Background: The neuroblastoma oncogene MYCN and the PRC2 members EZH2 and SUZ12 are regulators of gene transcription.
Results: MYCN and PRC2 form a repressive complex on the promoter of the tumor suppressor gene CLU.
Conclusion: PRC2 members are recruited by MYCN to repress gene expression and induce tumorigenesis.
Significance: Reactivation of MYCN-PRC2-repressed genes by epigenetic drugs could be of clinical value in neuroblastoma.
Keywords: Epigenetics, Myc, Neuroblastoma, Polycomb, Transcription, Transcription Target Genes
Abstract
CLU (clusterin) is a tumor suppressor gene that we have previously shown to be negatively modulated by the MYCN proto-oncogene, but the mechanism of repression was unclear. Here, we show that MYCN inhibits the expression of CLU by direct interaction with the non-canonical E box sequence CACGCG in the 5′-flanking region. Binding of MYCN to the CLU gene induces bivalent epigenetic marks and recruitment of repressive proteins such as histone deacetylases and Polycomb members. MYCN physically binds in vitro and in vivo to EZH2, a component of the Polycomb repressive complex 2, required to repress CLU. Notably, EZH2 interacts with the Myc box domain 3, a segment of MYC known to be essential for its transforming effects. The expression of CLU can be restored in MYCN-amplified cells by epigenetic drugs with therapeutic results. Importantly, the anticancer effects of the drugs are ablated if CLU expression is blunted by RNA interference. Our study implies that MYC tumorigenesis can be effectively antagonized by epigenetic drugs that interfere with the recruitment of chromatin modifiers at repressive E boxes of tumor suppressor genes such as CLU.
Introduction
Epigenetic modification of the DNA or associated proteins is thought to be involved in the control of gene expression in normal and pathological contexts. In cancer, many genes appear to be deregulated by aberrant rearrangements of the chromatin, which frequently occurs during neoplastic transformation. For example, methylation of the DNA is generally increased in cancer cells, leading to the silencing of tumor suppressor genes (1). Deacetylation of the histones, proteins tightly associated to the DNA, is also an important mechanism by which cancer cells reduce or silence the expression of genes that restrict cell proliferation or increase cell death (2, 3)
MYCN is a member of the MYC family of transcription factors frequently amplified in neuroblastoma, a childhood cancer originating from the peripheral nervous system (4). Amplification of MYCN is a poor prognostic factor in cancer patients, and transgenic expression of MYCN in the neuroectoderm causes neuroblastoma with high penetrance in mice (4, 5). Conversely, ablating the expression of MYCN in human neuroblastoma cell lines causes inhibition of their proliferation and stimulates apoptosis (6–8). Thus, in neuroblastoma patients carrying amplification of the gene, MYCN is likely to be required and sufficient to cause a fatal form of the disease.
It has been hypothesized that one of the mechanisms by which increased expression of MYCN drives tumorigenesis is by increasing the expression of cell cycle-related genes such as ornitine decarboxylase via direct promoter interaction and transactivation (9). More recently, it was observed that MYC proteins can also suppress gene expression indirectly, by interacting with sequence-specific transcription factors such as SP1 and MIZ1 and bringing transcriptional co-repressors near the transcription initiation site of the growth suppressor gene p21 (10–12). Using this mechanism, MYCN could induce transcriptional silencing of genes involved in negative regulation of cell proliferation and transformation. A further mechanism by which MYCN could mediate its oncogenic effects is by modulating microRNAs. We and others have shown that MYCN can induce the expression of transcripts of the 17–92 cluster of microRNAs. Among the targets of the 17–92 microRNA cluster, p21, BIM, and CLU appear to be critically involved in MYCN tumorigenesis (13–15). A sensible hypothesis is that the aberrant expression of MYCN could modify gene expression both via direct and indirect mechanisms.
