Background: How the cathelicidin LL-37 enhances Toll-like receptor 3 signaling is poorly understood.
Results: We determined that LL-37-poly(I:C) complex can localize to early endosomes with TLR3 through the FPRL1 receptor, whereas poly(I:C) localizes to endosomes by a different mechanism.
Conclusion: Double-stranded RNAs can localize with TLR3 by multiple trafficking pathways.
Significance: This work establishes a mechanism for cross-talk between LL-37, double-stranded RNA, and TLR3.
Keywords: Endosomes, Innate Immunity, Peptides, Signal Transduction, Toll-like Receptors (TLR), Cathelicidin LL-37, Formyl Peptide Receptor-like 1
Abstract
LL-37 is an antimicrobial peptide produced by human cells that can down-regulate the lipopolysaccharide-induced innate immune responses and up-regulate double-stranded (ds) RNA-induced innate responses through Toll-like receptor 3 (TLR3). The murine LL-37 ortholog, mCRAMP, also inhibited lipopolysaccharide-induced responses, but unlike LL-37, it inhibited viral-induced responses in mouse cells. A fluorescence polarization assay showed that LL-37 was able to bind dsRNA better than mCRAMP. In the human lung epithelial cell line BEAS-2B, LL-37, but not mCRAMP, colocalized with TLR3, and the colocalization was increased in the presence of dsRNA. The presence of poly(I:C) increased the accumulation of LL-37 in Rab5 endosomes. Signaling by cells induced with both LL-37 and poly(I:C) was sensitive to inhibitors that affect clathrin-independent trafficking, whereas signaling by poly(I:C) alone was not, suggesting that the LL-37-poly(I:C) complex trafficked to signaling endosomes by a different mechanism than poly(I:C) alone. siRNA knockdown of known LL-37 receptors identified that FPRL1 was responsible for TLR3 signaling induced by LL-37-poly(I:C). These results show that LL-37 and mCRAMP have different activities in TLR3 signaling and that LL-37 can redirect trafficking of poly(I:C) to effect signaling by TLR3 in early endosomes in a mechanism that involves FPRL1.
Introduction
Cathelicidins are multifunctional peptides produced by many mammals to act as bactericides, to promote wound repair and angiogenesis, and to modulate innate immune responses (1, 2). LL-37 is a 37-residue cathelicidin that is released by proteolytic cleavage from the C-terminal fragment of the protein hCAP-18. Elevated levels of LL-37 in human tissues associated with autoimmune diseases such as psoriasis (2–5).
LL-37 can bind several ligands that are also recognized by innate immune receptors (6). LL-37 binding to lipopolysaccharides (LPS) can down-regulate signaling by Toll-like receptor 4 (TLR4),2 binding to ssRNA can up-regulate TLR7 and TLR8 signaling, and interaction with flagellin can activate TLR5 signaling (3, 7, 8). Most relevant to this work, LL-37 binds double-stranded (ds) RNA, including poly(I:C) to enhance TLR3 signaling above the level observed with poly(I:C) alone (7, 9, 10). How LL-37 functions in concert with dsRNA to affect TLR3 signaling remains to be better understood.
LL-37 consists of an N-terminal α-helical structure followed by more flexible sequence that may be intrinsically disordered until it binds to ligands (11). Multiple LL-37 molecules may also form higher order structures with TLR ligands. The mouse ortholog of LL-37, mCRAMP, is believed to have a similar structure. Both LL-37 and mCRAMP are secreted from neutrophil granules upon wounding (8, 12). Unlike LL-37, however, mCRAMP inhibited TLR3 signaling in mouse cells (13). It is unclear whether these effects are due to the species of the cell lines examined, the tissues from which the cell lines have been derived, and/or sequence/structural differences in the two peptides.
In this study we document that LL-37 can activate TLR3 signaling in three human cell lines derived from different tissues but inhibit signaling in three murine cell lines. In contrast, mCRAMP inhibited dsRNA-dependent signaling in both human and murine cells. The differences in the activities of LL-37 and mCRAMP are correlated with their abilities to bind poly(I:C) and their localization to early endosomes. We also observed that signaling in the presence of both LL-37 and poly(I:C) was sensitive to inhibitors of clathrin-independent endocytosis, whereas signaling by poly(I:C) alone was sensitive to the clathrin-dependent endocytosis inhibitor. siRNA knockdown of the LL-37 receptor, formyl peptide receptor-like 1 (FPRL1) (14), prevented enhancement of signaling by the poly(I:C)-LL-37 complex.
MATERIALS AND METHODS
Reagents
The BEAS-2B, A549, RAW264.7, and NK-92MI cell lines were from ATCC. Mouse embryonic fibroblasts and L929 are the generous gifts of Pranav Danthi of Indiana University, Bloomington. Cell culture media were from Invitrogen. Endocytosis inhibitors methyl-β-cyclodextrin, genistein, chloropromazine, and bafilomycin A1 were from Sigma and dissolved in either water or DMSO according to its solubility. Poly(I:C) and LPS were from Invivogen. Reovirus (RV) dsRNA S4 were prepared as described in Lai et al. (9). Antimicrobial peptides with or without covalently attached fluorophores were custom-synthesized to greater than 95% purity by AnaSpec Inc.
Antibodies to detect the TLR3 ectodomain were from R&D systems (catalog #AF1487). Antibodies to detect Rab5, Rab7, and Rab11 were from Cell Signaling Inc. (catalog #9385). The antibody to detect mouse anti-LAMP1 was from Santa Cruz Biotechnology (catalog #Sc-20011). The antibody to detect FPRL1 was from Novus Biologicals (catalog #NLS1878). Secondary antibodies conjugated to Alexa Fluor 488 or 594 were from Invitrogen. Alexa Fluor-conjugated cholera toxin B subunit (catalog #C-34777) and human transferrin (catalog #T-13343) were from Invitrogen. An HRP-conjugated secondary antibodies that recognized rabbit IgGs was from Santa Cruz Biotechnologies.
Cell Culture
RAW264.7, L929, mouse embryonic fibroblasts, and A549 were cultured in DMEM, high glucose, GlutaMAX (Life Technology, Inc.) supplemented with 10% FBS at 37 °C with 5% CO2. BEAS-2B cells were cultured in BEGM media with its supplements (Lonza Inc.) (9, 17). NK-92MI cells were propagated in α-minimum essential medium with final concentrations of 2 mm l-glutamine, 0.2 mm inositol, 0.1 mm 2-mercaptoethanol, 0.02 m folic acid, 12.5% horse serum, and 12.5% FBS.
Quantification of Cytokine Production
A typical assay used 2 × 104 BEAS-2B cells/well grown for 24 h in flat-bottom 96-well plates. At that point, the media were removed and replaced with fresh media amended with TLR ligands and/or peptides. Human cell lines were treated with LPS at a final concentration of 1 μg/ml, poly(I:C) at 0.13 μg/ml, and the peptides at a final concentration of 3 μm. RAW264.7 cells were plated at 2 × 104 cells per ml and grown for 24 h. At that time, the media were removed and replaced with fresh media containing LPS at a final concentration of 1 μg/ml, poly(I:C) at 1 μg/ml, and/or the peptides at 3 μm. TLR ligand and peptides were not preincubated before their addition to the culture medium.
IL-6 was quantified by ELISA using the human or mouse BD OptEIATM kit (BD Biosciences) from cell media clarified by centrifugation at 1000 × g for 2 min. All ELISA results shown were performed in triplicate and in at least three independent experiments.
Messenger RNAs for human and mouse IL-6, IL-1β, TNF-α, and CCL5/RANTES were quantified by real-time RT-PCR using a primer(s) whose sequences will be made available upon request. Briefly, RNA was isolated using the Qiagen RNEasy kit (Qiagen Inc.). Moloney murine leukemia virus reverse transcriptase (New England Biolabs) was used to synthesize the cDNA. Each experiment was performed in triplicate in 96-well plates using 1× SYBR Green master mix (Bio-Rad) and 2 μg of the cDNA in a final volume of 25 μl. Amplifications were performed with each cycle consisting of 95 °C for 3 min followed by 50 cycles of 94 °C for 10 s and 60 °C for 30 s and 72 °C for 30 s. The data were analyzed as described in Qi et al. (15).
siRNA Knockdown
BEAS-2B cells were seeded at 2 × 106 cells per 6-well plate in BEGM amended with supplements (Lonza). 6 h later the cells were transfected with 30 nm concentrations of a pool of three siRNAs from Santa Cruz Biotechnology specific to TLR3 (catalog #sc-36685), FPRL1 (sc-40123), IGF-IRα/β (sc-29358), epidermal growth factor receptor (EGFR; sc-29301), or a nonspecific control siRNA (catalog #sc-37007). Transfection of the siRNAs used Lipofectamine RNAiMax (Life Technology Inc.) according to the manufacturer's protocol. The cells were transfected with siRNA then incubated for 48 h before transfection of plasmids to express proteins. The effects of the siRNAs on target mRNA levels were analyzed using real-time quantitative RT-PCR. Effects of the knockdown on IL-6 production were assessed in culture media collected 24 h after the siRNA transfection. The cells were also fixed at that time to allow localization of LL-37 and poly(I:C) using confocal microscopy.
