Background: Assembly of cytochrome c oxidase (COX), complex IV of the respiratory chain, requires a great number of accessory proteins known as assembly factors.
Results: hCOA3 interacts with newly synthesized COX1 and promotes its assembly with subsequent COX subunits.
Conclusion: hCOA3 participates in COX biogenesis in humans.
Significance: hCOA3 is a new candidate gene to screen in patients with COX deficiency.
Keywords: Energy Metabolism, Mitochondria, Mitochondrial Diseases, Molecular Cell Biology, Respiratory Chain, CCDC56, Cytochrome c Oxidase, Oxidative Phosphorylation, hCOA3
Abstract
Cytochrome c oxidase (COX) or complex IV of the mitochondrial respiratory chain plays a fundamental role in energy production of aerobic cells. In humans, COX deficiency is the most frequent cause of mitochondrial encephalomyopathies. Human COX is composed of 13 subunits of dual genetic origin, whose assembly requires an increasing number of nuclear-encoded accessory proteins known as assembly factors. Here, we have identified and characterized human CCDC56, an 11.7-kDa mitochondrial transmembrane protein, as a new factor essential for COX biogenesis. CCDC56 shares sequence similarity with the yeast COX assembly factor Coa3 and was termed hCOA3. hCOA3-silenced cells display a severe COX functional alteration owing to a decreased stability of newly synthesized COX1 and an impairment in the holoenzyme assembly process. We show that hCOA3 physically interacts with both the mitochondrial translation machinery and COX structural subunits. We conclude that hCOA3 stabilizes COX1 co-translationally and promotes its assembly with COX partner subunits. Finally, our results identify hCOA3 as a new candidate when screening for genes responsible for mitochondrial diseases associated with COX deficiency.
Introduction
Mitochondria produce most cellular ATP by the process of oxidative phosphorylation (OXPHOS)5 carried out by the four complexes of the respiratory chain and the F1F0 ATPase. Cytochrome c oxidase (COX) or complex IV, the terminal enzyme of the respiratory chain, catalyzes the oxidation of cytochrome c by transferring its electrons to molecular oxygen (1). Mammalian complex IV is composed of 13 subunits. Three subunits forming the catalytic core, COX1, COX2, and COX3, are encoded in the mitochondrial DNA (mtDNA), whereas the remaining ten subunits (COX4, COX5a, COX5b, COX6a, COX6b, COX6c, COX7a, COX7b, COX7c, and COX8) are encoded in the nuclear genome (2, 3). The enzyme contains four redox-active metal centers: two iron centers, heme a and heme a3, and two copper centers, CuA and CuB, which are essential for its assembly and function (4).
COX assembly is thought to be a linear process in which its subunits and cofactors are incorporated in an ordered manner (5). More than 30 COX assembly factors have been identified in model organisms, mainly the yeast Saccharomyces cerevisiae, and in human patients with COX deficiencies. They are required for every step of the formation of the complex, from translation of the individual subunits to incorporation of the prosthetic groups and assembly of the holoenzyme (6). COX biogenesis must be tightly regulated to prevent the accumulation of pro-oxidant assembly intermediates. We and others (6, 7) have recently shown that in S. cerevisiae, there is a negative feedback translational regulatory system that serves to coordinate the synthesis in mitochondrial ribosomes of COX1 a subunit containing heme a and copper centers, with its assembly into the holoenzyme. The mechanism involves Mss51, a translational activator that additionally acts as a COX1 chaperone, as well as Cox14 and Coa3, two COX1 stability/assembly factors. Whether this regulatory mechanism is conserved in humans remains to be fully disclosed.
In recent years, COX biogenesis gained much interest because defects in the assembly of the enzyme are a major cause of mitochondrial disorders in humans and may play an important role in aging and degenerative diseases (8–10). In fact, COX deficiency is one of the most frequent causes of mitochondrial encephalomyopathies in humans (11). Mutations in the three mtDNA-encoded COX subunits and nuclear-encoded subunits COX4, COX6b, and COX7b have been identified in patients (12–17), although these defects are relatively rare. Most COX deficiencies described to date are caused by mutations in assembly factors (8, 11). However, in the majority of patients, the molecular basis of their autosomal recessive COX deficiency remains unknown. Although the list of COX assembly genes identified in yeast is likely close to completion, identifying the respective human homologues is not simple in some cases due to their divergent primary sequences. Recent studies based on iterative orthology prediction have identified new human homologues of yeast COX assembly factors (18). Among them are some key proteins involved in the COX assembly regulatory process described earlier (Cox14, Coa3, and Mss51), which suggests that such a regulatory mechanism could actually exist in human cells. Notably, mutations in C12orf62, the human homologue of yeast Cox14, have been recently shown to produce COX deficiency in a patient with severe congenital lactic acidosis (19). C12orf62 was proposed to couple COX1 synthesis and cytochrome c oxidase assembly as its yeast counterpart (19).
We have recently identified a mitochondrial protein in Drosophila melanogaster, CCDC56, encoded on a bicistronic mRNA with mitochondrial translation factor B1 (20). This protein is essential for COX activity in Drosophila and is highly conserved throughout evolution. It has recently become evident that CCDC56 is actually the fly homologue of yeast Coa3. Here, we demonstrate that human CCDC56, hereafter termed hCOA3, is involved in the early steps of COX biogenesis by securing the stability and promoting the assembly of newly synthesized COX1. These results suggest that hCOA3 could play a role in coordinating COX1 synthesis and assembly. Finally, our results identify hCOA3 as a new candidate when screening for genes responsible for mitochondrial diseases associated with COX deficiency.
EXPERIMENTAL PROCEDURES
siRNAs, Taqman Probes, and Constructs
Three Silencer Select predesigned siRNAs against hCOA3 (s26294, s26295, and s26296), siRNA Silencer negative control 2 (Ki 2) and Taqman assays for CCDC56 mRNA (Hs00360235_m1) and 18 S rRNA (Hs99999901_s1) were purchased from Applied Biosystems.
For the FLAG-tagged hCOA3 construct, hCOA3 cDNA was amplified using the cDNA clone BX280424 (ImaGenes) as a template, with specific primers carrying in-frame the FLAG tag and recognition sites for AflII and NotI, respectively, (5′-TTCTTAAGATGGCGTCTTCGGGAGCTGGTG-3′, 5′-ATGCGGCCGCTTACTTGTCGTCATCGTCTTTGTAGTCGGACCCTGACGCCCTTGCCAGAGCTCG-3′). The PCR product was digested and cloned into pIRESpuro2 (Clontech). Fidelity of the clones was confirmed by sequencing.
Antibodies
To express hCOA3 in E. coli, a PCR fragment encoding the full hCOA3 cDNA was cloned in pRSET-B (Invitrogen). Polyclonal antibody was generated using standard procedures. Polyclonal antibody anti-Mn-SOD was obtained from Millipore. Monoclonal antibodies against COX1, COX2, COX3, COX4, COX5a, NDUFA9, and Porin were purchased from Mitosciences. Monoclonal antibodies against CII 70 kDa and CIII Core2 and secondary antibody coupled to Alexa Fluor 488 were purchased from Molecular Probes (Invitrogen). Anti-FLAG M2 monoclonal antibody was obtained from Stratagene. Anti-SLIRP, anti-mt-EFTu, anti-LRPPRC, anti-NDUFV1, and anti-NDUFS1 antibodies were obtained from Abcam. Secondary antibodies coupled to horseradish peroxidase were obtained from Santa Cruz Biotechnology. Anti-β-ATPase polyclonal antibodies had been previously generated in the lab (21).
Quantitative RT-PCR of Mitochondrial mRNAs
Mitochondrial mRNAs were quantified using SYBR Green (Applied Biosystems) and specific primer pairs for each gene following standard procedures. Primers used in this study were: COX1: 5′-CTCTTCGTCTGATCCGTCCT-3′, 5′-ATTCCGAAGCCTGGTAGGAT-3′; COX2: 5′-ACGAGTACACCGACTACGGC-3′, 5′-CGGGAATTGCATCTGTTTTT-3′; COX3: 5′-CCCACCAATCACATGCCTAT-3′, 5′-GTGGCCTTGGTATGTGCTTT-3′; ND5: 5′-AAACAACCCAGCTCTCCCTAA-3′, 5′-AGAAGGATATAATTCCTACG-3′; CytB: 5′-TGAAACTTCGGCTCACTCCT-3′, 5′-AGAATATTGAGGCGCCATTG-3′; ATP6: 5′-TTTCCCCCTCTATTGATCCC-3′, 5′-TGGGTGGTTGGTGTAAATGA-3′ and RNR1: 5′-CGATCAACCTCACCACCTCT-3′, 5′-TGCTAAATCCACCTTCGACC-3′. rRNA 18 S was used as a control: 5′-CCAGTAAGTGCGGGTCATAAGC-3′, 5′-CCTCACTAAACCATCCAA TCGG-3′.