In this study, we focus our attention on one of the MYCN-regulated genes, the putative tumor suppressor gene CLU. In a previous work, we showed that CLU is a suppressor of MYCN tumorigenesis in vivo, and silencing of CLU is required by MYCN to exert its malignant behavior (13). We noticed the presence of potential MYC-binding sites (also known as E box) in the human CLU promoter, so we wondered whether MYCN could modulate the expression of CLU via a direct mechanism.
EXPERIMENTAL PROCEDURES
Cell Culture
The human neuroblastoma cell lines SH-SY5Y, IMR-32, and HEK-293 cells were obtained from the American Type Culture Collection (ATCC). The human neuroblastoma cell lines LAN-1, Kelly, and Tet-Off 21N (stably transfected with a tetracycline-controlled transactivator protein and an expression vector encoding MYCN cDNA, which is switched off in the presence of Tet)4 were described previously (16–18). Primary neuroblastoma cells were obtained by mechanically disaggregating a tumor resection from a patient with a MYCN-amplified, relapsing tumor. Written consent for the utilization of the tumor material was obtained from the family. All cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% fetal calf serum, 2 mm l-glutamine and 50 μg/ml gentamicin, with the exception of Kelly, which were cultured in RPMI 1640 (Invitrogen) supplemented with 10% fetal calf serum, 1% l-glutamine, and 1% penicillin-streptomycin. For growth assays, cells were plated at a density of 1.5 × 105 cells/well in six-well plates. Cells were counted with a hemocytometer, and cell death was scored by trypan blue dye staining. For epigenetic drug treatments, cells were plated at a density of 5 × 104 cells/well in six-well plates. After 24 h, cells were exposed to trichostatin A (1 mm), valproic acid (1.5 mm), or 5-aza-2′-deoxycytidine (10 mm) for 24 or 48 h.
Plasmid Construction and Transfections
To generate the pGL2 clusterin WT reporter vector, the CLU human promoter region containing the putative MYCN binding site (E box) was first amplified by PCR from genomic DNA (the primers used were as follows: 5′-cgactggtaccctgtgtgtgctctcttctccagca-3′ (forward) and 5′-ttcgatcgaattggggctggctgcaaacctg-3′ (reverse)) and ligated in the TOPO vector using the TOPO TA cloning kit (Invitrogen). The CLU promoter segment was cut with KpnI and HindIII and subcloned into the luciferase pGL2 promoter vector (Promega Biosciences, Promega Corp., San Luis Oispo, CA). The pGL2 clusterin MUT reporter vector was obtained by mutation of the E box sequence located at −482 from the transcription start site (wt, cacgcg; mutant, TCTGCT) by site-directed mutagenesis (QuikChange multisite-directed mutagenesis kit, Stratagene). The MYCN expression vector lacking the MYC box domain 3 (MB3) domain (amino acids 187–241) was obtained by amplifying the plasmid pCMV14-MYCN-3Xflag (19) by PCR using the following primers: forward, ACCAGCGGCGGCGACCACAA; reverse, GCACTCGGCGGCCGGGTGGG. The PCR product was ligated and verified by sequencing. The pGL2 clusterin WT or the pGL2 clusterin MUT reporter vectors were transiently transfected in the presence or absence of CMV-MYCN into SH-SY5Y cells using Lipofectamine 2000 reagent (Invitrogen). Luciferease assays were carried out 36 h post transfections. PcDNA 3.1(+)/His- MYCN, pSUPER.gfp+neo. CLU and pSUPER.gfp+neo.Scr have been described previously (13). SH-SY5Y cells were transfected with pcDNA 3.1(+)/His-MYCN, and a mix population was obtained after selection in medium containing 0.8 mg/ml G418 (Invitrogen) for 3 weeks. IMR-32 and LAN-1 cells were transfected with pSUPER.gfp+neo.CLU or pSUPER.gfp+neo.Scr plasmids using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. Cells were maintained in selection medium containing 1.0 mg/ml G418 for 3 weeks to select individual clones with or without expression of CLU.