Confocal Microscopy
Cells were grown on poly-l-lysine-coated coverslips to 60% confluency. 12 h after treatment with poly(I:C) or LPS in the absence or presence of antimicrobial peptide(s), the cells were fixed with 4% paraformaldehyde for 15 min at room temperature and permeabilized for 30 min on ice in T buffer (0.5% Triton X-100 in PBS with 1% normal goat serum). Nonspecific antibody binding sites were blocked with 2% BSA in TBS-T (Tris-buffered saline, pH 7.4 with 0.5% Triton X-100) for 1 h before an overnight incubation at 4 °C with primary antibodies in TBS-T containing 2% BSA. The cells were then washed three times with TBS-T and incubated with secondary antibodies for 1 h at room temperature. After three additional washes with TBS-T, the coverslips were mounted on glass slides with anti-fade mounting medium with DAPI and dried overnight in the dark. Micrographs were acquired with a Leica TCS SP5 confocal inverted-base microscope with a 63× oil objective. Images were analyzed by Leica LAS AF and Image J software. At least 20 cells were analyzed for each treatment, and each experiment was repeated at least twice. Colocalization of fluorophores was quantified using the ImageJ plug-in tool, JACoP (16).
Labeling of dsRNAs
Rhodamine-labeled poly(I:C) was from Invitrogen. RV S4 dsRNA was labeled with Alexa Fluor 594-conjugated Universal Linkage System (Invitrogen) according to the manufacturer's instructions. 1 μg of RNA was heated at 95 °C for 5 min. Conjugated Alexa Fluor 594 was added to denatured RNA, and the reaction was incubated at 90 °C for 10 min. The labeling reaction was then cooled on ice, and unbound labels were removed by using a gel filtration-based spin column. RNA concentration was quantified by spectrophotometry.
Fluorescence Polarization Assay
LL-37 or mCRAMP binding to poly(I:C) was analyzed by a fluorescence polarization assay in a microplate reader (BioTek). Fluorescein-labeled LL-37 or mCRAMP (10 nm) was mixed with poly(I:C) in a 100-μl solution of buffer P (25 mm Tris and 50 mm NaCl, pH 7.4) before analysis. All binding assays were performed in triplicate, and the experiment was reproduced in two additional, independent assays to assess consistency of the results.
Western Blots
Approximately 5 × 105 cells were solubilized in NET buffer (150 mm NaCl, 5 mm EDTA, 50 mm Tris-Cl, pH 6.5, and 1% Nonidet P-40) amended with protease inhibitor mixture (Sigma). The lysate was clarified from insoluble materials by a brief centrifugation at 2000 × g for 5 min and denatured with an SDS-containing solution for 5 min at 70 °C. The proteins were separated by gel electrophoresis in a 4–12% Bis-Tris gel and processed for Western blot analysis as described by Qi et al. (15).
Statistical Analysis
All data shown are the means and ranges for one S.E. for a minimal of three independent samples. Data sets were compared using Student's t test calculated with GraphPad Prism 5 software.
RESULTS
LL-37 and mCRAMP Have Different Effects on dsRNA-induced TLR3 Signaling
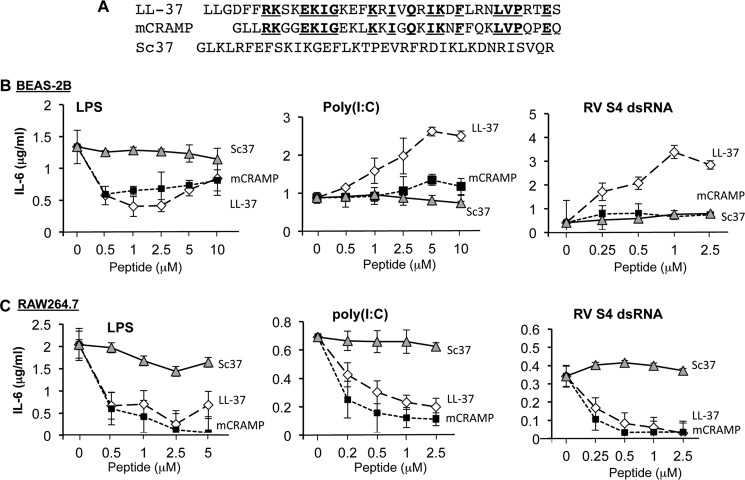
Discrepant observations of the activities of LL-37 and mCRAMP on dsRNA-induced signaling in human and mouse cells prompted us to better document the activities of the peptides (9, 17). The sequences of the peptides LL-37, mCRAMP, and a scrambled version of LL-37 named Sc37 are shown in Fig. 1A. The human bronchial epithelial cell line, BEAS-2B, and the mouse macrophage RAW264.7 cell line were selected for the initial analysis because they have been characterized for signaling by TLR3 and TLR4 (9, 18). TLR3 and TLR4 signaling can be assessed by the release of the cytokine IL-6 upon the addition of the agonists for the receptors to the cell culture medium (7, 9, 17). In both cell lines the addition of LPS, a ligand for TLR4, or poly(I:C), a ligand for TLR3, results in IL-6 production routinely being increased by 5-fold or higher. Furthermore, the addition of LL-37 or mCRAMP to either BEAS-2B or RAW264.7 cells inhibited LPS-induced IL-6 production in a concentration-dependent manner, whereas Sc37 caused little effect (left panels, Fig. 1, B and C). These results validate that the LL-37 and mCRAMP peptides used in our assays are effective for modulating innate immune signaling.
FIGURE 1.
IL-6 production induced by poly(I:C) or RV dsRNA in the presence of antimicrobial peptides. A, sequence of human antimicrobial peptide (LL-37), mouse antimicrobial peptide (mCRAMP), and Scrambled peptide (Sc37) is shown. The residues that are identical in LL-37 and mCRAMP are in bold and underlined. B, changes in IL-6 production in the human BEAS-2B cells in response to the peptides along with LPS, poly(I:C), or the RV S4 dsRNA are shown. ELISA was used to quantify the amounts of IL-6 present in the cell culture media. C, changes in IL-6 production in the mouse macrophage cell line, RAW264.7, are shown. Where present, LPS, poly(I:C), and S4 dsRNA were added to the cell culture media to, respectively, a final concentration of 1, 0.13, and 0.13 μg/ml. All the ELISA data in this work are represented as the mean ± S.D. of triplicates. The data were repeated twice after the initial experiment.
In BEAS-2B cells, LL-37 added to the media along with poly(I:C) increased IL-6 production (middle panel, Fig. 1B). A similar enhancement was observed with cells treated with LL-37 and RV S4 dsRNA (right panel, Fig. 1B). The addition of mCRAMP or Sc37 with either poly(I:C) or S4 resulted in little or no enhancement of IL-6 production in BEAS-2B cells (Fig. 1B). In RAW264.7 cells, the addition of either LL-37 or mCRAMP inhibited poly(I:C)-induced IL-6 production (Fig. 1C), consistent with the observations of Hasan et al. (13) in mouse cells. Altogether, results in BEAS-2B and RAW264.7 cells show that LL-37 and mCRAMP share the ability to inhibit LPS-induced signaling but differ in their effects on dsRNA-induced signaling.
To further examine the effects of LL-37 and mCRAMP on poly(I:C)-induced TLR3 signaling, real-time RT-PCR was used to quantify the mRNA levels of TNF-α, IL-1β, CCL5/RANTES, and IL-6, cytokines and chemokines produced in response to TLR3 activation (19, 20). The mRNAs were normalized to the levels of GAPDH message in the same samples. BEAS-2B cells treated with poly(I:C) and LL-37 increased TNF-α, IL-1β, IL-6, and CCL5/RANTES mRNAs by 68-, 3-, 17-, and 663-fold, respectively, relative to the signal from poly(I:C) alone (Table 1). The addition of mCRAMP along with poly(I:C) to BEAS-2B cells had either no effect or caused only a modest increase in the four cytokine messages (Table 1). In RAW264.7 cells, the amount of poly(I:C)-induced TNF-α, IL-1β, and CCL5/RANTES mRNAs were all inhibited by the addition of either LL-37 or mCRAMP but not by Sc37. These results are consistent with those from ELISA assays for IL-6 levels. All of the assays for cytokine and chemokine production show that LL-37 and mCRAMP have different effects on dsRNA-induced signaling in human BEAS-2B and mouse RAW264.7 cells.
TABLE 1.
