In contrast, the free RING domain generated by in situ thrombin processing catalyzes only automonoubiquitination, suggesting oligomerization is required for chain formation (Figs. 2, B, lane 6, and 4C). When the initial rate of monoubiquitination is exploited as a functional readout of free RING domain catalytic activity, this module exhibits hyperbolic kinetics with respect to [Ubc5A]o (Km = 630 ± 286 nm and kcat = 8.2 ± 1.9 × 10−6 s−1), confirmed by the linearity of the corresponding double-reciprocal plot (Fig. 4D, inset).
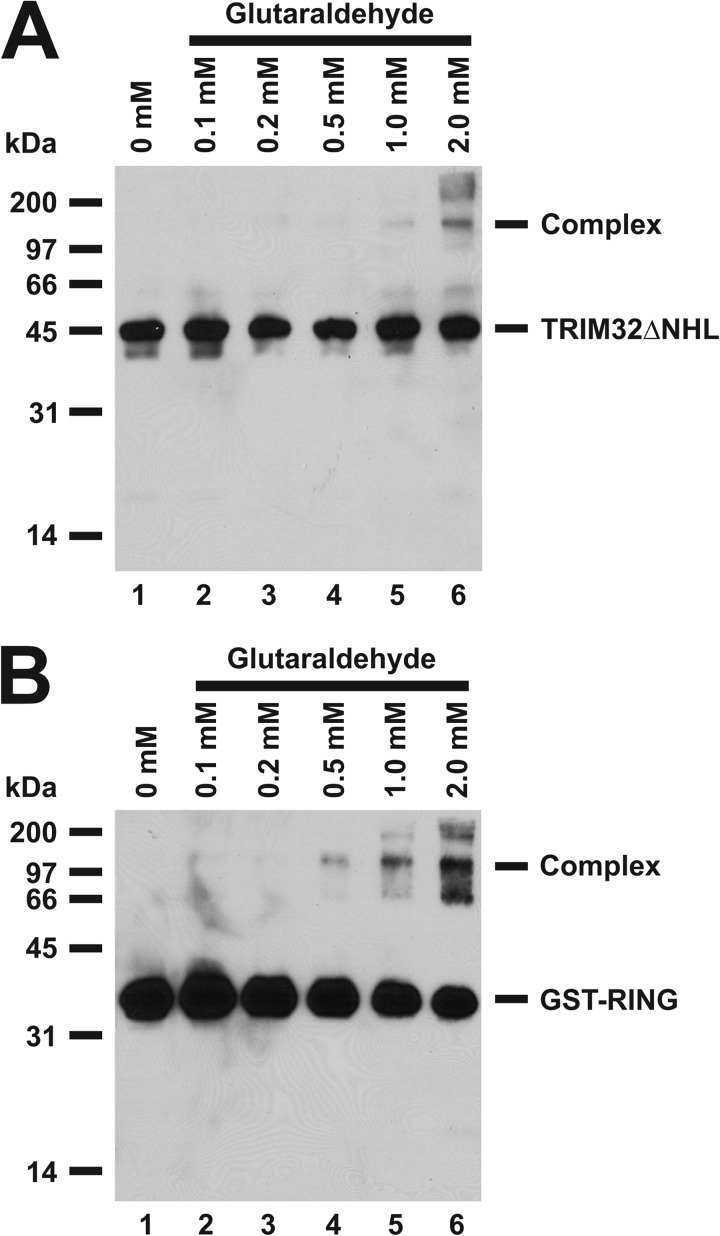
Glutaraldehyde cross-linking experiments were performed on Trim32ΔNHL and GST-RING proteins to determine the number of subunits present in the active E3. Consistent with previous observations for TRIM5α, TRIM28/KAP1, and TRIM33/TIF1α/γ (51, 52), glutaraldehyde cross-linking experiments demonstrate a fraction of the TRIM32ΔNHL and GST-RING proteins can be covalently cross-linked (Fig. 5). Previously, Li et al. (51) observed cross-linking that they described as consistent with a trimer, whereas Peng et al. (52) observed even higher oligomers. In the present studies, cross-linking of TRIM32ΔNHL or GST-RING yield bands with relative molecular weights consistent with dimers, tetramers, and higher order oligomers (Fig. 5), supporting levels of oligomerization of TRIM32 predicted by the observed Hill coefficient (Table 1).
FIGURE 5.
Glutaraldehyde cross-linking confirms oligomerization of TRIM32ΔNHL and GST-RING. Glutaraldehyde cross-linking reactions were conducted with either TRIM32ΔNHL (panel A) or GST-RING (panel B) proteins as described under “Materials and Methods” then resolved by 12% (w/v) SDS-PAGE before visualization by Western blotting. Migration positions for relative molecular weight standards are shown to the left.