Cell Lines, Cell Culture, and Transfection
HeLa cells were cultured in high-glucose DMEM (Invitrogen) supplemented with 10% fetal bovine serum, uridine, and antibiotics at 37 °C in a 5% CO2 atmosphere. Cells were transfected with 30 nm siRNAs or 5 μg of the constructs using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. For hCOA3 silencing experiments, cells were harvested 96 h after transfection. To generate the cell lines stably overexpressing hCOA3-FLAG, 1.5 μg/ml of puromycin were added to the medium 2 days after transfection. The resultant cell line was grown in the presence of the antibiotic.
Mitochondrial Subfractionation and Proteinase K Protection Assays
Mitochondria were purified from HeLa cells expressing hCOA3-FLAG as described previously (20). Mitochondria were ruptured by sonication. Soluble and insoluble fractions were obtained by centrifugation at 50,000 × g for 15 min at 4 °C. The membrane pellet was resuspended in 0.1 m Na2CO3, pH 11. After 30 min on ice, the sample was centrifuged at 50,000 × g for 15 min at 4 °C to separate the soluble membrane extrinsic from the insoluble intrinsic membrane proteins.
Purified mitochondria from hCOA3-FLAG were resuspended in 10 mm Tris-HCl, pH 7, 10 mm KCl, 0.15 mm MgSO4 buffer either containing 0.25 m sucrose or devoid of sucrose to allow mitochondria swelling and conversion to mitoplasts. Samples were treated where indicated with 0.62 μg/ml proteinase K for 20 min in ice. Mitochondria and mitoplasts were recovered by centrifugation at 50,000 × g for 15 min at 4 °C and analyzed by Western blotting.
To selectively solubilize the outer mitochondrial membrane, purified mitochondria from hCOA3-FLAG-overexpressing cells were resuspended in 10 mm Tris-HCl, pH 7, 10 mm KCl, 0.15 mm MgSO4, 0.25 m sucrose buffer, and treated with 1% digitonin for 30 min in ice. Membranes and the soluble fraction were separated by centrifugation at 50,000 × g for 15 min at 4 °C and analyzed by Western blotting.
Activity of the OXPHOS Complexes
The activity of the OXPHOS complexes and the enzyme citrate synthase were determined as described previously (20).
BN-PAGE and Two-dimensional BN/SDS-PAGE
Blue native electrophoresis and two-dimensional BN/SDS electrophoresis of the OXPHOS complexes was performed as described previously (22). For two-dimensional BN/SDS-PAGE experiments, transfected cells were treated with 10 μg/ml chloramphenicol for 2 days to deplete the respiratory chain complexes and retransfected with siRNAs 48 h after the first transfection. Cells were collected at 96 h of hCOA3 interference and 48 h of recovery after chloramphenicol treatment.
Pulse Labeling of Mitochondrial Translation Products
In vivo labeling of the mitochondrial translation products was performed as described previously (23). Briefly, cells were labeled for 30 min in methionine/cysteine-free DMEM containing 200 μCi/ml of [35S]methionine (PerkinElmer Life Sciences) and 100 μg/ml of either emetine (pulse experiments) or anisomycin (pulse-chase experiments). After the incubation, cells were washed and harvested (pulse) or incubated in regular DMEM for 2, 4, and 7 h (chase). 100 μg of whole protein extracts were loaded on 15–20% SDS-PAGE gradient gels, and the labeled mitochondrial translation products were detected through direct autoradiography or quantified using a Typhoon PhosphorImager system (GE Healthcare).
Immunocytochemistry
HeLa cells expressing hCOA3-FLAG were plated on coverslips and grown overnight at 37 °C. Cells were stained for 30 min with 50 nm Mitotracker red (Molecular Probes, Invitrogen), fixed with 2% paraformaldehyde and treated with methanol before the incubation with an anti-FLAG M2 primary antibody (Stratagene) in 2% BSA. Secondary antibody Alexa Fluor 488 (Molecular Probes, Invitrogen) was used for immunofluorescence detection. Images were obtained in a Leica SP5 confocal microscope (DMI6000 stand), Leica plan apochromat 63×/1.4 numerical aperture objective lens, 60-nm XY pixel size (Nyquist criterion optimized), 2-frame averaging, and sequential scan mode. For Alexa Fluor 488, Argon ion laser, PMT gain 600, 510–540-nm emission band; for Mitotracker Red, 561 nm laser, PMT gain 600, and 580–630-nm emission band.
Immunoprecipitation and Proteomic Analysis
Mitochondria were purified from HeLa cells expressing hCOA3-FLAG or containing empty pIRESpuro2 vector as described previously (20). Equal amounts of mitochondrial protein of each cell line were lysed using 1% DDM in 50 mm Tris HCl pH 7.4, 150 mm NaCl and 1 mm EDTA. hCOA3-FLAG was immunoprecipitated using FLAG immunoprecipitation kit (Sigma-Aldrich) following the manufacturer's instructions. Proteins were eluted using a FLAG peptide and analyzed though mass spectrometry and immunoblotting.
Protein extracts were applied onto a SDS-PAGE gel, and the run was stopped as soon as the front entered into the resolving gel. The unseparated protein bands were visualized by Coomassie staining, excised, destained in acetonitrile:water (1:1), and digested in situ with sequencing grade trypsin as described by Shevchenko et al. (24) with minor modifications. The dried gel pieces were reswollen in 50 mm ammonium bicarbonate with 60 ng/μl trypsin (Promega) at a 5:1 protein:trypsin (w/w) ratio in 50 mm ammonium bicarbonate, pH 8.8. The tubes were kept in ice for 2 h and incubated at 37 °C for 12 h. Whole supernatants were dried down and desalted onto Omix tips. The desalted protein digest was dried, resuspended in 0.1% formic acid and analyzed by reverse phase-liquid chromatography MS/MS (RP-LC-MS/MS) in an Agilent 1100 system coupled to a linear ion trap LTQ-Velos mass spectrometer (Thermo Fisher Scientific). Peptides were detected in survey scans from 400 to 1600 atomic mass units (1 μscan), followed by 15 data-dependent MS/MS scans (Top 15), using an isolation width of 2 units (mass-to-charge ratio units), normalized collision energy of 35%, and dynamic exclusion applied during 30-s periods. Peptide identification from raw data was carried out using the SEQUEST algorithm (Proteome Discoverer 1.3, Thermo Fisher Scientific). Database search was performed against uniprot-homo.fasta. The following constraints were used for the searches: tryptic cleavage after Arg and Lys, up to two missed cleavage sites, and tolerances of 1 Da for precursor ions and 0.8 Da for MS/MS fragment ions and the searches were performed allowing optional Met oxidation and Cys carbamidomethylation. Search against decoy database (integrated decoy approach) was performed using false discovery rate < 0.01.
Statistical Analysis
Graphs represent mean values of at least three independent measurements. Error bars represent S.D. One-way or two-way analyses of variance were performed using GraphPad Prism (version 4.00 for Windows, GraphPad Software, San Diego, CA).
RESULTS
hCOA3 Is a Transmembrane Protein of the Inner Mitochondrial Membrane
We have recently shown CCDC56 to be a mitochondrial protein essential for COX activity in D. melanogaster (20), although its precise function remained to be elucidated. BLAST alignments showed this protein is well conserved from Drosophila to humans, and more recent studies have suggested CCDC56 is the homologue of yeast Coa3 (18). Human COA3 is located on chromosome 17 (17q.21.31) and encodes an 11.7-kDa protein with predicted transmembrane and coiled-coil domains.
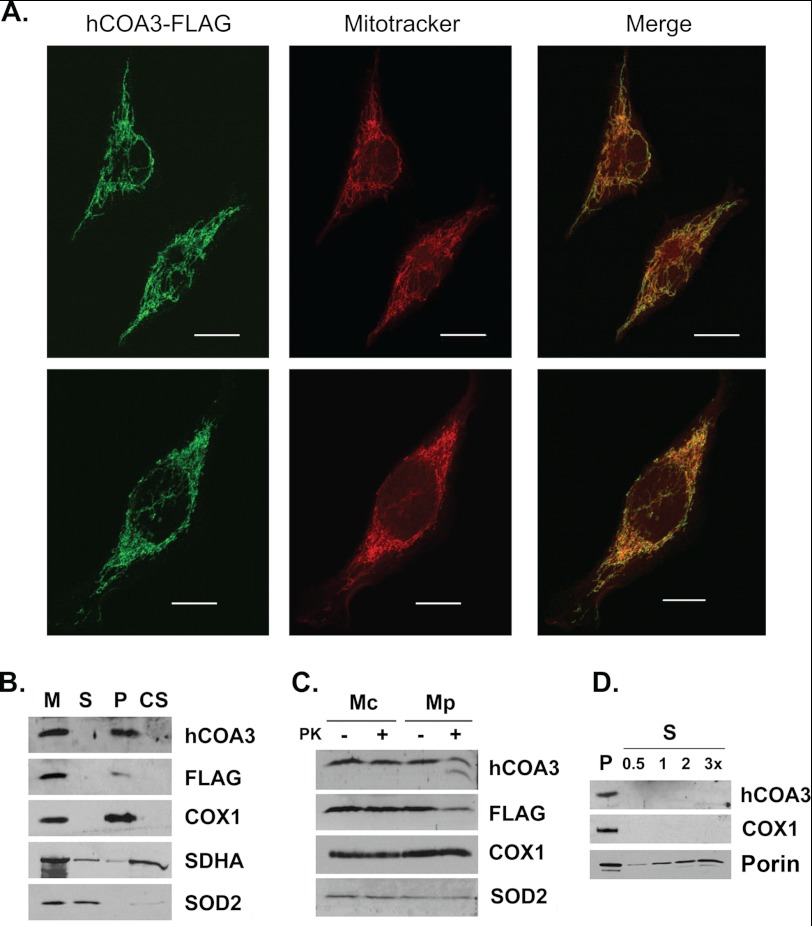
To analyze whether hCOA3 is a mitochondrial protein as described for its D. melanogaster orthologue (20), we overexpressed a C-terminal FLAG-tagged version of the protein in HeLa cells. Immunocytochemical experiments showed this protein to be localized exclusively to mitochondria (Fig. 1A). Sonication of mitochondria followed by centrifugation to separate soluble and insoluble proteins showed hCOA3-FLAG behaves as a mitochondrial membrane protein (Fig. 1B). hCOA3-FLAG is not extracted from the mitochondrial membrane by sodium carbonate treatment, indicating hCOA3 is an intrinsic membrane protein, as expected from its predicted transmembrane domain (Fig. 1B).