Real time quantitative PCR analysis of the effects of LL-37 and mCRAMP on cytokines produced by poly(I:C)-induced TLR3 signaling
The -fold of the results is relative to those from cells treated with only poly(I:C).
| Ligand(s) | BEAS-2B |
RAW264.7 |
||||||
|---|---|---|---|---|---|---|---|---|
| TNFα | IL-1β | IL-6 | RANTES | TNFα | IL-1β | IL-6 | RANTES | |
| Poly(I:C) | 47.2 (−8.4, +10.9)a | 1.6 (−0.7, +1.2) | 358.4 (−54.3, +66.7) | 4.59 (−1.1, +1.8) | 1.8 (−0.5, +0.8) | 1354.3 (−491, +771) | 3897.5 (−260, +279) | 244.4 (−29, +32.) |
| Poly(I:C)+LL-37 | 3210 (−756, +1220) (68-fold) | 5.1 (−1.2, +1.7) (3.2-fold) | 5937 (−1158, + 1577) (16.6-fold) | 3044 (−627, +883) (663-fold) | 0.9 (−0.3, +0.5) (0.5-fold) | 4.6 (−2.7, +6.6) (0.003-fold) | 34.4 (−5.0, +5.8) (0.009-fold) | 4.5 (−0.64, +0.75) (0.02-fold) |
| Poly(I:C)+mCRAMP | 209.0 (−23.5, +27.0) (4.4-fold) | 2.1 (−0.7, +0.9) (1.3-fold) | 462.4 (−48.9, +55.6) (1.3-fold) | 6.6 (−0.6, +0.7) (1.4-fold) | 0.9 (−0.1, +0.1) (0.5-fold) | 1.5 (−0.9, +2.0) (0.001-fold) | 6.1 (−0.5, +0.5) (0.002-fold) | 4.5 (−0.6, +0.7) (0.02-fold) |
| Poly(I:C)+Sc37 | 107.2 (−19.4, +25.3) | 1.3 (−0.3, +0.4) | 423.6 (−72.2, +92.3) | 383.6 (−53.3, +4.0) | 1.6 (−0.4, +0.6) | 1758 (−258, +303) | 4225 (−236, +250) | 266. (−21.0, +23) |
| LPS | 146.8 (−19.5, +23.2) | 3.5 (−0.9, +1.2) | 37.7 (−8.0, +11.4) | 715.3 (−41.0, +67.2) | 1.9 (−41.0, +67.2) | 1897.7 (−0.2, +0.2) | 4798 (−1311, +1804) | 347 (−87, +116) |
a 1 S.D calculated from the ΔΔCt is shown in parentheses.
Two additional human cell lines were tested to determine whether LL-37 and mCRAMP have different effects on signaling by TLR3. Human cell lines NK-93MI (natural killer origin) and A549 (alveolar adenocarcinoma origin) both showed increased IL-6 production when treated with LPS. The addition of LL-37 and mCRAMP inhibited the LPS-induced IL-6 production by more than 3-fold, whereas Sc37 had no effect (Table 2). When treated with poly(I:C), the two cell lines produced between 5- and 6-fold higher IL-6. The addition of LL-37 with poly(I:C) resulted in another 2-fold increase in IL-6 production. In contrast, mCRAMP added along with poly(I:C) reduced IL-6 production in NK-93MI and A549 cells (Table 2). Thus, in three human cells lines derived from different tissues, LL-37 consistently increased TLR3 signaling by dsRNA, whereas mCRAMP inhibited signaling.
TABLE 2.
LL-37 and mCRAMP on IL-6 production in different human and mouse cell lines
All means and S.E. were derived from a minimum of six independently processed samples.
| Ligands | Human IL-6 (mean ± S.D) |
Mouse IL-6 (mean ± S.D) |
||
|---|---|---|---|---|
| A549 | NK-92MI | MEFs | L929 | |
| μg/ml | μg/ml | |||
| Mock | 0.19 ± 0.09 | 0.08 ± 0.08 | 0.09 ± 0.07 | 0.08 ± 0.06 |
| LL-37 | 0.25 ± 0.042 | 0.06 ± 0.07 | 0.09 ± 0.06 | 0.09 ± 0.06 |
| mCRAMP | 0.25 ± 0.021 | 0.05 ± 0.05 | 0.09 ± 0.07 | 0.098 ± 0.08 |
| Sc37 | 0.26 ± 0.051 | 0.05 ± 0.02 | 0.09 ± 0.07 | 0.08 ± 0.06 |
| LPS | 1.40 ± 0.95 | 0.61 ± 0.59 | 2.16 ± 1.25 | 1.42 ± 0.09 |
| LPS + LL-37 | 0.44 ± 0.29 (0.03a) | 0.17 ± 0.19 (0.08a) | 0.10 ± 0.08 (0.004a) | 0.08 ± 0.07 (0.01a) |
| LPS + mCRAMP | 0.43 ± 0.17 (0.07a) | 0.09 ± 0.11 (0.07a) | 0.21 ± 0.06 (0.01a) | 0.07 ± 0.06 (0.01a) |
| LPS + Sc37 | 1.40 ± 0.55 | 0.62 ± 0.43 | 2.19 ± 1.19 | 1.12 ± 1.6 |
| poly(I:C) | 0.96 ± 0.31 | 0.51 ± 0.40 | 1.11 ± 0.58 | 0.98 ± 0.66 |
| poly(I:C) + LL-37 | 1.98 ± 0.45 (0.001a) | 1.23 ± 0.90 (0.01a) | 0.17 ± 019 (0.003) | 0.14 ± 0.19 (0.01a) |
| poly(I:C) + mCRAMP | 0.57 ± 0.17 (0.07a) | 0.25 ± 0.21 (0.00a) | 0.18 ± 0.16 (0.002) | 0.19 ± 0.25 (0.01a) |
| poly(I:C)+Sc37 | 0.97 ± 0.39 | 0.51 ± 0.49 | 1.10 ± 0.58 | 1.03 ± 0.71 |
a The p values were determine between the sample that treated with LL-37 or mCRAMP to those treated with either only LPS or poly(I:C).
In mouse fibroblast cell lines L929 and mouse embryonic fibroblasts, LPS-induced IL-6 production was inhibited by LL-37 and mCRAMP, demonstrating that the peptides possess bioactivity with these two cell lines. Furthermore, poly(I:C)-induced IL-6 production increased by more than 10-fold in both cell lines. The addition of either LL-37 or mCRAMP along with poly(I:C) inhibited IL-6 production (Table 2). These results are consistent with those from RAW264.7 cells and demonstrate that multiple mouse cell lines have a consistently different response to LL-37 and mCRAMP when compared with human cell lines.
LL-37 and mCRAMP Differ in Their Ability to Bind Poly(I:C)
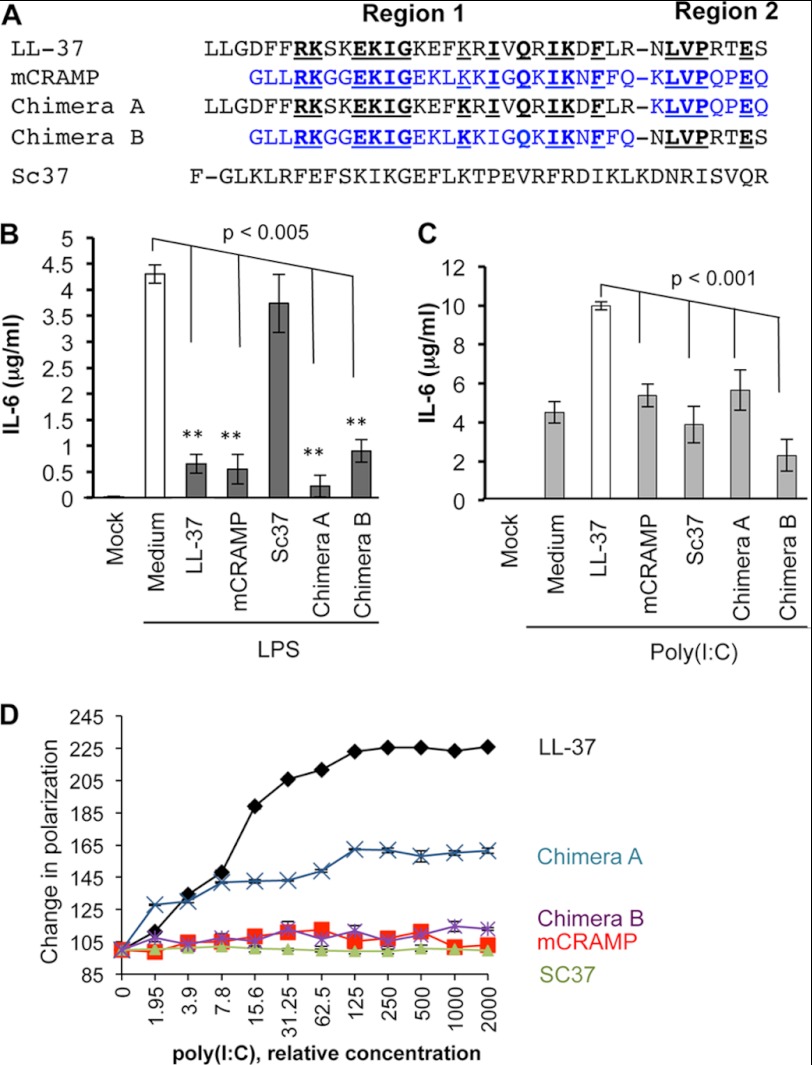
We seek to better understand the basis for the distinct effects of LL-37 and mCRAMP. We hypothesize that the difference in dsRNA-induced signaling could be due to LL-37 and mCRAMP having different abilities to bind dsRNAs. To test this, LL-37 and mCRAMP were synthesized with N-terminal fluorescein. Both labeled peptides modulated LPS- and poly(I:C)-induced IL-6 production in BEAS-2B cells, indicating that the addition of the fluorophore did not alter the ability of the peptides to affect LPS or poly(I:C) signaling (Fig. 2, B and C). However, when the peptides were assessed for poly(I:C) binding using a fluorescence polarization assay, the change in the LL-37 polarization was increased by poly(I:C) in a concentration-dependent manner, whereas that of mCRAMP was not (Fig. 2D). These results show that LL-37 is better at binding poly(I:C) than is mCRAMP.