In Vitro Invasion and Cell Viability Assays
In vitro invasion assay was carried out as reported previously (13). Briefly 2.5 × 104 neuroblastoma cells were resuspended in serum-free medium and seeded on the top of the invasion assay chamber following manufacturer's instructions (BD Biosciences). Medium containing 10% fetal calf serum served as chemo-attractant in the bottom chamber.
Electrophoretic Mobility Shift Assay
Nuclei were prepared from SHSY-5Y cells stably transfected with empty or MYCN expressing CMV vector. Cells were harvested in hypotonic buffer (10 nm Tris-HCl, pH 7.8, 5 mm KCl, 2 mm MgCl2, 1 mm DTT, protease inhibitors) and subsequently lysed in the hypotonic buffer supplemented with 0.25% Nonidet P-40 for 5 min on ice. Nuclei were pelleted, washed twice with hypotonic buffer, and suspended in 2 volumes of hypotonic buffer containing 0.3 m NaCl. The suspension was kept on ice for 30 min and centrifuged, and the supernatant was used directly for EMSA. Double-stranded DNA oligomers containing the wild type (5′-GGGGCTCCCAGATGGGCACGCGAGTTCAGGCTCTTCC-3′) or mutant (5′-GGGGCTCCCAGATGGGCTTAGGAGTTCAGGCTCTTCC-3′) putative MYCN-binding site (underlined) were labeled with [γ-32P]dATP and used as probes for gel shift analysis. Nuclear cell extract (5 μg) and 1 ng of probes were mixed for 30 min at room temperature in 25 μl of binding buffer (10 mm Tris-HCl, pH 8.0, 150 mm KCl, 0.5 mm EDTA, 0.1% Triton X-100, 4% Ficoll 400 (v/v), 1 mm DTT, 1.5 μg poly(dI-dC)). Shifted protein complexes were resolved by electrophoresis in a 5% native polyacrylamide gel.
Standard ChIP and Dual Chromatin Immunoprecipitation Assays
Standard ChIP and dual ChIP assays were performed as described (19). Antibodies used in this study were as follows: IgG (sc-2027, Santa Cruz Biotechnology, Santa Cruz, CA), GAL4 (IgG2A negative control) (Santa Cruz Biotechnology), MYCN (sc53993, Santa Cruz Biotechnology), Max (sc197, Santa Cruz Biotechnology), HDAC1 (Ab7028, Abcam), HDAC2 (05-814, Upstate), Bmi1 (Ab 14389), EZH2 (5246, Cell Signaling), SUZ12 (Ab12073, Abcam), AcH3 (07-599, Upstate, H3K4-2Me (07-030, Upstate), H3K9-3Me (07-442, Upstate), H3K27-3Me (07-449, Upstate), H4K20-3Me (07-463, Upstate). The primers used were as follows: BS2 (E box), 5′-ttgtgtcttggactgggaca-3′ (forward) and 5′-gaaagcaaggggagctttct-3′ (reverse); −1000, 5′-tccatagtcctgatcctgaactg-3′ (forward) and 5′-tttggagccagggatgtttaag-3 (reverse); +1000, 5′-gtggagcattgggcacaactg-3′ (forward) and 5′-ccagaggcaaaggttagcactg-3′ (reverse); +3000, 5′-ccttgttaatgtgctacttgagtgtc-3′ (forward) and 5′-tcacagaaccaacaggacgatg-3′ (reverse).
GST Pulldown Assay
The different MYCN segments were cloned into the pGEX-3X plasmid, in frame with GST. Recombinant proteins were expressed in BL21 Escherichia coli cells, purified, and immobilized onto glutathione-agarose beads (Sigma-Aldrich). GST beads were then incubated with in vitro-translated EZH2 protein (TNT® Quick Coupled Transcription/Translation System, Promega, Madison, WI) pretreated with DNase (GE Healthcare, Waukesha, WI). Purified complexes were separated on SDS-PAGE and analyzed by Western blotting using anti-EZH2 antibodies (5246, Cell Signaling).