FIGURE 1.
hCOA3 is a transmembrane protein of the inner mitochondrial membrane. A, immunocytochemistry of HeLa cells overexpressing hCOA3-FLAG (hCOA3-FLAG). hCOA3-FLAG was visualized using anti-FLAG M2 antibodies. Mitochondria were stained using Mitotracker Red. Images were obtained in a Leica SP5 confocal microscope with a Leica plan apochromat 63×/1.4 numerical aperture objective lens. Scale bars represent 10 μm. B, purified mitochondria from hCOA3-FLAG-overexpressing cells (hCOA3-FLAG) were sonically irradiated and centrifuged to separate the soluble (S) and membrane (M) proteins. The membrane pellet was treated with 0.1 m Na2CO3 and centrifuged to separate the soluble membrane extrinsic (CS) from the insoluble intrinsic membrane proteins (P). Equivalent volumes of each fraction were analyzed by Western blotting. C, mitochondria (Mc) and mitoplasts (Mp) from hCOA3-FLAG-overexpressing cells were treated with proteinase K where indicated. After proteinase K treatment, mitochondria and mitoplasts were recovered by centrifugation and analyzed by Western blotting. D, purified mitochondria from hCOA3-FLAG overexpressing cells were treated with 1% digitonin to selectively solubilize the mitochondrial outer membrane. Membranes (P) and the soluble fraction (S) were separated by centrifugation and analyzed by Western blotting. 0.5, 1, 2, and 3 times of equivalent soluble fraction were loaded on the gel.
hCOA3-FLAG is protected from proteinase K digestion in isolated mitochondria but is partially digested in mitoplasts where the outer membrane is disrupted (Fig. 1C). Interestingly, anti-hCOA3 antibodies but not anti-FLAG M2 antibodies detect a polypeptide of smaller size in mitoplasts treated with proteinase K, probably hCOA3 after the FLAG tag has been digested (Fig. 1C). In addition, hCOA3-FLAG behaves as an inner mitochondrial membrane protein when the outer mitochondrial membrane is solubilized by digitonin treatment (Fig. 1D). Taken together, these data demonstrate that hCOA3 is an intrinsic protein of the mitochondrial inner membrane with the C terminus exposed to the intermembrane space.
hCOA3 Is Essential for Cytochrome c Oxidase Activity and Holocomplex Accumulation
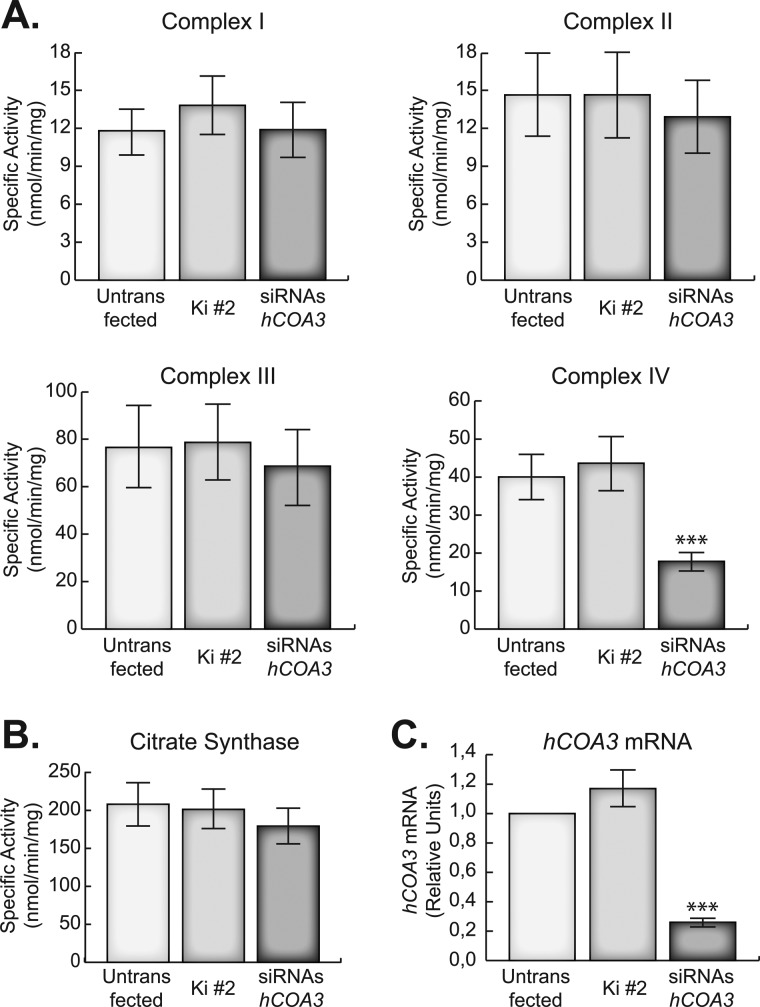
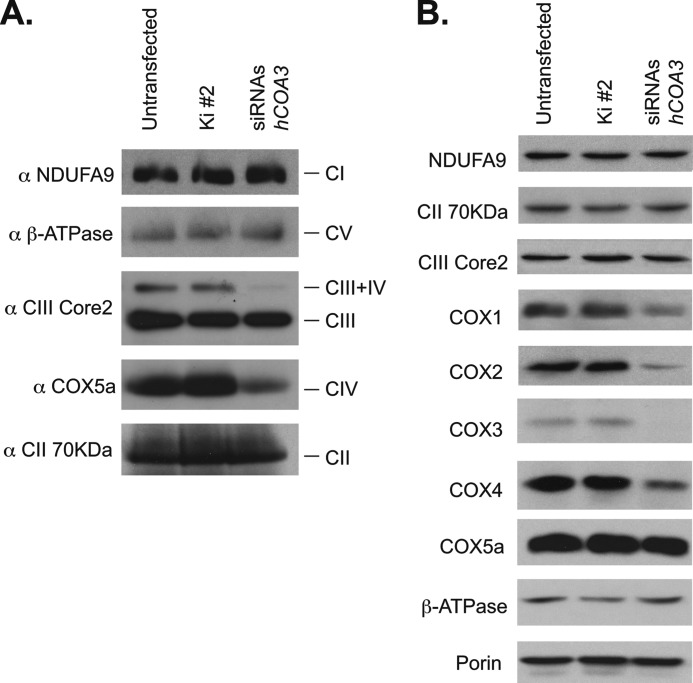
To characterize the function of hCOA3 in human mitochondria, we have silenced its expression using siRNAs. For this purpose, three specific siRNAs against hCOA3 and a non-interfering control siRNA were transiently transfected into HeLa cells. Four days after transfection, the cells were harvested and analyzed for the activity of the OXPHOS complexes (Fig. 2, A and B) and hCOA3 mRNA levels using quantitative RT-PCR (Fig. 2C). hCOA3 interfered cells present a specific defect in COX activity, which was reduced to 30% of controls, whereas the activity of all other respiratory chain complexes, as well as citrate synthase activity, remained unchanged (Fig. 2, A and B). This decrease in COX activity correlated with a similar decrease in the steady-state levels of fully assembled complex IV, as observed in blue native gel electrophoresis (BN-PAGE) experiments (Fig. 3A). Although our extraction conditions (2% dodecyl maltoside, DDM) are known to significantly disrupt the macromolecular assemblies of respiratory chain complexes into supercomplexes (25), we were able to also detect a decrease in complex CIII/CIV in silenced cells. There was a perfect correlation between the activity and the levels of fully assembled complex in each experiment, suggesting that the residual enzyme present in the interfered cells is fully active and the respiratory defect results from an impairment of COX assembly. These data point toward an involvement of hCOA3 in COX biogenesis rather than in the regulation of its activity.