FIGURE 2.
Antimicrobial peptides binding to poly(I:C) and their abilities to affect IL-6 production. A, shown are sequences of the peptides used in this study. All peptides were synthesized to contain a fluorescein at the N terminus. Residues from mCRAMP are shown in blue. The residues in LL-37 and mCRAMP that are identical are in bold and underlined. Regions 1 and region 2 were identified by predictions of the secondary structures of LL-37. Region 1 has a propensity to fold into an α-helix, whereas region 2 is unstructured. B, biological activities of the fluorescein-tagged peptides are shown. Each peptide was added to a final concentration of 3 μm. The ability of the peptides to inhibit IL-6 production in BEAS-2B cells that were also treated with LPS (1 μg/ml) was quantified by ELISA. IL-6 production in cells treated with IL-6 was statistically significant to those treated with mCRAMP, Chimera A, and Chimera B in the Student t test by p values of >0.05. C, the ability of the peptides to enhance IL-6 production in BEAS-2B cells is shown. Poly(I:C) was added to a final concentration of 0.13 μg/ml. IL-6 levels were quantified by ELISA. D, fluorescence polarization assay for the peptides in the presence of increasing concentrations of poly(I:C) is shown. Fluorescently labeled chimera peptides (10 nm) were titrated with increasing concentrations of poly(I:C) in a 100-μl solution of buffer P (25 mm Tris and 50 mm NaCl, pH 7.4).
LL-37 and mCRAMP are predicted to form an α-helical structure that starts at the N terminus (region 1), whereas the C-terminal eight residues is more flexible (region 2) (Fig. 2A; Ref. 31). Region 1 of mCRAMP is shorter by three residues than that of LL-37 but possesses a central portion that is highly similar in sequence to that of LL-37. The respective region 2s of LL-37 and mCRAMP have four residues of eight that are identical (Fig. 2A). To better define the regions in LL-37 that contribute to dsRNA binding, we synthesized two chimeras with mixed and matched sequences from the two regions (Fig. 2A). Both chimeras inhibited LPS-dependent signaling by TLR4 (Fig. 2B) but failed to significantly induce IL-6 production in response to poly(I:C) (Fig. 2C). In the fluorescence polarization assay, Chimera A that contained region 1 from LL-37 could bind poly(I:C) at a reduced rate relative to that of LL-37 (Fig. 2D). Chimera B that contained region 1 from mCRAMP failed to bind RNA. These results show that region 1 of LL-37 retained some ability to interact with poly(I:C) but that this is insufficient to enhance TLR3 signaling. Furthermore, both regions 1 and 2 contribute to optimal dsRNA binding and enhancement of TLR3 signaling.
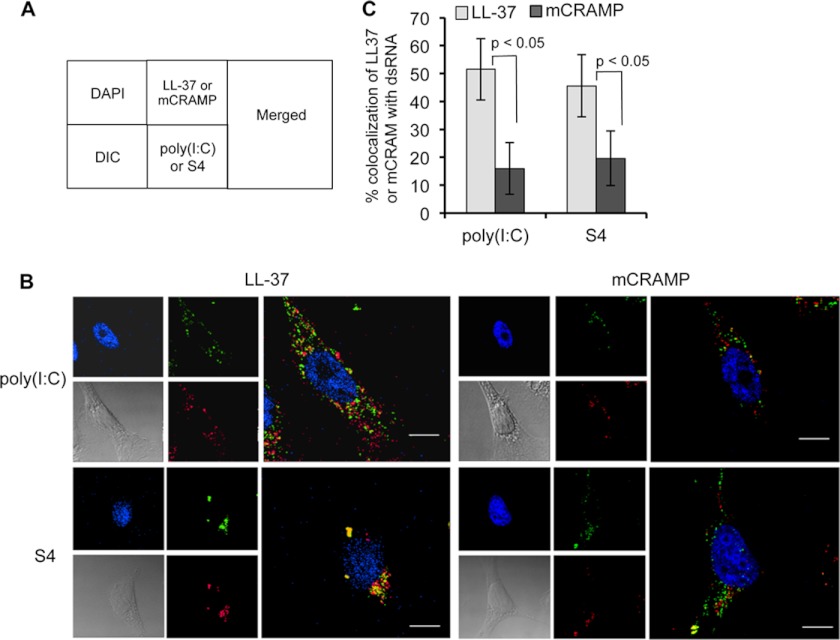
LL-37, But Not mCRAMP, Preferentially Colocalizes with Poly(I:C) in Cells
We examined whether fluorescently labeled poly(I:C) or S4 could colocalize with LL-37 and/or mCRAMP in cells. A schematic of the collage of the confocal micrographs is shown in Fig. 3A. In BEAS-2B cells, the dsRNAs and the peptides were localized to punctate structures that are likely to be endosomes (Fig. 3B). However, although 50% of LL-37 colocalized with poly(I:C), only 18% of mCRAMP did so (Fig. 3C). Similar results were observed with the colocalization of the peptides with S4 dsRNA (Fig. 3, B and C). These results demonstrate that in human cells, LL-37 and mCRAMP differ in their intracellular localization with respect to dsRNAs, likely as a consequence to their different abilities to associate with dsRNA.
FIGURE 3.
Colocalization of LL-37 or mCRAMP with poly(I:C) in BEAS-2B cells. A, shown is a schematic of the confocal images shown in panel B. DIC, differential interference contrast. B, shown are confocal microscopic images of the localizations of fluorescently labeled poly(I:C) and either LL-37 or mCRAMP. The antimicrobial peptides are pseudocolored green, and poly(I:C) is pseudocolored red. The cell nuclei were counter-stained with DAPI, and the cell is visualized in a differential interference contrast image. Each scale bar represents a length of 10 μm. C, quantification of the degree of colocalization of poly(I:C) or S4 with the two antimicrobial peptides. The percent colocalization was calculated using ImageJ software plug-in JACoP. All the data are represented as the mean ± S.D. of percent colocalization from at least 20 independent cells in each treatment. Statistical analysis of the colocalization of LL-37 or mCRAMP with the dsRNAs were determined by use of Student's t test.
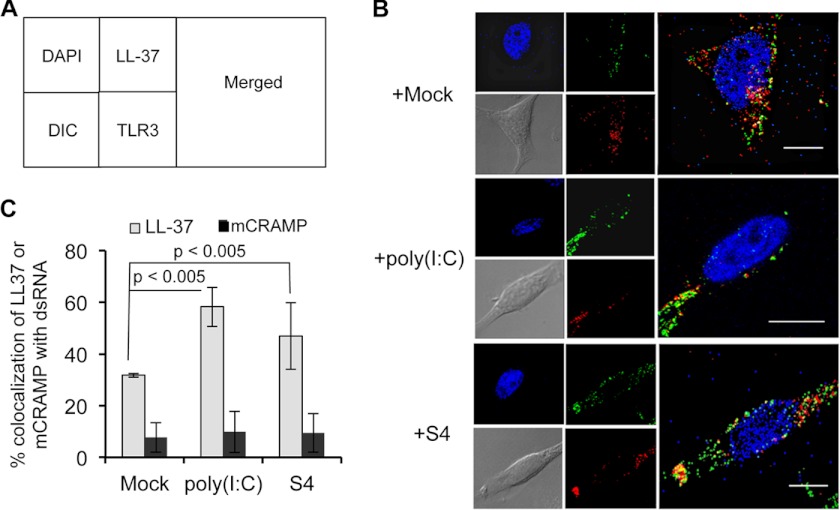
Colocalization of LL-37 and mCRAMP with TLR3
We examined whether fluorescently labeled LL-37 and mCRAMP will colocalize with TLR3 in the absence or presence of poly(I:C) or RV S4 dsRNA. TLR3 was detected by immunostaining using a fluorescently labeled mAb. Approximately 33% of LL-37 colocalized with TLR3 in endosomes in the absence of dsRNA (Fig. 4). When added to cells along with poly(I:C), 60% of LL-37 colocalized with TLR3 (Fig. 4C). The presence of S4 also increased the colocalization of LL-37 with TLR3.
FIGURE 4.