TRIM32 Binds Ubc5A via Two Distinct Surfaces on the E2
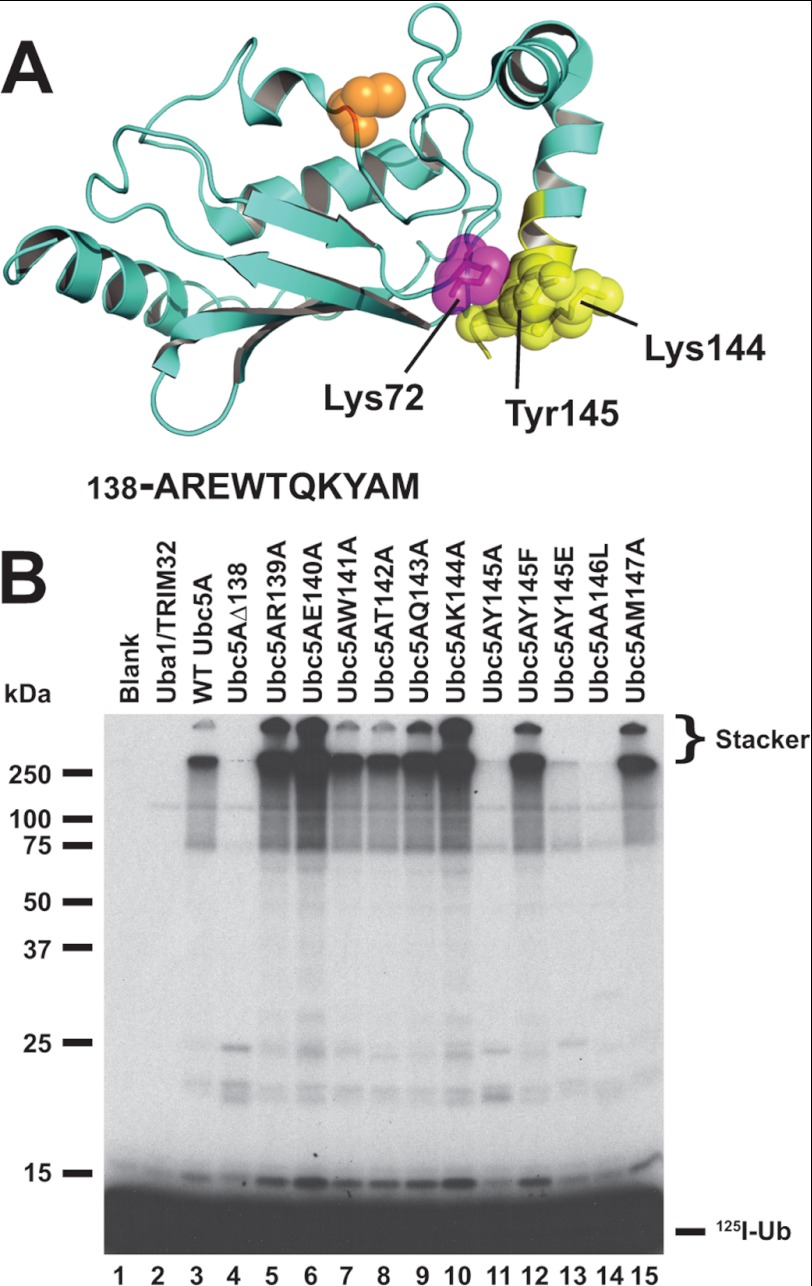
Ubiquitin carrier proteins bind RING domains such as those present on the TRIM ligases through the Loop 1/2 surface encompassing the N-terminal helix of the E2, as originally shown for UbcH7 binding to Cbl (53). The RING binding interface overlaps that for E1, precluding E1-E2-E3 complexes and requiring dissociation of the E2-ubiquitin thioester intermediate from E1 before binding to the cognate E3 is possible (29, 54). In contrast, we found that truncating the last 10 residues of UbcH5A comprising the C-terminal α-helix, highlighted in yellow in Fig. 6A, abrogates chain formation (Fig. 6B, lane 3 versus 4). Loss of function was not due to disruption of the E2-ubiquitin thioester formation because Uba1 ternary complex binding occurs at the opposite end of the uncharged E2 (29) and Ubc5AΔ138 supports Uba1-catalyzed transthiolation (not shown). In addition, the concentration dependence for Ubc5AΔ138 transthiolation by Uba1 showed wild type kinetics for Km and kcat (not shown), confirming that the truncation does not obviate correct folding of the protein. Alanine scanning mutagenesis within the 10 truncated residues indicates that some residues have no effect on polyubiquitin chain formation (W141A and T142A; Fig. 6B, lanes 7 and 8), whereas other positions qualitatively enhance the rate of chain formation (R139A, E140A, Q143A, K144A, and M147A; Fig. 6B, lanes 5, 6, 9, 10, and 15). However, mutation of Tyr145 to alanine or Ala146 to a more bulky leucine ablates chain formation (Fig. 6B, lanes 11 and 14). These qualitative results were confirmed by quantitative kinetics for the E2 concentration dependence for 125I-ubiquitin polyubiquitin chain formation determined similar to the study of Fig. 4B and Table 2.
FIGURE 6.
Mutations in the C-terminal α-helix of Ubc5A abrogate polyubiquitin chain formation. A, structure of Ubc5A with the active cysteine shown in orange spheres and the last 10 residues of the protein within the C-terminal α-helix are highlighted in yellow. The packing of Tyr145 between Lys72 and Lys146 is highlighted. The sequence for the C-terminal 10 residues is shown below the structure. The image was created from PDB file 2C4P (Dodd, R. B., and Read, R. J., unpublished coordinates) using PyMol (Schrodinger). B, autoradiogram comparing wild type Ubc5A (lane 3) and the indicated C-terminal point mutants (lanes 4–15) in their ability to support TRIM32-catalyzed 125I-ubiquitin chain formation. Reactions contained 64 nm Uba1, 100 nm active E2, and 200 nm TRIM32 protein, as indicated. Migration positions for relative molecular weight standards are shown to the left. The position of the 5% stacking gel and free 125I-ubiquitin are shown to the right.
TABLE 2.
Summary of kinetic constants for Ubc5A C-terminal mutants
| kcata | [S]0.5 | nH | |
|---|---|---|---|
| s−1 | nm | ||
| Ubc5A (wt)b | 4.5 ± 0.8 × 10−3 | 410 ± 10 | 5.1 ± 0.3 |
| Ubc5AΔ138 | NAc | NA | NA |
| Ubc5AR139A | 8.3 ± 0.1 × 10−3 | 302 ± 11 | 4.6 ± 0.7 |
| Ubc5AE140A | 4.9 ± 0.1 × 10−3 | 174 ± 6 | 4.6 ± 0.6 |
| Ubc5AW141A | 4.7 ± 0.1 × 10−3 | 480 ± 25 | 3.3 ± 0.3 |
| Ubc5AT142A | 1.8 ± 0.1 × 10−3 | 240 ± 2 | 5.2 ± 0.2 |
| Ubc5AQ143A | 4.6 ± 0.1 × 10−3 | 240 ± 10 | 5.1 ± 0.3 |
| Ubc5AK144A | 6.9 ± 0.1 × 10−3 | 190 ± 6 | 4.6 ± 0.6 |
| Ubc5AY145A | NA | NA | NA |
| Ubc5AA146L | 0.10 ± 0.01 × 10−3 | 570 ± 380 | 3.4 ± 1.4 |
| Ubc5AM147A | 4.3 ± 0.1 × 10−3 | 249 ± 6 | 5.7 ± 0.8 |
a Values of kcat determined as Vmax/[E3]o, where [E3]o is calculated from total TRIM protein.
b Values from Table I.
c NA, no activity.