FIGURE 2.
hCOA3 knockdown causes a defect in COX activity in HeLa cells. A, specific activity of the OXPHOS complexes. B, citrate synthase-specific activity. C, quantitative RT-PCR of hCOA3 mRNA. All experiments were performed in untransfected HeLa cells (untransfected), HeLa cells transfected with non-interfering siRNAs (Ki #2) or siRNAs against hCOA3 (siRNAs hCOA3) 96 h after transfection. Data represent mean values ± S.D. ***, p < 0.001 (n = 6).
FIGURE 3.
hCOA3 knockdown causes a decrease of fully assembled COX in HeLa cells. BN-PAGE of the OXPHOS complexes (A) and steady-state levels of the OXPHOS subunits analyzed by Western blotting (B) in untransfected HeLa cells (untransfected), HeLa cells transfected with non-interfering siRNAs (Ki #2) or siRNAs against hCOA3 (siRNAs hCOA3) 96 h after transfection.
We next investigated whether the levels of COX structural subunits were also affected in hCOA3 knockdown cells. Consistently with the COX activity and BN-PAGE results, we observed hCOA3 interfered cells have lower steady-state levels of COX1, COX2, COX3, and COX4 subunits, whereas the levels of complex I, II, III, or V subunits remained unchanged when compared with controls (Fig. 3B). Noticeably, COX5a showed almost similar levels in knockdown and control cells, suggesting this protein has a longer-half-life than other COX structural subunits.
COX1 Is Rapidly Degraded in hCOA3-silenced Cells
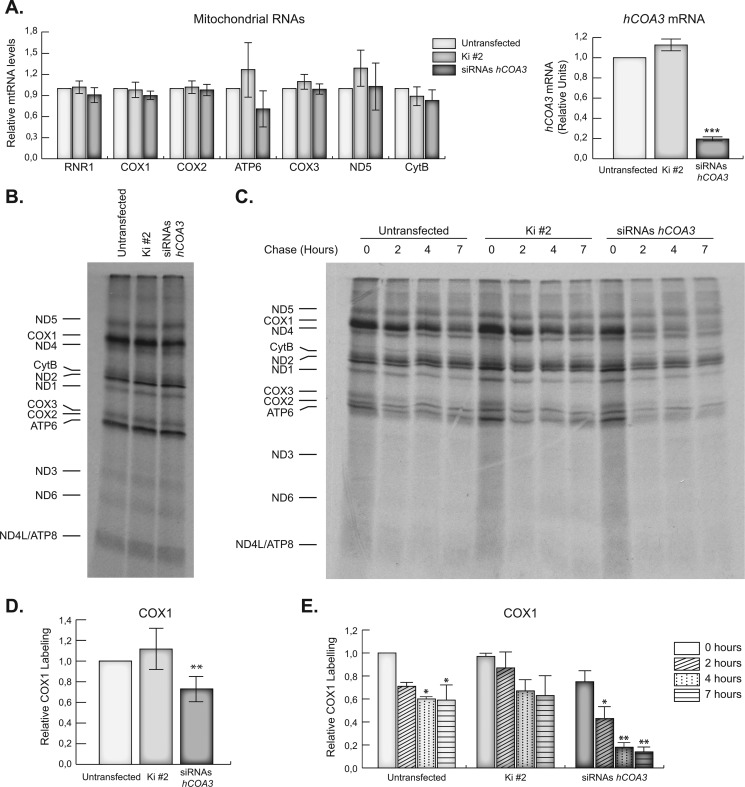
To systematically investigate the role of hCOA3 in COX assembly, we started by assessing whether the decrease in COX subunits was due to defective mitochondrial transcription. We measured the levels of their transcripts using quantitative RT-PCR and specific primer pairs for each gene and failed to find any differences in steady-state levels of COX1, COX2, and COX3 mRNAs or in the levels of other mitochondrial mRNAs and rRNAs when compared with controls (Fig. 4A), indicating mitochondrial transcription is not affected in the absence of hCOA3.
FIGURE 4.
hCOA3 knockdown induces a rapid degradation of the newly synthesized COX1. A, quantitative RT-PCR of mitochondrial mRNAs (left panel) and hCOA3 mRNA (right panel) in untransfected HeLa cells (untransfected), HeLa cells transfected with non-interfering siRNAs (Ki #2) or siRNAs against hCOA3 (siRNAs hCOA3) 96 h after transfection. Data represent mean values ± S.D. ***, p < 0.001 (n = 5). B, newly synthesized mitochondrial polypeptides were labeled with [35S]methionine for 30 min. Mitochondrial translation products were detected through direct autoradiography of the gels. C, newly synthesized mitochondrial polypeptides were labeled with [35S]methionine for 30 min. Cells were then washed and incubated in regular DMEM for 2, 4, or 7 h. Mitochondrial translation products were detected through direct autoradiography of the gels. D, quantification of COX1 labeling in pulse experiments. Data represent mean values ± S.D. **, p < 0.01 (n = 4). E, quantification of COX1 labeling in pulse-chase experiments. Data represent mean values ± S.D. *, p < 0.05; **, p < 0.01 (n = 4).
We next determined whether hCOA3 interfered cells had altered synthesis or turnover rates of any of the mtDNA-encoded COX subunits. To this end, we pulse-labeled mitochondrial translation products with [35S]methionine for 30 min in the presence of an inhibitor of cytoplasmic translation, and the labeled polypeptides were subsequently chased for 2, 4, or 7 h. Interfered cells showed a specific mild decrease in the amount of pulse-labeled COX1, whereas the rest of the mtDNA-encoded polypeptides were synthesized at rates similar than in control cells (Fig. 4, B and D). Pulse-chase experiments showed newly synthesized COX1 is rapidly degraded in hCOA3-deficient cells (Fig. 4, C and E). On the contrary, the stability of COX2, COX3, or any other mtDNA-encoded polypeptides was not markedly different in control and hCOA3-deficient cells during the chase periods analyzed (data not shown).
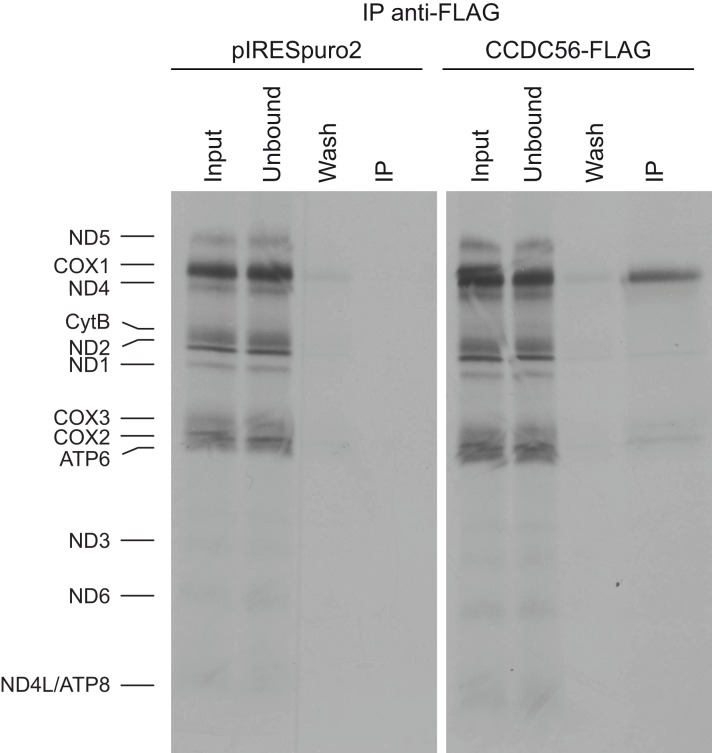
Even though newly synthesized COX2 and COX3 are more stable than COX1, Western blot analyses revealed low levels of the three catalytic core subunits. The three mtDNA-encoded COX subunits are preferentially degraded when COX fails to assemble and if one of the subunits is particularly labile, the other two become also unstable (26–28). To clarify whether hCOA3 plays a role on the stability or biogenesis of one or several of the mtDNA-encoded COX subunits, we labeled mitochondrial translation products for 30 min with [35S]methionine and performed immunoprecipitation experiments with anti-FLAG M2 antibodies in HeLa cells either overexpressing hCOA3-FLAG or containing an empty pIRESpuro2 vector. As shown in Fig. 5, newly synthesized COX1 is greatly enriched in the eluate from hCOA3-FLAG overexpressing cells, demonstrating that hCOA3 interacts with newly synthesized COX1. These results indicate that hCOA3 is required for COX1 stability and point toward the loss of COX1 in hCOA3-depleted cells as the cause of the degradation of the remaining COX catalytic core subunits.