Colocalization of antimicrobial peptides with TLR3 in the presence or absence of either poly(I:C) or S4 dsRNA. A, shown is a schematic of the confocal images of panel B. B, shown is colocalization of TLR3 with antimicrobial peptides. Representative confocal images of BEAS-2B cells are shown, and the antimicrobial peptides are colored green. Goat anti-TLR3 antibody and anti-goat Texas Red were used to stain TLR3. DAPI used to stain nucleus and cells are visualized in differential interference contrast (DIC). Scale bars are represented as 10 μm. C, shown are quantifications of percent colocalization of antimicrobial peptide with TLR3 either in the presence or absence of poly(I:C)/S4dsRNA in BEAS-2B cells. The percent colocalization was calculated using ImageJ software plug-in JACoP. All the data are represented as the mean ± S.D. of percent colocalization from at least 20 independent cells in each treatment. p values were determined by Student's t test.
The situation was different with mCRAMP. In the absence of poly(I:C), only 10% of mCRAMP colocalized with TLR3, and colocalization was not affected by the addition of either poly(I:C) or S4 dsRNA (Fig. 4C). These results suggest that LL-37 can colocalize to endosomes containing TLR3 even in the absence of exogenously provided dsRNA. However, the targeting of LL-37 in endosomes containing TLR3 will be increased.
LL-37 and mCRAMP Localize to Different Subsets of Endosomes
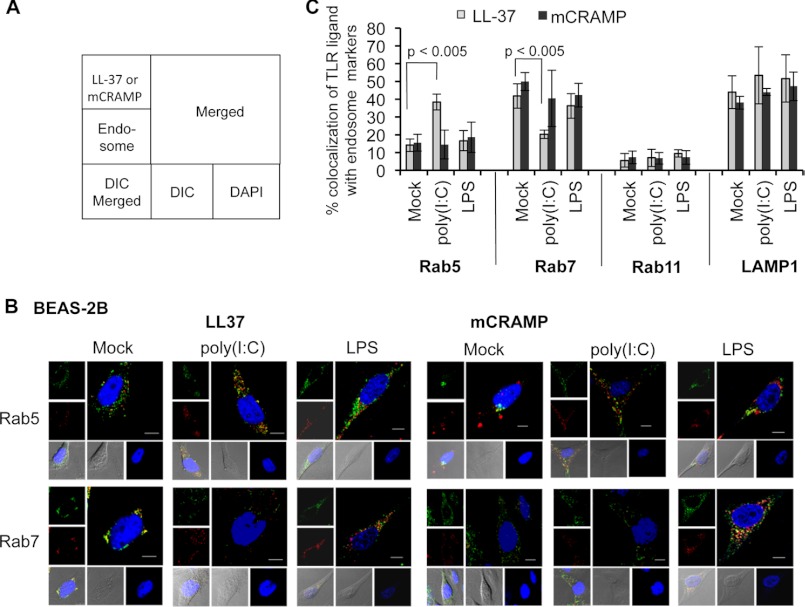
We next sought to determine whether LL-37 and mCRAMP were associated with specific subpopulations of endosomes. The localization of fluorescently labeled LL-37 and mCRAMP in endosomes marked with Rab5 or Rab7 was of interest as TLR3 signaling was associated with Rab5 endosomes (15). The early (Rab5), late (Rab7), and recycling endosomes (Rab11) as well as lysosomes (LAMP1) were detected with fluorescently labeled mAb specific to the endosomal markers (Fig. 5, A and B, and data not shown). Approximately half of the LL-37 and mCRAMP localized with LAMP1 in BEAS-2B cells, and the degree of this localization was not much changed by the presence of poly(I:C) or LPS (Fig. 5C). Less than 10% of LL-37 or mCRAMP localized to Rab11 endosomes, and this was also largely unaffected by the presence of poly(I:C) or LPS (Fig. 5C and data not shown).
FIGURE 5.
Colocalization of antimicrobial peptides with different endosomal markers in the presence or absence of TLR ligands. A, a schematic of the confocal images of B is shown. DIC, differential interference contrast. B, colocalization of LL-37 or mCRAMP with endosomal markers in the presence of mock or poly(I:C) or LPS in BEAS-2B cells is shown. All the confocal images are represented with pseudocolor green as antimicrobial peptides, red as endosomal markers, and blue as DAPI-stained nucleus. Scale bars are represented as 10 μm. C, quantifications of percent colocalization of LL-37 or mCRAMP with early endosome (Rab5), late endosome (Rab7), recycling endosome (Rab11), and lysosome (LAMP1) in the presence or absence of ligands. Each bar represents the means and 1 S.D. of 20 cells in each group. The localization of LL-37 in RAB5 or RAB7 endosomes was only statistically significant to p values of <0.05 in cells treated with poly(I:C). The calculated p values for these samples are shown above the bars.
Localization with the Rab5 and Rab7 endosomes showed more substantive changes. In the absence of poly(I:C), ∼12–14% of LL-37 and mCRAMP localized with Rab5-positive endosomes and 45% localized to Rab7 endosomes (Fig. 5). In the presence of poly(I:C), LL-37 colocalization with Rab5 endosomes was increased by 2-fold or more in six independent experiments, and LL-37 colocalization with Rab7 endosomes was decreased by a comparable amount (Fig. 5C). In contrast, mCRAMP localization to Rab5 or Rab7 endosomes was unaffected by poly(I:C), consistent with the observation that mCRAMP does not interact with poly(I:C) (Fig. 2).
Effect of Endocytosis Inhibitors on Activities of Antimicrobial Peptides
We sought to determine whether a clathrin-dependent or clathrin-independent mechanism was responsible for LL-37 enhancement of TLR3 signaling. Given that numerous differences between LL-37 and mCRAMP have already been established, we will focus on LL-37 for the remainder of this work.
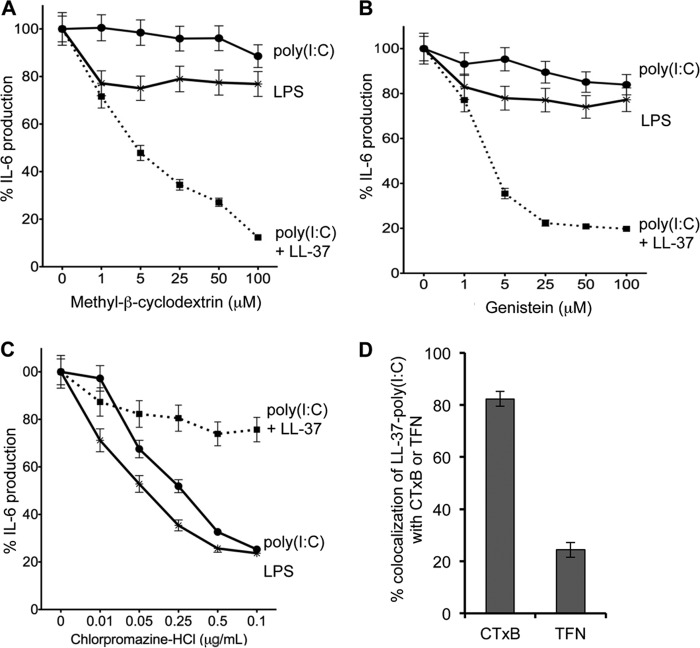
BEAS-2B cells were assessed for the effects of endocytosis inhibitors (21) on poly(I:C)-induced IL-6 production. Methyl-β-cyclodextrin, which removes cholesterols from cells to inhibit clathrin-independent endocytosis (21), caused only a modest change in both LPS and poly(I:C)-induced IL-6 production (Fig. 6A). However, methyl-β-cyclodextrin profoundly inhibited the LL-37-mediated enhancement of IL-6 production in cells treated with both poly(I:C) and LL-37 (Fig. 6A). The differential effects of methyl-β-cyclodextrin on cells treated with poly(I:C) and poly(I:C)-LL-37 suggest that LL-37 can direct poly(I:C) to enter cells by a different mechanism than used by poly(I:C) alone. To confirm this, we used genistein, another clathrin-independent endocytosis inhibitor (Fig. 6B). Again, genistein did not alter the uptake of LPS or poly(I:C) but did inhibit IL-6 production when the cells were induced by poly(I:C) and LL-37. These results suggest that a clathrin-independent mechanism for endocytosis is responsible for uptake of a complex of poly(I:C)-LL-37.
FIGURE 6.
Effects of endocytosis inhibitors on IL-6 production by BEAS-2B cells induced by LPS or poly(I:C) in the absence of presence of LL-37. A, shown are the effects of increasing concentration of clathrin-independent endocytosis inhibitor methyl-β-cyclodextrin. B, shown are the effects of increasing concentrations of clathrin-independent endocytosis inhibitor genistein. C, shown are the effects of increasing concentrations of clathrin-dependent endocytosis inhibitor chloropromazine-HCl. In the results for all three graphs, the inhibitor of endocytosis was added 6 h before the addition of the TLR ligand and the antimicrobial peptides. IL-6 production was quantified by ELISA, and the data are represented as percent changes in IL-6 production normalized to IL-6 production without inhibitor. D, colocalization of LL-37 and poly(I:C) with either cholera toxin-B (CTxB) or transferrin (TFN). LL-37 was labeled with fluorescein in this experiment, and cholera toxin-B and transferrin were labeled with Alexa Fluor 594. Poly(I:C) was added to a final concentration of 0.13 μg/ml but was not labeled. All the data are represented as mean and 1 S.D. of the percent colocalization of LL-37 with cholera toxin-B or transferrin from at least 20 cells.