Tyrosine 145 packs between Lys72 and Lys144 (Fig. 6A) so that mutation to alanine would deshield the charge repulsion otherwise present between these resides. Reconstitution of the aromatic nature of the side chain with Ubc5AY145F rescues polyubiquitin chain formation (Fig. 6B, lane 12), consistent with the model for deshielding by the aromatic ring. In contrast, mimicking the phenolate charge by substitution with glutamate does not restore chain formation (Fig. 6B, lane 13). Alanine 146 is buried in Ubc5 and substitution with leucine presumably disrupts packing of the C-terminal helix to the catalytic core of the E2. These results suggest that the C-terminal face of Ubc5A comprises a noncanonical E3 binding surface composed in part of the C-terminal α-helix and that this interaction requires native packing of this helix.
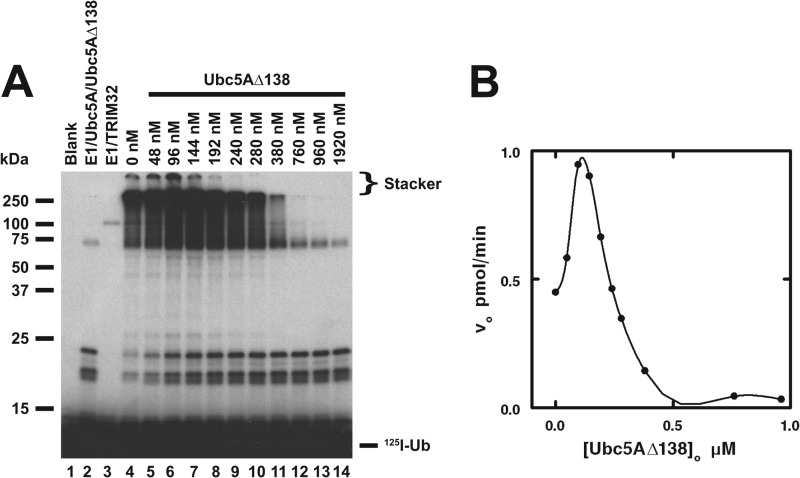
Although the Ubc5AΔ138 mutant does not support TRIM32-catalyzed chain formation, the data of Fig. 7 indicate that the mutant nonetheless binds TRIM32. The Ubc5AΔ138 mutant, present as its corresponding 125I-ubiquitin thioester due to its ability to be charged by Uba1 with wild type kinetics, stimulates Ubc5A-dependent chain formation at concentrations below 100 nm (Fig. 7), consistent with the allosteric cooperativity observed for the wild type E2 (Fig. 4). At higher concentrations, Ubc5AΔ138 displays the same concentration-dependent substrate inhibition observed for wild type protein. The latter is not due to inhibition of the Uba1 step because doubling [Uba1]o in parallel control incubations had no effect on rates of chain formation, confirming TRIM32-limiting conditions.
FIGURE 7.
Ubc5AΔ138 supports allosteric cooperative activation of TRIM32. A, autoradiogram of the dependence of TRIM32-catalyzed polyubiquitin chain formation on the Ubc5AΔ138 concentration (lanes 5–14) in incubations containing 72 nm Uba1, 135 nm wild type Ubc5A, the indicated concentrations of Ubc5AΔ138, and 2.4 μm TRIM32 protein as indicated. The positions of the 5% stacking gel and free 125I-ubiquitin are shown to the right. Migration positions for relative molecular weight standards are shown to the left. B, plot of the initial velocity for TRIM32-catalyzed 125I-ubiquitin chain formation versus [Ubc5AΔ138]o for the incubations in panel A as described under “Materials and Methods.”
TRIM32 Assembles Thioester-linked Polyubiquitin Chains on Ubc5A
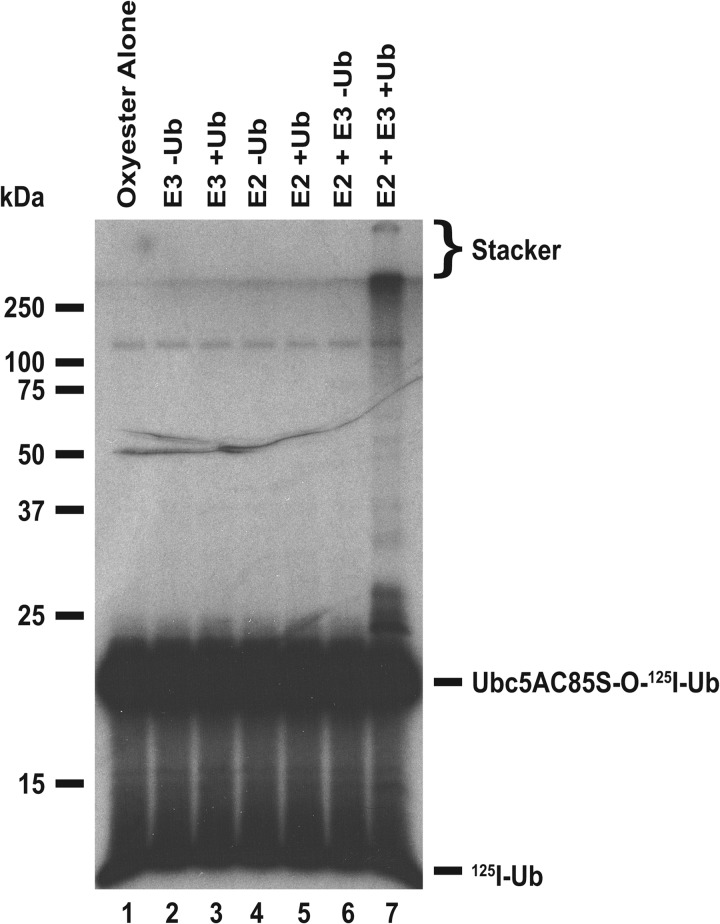
The mechanism of polyubiquitin chain formation is generally thought to proceed by sequential addition of successive ubiquitin molecules to the distal end of a growing chain attached to the target substrate (9). However, the ability of many ubiquitin ligases to assemble free polyubiquitin chains suggests a mechanism in which the chains are first formed then transferred to the target protein (10), as has been shown for the ER-associated ligase gp78 for which chains are assembled on its cognate Ube2g2/Ubc7 E2 carrier protein as a thioester (12). To test this for TRIM32, we exploited the ability of Uba1 to form a Ubc5AC85S-125I-ubiquitin oxyester from the corresponding active site point mutant then purified this inert substrate analog by gel filtration chromatography (33, 55). Such oxyesters are incapable of supporting subsequent E3-catalyzed conjugation reactions due to the stability of the E2-ubiquitin oxyester bond (33, 55); therefore, if TRIM32 is capable of preforming a polyubiquitin chain on Ubc5A as a thioester, one should observe similar chain formation with unlabeled ubiquitin to the radiolabeled E2-ubiquitin oxyester in a pulse-chase incubation.