Co-immunoprecipitations
For co-immunoprecipitation experiments, we used IMR32 because this cell line showed the strongest expression of Polycomb repressive complex 2 members compared with other MYCN-amplified neuroblastoma cell lines. Cells were washed twice in PBS and lysed in 3.2 ml of IP buffer (20 ml of HEPES, 175 mm NaCl, 10% glycerol, 0,5% Nonidet P-40, 1 mm EDTA, 1 mm DTT, protease, and phosphatase inhibitors) and kept on ice for 30 min. Cell debris were eliminated by centrifugation, and the supernatant was collected and incubated with 10 mg of the following antibodies: SUZ 12 (ab 12073, Abcam), EZH2 (clone AC22, Upstate, Millipore), MYCN (sc-53993, Santa Cruz Biotechnology). As control antibodies, we used Fyn (sc-434, Santa Cruz Biotechnology) and TRAF2 (N-19, Santa Cruz Biotechnology). After six washes in 1× PBS, immunoprecipitated samples were run onto a SDS-polyacrylamide gel and transferred to nitrocellulose. The nitrocellulose filter was blocked in PBS containing 5% dry milk and 0.1% Nonidet P-40 and incubated with the different antibodies overnight at 4 °C. The filters were then incubated with the appropriate secondary antibody labeled with horseradish peroxidase, followed by incubation with a chemiluminescent substrate (ECL, Amersham Biosciences). Proteins were detected by exposing the nitrocellulose filter to an autoradiographic film. All experiments were repeated for at least three times in duplicates. All data for statistical analysis were calculated as means ± S.E. Differences among groups were determined with an unpaired t test. A probability value of p = 0.05 or less was considered significant.
Analysis of Acetylated Histones
LAN-1 cells were treated with 300 nm TSA, 3 mm valproic acid or a combination of both drugs for 24 and 48 h. After treatment, nuclei were obtained by resuspending cells in low salt buffer (10 mm HEPES, pH 8, 50 mm NaCl, 1 mm EDTA, 1 mm DTT, 1 mm sodium pyrophosphate, 1 mm sodium orthovanadate, 1 mm sodium flourophosphate, 1 mm PMSF, and protease inhibitor mixtures). After centrifugation, nuclei were lysed in high salt buffer (10 mm HEPES, pH 8, 450 mm NaCl, 1 mm EDTA, 10% glycerol, 1 mm sodium pyrophosphate, 1 mm sodium orthovanadate, 1 mm sodium fluorophosphate, 1 mm PMSF, and protease inhibitor mixture). 20 μg of nuclear extracts were loaded onto a SDS-PAGE and analyzed by Western blot analysis. AcH3 (07-599) and H1 (05-547) antibodies were from Millipore.
RESULTS
MYCN Binds to the 5′-Flanking Sequence of the CLU Gene in Vitro and in Vivo
Scanning of the human CLU gene revealed two potential E boxes (named BS1 and BS2) in close proximity of the transcription start site (Fig. 1A). We firstly investigated whether MYCN could interact with the putative E boxes in vitro. We quickly ruled out BS1 as a genuine E box sequence (data not shown), but a probe containing BS2 could be shifted by cell lysates containing exogenously expressed MYCN protein. The interaction was specific because it was ablated by point mutations in BS2 (Fig. 1B). We next assessed whether MYCN was bound to the CLU promoter in neuroblastoma cells in vivo. Chromatin IP analysis was carried out using the MYCN amplified LAN-1 and Kelly neuroblastoma cell lines or non-amplified SH-SY5Y cells. This assay demonstrated that endogenously expressed MYCN binds to the CLU promoter in neuroblastoma cells in vivo (Fig. 1C). As a control, we used primers that amplify a region of the CLU promoter that does not contain the E box. As expected, no amplification was observed using these primers after immunoprecipitation with the MYCN antibody (supplemental Fig. S1).
FIGURE 1.