The involvement of a clathrin-independent trafficking mechanism for LL-37-poly(I:C) complex does not preclude the use of a clathrin-dependent trafficking mechanism (22). Therefore, IL-6 production was quantified in the presence of chlorpromazine-HCl, an inhibitor of clathrin-dependent trafficking. Chlorpromazine-HCl inhibited the effects of poly(I:C) on IL-6 production in a concentration-dependent manner (Fig. 6C). It also inhibited the effects of LPS. Because LPS has been reported to signal from endosomes (9, 23), these results suggest that LPS and poly(I:C) are internalized by some clathrin-dependent mechanisms, consistent with previous observations (23, 24). Interestingly, chlorpromazine-HCl had only a mild effect on IL-6 production induced by poly(I:C)-LL-37, consistent with our notion that LL-37-poly(I:C) complex enters cells by a mechanism different than that used by poly(I:C).
Should LL-37-poly(I:C) enter cells through a clathrin-independent endocytosis, it is likely that the complex would colocalize with cholera toxin B, which also enters cells using a clathrin-independent mechanism (2, 25). As a control, we examined poly(I:C)-LL-37 colocalization with the human transferrin, which uses a clathrin-dependent mechanism (22). In BEAS-2B cells, 82% of poly(I:C)-LL-37 colocalized with cholera toxin B, whereas only 22% colocalized with transferrin. These results support that the mechanism for poly(I:C) uptake differs from that used by poly(I:C)-LL-37.
Receptor for Poly(I:C)-LL-37 Signaling and Endocytosis
LL-37 has been reported to traffic into cells using three receptors, EGFR, IGF-1R, and FPRL1 (12, 14, 26, 27). To determine whether one or more of these receptors was involved in trafficking poly(I:C)-LL-37, siRNAs specific to each receptor were used to knock down their expression. Real-time RT-PCR was used to monitor the efficiency of knockdown. All three sets of siRNAs achieved a >70% reduction in the mRNA levels for their respective receptors in BEAS-2B cells (Fig. 7A). The effects of EGFR, IGF-1R, and FPRL1 knockdowns on IL-6 production were determined in cells stimulated with LPS, LL-37-LPS, poly(I:C), and poly(I:C)-LL-37. Knockdowns of EGFR and IGF-1R decreased the responses to LPS but did not affect the response to poly(I:C) (Fig. 7B, upper two panels). In the presence of LL-37, a reduction in IL-6 production was observed when LPS was added to cells with EGFR and IGR-1R knocked down.
FIGURE 7.
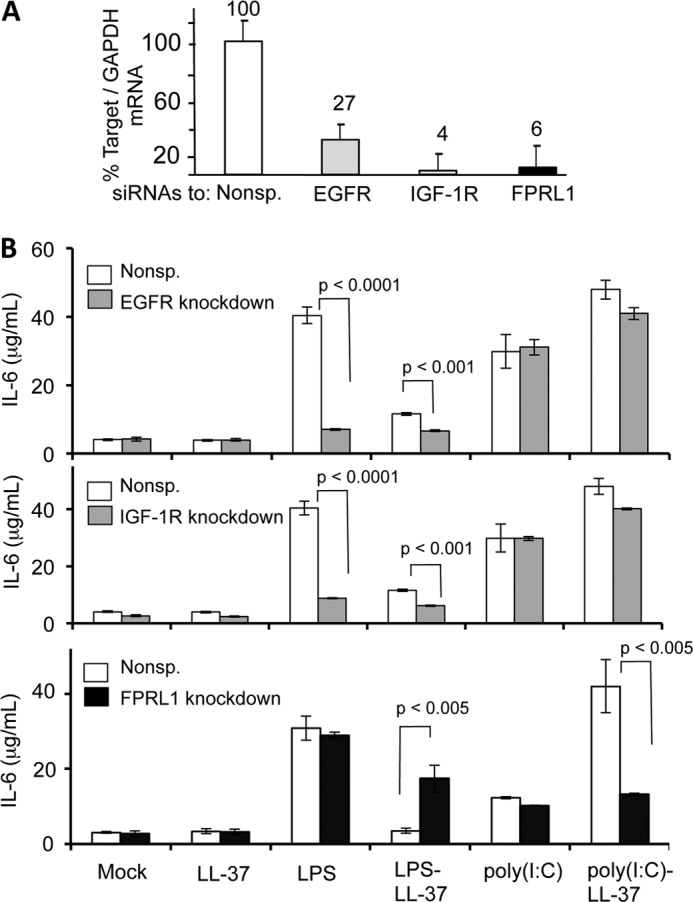
Identification of the LL-37 receptor required for IL-6 production in response to poly(I:C)-LL-37. A, shown is quantification of the siRNA knockdown of EGFR, IFG-1R, and FPRL1. The amount of the mRNAs was quantified by real-time RT-PCR. The data are normalized to the level of GAPDH message in each sample. Each result represents the mean and S.D. of three plates of BEAS-2B cells that were independently transfected with siRNA. B, shown are the effects of siRNA knockdown of EGFR, IGF-1R, and FPRL1 on IL-6 production induced by various ligands. White bars are the results from cells transfected with a nonspecific siRNA control (Nonsp.). Where present, LPS was added to a final concentration of 1 μg/ml, poly(I:C) was at 0.13 μg/ml, and LL-37 was at 3 μm. In the three graphs, only the samples that yielded p values of less than 0.05 are shown. The actual calculated p values are the samples that were compared.
In contrast, knockdown of FPRL1 did not affect the response to LPS. When LPS was added to the culture media along with LL-37, FPRL1 knockdown actually increased IL-6 production, possibly due to some cross-talk between FPRL1 and other LPS-signaling pathways. Importantly, the knockdown of FPRL1 had only a minimal effect on the response to poly(I:C) but severely reduced the response induced by poly(I:C)-LL-37 (Fig. 7B). These results suggest that FPRL1 is responsible for the trafficking of poly(I:C)-LL-37 but that poly(I:C) entered cells by an independent mechanism.
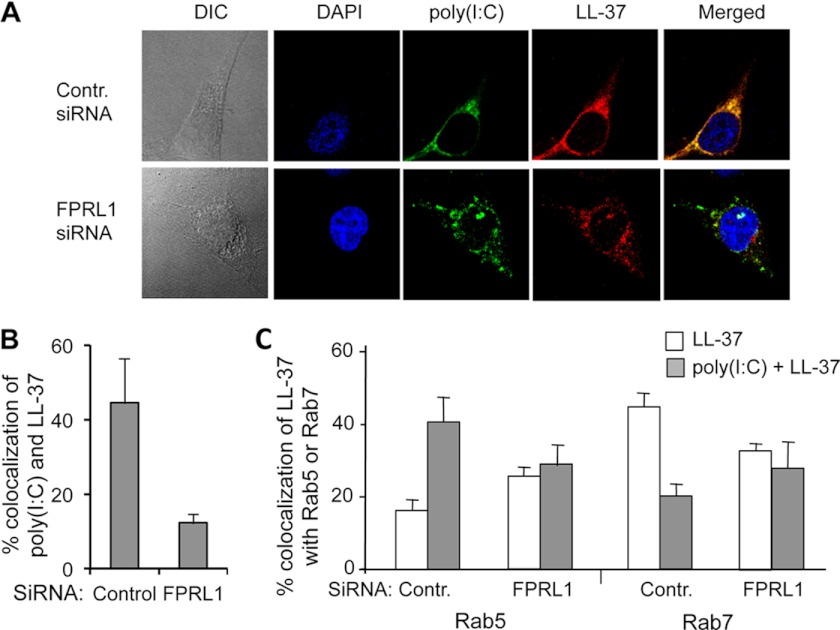
Confocal microscopy was used to examine the effects of a knockdown in FPLR1 on LL-37 localization. BEAS-2B cells treated with control siRNAs had extensive colocalization of fluorescently labeled poly(I:C) and LL-37 (Fig. 8, A and B). However, cells with decreased FPRL1 expression showed significantly reduced LL-37 internalization and also the colocalization of poly(I:C) and LL-37 (Fig. 8, A and B). The preferential accumulation of LL-37 in Rab5 endosomes (versus Rab7 endosomes) was also not observed with cells knocked down for FPRL1. These results confirmed that FPRL1 participates in the trafficking of the poly(I:C)-LL-37 complex in BEAS-2B cells.
FIGURE 8.
FPRL1 knockdown affected the localization of LL-37 with poly(I:C) or with early endosomes in BEAS-2B cells. A, confocal micrographs show the locations of fluorescently labeled LL-37, poly(I:C). Images in the upper row contained BEAS-2B cells transfected with a control siRNA. The images from the lower row were of BEAS-2B cells transfected to knockdown the FPRL1 message. DIC, differential interference contrast. B, shown is quantification of the amount of colocalization of LL-37 and poly(I:C) upon treatment to knockdown FPRL1. C, shown are the effects of FPRL1 knockdown on the localization of LL-37 or poly(I:C)-LL-37 complex in Rab5 and Rab7 endosomes. In panels C and D, each bar represents the means and 1 S.D. of colocalized signals from a minimal of 20 cells.