The autoradiogram of Fig. 8 demonstrates that this is indeed the case because only in the presence of the complete ligation reaction and unlabeled ubiquitin does one observe Ubc5AC85S-125I-ubiquitin oxyester, present at 80 nm, promoted to higher molecular weight forms (Fig. 8, lane 7). Presumably, the concentration of free 125I-ubiquitin in the incubation of Fig. 8, lane 6, resulting from slow hydrolysis of the oxyester, is too low to support the activation reaction of Uba1. The free 125I-ubiquitin present in the incubation of lane 6 was quantitated by directly determining associated radioactivity in the excised lane below the pronounced band of the oxyester. The resulting concentration of free 125I-ubiquitin of 16 nm was significantly below the Km of 0.8 μm for free ubiquitin shown previously for human Uba1 and predicts a rate of 2% of Vmax (33). Addition of 5 μm unlabeled ubiquitin to the incubation of lane 7 reduces the specific radioactivity of the polypeptide to 0.3% of that in lane 6; therefore, high molecular weight conjugates observed in lane 7 must result from conjugation of unlabeled polypeptide to the preformed Ubc5AC85S-125I-ubiquitin oxyester. Determination of associated radioactivity for Ubc5AC85S-125I-ubiquitin oxyester from lanes 6 and 7 show a 0.3 pmol (15%) loss from lane 6 that matched exactly with a 0.3 pmol gain in higher molecular conjugates in lane 7 above the band of the oxyester. In contrast, free ubiquitin-associated radioactivity was unchanged between lanes 6 (0.4 pmol) and 7 (0.4 pmol). Interestingly, TRIM32 is catalytically capable of assembling polyubiquitin chains on Ubc5AΔ138-125I-ubiquitin oxyester (not shown), consistent with the ability of the truncation to bind the ligase (Fig. 7B). The presence of preformed polyubiquitin chains on Ubc5 as the corresponding thioester provides a mechanism for free chain formation upon transfer of the adduct to water or DTT present in the reactions.
FIGURE 8.
TRIM32 assembles polyubiquitin chains on Ubc5A. Autoradiogram of 80 nm purified Ubc5AC85S-125I-ubiquitin oxyester (lanes 1–7) incubated with 80 nm Uba1 (lanes 2–7) and, where indicated, 100 nm wild type Ubc5A, and 5 μm unlabeled ubiquitin for 15 min at 37 °C before quenching with SDS sample buffer and SDS-PAGE resolution as described under “Materials and Methods.” Migration positions for relative molecular weight standards are shown to the left. The position of the 5% stacking gel, Ubc5AC85S-125I-ubiquitin oxyester, and free 125I-ubiquitin are shown to the right.
DISCUSSION
Conjugation of ubiquitin to protein targets is a critical mechanism for cellular regulation in eukaryotes and many of these pathways rely on the assembly of polyubiquitin chains on the targets as the committed step for their degradation by the 26S proteasome or as a scaffold for assembly of multimeric complexes (56, 57). Target protein specificity among these post-translational modifications is defined by a diverse subset of ubiquitin ligases (58) of which the TRIM superfamily constitutes more than 100 paralogs that serve important functions in cellular homeostasis and, more critically, in the innate immune response of cells to viral and bacterial pathogens (13, 14). The current study is the first to investigate the mechanism of polyubiquitin chain formation by TRIM32 and related paralogs, using initial rates for formation of this degradation signal as a functional readout. The results suggest a common catalytic mechanism among members of the superfamily, consistent with their marked conservation in sequence and domain architecture.
The identification of the cognate E2 for a ubiquitin ligase is an essential step in its characterization, yet conservation within the E2 superfamily and the qualitative assays typically used to assess E2 specificity makes this task technically challenging (10). The present biochemically defined quantitative E2 screens unambiguously identify Ubc5 as the sole cognate E2 paralog supporting TRIM32-, TRIM25-, and TRIM5α-catalyzed polyubiquitin chain formation (Fig. 1). The current approach has the advantage that E2 species are compared in parallel at equivalent concentrations determined in functional stoichiometric assays, rather than by total protein, and that activity is assayed under initial velocity conditions so that the signal is proportional to catalytic competence (10). Identification of the ability of the Ubc5 isozymes generally to support TRIM ligase activity agrees with previous observations (36, 59–61); however, earlier reports that TRIM32 (36) and TRIM25 (40, 41, 62) also function with UbcH6 likely reflect the intrinsic ability of the E2 to form E3-independent polyubiquitin chains because the protein fails to support the activity of the TRIM paralogs tested here (10). More recently, high throughput methods involving two-hybrid screens have been exploited in an attempt to provide a comprehensive understanding of E2-E3 interactions and have suggested that TRIM32 functions with a broad array of E2 species, including UbcH7 and UbcH8, among others (37). However, such approaches fail to recognize that the actual substrate for an E3 is the cognate E2-ubiquitin thioester, which cannot be replicated in such interaction screens, and that both binding and geometry of the resulting Michaelis complex define catalytic competence. Thus, whereas other uncharged E2 paralogs may bind TRIM32 with low affinity, they can potentially lack the correct orientation to support chain formation as their corresponding ubiquitin thioesters, as appears to be the case for UbcH7 and UbcH8.