FIGURE 5.
hCOA3 interacts with newly synthesized COX1. In vivo labeling of mitochondrial translation products followed by immunoprecipitation using anti-FLAG M2 antibodies in HeLa cells containing pIRESpuro2 (pIRESpuro2) and HeLa cells stably overexpressing hCOA3-FLAG (hCOA3-FLAG). 10% of the corresponding volume of input, unbound, and wash fractions was loaded on the gels.
hCOA3 Is a Cytochrome c Oxidase Assembly Factor
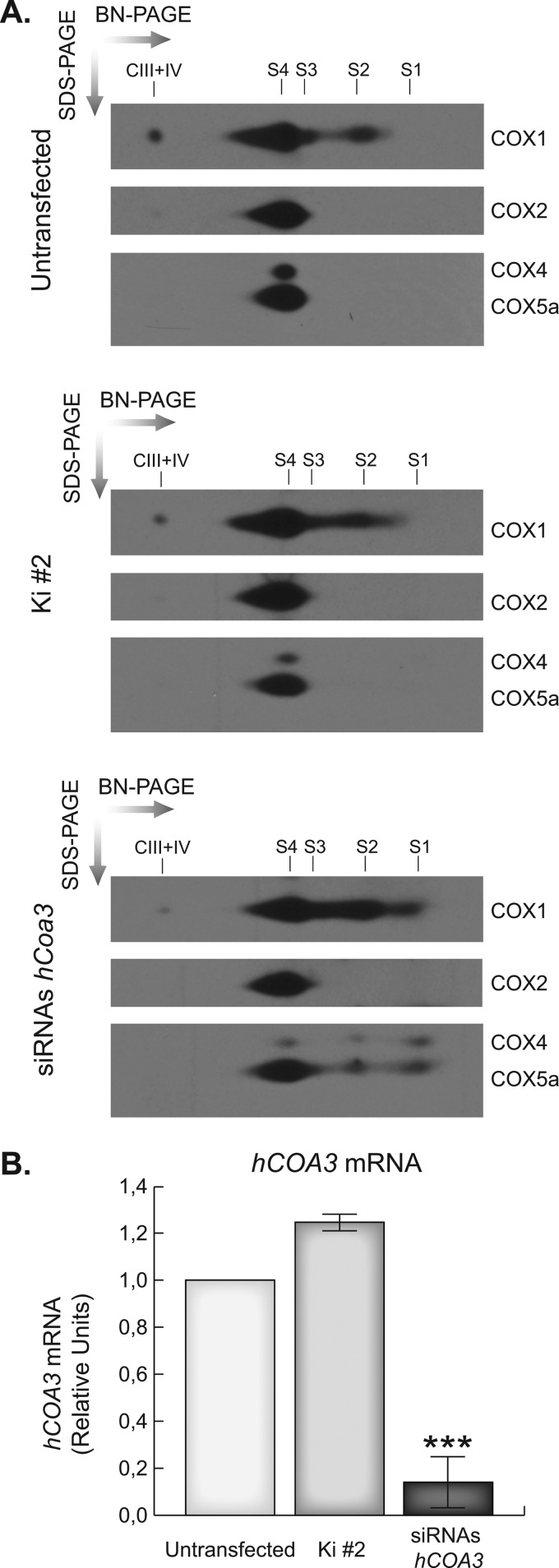
hCOA3 knockdown induces a rapid degradation of the newly synthesized COX1, which together with the decreased steady-state levels of COX2, COX3, and the nuclear DNA-encoded subunit COX4, suggest hCOA3-silenced cells may have an early defect in COX assembly most probably involving COX1. To gain insight into how COX assembly proceeds in silenced cells, we depleted the respiratory chain complexes using the mitochondrial translation inhibitor chloramphenicol, let the cells recover for 48 h, and performed two-dimensional BN/SDS-PAGE experiments to analyze the accumulation and distribution of the different subunits in each assembly intermediate. As shown in Fig. 6, hCOA3-deficient cells accumulate a subassembly intermediate that contains COX1, COX4, and COX5a. In the current model of human COX assembly, the first subassembly (S1) formed during the process exclusively contains COX1, which acts as a seed for sequential incorporation of COX subunits. In this model, the addition of subunits COX4 and COX5a to S1 results in the progression to the second assembly intermediate (S2) (5, 29, 30). The accumulated subassembly in interfered cells corresponds with subcomplex S2 and indicates that hCOA3 is participating in the early steps of COX assembly, most probably facilitating the addition of the subsequent COX subunits to COX1 containing intermediates.
FIGURE 6.
hCOA3 knockdown causes a defect in cytochrome c oxidase assembly. A, two-dimensional BN/SDS-PAGE of the OXPHOS complexes in untransfected HeLa cells (untransfected), HeLa cells transfected with non-interfering siRNAs (Ki #2) or siRNAs against hCOA3 (siRNAs hCOA3). S1, S2, and S3 indicate the three COX subassemblies, S4 indicates the COX monomer, and CIII+IV indicates supercomplex III+IV. B, quantitative RT-PCR of hCOA3 mRNA in untransfected HeLa cells (untransfected), HeLa cells transfected with non-interfering siRNAs (Ki #2) or siRNAs against hCOA3 (siRNAs hCOA3) 96 h after transfection. Data represent mean values ± S.D. ***, p < 0.001 (n = 4).
hCOA3-FLAG Overexpression Rescues the Cytochrome c Oxidase Defect
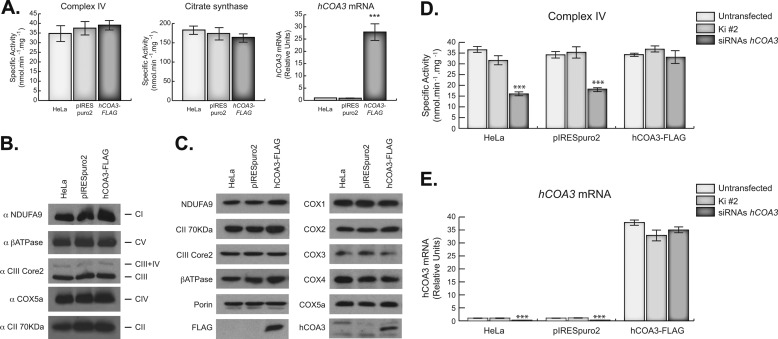
To ensure that the defects observed in interfered cells were exclusively due to hCOA3 silencing, we overexpressed in HeLa cells the hCOA3-FLAG construct described earlier. This construct does not include the 3′-UTR of hCOA3 mRNA that contains the regions of homology of the siRNAs, avoiding in this way the RNA interference machinery.
hCOA3-FLAG overexpression had no effect on COX activity or the levels of its subunits (Fig. 7A–C). Nevertheless, hCOA3-FLAG overexpression was able to fully rescue the COX defect caused by hCOA3 silencing (Fig. 7, C and D), confirming that the previously observed localization of the overexpressed protein in the inner mitochondrial membrane is correct, and the absence of hCOA3 is responsible for the phenotypes observed in interfered cells.
FIGURE 7.
hCOA3-FLAG overexpression fully rescues COX defect in interfered cells. A, COX activity (left panel), citrate synthase activity (middle panel), and quantitative RT-PCR of hCOA3 mRNA (right panel) in HeLa cells (HeLa), HeLa cells containing empty pIRESpuro2 vector (pIRESpuro2), and HeLa cells stably overexpressing hCOA3-FLAG (hCOA3-FLAG). Data represent mean values ± S.D. ***, p < 0.001 (n = 4). B, BN-PAGE of the OXPHOS complexes in HeLa cells (HeLa), HeLa cells containing empty pIRESpuro2 vector (pIRESpuro2), and HeLa cells stably overexpressing hCOA3-FLAG (hCOA3-FLAG). C, steady-state levels of the OXPHOS subunits analyzed by Western blotting in HeLa cells (HeLa), HeLa cells containing empty pIRESpuro2 vector (pIRESpuro2), and HeLa cells stably overexpressing hCOA3-FLAG (hCOA3-FLAG). COX-specific activity (D) and hCOA3 mRNA levels (E) in HeLa cells (HeLa), HeLa cells containing pIRESpuro2 (pIRESpuro2), and HeLa cells stably overexpressing hCOA3-FLAG (hCOA3-FLAG). Each cell line was transiently transfected with non-interfering siRNAs (Ki #2) and siRNAs against hCOA3 (siRNAs hCOA3). COX activity and hCOA3 mRNA were measured 96 h after transfection. Data represent mean values ± S.D. ***, p < 0.001 (n = 3).
hCOA3 Interacts with COX Subunits Promoting Cytochrome c Oxidase Assembly
To further investigate the function of hCOA3 in the early steps of COX biogenesis, we asked whether hCOA3 physically interacts with COX subassemblies. Two-dimensional BN/SDS-PAGE analysis of hCOA3-FLAG-overexpressing cells extracted with 2% DDM revealed that hCOA3-FLAG is part of several complexes, ranging in size from monomeric hCOA3-FLAG to almost 1 MDa (Fig. 8A), indicating its potential interaction with other proteins that could constitute COX subassemblies. Because the overexpressed FLAG-tagged version of hCOA3 is fully functional and is forming several high molecular mass complexes, we performed immunoprecipitation experiments using anti-FLAG M2 antibodies in overexpressing and control cells to identify its binding partners. Three independent immunoprecipitation experiments followed by mass spectrometry analysis identified several COX structural subunits present in the eluate, supporting previous observations that indicate a role for hCOA3 in COX assembly. Moreover, this analysis showed hCOA3-FLAG coimmunoprecipitates with the mitochondrial translation elongation factor Tu (mt-EfTu) and SLIRP (stem-loop RNA binding protein), both involved in mitochondrial translation (supplemental Table 1).