Given that FPRL1 has a role in LL-37 trafficking and enhancement of TLR3 expression, we determined whether the murine cells that do not respond to LL-37 and mCRAMP to affect TLR3 signaling, expressed FPRL1. Western blot analysis was performed with lysates from four human cell lines and the three murine cell lines we characterized for LL-37 and mCRAMP signaling (Fig. 1 and Table 1). FPRL1 in human and mouse have calculated molecular masses of 39–40 kDa, and bands of approximately these masses was observed in lysates from all of the cell lines (Fig. 9). Furthermore, siRNA targeting the FPRL1 message in HEK293T cells caused an obvious decrease in the level of the FPRL1 protein. We note that in the murine cell lines the abundance of a higher molecular weight form of FPRL1, likely a glycosylated form of FPRL1, appear to be severely reduced in comparison to the FPRL1 from the four human cell lines (Fig. 9).
FIGURE 9.
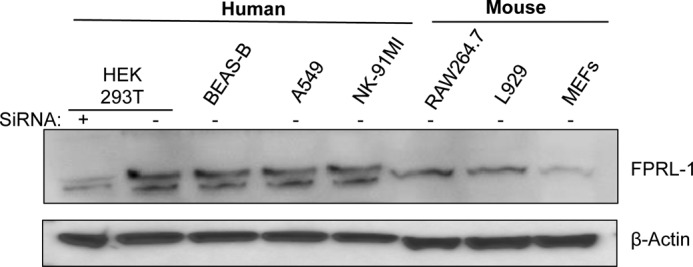
FPRL1 is expressed in the human and mouse cell lines used in this study. The image is of a Western blot probed with a polyclonal antibody that recognizes both the human and mouse FRLR1. The sample in the left-most lane was treated with siRNA to knock down the FPRL1 message and, thus, provides an independent confirmation of the identity of the FPRL1 band. The same blot was probed to detect the β-actin protein, which served as a loading control.
DISCUSSION
Cathelicidins have been isolated from many mammalian species and are demonstrated to have a range of activities from modulating wound repair responses to regulation of innate immune responses. In this work we showed that LL-37 could bind dsRNA and up-regulate TLR3 signaling, likely by trafficking to endosomes where TLR3 is resident. In addition, although the mouse CRAMP peptide is an ortholog to the human LL-37, mCRAMP does not bind dsRNA as well as LL-37 and cannot activate TLR3 signaling. Localization of LL-37 in endosomes with TLR3 in BEAS-2B cells was increased by the presence of poly(I:C) or S4 dsRNA (Fig. 3, B and C). These observations suggest that LL-37 binds to poly(I:C) and that the complex is delivered to endosomes for TLR3 signaling. Perhaps the most important observation for TLR3 signaling in this work is that the poly(I:C)-LL-37 complex is trafficked into BEAS-2B cells by a different mechanism used by poly(I:C). Poly(I:C) trafficking primarily uses a clathrin-dependent mechanism, whereas poly(I:C)-LL-37 used a clathrin-independent mechanism. siRNA knockdown of FPRL1, a known LL-37 receptor (14), showed that FPRL1 was required to traffic the poly(I:C)-LL-37 complex into Rab5 endosomes of BEAS-2B cells.
Although both LL-37 and mCRAMP can inhibit signaling by LPS, we and others have observed that their activities do not overlap with regard to dsRNA-induced signaling. In three independently derived human cell lines, LL-37 enhanced TLR3 signaling, but mCRAMP did not (Fig. 1B, Table 1, Table 2). We note, however, that LL-37 may not always enhance TLR3 signaling, as it was shown to reduce IL-6 production in human gingival fibroblasts (28). A difference in the activities of LL-37 and mCRAMP could be due to mCRAMP having reduced ability to bind dsRNA when compared with LL-37 (Figs. 2D and 3B).
The activities of LL-37 and mCRAMP was also different between human and mouse cell lines. Both LL-37 and mCRAMP inhibited signaling by LPS and by dsRNA in murine cell lines (Fig. 1C, Tables 1 and 2). Although the presence of poly(I:C) or viral dsRNA increased the localization of LL-37 in Rab5 endosomes in human cells, this was not observed in the murine RAW264.7.3 Thus, trafficking of either LL-37 and/or dsRNA is likely to be significantly different in mouse cells than in human cells. We note that all three murine cell lines we analyzed appear to express FPRL1 (Fig. 9), consistent with previous reports (29, 30). However, there may be differences in the post-translational modifications of the murine proteins that could affect the ability to interact with the antimicrobial peptides. There are also a number of differences in the sequences of the two proteins that could affect the functions of the human and mouse FPRL1 (supplemental Fig. 1). The differences in the murine and human FPRL1 molecules will require more thorough analysis. Nonetheless, it is worth noting that the innate immune systems of the mouse have a number of significant differences from those in human (31). Studies of LL-37 may thus require other animal models.
Several lines of evidence suggest that trafficking of the poly(I:C)-LL-37 complex and poly(I:C) in human cells function by different mechanisms. First, an inhibitor of clathrin-dependent endocytosis, chlorpromazine, inhibited poly(I:C)-mediated signaling but was much less effective in inhibiting signaling by the poly(I:C)-LL-37 complex (Fig. 6C). Second, poly(I:C)-LL-37 was sensitive to clathrin-independent inhibitors, whereas poly(I:C)-dependent signaling was not. Third, siRNA knockdown of the FPRL1 receptor known to traffic LL-37 was able to inhibit LL-37-poly(I:C)-induced signaling and localization to early endosomes (Figs. 7 and 8). These results further extend the previous observations of Limmon et al. (32) and Itoh et al. (24), who showed that poly(I:C) binds to Class A scavenger receptors and traffics to endosomes through clathrin-mediated endocytosis. The demonstration that poly(I:C)-LL-37 complexes enter cells using a different endocytic mechanism than poly(I:C) and the increased colocalization of the complex with TLR3 could provide a mechanism for LL-37 enhancing poly(I:C)-induced signaling by TLR3. Our results also suggest that in BEAS-2B cells, LPS and poly(I:C) trafficking could use distinguishable pathways in the presence of LL-37. Signaling by cells incubated with LL-37 and LPS is affected by siRNA knockdown of EGFR and IGF-1R but not by knockdown of FPRL1. In turn, signaling by cells incubated with LL-37 and poly(I:C) was largely unaffected by knockdowns of EGFR and IGF-1R but severely affected by knockdown of FPRL1. Interestingly, signaling in response to LPS by BEAS-2B cells was affected by knockdowns of EGFR and IGF-R1, demonstrating that there are distinct endocytic mechanisms for the ligands of TLR3 and TLR4.
These results suggest that recognition of LL-37 by the three receptors could be modulated by either LPS or dsRNA complexed to LL-37. Furthermore, because LL-37, mCRAMP, and the two chimeric peptides contained swaps within the two regions of the peptides could inhibit LPS-induced signaling, it is likely that binding to LPS by these cathelicidin peptides have less stringent requirements than the binding of double-stranded RNA.
Poly(I:C) is a potential adjuvant for vaccines (33). Poly(I:C) complexed with LL-37 could enhance the responses to increase the efficacy of vaccines. In addition, as high levels of LL-37 have been observed in the context of certain inflammatory diseases, such as psoriasis and rheumatoid arthritis (34), antagonizing FPRL1 could represent a means of decreasing certain inflammatory responses.
Acknowledgments
We are grateful to Y. Lai and members of the Kao laboratory at Indiana University for numerous helpful discussions. We thank L. Kao for editing the manuscript and Jim Powers and the Indiana University Light Microscopy center for use of the confocal microscopy facility, which is supported by an Indiana METACyt Initiative funded in part through a grant from the Lilly Endowment, Inc.
This work was supported in part by a grant from the Indiana Economic Development Corporation (to C. K.).

This article contains supplemental Fig. S1.
D. Singh, unpublished observation.
- TLR4
- Toll-like receptor 4
- FPRL1
- formyl peptide receptor-like 1
- RV
- reovirus
- Bis-Tris
- bis(2-hydroxyethyl)iminotris(hydroxymethylmethane)
- EGFR
- epidermal growth factor receptor
- IGF-1R
- Insulin-like growth factor 1 receptor
- RANTES
- regulated on activation normal T cell expressed and secreted.