Previously we have shown that E2-ubiquitin thioesters bind their cognate ligase with hyperbolic kinetics, from which the intrinsic affinity (Kd) could be determined from the corresponding Km (10, 30, 33). In contrast, the TRIM ligases exhibit cooperative allosteric kinetics with respect to Ubc5-ubiquitin thioester concentration, requiring binding-dependent conformational cross-talk among the subunits of the ligase (Fig. 4). This is the first evidence of cooperativity for a ubiquitin ligase and provides important insights into the mechanism of polyubiquitin chain formation because deletion of the B box-coiled coil oligomerization domain abrogates chain formation unless association of the catalytic RING domains is rescued by linkage to an artificial dimerization domain such as GST (Fig. 3 and Table 1). Similar rescue of activity through fusion to GST has been reported previously for the RING domains of c-Cbl (63), APC11 (64, 65), BRCA1 (66), ROC1 and -2 (67), and Bcr-Abl protein (34), although the mechanism by which the GST moiety restores activity was not pursued. Mounting evidence suggests homo- and heterodimerization is required for the activity of many ligases (68–72) (reviewed in Ref. 73). The current kinetic studies suggest a mechanistic context for oligomerization as a necessary component of polyubiquitin chain formation.
The current kinetic data require that conformational communication among the multiple E2-ubiquitin binding sites occurs across the oligomer and that the portion of TRIM32 from the N terminus to the B-box mediates this mechanical coupling, evidenced by the loss of cooperativity in favor of hyperbolic kinetics with respect to [Ubc5A]o for the isolated RING-catalyzed automonoubiquitination (Fig. 4, C and D). From the theoretical sigmoidal curve we can calculate the Hill coefficient (n), a direct measure of the lower limit to the number of substrate binding sites (74). The range of n values for TRIM32 and TRIM32ΔNHL indicate that there are several binding sites on the E3; however, limitations imposed by substrate inhibition at higher concentrations of the Ubc5-ubiquitin thioester precludes accurate estimates of the intrinsic Hill coefficient. That the GST-RING displays a lower n value may indicate a diminished number of substrate binding sites, but more likely results from ablated conformational coupling of the subunits when linked artificially through GST. Despite this limitation, the data indicate that there are at least four Ubc5-ubiquitin thioester binding sites present on wild type TRIM32. Furthermore, an n value equal to two for GST-RING requires more than two binding sites for Ubc5-ubiquitin thioester, recapitulating the four binding sites of the wild type protein, but with diminished conformational coupling.
Nonlinear regression analysis allows calculation of values for [S]0.5 that are comparable (but not formally equivalent) to the Km for hyperbolic enzymes. The range of [S]0.5 values for the TRIM paralogs examined are somewhat higher than the Km of 54 ± 18 nm previously reported for the interaction between human Ubc2b-ubiquitin thioester with E3α/Ubr1 (33). Values of Km frequently evolve to approximate the corresponding intracellular substrate concentration so that rates of the enzyme fluctuate with changes in metabolite concentration. Using quantitative Western blots, we estimate the intracellular concentration of Ubc5 in A549 cells to be ∼160 nm, somewhat lower than the [S]0.5 values for the three TRIM ligases tested (Table 1). However, because of the sharp increase in activity inherent in the sigmoidal cooperative concentration dependences observed here, small changes in Ubc5 concentration will result in large alterations in activity, rendering such enzymes molecular switches. In the case of the TRIM ligases, the observed substrate inhibition at higher concentrations of Ubc5-ubiquitin thioester further enforces a narrow range of activity.
Substrate inhibition at high E2 concentrations was previously observed with the E3α/Ubr1 RING ligase and the Hect ligase family (10, 33). In the present context, substrate inhibition is consistent with ordered binding of the Ubc5-ubiquitin thioester to distinct substrate sites on the TRIM ligases that differ in affinity. Previous structural work demonstrates that E2 proteins bind the RING domain through a specific surface near their N terminus encompassing the Loop 1/2 region (53). However, the current study suggests that TRIM32 also binds Ubc5-ubiquitin thioester independently through an additional noncanonical surface on the C-terminal α-helix because truncation of the last 10 Ubc5A residues abrogates free chain formation and this effect can be replicated by mutation of Tyr145 (Fig. 6 and Table 2). However, because the Ubc5AΔ138 mutant can stimulate the rate of polyubiquitin chain formation through allosteric cooperativity even though it cannot formally support the net reaction of chain elongation demonstrates that the substrate is capable of binding to TRIM32, presumably through the canonical RING binding interaction.
Overall, the current data support a model for TRIM32 and other members of this ligase superfamily in which oligomerization is essential for catalytic competence in formation of the resulting polyubiquitin degradation signal. Other observations suggest that polyubiquitin is first assembled as an enzyme-bound E2 thioester intermediate prior to transfer en bloc to the protein target, although the latter step has not been directly confirmed. Such a mechanism obviates the topological constraints posed by translating a catalytic site along the growing polyubiquitin chain, whereas target protein remains bound. Empirical evidence for distinct TRIM32 binding sites on E2, substrate inhibition by the corresponding E2-ubiquitin thioester, and the stoichiometry of the Hill coefficient relative to the oligomerization state of the ligase are consistent with multiple binding sites per subunit for the enzyme. Whether this mechanism conforms to a “seesaw” model proposed earlier (9) or an indexation model in which chains are built “inside out” remains to be determined. However, ordered E2-ubiquitin thioester binding to distinct nonsymmetric sites, indicated by substrate inhibition at higher concentrations, strongly favors the latter alternative. Nonetheless, the present observations provide the first detailed examination of the mechanism of TRIM ligase function.
Acknowledgments
We thank Dr. Keith Wilkinson (Emory University School of Medicine, Department of Biochemistry) for the generous gift of the IsoT expression plasmid and Dr. Ronald Luftig (Louisiana State University Medical School, Department of Microbiology, Immunology, and Parasitology) for the generous gift of the TRIM5α plasmid.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM0034009 (to A. L. H.).
- E1
- ubiquitin activating enzyme
- E3
- ubiquitin-protein isopeptide ligase
- E2/Ubc
- ubiquitin carrier protein or ubiquitin conjugating enzyme
- NHL
- protein domain from representative proteins NCL-1, HT2A, and LIN-41
- RING
- Really Interesting New Gene protein domain
- TRIM
- tripartite motif
- Uba1
- human ubiquitin activating enzyme.