FIGURE 8.
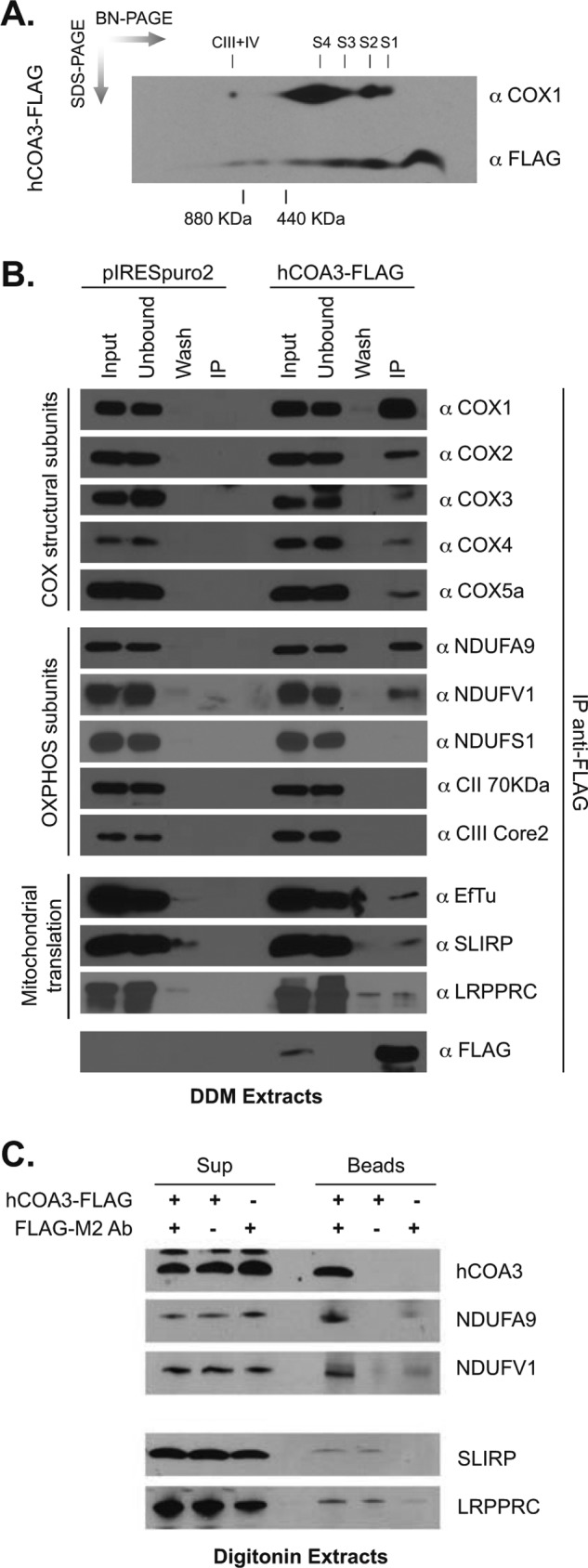
hCOA3 interacts with COX structural subunits and complex I. A, two-dimensional BN/SDS-PAGE of hCOA3-FLAG-overexpressing HeLa cells. S1, S2 and S3 indicate the three COX subassemblies, S4 indicates the COX monomer, and CIII+IV indicates supercomplex III+IV. B, immunoprecipitation using anti-FLAG M2 antibodies in control (pIRESpuro2) and hCOA3-FLAG-overexpressing (hCOA3-FLAG) mitochondria. Proteins were extracted with 1% DDM. 10% of the corresponding volume of input, unbound, and wash fractions was loaded on the gels. C, proteins were extracted from isolated mitochondria from HeLa cells and HeLa cells overexpressing hCOA3-FLAG with 1% digitonin. Extracts were incubated with protein A-Sepharose beads and anti-FLAG M2 antibodies where indicated. Beads and supernatant were recovered by centrifugation. Samples were analyzed by Western blotting.
To confirm these results, we performed additional immunoprecipitation experiments followed by immunoblotting (Fig. 8B). Anti-FLAG M2 antibodies efficiently immunoprecipitated COX1 in hCOA3-FLAG-overexpressing cells. COX structural subunits COX2, COX3, COX4, and COX5a are also coimmunoprecipitated with hCOA3-FLAG, although less efficiently than COX1. None of these COX subunits were detected in the eluate of control cells either using mass spectrometry or Western blot analysis. Western blot experiments also confirmed mt-EFTu coimmunoprecipitates with hCOA3-FLAG but showed a weak coimmunoprecipitation of SLIRP and hCOA3-FLAG (Fig. 8B). SLIRP forms a ribonucleoprotein complex with LRPPRC, a protein involved in the stabilization and polyadenylation of mitochondrial mRNAs (31–33). LRPPRC, however, was barely detected in the eluate of hCOA3-FLAG-overexpressing cells. To confirm these results, mitochondria from hCOA3-FLAG overexpressing cells were lysed using the milder detergent digitonin (1%). Neither SLIRP nor LRPPRC were specifically recovered after immunoprecipitation using anti-FLAG M2 antibodies (Fig. 8C), thus suggesting that these proteins do not form a stable complex. Our mass spectrometry analyses of hCOA3-FLAG eluates also detected several mitochondrial import receptors, namely the translocase of the inner membrane (TIM) proteins TIM22, TIM13, TIM23, and TIM23B (supplemental Table 1).
Focusing on respiratory chain complex subunits, when the extraction was performed with DDM, the CII 70 kDa (complex II) or CIII Core 2 (complex III) subunits were not immunoprecipitated using anti-FLAG M2 antibodies in hCOA3-FLAG-overexpressing cells, perhaps because the low abundance of the CIII-CIV supercomplex under these extraction conditions. Interestingly, however, mass spectrometry results revealed the presence of several complex I (CI) subunits in the eluate (supplemental Table 1) and immunoblot experiments confirmed the interaction of hCOA3 with NDUFA9 and NDUFV1, thus suggesting an early interaction of complex I subunits with COX assembly intermediates as reported recently (34). The same subunits were detected in hCOA3-FLAG immunoprecipitates from digitonin extracts (Fig. 8C). However, not all complex I subunits analyzed were recovered in the immunoprecipitate (see NDUFS1 in Fig. 8B), further supporting the concept that the interaction between complex I and complex IV subunits occurs before the complexes are fully assembled.
DISCUSSION
CCDC56 was recently identified by our group as a new protein essential for cytochrome c oxidase activity in Drosophila melanogaster, although its precise function remained to be elucidated (20). In this study, we have identified and characterized the function of the human homologue of CCDC56. Our results show human CCDC56 participates in the early steps of the assembly of the holoenzyme by promoting nascent COX1 stability and facilitating its assembly with other COX subunits, confirming the results by Szklarczyk and collaborators who proposed CCDC56 to be the human homologue of yeast Coa3 (18) and therefore the protein has been termed hCOA3.
hCOA3 is located on chromosome 17 (17q.21.31) and encodes an 11.7-kDa transmembrane protein of the inner mitochondrial membrane with a coiled-coil domain in its C terminus facing the intermembrane space. hCOA3-silenced cells present a severe defect in COX activity and a similar decrease in the levels of fully assembled complex. These data, together with the decrease in the steady-state levels of several COX subunits in silenced cells, pointed toward an involvement of hCOA3 in COX biogenesis. Silenced cells presented a mild but specific decrease in the amount of pulse-labeled COX1, suggesting its translation could be impaired in the absence of hCOA3. However, newly synthesized COX1 was rapidly degraded in interfered cells, indicating that the observed decrease in COX1 labeling could be due to increased degradation of the newly synthesized polypeptide rather than to defective translation. Synthesis of COX2 and COX3 and their stability over a 7-h chase were as in wild-type cells. Although these subunits have a longer half-life than COX1, they are known to be unstable when COX assembly is compromised (26, 35), and, accordingly, their steady-state levels in hCOA3 interfered cells were lowered markedly.
COX assembly is a linear process in which at least three subassemblies, S1, S2, and S3, can be detected, which probably represent the rate-limiting steps of the process (5). In this model, originally proposed by Nijtmans et al. (5), S1 is formed exclusively by COX1, and the addition of subunits COX4 and COX5a to S1 results in the progression to S2. We have observed that hCOA3-silenced cells presented accumulates of an assembly intermediate that contains COX1, COX4, and COX5a and corresponds to the S2 subassembly. The accumulation of this intermediate, however, is not very prominent probably due to remaining amounts of hCOA3 in silenced cells that allow COX assembly to proceed, resulting in the residual amounts of fully assembled holoenzyme observed in BN-PAGE experiments.