REFERENCES
- 1. Ganz T., Selsted M. E., Szklarek D., Harwig S. S., Daher K., Bainton D. F., Lehrer R. I. (1985) Defensins. Natural peptide antibiotics of human neutrophils. J. Clin. Invest. 76, 1427–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nizet V., Ohtake T., Lauth X., Trowbridge J., Rudisill J., Dorschner R. A., Pestonjamasp V., Piraino J., Huttner K., Gallo R. L. (2001) Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature 414, 454–457 [DOI] [PubMed] [Google Scholar]
- 3. Hancock R. E., Scott M. G. (2000) The role of antimicrobial peptides in animal defenses. Proc. Natl. Acad. Sci. U.S.A. 97, 8856–8861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lande R., Ganguly D., Facchinetti V., Frasca L., Conrad C., Gregorio J., Meller S., Chamilos G., Sebasigari R., Riccieri V., Bassett R., Amuro H., Fukuhara S., Ito T., Liu Y. J., Gilliet M. (2011) Neutrophils activate plasmacytoid dendritic cells by releasing self-DNA-peptide complexes in systemic lupus erythematosus. Sci. Transl Med. 3, 73ra19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lande R., Gregorio J., Facchinetti V., Chatterjee B., Wang Y. H., Homey B., Cao W., Wang Y. H., Su B., Nestle F. O., Zal T., Mellman I., Schröder J. M., Liu Y. J., Gilliet M. (2007) Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature 449, 564–569 [DOI] [PubMed] [Google Scholar]
- 6. Zhang X., Oglcka K., Sandgren S., Belting M., Esbjörner E. K., Nordén B., Gräslund A. (2010) Dual functions of the human antimicrobial peptide LL-37-target membrane perturbation and host cell cargo delivery. Biochim. Biophys. Acta 1798, 2201–2208 [DOI] [PubMed] [Google Scholar]
- 7. Filewod N. C., Pistolic J., Hancock R. E. (2009) Low concentrations of LL-37 alter IL-8 production by keratinocytes and bronchial epithelial cells in response to proinflammatory stimuli. FEMS Immunol. Med. Microbiol. 56, 233–240 [DOI] [PubMed] [Google Scholar]
- 8. Gilliet M., Lande R. (2008) Antimicrobial peptides and self-DNA in autoimmune skin inflammation. Curr. Opin. Immunol. 20, 401–407 [DOI] [PubMed] [Google Scholar]
- 9. Lai Y., Adhikarakunnathu S., Bhardwaj K., Ranjith-Kumar C. T., Wen Y., Jordan J. L., Wu L. H., Dragnea B., San Mateo L., Kao C. C. (2011) LL-37 and cationic peptides enhance TLR3 signaling by viral double-stranded RNAs. PLoS One 6, e26632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lai Y., Yi G., Chen A., Bhardwaj K., Tragesser B. J., Rodrigo A Valverde, Zlotnick A., Mukhopadhyay S., Ranjith-Kumar C. T., Kao C. C. (2011) Viral double-strand RNA-binding proteins can enhance innate immune signaling by toll-like Receptor 3. PLoS One 6, e25837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oren Z., Lerman J. C., Gudmundsson G. H., Agerberth B., Shai Y. (1999) Structure and organization of the human antimicrobial peptide LL-37 in phospholipid membranes. Relevance to the molecular basis for its non-cell-selective activity. Biochem. J. 341, 501–513 [PMC free article] [PubMed] [Google Scholar]
- 12. Wan M., Sabirsh A., Wetterholm A., Agerberth B., Haeggström J. Z. (2007) Leukotriene B4 triggers release of the cathelicidin LL-37 from human neutrophils. Novel lipid-peptide interactions in innate immune responses. FASEB J. 21, 2897–2905 [DOI] [PubMed] [Google Scholar]
- 13. Hasan M., Ruksznis C., Wang Y., Leifer C. A. (2011) Antimicrobial peptides inhibit polyinosinic-polycytidylic acid-induced immune responses. J. Immunol. 187, 5653–5659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Coffelt S. B., Tomchuck S. L., Zwezdaryk K. J., Danka E. S., Scandurro A. B. (2009) Leucine leucine-37 uses formyl peptide receptor-like 1 to activate signal transduction pathways, stimulate oncogenic gene expression, and enhance the invasiveness of ovarian cancer cells. Mol. Cancer Res. 7, 907–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Qi R., Singh D., Kao C. C. (2012) Proteolytic processing regulates Toll-like receptor 3 stability and endosomal localization. J. Biol. Chem. 287, 32617–32629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bolte S., Cordelières F. P. (2006) A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213–232 [DOI] [PubMed] [Google Scholar]
- 17. Kandler K., Shaykhiev R., Kleemann P., Klescz F., Lohoff M., Vogelmeier C., Bals R. (2006) The anti-microbial peptide LL-37 inhibits the activation of dendritic cells by TLR ligands. Int. Immunol. 18, 1729–1736 [DOI] [PubMed] [Google Scholar]
- 18. Qi R., Hoose S., Schreiter J., Sawant K. V., Lamb R., Ranjith-Kumar C. T., Mills J., San Mateo L., Jordan J. L., Kao C. C. (2010) Secretion of the human Toll-like receptor 3 ectodomain is affected by single nucleotide polymorphisms and regulated by Unc93b1. J. Biol. Chem. 285, 36635–36644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alexopoulou L., Holt A. C., Medzhitov R., Flavell R. A. (2001) Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413, 732–738 [DOI] [PubMed] [Google Scholar]
- 20. Ritter M., Mennerich D., Weith A., Seither P. (2005) Characterization of Toll-like receptors in primary lung epithelial cells. Strong impact of the TLR3 ligand poly(I:C) on the regulation of Toll-like receptors, adaptor proteins, and inflammatory response. J. Inflamm. (Lond) 2, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vercauteren D., Vandenbroucke R. E., Jones A. T., Rejman J., Demeester J., De Smedt S. C., Sanders N. N., Braeckmans K. (2010) The use of inhibitors to study endocytic pathways of gene carriers. Optimization and pitfalls. Mol. Ther. 18, 561–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pearse B. M., Robinson M. S. (1990) Clathrin, adaptors, and sorting. Annu. Rev. Cell Biol. 6, 151–171 [DOI] [PubMed] [Google Scholar]
- 23. Husebye H., Halaas Ø., Stenmark H., Tunheim G., Sandanger Ø., Bogen B., Brech A., Latz E., Espevik T. (2006) Endocytic pathways regulate Toll-like receptor 4 signaling and link innate and adaptive immunity. EMBO J. 25, 683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Itoh K., Watanabe A., Funami K., Seya T., Matsumoto M. (2008) The clathrin-mediated endocytic pathway participates in dsRNA-induced IFN-β production. J. Immunol. 181, 5522–5529 [DOI] [PubMed] [Google Scholar]
- 25. Limmongkon A., Giuliani C., Valenta R., Mittermann I., Heberle-Bors E., Wilson C. (2004) MAP kinase phosphorylation of plant profilin. Biochem. Biophys. Res. Commun. 324, 382–386 [DOI] [PubMed] [Google Scholar]
- 26. Girnita A., Zheng H., Grönberg A., Girnita L., Ståhle M. (2012) Identification of the cathelicidin peptide LL-37 as agonist for the type I insulin-like growth factor receptor. Oncogene 31, 352–365 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27. Tjabringa G. S., Aarbiou J., Ninaber D. K., Drijfhout J. W., Sørensen O. E., Borregaard N., Rabe K. F., Hiemstra P. S. (2003) The antimicrobial peptide LL-37 activates innate immunity at the airway epithelial surface by transactivation of the epidermal growth factor receptor. J. Immunol. 171, 6690–6696 [DOI] [PubMed] [Google Scholar]
- 28. Into T., Inomata M., Shibata K., Murakami Y. (2010) Effect of the antimicrobial peptide LL-37 on Toll-like receptors 2-, 3-, and 4-triggered expression of IL-6, IL-8. and CXCL10 in human gingival fibroblasts. Cell. Immunol. 264, 104–109 [DOI] [PubMed] [Google Scholar]
- 29. Gao J. L., Murphy P. M. (1993) Species and subtype variants of the N-formyl peptide chemotactic receptor reveal multiple important functional domains. J. Biol. Chem. 268, 25395–25401 [PubMed] [Google Scholar]
- 30. He H. Q., Liao D., Wang Z. G., Wang Z. L., Zhou H. C., Wang M. W., Ye R. D. (2013) Functional characterization of three mouse formyl peptide receptors. Mol. Pharmacol. 83, 389–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ranjith-Kumar C. T., Duffy K. E., Jordan J. L., Eaton-Bassiri A., Vaughan R., Hoose S. A., Lamb R. J., Sarisky R. T., Kao C. C. (2008) Single-stranded oligonucleotides can inhibit cytokine production induced by human toll-like receptor 3. Mol. Cell Biol. 28, 4507–4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Limmon G. V., Arredouani M., McCann K. L., Corn Minor R. A., Kobzik L., Imani F. (2008) Scavenger receptor class-A is a novel cell surface receptor for double-stranded RNA. FASEB J. 22, 159–167 [DOI] [PubMed] [Google Scholar]
- 33. McCartney S., Vermi W., Gilfillan S., Cella M., Murphy T. L., Schreiber R. D., Murphy K. M., Colonna M. (2009) Distinct and complementary functions of MDA5 and TLR3 in poly(I:C)-mediated activation of mouse NK cells. J. Exp. Med. 206, 2967–2976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hoffmann M. H., Bruns H., Backdahl L., Neregard P., Niederreiter B., Herrmann M., Catrina A. I., Agerberth B., Holmdahl R. (2012) The cathelicidins LL-37 and rCRAMP are associated with pathogenic events of arthritis in humans and rats. Ann. Rheum. Dis., in press [DOI] [PubMed] [Google Scholar]