REFERENCES
- 1. Chau V., Tobias J. W., Bachmair A., Marriott D., Ecker D. J., Gonda D. K., Varshavsky A. (1989) A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243, 1576–1583 [DOI] [PubMed] [Google Scholar]
- 2. Pickart C. M. (2001) Mechanism underlying ubiquitination. Annu. Rev. Biochem. 70, 503–533 [DOI] [PubMed] [Google Scholar]
- 3. Ravid T., Hochstrasser M. (2008) Diversity of degradation signals in the ubiquitin-proteasome system. Nat. Rev. Mol. Cell Biol. 9, 679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Deshaies R. J., Joazeiro C. A. (2009) RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 [DOI] [PubMed] [Google Scholar]
- 5. Haas A. L., Siepmann T. J. (1997) Pathways of ubiquitin conjugation. FASEB J. 11, 1257–1268 [DOI] [PubMed] [Google Scholar]
- 6. Streich F. C., Jr., Haas A. L. (2010) Activation of ubiquitin and ubiquitin-like proteins. Subcell. Biochem. 54, 1–16 [DOI] [PubMed] [Google Scholar]
- 7. Pickart C. M., Rose I. A. (1985) Functional heterogeneity of ubiquitin carrier proteins. J. Biol. Chem. 260, 1573–1581 [PubMed] [Google Scholar]
- 8. Ye Y., Rape M. (2009) Building ubiquitin chains. E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 10, 755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hochstrasser M. (2006) Lingering mysteries of ubiquitin-chain assembly. Cell 124, 27–34 [DOI] [PubMed] [Google Scholar]
- 10. Ronchi V. P., Haas A. L. (2012) Measuring rates of ubiquitin chain formation as a functional readout of ligase activity. Meth. Mol. Biol. 832, 197–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lorick K. L., Jensen J. P., Fang S., Ong A. M., Hatakeyama S., Weissman A. M. (1999) RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. U.S.A. 96, 11364–11369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li W., Tu D., Brunger A. T., Ye Y. (2007) A ubiquitin ligase transfers preformed polyubiquitin chains from a conjugating enzyme to a substrate. Nature 446, 333–337 [DOI] [PubMed] [Google Scholar]
- 13. Nisole S., Stoye J. P., Saïb A. (2005) TRIM family proteins. Retroviral restriction and antiviral defence. Nat. Rev. Microbiol. 3, 799–808 [DOI] [PubMed] [Google Scholar]
- 14. McNab F. W., Rajsbaum R., Stoye J. P., O'Garra A. (2011) Tripartite-motif proteins and innate immune regulation. Curr. Opin. Immunol. 23, 46–56 [DOI] [PubMed] [Google Scholar]
- 15. Munir M. (2010) TRIM proteins. Another class of viral victims. Sci. Signal. 3, jc2. [DOI] [PubMed] [Google Scholar]
- 16. Horn E. J., Albor A., Liu Y., El-Hizawi S., Vanderbeek G. E., Babcock M., Bowden G. T., Hennings H., Lozano G., Weinberg W. C., Kulesz-Martin M. (2004) RING protein Trim32 associated with skin carcinogenesis has anti-apoptotic and E3-ubiquitin ligase properties. Carcinogenesis 25, 157–167 [DOI] [PubMed] [Google Scholar]
- 17. Albor A., Kulesz-Martin M. (2007) Novel initiation genes in squamous cell carcinomagenesis. A role for substrate-specific ubiquitylation in the control of cell survival. Mol. Carcinog. 46, 585–590 [DOI] [PubMed] [Google Scholar]
- 18. Kano S., Miyajima N., Fukuda S., Hatakeyama S. (2008) Tripartite motif protein 32 facilitates cell growth and migration via degradation of Abl-interactor 2. Cancer Res. 68, 5572–5580 [DOI] [PubMed] [Google Scholar]
- 19. Liu Y., Lagowski J. P., Gao S., Raymond J. H., White C. R., Kulesz-Martin M. F. (2010) Regulation of the psoriatic chemokine CCL20 by E3 ligases Trim32 and Piasy in keratinocytes. J. Invest Dermatol. 130, 1384–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yokota T., Mishra M., Akatsu H., Tani Y., Miyauchi T., Yamamoto T., Kosaka K., Nagai Y., Sawada T., Heese K. (2006) Brain site-specific gene expression analysis in Alzheimer's disease patients. Eur. J. Clin. Invest. 36, 820–830 [DOI] [PubMed] [Google Scholar]
- 21. Kudryashova E., Wu J., Havton L. A., Spencer M. J. (2009) Deficiency of the E3 ubiquitin ligase TRIM32 in mice leads to a myopathy with a neurogenic component. Hum. Mol. Genet. 18, 1353–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schwamborn J. C., Berezikov E., Knoblich J. A. (2009) The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell 136, 913–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guglieri M., Magri F., Comi G. P. (2005) Molecular etiopathogenesis of limb girdle muscular and congenital muscular dystrophies. Boundaries and contiguities. Clin. Chim. Acta 361, 54–79 [DOI] [PubMed] [Google Scholar]
- 24. Muller J., Stoetzel C., Vincent M. C., Leitch C. C., Laurier V., Danse J. M., Helle S., Marion V., Bennouna-Greene V., Vicaire S., Megarbane A., Kaplan J., Drouin-Garraud V., Hamdani M., Sigaudy S., Francannet C., Roume J., Bitoun P., Goldenberg A., Philip N., Odent S., Green J., Cossée M., Davis E. E., Katsanis N., Bonneau D., Verloes A., Poch O., Mandel J. L., Dollfus H. (2010) Identification of 28 novel mutations in the Bardet-Biedl syndrome genes. The burden of private mutations in an extensively heterogeneous disease. Hum. Genet. 127, 583–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Baboshina O. V., Haas A. L. (1996) Novel multiubiquitin chain linkages catalyzed by the conjugating enzymes E2epf and Rad6 are recognized by the 26S proteasome subunit 5. J. Biol. Chem. 271, 2823–2831 [DOI] [PubMed] [Google Scholar]
- 26. Ciechanover A., Heller H., Elias S., Haas A. L., Hershko A. (1980) ATP-dependent conjugation of reticulocyte proteins with the polypeptide required for protein degradation. Proc. Natl. Acad. Sci. U.S.A. 77, 1365–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Haas A. L. (2005) Purification of E1 and E1-like enzymes. Methods Mol. Biol. 301, 23–35 [DOI] [PubMed] [Google Scholar]