A similar subassembly has been reported to accumulate in cells from patients suffering from mitochondrial diseases due to mutations in several COX assembly genes. For example, SCO1 and SCO2 mutant fibroblasts, both of which are required for COX2 maturation, accumulate the S2 intermediate (29, 36). In the hCOA3-silenced cells, the accumulation of the S2 intermediate indicates hCOA3 is a COX assembly factor involved in the first steps of the assembly process, probably facilitating the incorporation of COX subunits to COX1-containing intermediates.
We have observed hCOA3-FLAG is forming several high molecular weight complexes, which could constitute COX subassemblies as well as CIII-CIV supercomplexes, as has already been observed for its yeast homologue Coa3 (37, 38). The physical interaction with COX subunits was demonstrated by immunoprecipitation studies followed by mass spectrometric and immunoblot analyses. Interestingly, these analyses also showed hCOA3 interacts with some complex I subunits such as NDUFA9 and NDUFV1 but not others such as NDUFS1. It has been recently reported that complex I assembly takes place in the context of supercomplexes or respirasomes (26), which could explain the interaction between COX and complex I assembly intermediates.
Because COX contains highly reactive heme A and copper prosthetic groups, the biogenesis of this enzyme must be tightly regulated to prevent the accumulation of pro-oxidant assembly intermediates. Coordination of COX1 synthesis and COX assembly has been extensively studied in S. cerevisiae. Mss51, the central protein of a negative feedback translational regulatory system in yeast, has a dual function acting as a translational activator of COX1 mRNA and as a COX1 chaperone during the first steps of COX biogenesis (39). During and after synthesis, COX1 is bound to Mss51 in a complex stabilized by two small COX-specific chaperones, Cox14 (40) and Coa3 (37, 38) and the mitochondrial Hsp70 chaperone (41). When COX1 proceeds in the assembly process, Mss51 is released from these complexes and is available to activate new rounds of Cox1 synthesis (41–43).
For many years, the translational regulatory system was believed to be restricted to yeast because homologues of Mss51, Cox14 and Coa3 had not been identified. However, the recent discovery that the three proteins are conserved in humans opened the possibility that such a regulatory system could have been also conserved. So far, the human homologues of Cox14 (C12orf62; Ref. 19) and Coa3 (CCDC56/hCOA3; this work) have been shown to be COX assembly factors important for COX1 stability and assembly, similar to their yeast counterparts. Our mass spectrometry studies failed to detect C12orf62 as an hCOA3 interactor, although this could perhaps be justified by the small size of the protein (∼15 kDa). Importantly, our immunoprecipitation results have suggested hCOA3 interacts with mt-EfTu. A similar interaction was previously described for C12orf62 (19). hCOA3 interaction with newly synthesized COX1 and proteins involved in translation, together with our pulse-labeling results, could reflect an early interaction of hCOA3 with COX1 during translation, required to stabilize the protein and prevent its degradation. mt-EfTu, although primarily acts as a mitochondrial translation elongation factor, can also function as a chaperone promoting the folding of newly synthesized polypeptides (44) and could therefore have a role in quality control of the nascent COX enzyme subunits and assembly intermediates through the binding of C12orf62 and hCOA3. It is conceivable that hCOA3 interacts early with newly synthesized COX1 polypeptide, perhaps during its elongation following its co-translational membrane insertion. Altogether, our results support that hCOA3 associates directly or indirectly with the COX1 mRNA translational machinery and interacts with the newly synthesized polypeptide providing stability and facilitating its assembly with other COX subunits to form the mature holoenzyme.
Finally, the identification of hCOA3 as a new COX assembly factor in humans is not only relevant from a biological point of view but also from a biomedical perspective. Most isolated COX deficiencies described to date in patients with mitochondrial diseases are caused by mutations in COX assembly factors, rather than mutations in structural subunits of the complex. Expanding the list of COX assembly factors responsible for human diseases, a recent report has identified mutations in c12orf62 in a patient with fatal neonatal lactic acidosis (19). Our data indicates that hCOA3 is a new candidate gene to screen in patients suffering from mitochondrial diseases associated with isolated COX deficiency. Increasing our knowledge of COX biogenesis and its regulation is expected to contribute to our understanding of mechanisms underlying these disorders.
Acknowledgments
We thank Rosana Hernandez-Sierra for excellent technical assistance. The proteomics analysis was carried out in the Centro de Biología Molecular “Severo Ochoa”, Protein Chemistry Facility, a member of ProteoRed network.
Addendum
These results are relevant because during the preparation of the revised version of this manuscript, it has been reported that TIM21, a component of the TIM23 transport machinery, is also present in respiratory chain subassemblies containing newly mitochondria-synthesized and imported respiratory chain subunits as well as some assembly factors such as CCDC56 (45). The authors have proposed that in this way, the transfer of newly imported proteins from the presequence translocase is coordinated with the assembly of respiratory chain complexes (45).
This work was supported by Instituto de Salud Carlos III Grant PI 10/0703 (to R. G.), Comunidad Autónoma de Madrid Grant S2010/BMD-2402 (to R. G.), Muscular Dystrophy Association Grants 158547 (to F. F). and 158547 (to A. B.), and National Institutes of Health Grant GM071775A (to A. B.).

This article contains supplemental Table 1.
- OXPHOS
- oxidative phosphorylation
- BN-PAGE
- blue native PAGE
- DDM
- dodecyl maltoside
- mt-EfTu
- mitochondrial translation elongation factor Tu
- TIM
- translocase of the inner membrane.
REFERENCES
- 1. Wallace D. C. (2005) A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 39, 359–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F., Schreier P. H., Smith A. J., Staden R., Young I. G. (1981) Sequence and organization of the human mitochondrial genome. Nature 290, 457–465 [DOI] [PubMed] [Google Scholar]
- 3. Tsukihara T., Aoyama H., Yamashita E., Tomizaki T., Yamaguchi H., Shinzawa-Itoh K., Nakashima R., Yaono R., Yoshikawa S. (1996) The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science 272, 1136–1144 [DOI] [PubMed] [Google Scholar]
- 4. Tsukihara T., Aoyama H., Yamashita E., Tomizaki T., Yamaguchi H., Shinzawa-Itoh K., Nakashima R., Yaono R., Yoshikawa S. (1995) Structures of metal sites of oxidized bovine heart cytochrome c oxidase at 2.8 A. Science 269, 1069–1074 [DOI] [PubMed] [Google Scholar]
- 5. Nijtmans L. G., Taanman J. W., Muijsers A. O., Speijer D., Van den Bogert C. (1998) Assembly of cytochrome c oxidase in cultured human cells. Eur. J. Biochem. 254, 389–394 [DOI] [PubMed] [Google Scholar]
- 6. Soto I. C., Fontanesi F., Liu J., Barrientos A. (2012) Biogenesis and assembly of eukaryotic cytochrome c oxidase catalytic core. Biochim. Biophys. Acta 1817, 883–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mick D. U., Fox T. D., Rehling P. (2011) Inventory control: cytochrome c oxidase assembly regulates mitochondrial translation. Nat. Rev. Mol. Cell Biol. 12, 14–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shoubridge E. A. (2001) Cytochrome c oxidase deficiency. Am. J. Med. Genet. 106, 46–52 [DOI] [PubMed] [Google Scholar]
- 9. Zee J. M., Glerum D. M. (2006) Defects in cytochrome oxidase assembly in humans: lessons from yeast. Biochem. Cell Biol. 84, 859–869 [DOI] [PubMed] [Google Scholar]
- 10. Barrientos A., Gouget K., Horn D., Soto I. C., Fontanesi F. (2009) Suppression mechanisms of COX assembly defects in yeast and human: insights into the COX assembly process. Biochim. Biophys. Acta 1793, 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Diaz F. (2010) Cytochrome c oxidase deficiency: patients and animal models. Biochim. Biophys. Acta 1802, 100–110 [DOI] [PubMed] [Google Scholar]
- 12. Comi G. P., Bordoni A., Salani S., Franceschina L., Sciacco M., Prelle A., Fortunato F., Zeviani M., Napoli L., Bresolin N., Moggio M., Ausenda C. D., Taanman J. W., Scarlato G. (1998) Cytochrome c oxidase subunit I microdeletion in a patient with motor neuron disease. Ann. Neurol. 43, 110–116 [DOI] [PubMed] [Google Scholar]