- 28. Haas A. L., Bright P. M. (1988) The resolution and characterization of putative ubiquitin carrier protein isozymes from rabbit reticulocytes. J. Biol. Chem. 263, 13258–13267 [PubMed] [Google Scholar]
- 29. Tokgöz Z., Siepmann T. J., Streich F., Jr., Kumar B., Klein J. M., Haas A. L. (2012) E1-E2 interactions in the ubiquitin and Nedd8 ligation pathways. J. Biol. Chem. 287, 311–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kumar B., Lecompte K. G., Klein J. M., Haas A. L. (2010) Ser120 of Ubc2/Rad6 regulates ubiquitin-dependent N-end rule targeting by E3α/Ubr1. J. Biol. Chem. 285, 41300–41309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Russell N. S., Wilkinson K. D. (2005) Deubiquitinating enzyme purification, assay inhibitors, and characterization. Methods Mol. Biol. 301, 207–219 [DOI] [PubMed] [Google Scholar]
- 32. Haas A. L., Rose I. A. (1981) Hemin inhibits ATP-dependent ubiquitin-dependent proteolysis. Role of hemin in regulating ubiquitin conjugate degradation. Proc. Natl. Acad. Sci. U.S.A. 78, 6845–6848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Siepmann T. J., Bohnsack R. N., Tokgöz Z., Baboshina O. V., Haas A. L. (2003) Protein interactions within the N-end rule ubiquitin ligation pathway. J. Biol. Chem. 278, 9448–9457 [DOI] [PubMed] [Google Scholar]
- 34. Maru Y., Afar D. E., Witte O. N., Shibuya M. (1996) The dimerization property of glutathione S-transferase partially reactivates Bcr-Abl lacking the oligomerization domain. J. Biol. Chem. 271, 15353–15357 [DOI] [PubMed] [Google Scholar]
- 35. Winn P. J., Religa T. L., Battey J. N., Banerjee A., Wade R. C. (2004) Determinants of functionality in the ubiquitin conjugating enzyme family. Structure 12, 1563–1574 [DOI] [PubMed] [Google Scholar]
- 36. Kudryashova E., Kudryashov D., Kramerova I., Spencer M. J. (2005) Trim32 is a ubiquitin ligase mutated in limb girdle muscular dystrophy type 2H that binds to skeletal muscle myosin and ubiquitinates actin. J. Mol. Biol. 354, 413–424 [DOI] [PubMed] [Google Scholar]
- 37. Napolitano L. M., Jaffray E. G., Hay R. T., Meroni G. (2011) Functional interactions between ubiquitin E2 enzymes and TRIM proteins. Biochem. J. 434, 309–319 [DOI] [PubMed] [Google Scholar]
- 38. Hershko A., Heller H. (1985) Occurrence of a polyubiquitin structure in ubiquitin-protein conjugates. Biochem. Biophys. Res. Commun. 128, 1079–1086 [DOI] [PubMed] [Google Scholar]
- 39. Wilkinson K. D., Tashayev V. L., O'Connor L. B., Larsen C. N., Kasperek E., Pickart C. M. (1995) Metabolism of the polyubiquitin degradation signal. Structure, mechanism, and role of isopeptidase T. Biochemistry 34, 14535–14546 [DOI] [PubMed] [Google Scholar]
- 40. Urano T., Saito T., Tsukui T., Fujita M., Hosoi T., Muramatsu M., Ouchi Y., Inoue S. (2002) Efp targets 14-3-3σ for proteolysis and promotes breast tumour growth. Nature 417, 871–875 [DOI] [PubMed] [Google Scholar]
- 41. Zou W., Wang J., Zhang D. E. (2007) Negative regulation of ISG15 E3 ligase EFP through its autoISGylation. Biochem. Biophys. Res. Commun. 354, 321–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gack M. U., Shin Y. C., Joo C. H., Urano T., Liang C., Sun L., Takeuchi O., Akira S., Chen Z., Inoue S., Jung J. U. (2007) TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446, 916–920 [DOI] [PubMed] [Google Scholar]
- 43. Zeng W., Sun L., Jiang X., Chen X., Hou F., Adhikari A., Xu M., Chen Z. J. (2010) Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell 141, 315–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nakajima A., Maruyama S., Bohgaki M., Miyajima N., Tsukiyama T., Sakuragi N., Hatakeyama S. (2007) Ligand-dependent transcription of estrogen receptor α is mediated by the ubiquitin ligase EFP. Biochem. Biophys. Res. Commun. 357, 245–251 [DOI] [PubMed] [Google Scholar]
- 45. Langelier C. R., Sandrin V., Eckert D. M., Christensen D. E., Chandrasekaran V., Alam S. L., Aiken C., Olsen J. C., Kar A. K., Sodroski J. G., Sundquist W. I. (2008) Biochemical characterization of a recombinant TRIM5α protein that restricts human immunodeficiency virus type 1 replication. J. Virol. 82, 11682–11694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Reymond A., Meroni G., Fantozzi A., Merla G., Cairo S., Luzi L., Riganelli D., Zanaria E., Messali S., Cainarca S., Guffanti A., Minucci S., Pelicci P. G., Ballabio A. (2001) The tripartite motif family identifies cell compartments. EMBO J. 20, 2140–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Parker M. W., Lo Bello M., Federici G. (1990) Crystallization of glutathione S-transferase from human placenta. J. Mol. Biol. 213, 221–222 [DOI] [PubMed] [Google Scholar]
- 48. Ji X., Zhang P., Armstrong R. N., Gilliland G. L. (1992) The three-dimensional structure of a glutathione S-transferase from the mu gene class. Structural analysis of the binary complex of isoenzyme 3-3 and glutathione at 2.2-Å resolution. Biochemistry 31, 10169–10184 [DOI] [PubMed] [Google Scholar]
- 49. Baboshina O. V., Crinelli R., Siepmann T. J., Haas A. L. (2001) N-end rule specificity within the ubiquitin/proteasome pathway is not an affinity effect. J. Biol. Chem. 276, 39428–39437 [DOI] [PubMed] [Google Scholar]
- 50. Vargo M. A., Nguyen L., Colman R. F. (2004) Subunit interface residues of glutathione S-transferase A1-1 that are important in the monomer-dimer equilibrium. Biochemistry 43, 3327–3335 [DOI] [PubMed] [Google Scholar]