- 13. Clark K. M., Taylor R. W., Johnson M. A., Chinnery P. F., Chrzanowska-Lightowlers Z. M., Andrews R. M., Nelson I. P., Wood N. W., Lamont P. J., Hanna M. G., Lightowlers R. N., Turnbull D. M. (1999) An mtDNA mutation in the initiation codon of the cytochrome C oxidase subunit II gene results in lower levels of the protein and a mitochondrial encephalomyopathy. Am. J. Hum. Genet. 64, 1330–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Manfredi G., Schon E. A., Moraes C. T., Bonilla E., Berry G. T., Sladky J. T., DiMauro S. (1995) A new mutation associated with MELAS is located in a mitochondrial DNA polypeptide-coding gene. Neuromuscul. Disord. 5, 391–398 [DOI] [PubMed] [Google Scholar]
- 15. Shteyer E., Saada A., Shaag A., Al-Hijawi F. A., Kidess R., Revel-Vilk S., Elpeleg O. (2009) Exocrine pancreatic insufficiency, dyserythropoeitic anemia, and calvarial hyperostosis are caused by a mutation in the COX4I2 gene. Am. J. Hum. Genet. 84, 412–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Massa V., Fernandez-Vizarra E., Alshahwan S., Bakhsh E., Goffrini P., Ferrero I., Mereghetti P., D'Adamo P., Gasparini P., Zeviani M. (2008) Severe infantile encephalomyopathy caused by a mutation in COX6B1, a nucleus-encoded subunit of cytochrome c oxidase. Am. J. Hum. Genet. 82, 1281–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Indrieri A., van Rahden V. A., Tiranti V., Morleo M., Iaconis D., Tammaro R., D'Amato I., Conte I., Maystadt I., Demuth S., Zvulunov A., Kutsche K., Zeviani M., Franco B. (2012) Mutations in COX7B cause microphthalmia with linear skin lesions, an unconventional mitochondrial disease. Am. J. Hum. Genet. 91, 942–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Szklarczyk R., Wanschers B. F., Cuypers T. D., Esseling J. J., Riemersma M., van den Brand M. A., Gloerich J., Lasonder E., van den Heuvel L. P., Nijtmans L. G., Huynen M. A. (2012) Iterative orthology prediction uncovers new mitochondrial proteins and identifies C12orf62 as the human ortholog of COX14, a protein involved in the assembly of cytochrome c oxidase. Genome Biol. 13, R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Weraarpachai W., Sasarman F., Nishimura T., Antonicka H., Auré K., Rötig A., Lombès A., Shoubridge E. A. (2012) Mutations in C12orf62, a Factor that Couples COX I Synthesis with Cytochrome c Oxidase Assembly, Cause Fatal Neonatal Lactic Acidosis. Am. J. Hum. Genet. 90, 142–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Peralta S., Clemente P., Sánchez-Martínez A., Calleja M., Hernández-Sierra R., Matsushima Y., Adán C., Ugalde C., Fernández-Moreno M. Á., Kaguni L. S., Garesse R. (2012) Coiled-coil domain-containing protein 56 (CCDC56) is a novel mitochondrial protein essential for cytochrome c oxidase function. J. Biol. Chem. 287, 24174–24185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Peña P., Garesse R. (1993) The β subunit of the Drosophila melanogaster ATP synthase: cDNA cloning, amino acid analysis and identification of the protein in adult flies. Biochem. Biophys. Res. Commun. 195, 785–791 [DOI] [PubMed] [Google Scholar]
- 22. Calvaruso M. A., Smeitink J., Nijtmans L. (2008) Electrophoresis techniques to investigate defects in oxidative phosphorylation. Methods 46, 281–287 [DOI] [PubMed] [Google Scholar]
- 23. Chomyn A. (1996) In vivo labeling and analysis of human mitochondrial translation products. Methods Enzymol. 264, 197–211 [DOI] [PubMed] [Google Scholar]
- 24. Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 25. Schägger H., Pfeiffer K. (2000) Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 19, 1777–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Antonicka H., Leary S. C., Guercin G. H., Agar J. N., Horvath R., Kennaway N. G., Harding C. O., Jaksch M., Shoubridge E. A. (2003) Mutations in COX10 result in a defect in mitochondrial heme A biosynthesis and account for multiple, early-onset clinical phenotypes associated with isolated COX deficiency. Hum. Mol. Genet. 12, 2693–2702 [DOI] [PubMed] [Google Scholar]
- 27. Tam E. W., Feigenbaum A., Addis J. B., Blaser S., Mackay N., Al-Dosary M., Taylor R. W., Ackerley C., Cameron J. M., Robinson B. H. (2008) A novel mitochondrial DNA mutation in COX1 leads to strokes, seizures, and lactic acidosis. Neuropediatrics 39, 328–334 [DOI] [PubMed] [Google Scholar]
- 28. Weraarpachai W., Antonicka H., Sasarman F., Seeger J., Schrank B., Kolesar J. E., Lochmüller H., Chevrette M., Kaufman B. A., Horvath R., Shoubridge E. A. (2009) Mutation in TACO1, encoding a translational activator of COX I, results in cytochrome c oxidase deficiency and late-onset Leigh syndrome. Nat. Genet. 41, 833–837 [DOI] [PubMed] [Google Scholar]
- 29. Williams S. L., Valnot I., Rustin P., Taanman J. W. (2004) Cytochrome c oxidase subassemblies in fibroblast cultures from patients carrying mutations in COX10, SCO1, or SURF1. J. Biol. Chem. 279, 7462–7469 [DOI] [PubMed] [Google Scholar]
- 30. Stiburek L., Vesela K., Hansikova H., Hulkova H., Zeman J. (2009) Loss of function of Sco1 and its interaction with cytochrome c oxidase. Am. J. Physiol. Cell Physiol. 296, C1218–1226 [DOI] [PubMed] [Google Scholar]
- 31. Sasarman F., Brunel-Guitton C., Antonicka H., Wai T., Shoubridge E. A. (2010) LRPPRC and SLIRP interact in a ribonucleoprotein complex that regulates posttranscriptional gene expression in mitochondria. Mol. Biol. Cell 21, 1315–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bratic A., Wredenberg A., Grönke S., Stewart J. B., Mourier A., Ruzzenente B., Kukat C., Wibom R., Habermann B., Partridge L., Larsson N. G. (2011) The bicoid stability factor controls polyadenylation and expression of specific mitochondrial mRNAs in Drosophila melanogaster. PLoS Genet. 7, e1002324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ruzzenente B., Metodiev M. D., Wredenberg A., Bratic A., Park C. B., Cámara Y., Milenkovic D., Zickermann V., Wibom R., Hultenby K., Erdjument-Bromage H., Tempst P., Brandt U., Stewart J. B., Gustafsson C. M., Larsson N. G. (2012) LRPPRC is necessary for polyadenylation and coordination of translation of mitochondrial mRNAs. EMBO J. 31, 443–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moreno-Lastres D., Fontanesi F., García-Consuegra I., Martín M. A., Arenas J., Barrientos A., Ugalde C. (2012) Mitochondrial complex I plays an essential role in human respirasome assembly. Cell Metab. 15, 324–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nijtmans L. G., Artal Sanz M., Bucko M., Farhoud M. H., Feenstra M., Hakkaart G. A., Zeviani M., Grivell L. A. (2001) Shy1p occurs in a high molecular weight complex and is required for efficient assembly of cytochrome c oxidase in yeast. FEBS Lett. 498, 46–51 [DOI] [PubMed] [Google Scholar]
- 36. Stiburek L., Vesela K., Hansikova H., Pecina P., Tesarova M., Cerna L., Houstek J., Zeman J. (2005) Tissue-specific cytochrome c oxidase assembly defects due to mutations in SCO2 and SURF1. Biochem. J. 392, 625–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mick D. U., Vukotic M., Piechura H., Meyer H. E., Warscheid B., Deckers M., Rehling P. (2010) Coa3 and Cox14 are essential for negative feedback regulation of COX1 translation in mitochondria. J. Cell Biol. 191, 141–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fontanesi F., Clemente P., Barrientos A. (2011) Cox25 teams up with Mss51, Ssc1, and Cox14 to regulate mitochondrial cytochrome c oxidase subunit 1 expression and assembly in Saccharomyces cerevisiae. J. Biol. Chem. 286, 555–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Perez-Martinez X., Broadley S. A., Fox T. D. (2003) Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J. 22, 5951–5961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Barrientos A., Zambrano A., Tzagoloff A. (2004) Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J. 23, 3472–3482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fontanesi F., Soto I. C., Horn D., Barrientos A. (2010) Mss51 and Ssc1 facilitate translational regulation of cytochrome c oxidase biogenesis. Mol. Cell Biol. 30, 245–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Perez-Martinez X., Butler C. A., Shingu-Vazquez M., Fox T. D. (2009) Dual functions of Mss51 couple synthesis of Cox1 to assembly of cytochrome c oxidase in Saccharomyces cerevisiae mitochondria. Mol. Biol. Cell 20, 4371–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Soto I. C., Fontanesi F., Myers R. S., Hamel P., Barrientos A. (2012) A heme-sensing mechanism in the translational regulation of mitochondrial cytochrome c oxidase biogenesis. Cell Metab. 16, 801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Suzuki H., Ueda T., Taguchi H., Takeuchi N. (2007) Chaperone properties of mammalian mitochondrial translation elongation factor Tu. J. Biol. Chem. 282, 4076–4084 [DOI] [PubMed] [Google Scholar]
- 45. Mick D. U., Dennerlein S., Wiese H., Reinhold R., Pacheu-Grau D., Lorenzi I., Sasarman F., Weraarpachai W., Shoubridge E. A., Warscheid B., Rehling P. (2012) MITRAC Links Mitochondrial Protein Translocation to Respiratory-Chain Assembly and Translational Regulation. Cell 151, 1528–1541 [DOI] [PubMed] [Google Scholar]