- 51. Li X., Li Y., Stremlau M., Yuan W., Song B., Perron M., Sodroski J. (2006) Functional replacement of the RING, B-box 2, and coiled-coil domains of tripartite motif 5α (TRIM5α) by heterologous TRIM domains. J. Virol. 80, 6198–6206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Peng H., Feldman I., Rauscher F. J., 3rd (2002) Hetero-oligomerization among the TIF family of RBCC/TRIM domain-containing nuclear cofactors. A potential mechanism for regulating the switch between coactivation and corepression. J. Mol. Biol. 320, 629–644 [DOI] [PubMed] [Google Scholar]
- 53. Zheng N., Wang P., Jeffrey P. D., Pavletich N. P. (2000) Structure of a c-Cbl-UbcH7 complex. RING domain function in ubiquitin-protein ligases. Cell 102, 533–539 [DOI] [PubMed] [Google Scholar]
- 54. Eletr Z. M., Huang D. T., Duda D. M., Schulman B. A., Kuhlman B. (2005) E2 conjugating enzymes must disengage from their E1 enzymes before E3-dependent ubiquitin and ubiquitin-like transfer. Nat. Struct. Mol. Biol. 12, 933–934 [DOI] [PubMed] [Google Scholar]
- 55. Sullivan M. L., Vierstra R. D. (1993) Formation of a stable adduct between ubiquitin and the Arabidopsis ubiquitin-conjugating enzyme, AtUBC1. J. Biol. Chem. 268, 8777–8780 [PubMed] [Google Scholar]
- 56. Ikeda F., Dikic I. (2008) Atypical ubiquitin chains. New molecular signals. EMBO Rep. 9, 536–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kim I., Rao H. (2006) What's Ub chain linkage got to do with it? Sci. STKE 2006, e18. [DOI] [PubMed] [Google Scholar]
- 58. de Bie P., Ciechanover A. (2011) Ubiquitination of E3 ligases. Self-regulation of the ubiquitin system via proteolytic and non-proteolytic mechanisms. Cell Death. Differ. 18, 1393–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yamauchi K., Wada K., Tanji K., Tanaka M., Kamitani T. (2008) Ubiquitination of E3 ubiquitin ligase TRIM5α and its potential role. FEBS J. 275, 1540–1555 [DOI] [PubMed] [Google Scholar]
- 60. Urano T., Usui T., Takeda S., Ikeda K., Okada A., Ishida Y., Iwayanagi T., Otomo J., Ouchi Y., Inoue S. (2009) TRIM44 interacts with and stabilizes terf, a TRIM ubiquitin E3 ligase. Biochem. Biophys. Res. Commun. 383, 263–268 [DOI] [PubMed] [Google Scholar]
- 61. Chen D., Gould C., Garza R., Gao T., Hampton R. Y., Newton A. C. (2007) Amplitude control of protein kinase C by RINCK, a novel E3 ubiquitin ligase. J. Biol. Chem. 282, 33776–33787 [DOI] [PubMed] [Google Scholar]
- 62. Horie-Inoue K., Inoue S. (2006) Epigenetic and proteolytic inactivation of 14-3-3σ in breast and prostate cancers. Semin. Cancer Biol. 16, 235–239 [DOI] [PubMed] [Google Scholar]
- 63. Joazeiro C. A., Wing S. S., Huang H., Leverson J. D., Hunter T., Liu Y. C. (1999) The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science 286, 309–312 [DOI] [PubMed] [Google Scholar]
- 64. Leverson J. D., Joazeiro C. A., Page A. M., Huang Hk., Hieter P., Hunter T. (2000) The APC11 RING-H2 finger mediates E2-dependent ubiquitination. Mol. Biol. Cell 11, 2315–2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gmachl M., Gieffers C., Podtelejnikov A. V., Mann M., Peters J. M. (2000) The RING-H2 finger protein APC11 and the E2 enzyme UBC4 are sufficient to ubiquitinate substrates of the anaphase-promoting complex. Proc. Natl. Acad. Sci. U.S.A. 97, 8973–8978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ruffner H., Joazeiro C. A., Hemmati D., Hunter T., Verma I. M. (2001) Cancer-predisposing mutations within the RING domain of BRCA1. Loss of ubiquitin protein ligase activity and protection from radiation hypersensitivity. Proc. Natl. Acad. Sci. U.S.A. 98, 5134–5139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Furukawa M., Ohta T., Xiong Y. (2002) Activation of UBC5 ubiquitin-conjugating enzyme by the RING finger of ROC1 and assembly of active ubiquitin ligases by all cullins. J. Biol. Chem. 277, 15758–15765 [DOI] [PubMed] [Google Scholar]
- 68. Barbash O., Zamfirova P., Lin D. I., Chen X., Yang K., Nakagawa H., Lu F., Rustgi A. K., Diehl J. A. (2008) Mutations in Fbx4 inhibit dimerization of the SCFFbx4 ligase and contribute to cyclin D1 overexpression in human cancer. Cancer Cell 14, 68–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Brzovic P. S., Rajagopal P., Hoyt D. W., King M. C., Klevit R. E. (2001) Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat. Struct. Biol. 8, 833–837 [DOI] [PubMed] [Google Scholar]
- 70. Buchwald G., van der Stoop P., Weichenrieder O., Perrakis A., van Lohuizen M., Sixma T. K. (2006) Structure and E3-ligase activity of the Ring-Ring complex of Polycomb proteins Bmi1 and Ring1b. EMBO J. 25, 2465–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Linke K., Mace P. D., Smith C. A., Vaux D. L., Silke J., Day C. L. (2008) Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ. 15, 841–848 [DOI] [PubMed] [Google Scholar]
- 72. Mace P. D., Linke K., Feltham R., Schumacher F. R., Smith C. A., Vaux D. L., Silke J., Day C. L. (2008) Structures of the cIAP2 RING domain reveal conformational changes associated with ubiquitin-conjugating enzyme (E2) recruitment. J. Biol. Chem. 283, 31633–31640 [DOI] [PubMed] [Google Scholar]
- 73. Knipscheer P., Sixma T. K. (2007) Protein-protein interactions regulate Ubl conjugation. Curr. Opin. Struct. Biol. 17, 665–673 [DOI] [PubMed] [Google Scholar]
- 74. Segal I. A. (1975) Enzyme Kinetics, Wiley-Interscience, New York [Google Scholar]