MYCN binds to a non-canonical, repressive E box in the 5′-flanking region of CLU. A, the promoter region of the human CLU gene proximal to the transcription start site (indicated by the arrow) is shown. Two E box-like sequences and putative binding sequences for MYCN are indicated by BS1 and BS2. B, gel shift assay. Cell lysates from SH-SY5Y cells stably transfected with empty CMV or MYCN vectors were mixed with radioactive probes encompassing the wild type (WT) or mutated (Mut) binding sequence BS2 as indicated. A specific complex containing MYCN is indicated by the bracket, and free indicates the probe without lysates. Note that naïve SH-SY5Y cells do express a small amount of endogenous MYCN, explaining the faint band shift observed in empty vector (CMV)-transfected cells. C, chromatin immunoprecipitation assay. The SH-SY5Y, LAN-1 and Kelly neuroblastoma cell lines were subjected to chromatin IP assays with MYCN or GAL4 (control) antibodies. A specific product (indicated by the arrow) is amplified with 10% of the input chromatin after immunoprecipitation with the MYCN, but not GAL4, antibody. D, a segment of the CLU promoter containing the putative BS2 binding site characterized in the gel shift analysis was subcloned upstream of the luciferase gene, in the wild type or mutant conformations, and co-transfected into non-MYCN amplified SH-SY5Y cells with or without the CMV-MYCN expression vector, as indicated. The activity of the promoters in the absence of MYCN was set to 100% and is indicated in the Y axis. S.D. are indicated by the error bars. Statistical significance (indicated by the asterisk) was assessed using the Student's t test (p = 0.0413; n = 4).
Bioinformatic Assessment of the BS2 E Box Sequence in Human Neuroblastomas
To verify whether BS2 could be found in other MYCN-regulated genes, we carried out a bioinformatic analysis asking the following question: is the sequence CACGCG enriched in the flanking region of genes whose expression is regulated with, and possibly by, MYCN in neuroblastoma tumors? We observed a significant and robust enrichment of the non-canonical E box BS2 in MYCN co-regulated genes in a large neuroblastoma data set (neuroblastoma prognosis database -Oberthuer Lab-) in the Oncogenomics repository (http://pob.abcc.ncifcrf.gov/cgi-bin/JK). The enrichment was equal or superior to that observed with the canonical E box sequence CACGTG (Table 1). Interestingly, only the non-canonical E box showed a slight but statistically significant enrichment in the promoter of genes whose expression is anti-regulated with that of MYCN (i.e. genes that have an expression pattern similar to CLU) (Table 2). Overall, the bioinformatic analysis indicates that the non-canonical E box BS2 is, similar to the canonical, involved in MYCN transcriptional activation of target genes. However, BS2 but not the canonical E box motif could also mediate MYCN repression of a set of genes in human neuroblastomas (the complete list of genes correlated and anticorrelated with BS2 is shown in supplemental Table S1).
TABLE 1.
E box motif enrichment in the MYCN co-regulated set of the Obertheuer data set
The canonical and BS2 sequence were scored in at least one (>0), 10% (>0.1), or 50% (>0.5) of the transcripts originating from MYCN co-regulated genes. The noncanonical sequence was scored in both orientations (indicated by +/−), whereas the canonical sequence was only scored in one orientation because it is a palindrome. Significant p values are shown in boldface type.
| p value | Enrichment | |
|---|---|---|
| CACGTG | ||
| >0 | 8.55 × 10−3 | 1.24 |
| >0.1 | 1.31 × 10−3 | 1.32 |
| >0.5 | 1.13 × 10−4 | 1.65 |
| CACGCG (+/−) | ||
| >0 | 6.98 × 10−5 | 1.35 |
| >0.1 | 1.55 × 10−5 | 1.40 |
| >0.5 | 1.07 × 10−7 | 1.74 |
TABLE 2.
E box motif enrichment in the MYCN anti-correlated set of the Oberthuer data set
Significant p values are shown in boldface type.
| p value | Enrichment | |
|---|---|---|
| CACGTG | ||
| >0 | 0.210 | 1.15 |
| >0.1 | 0.854 | 0.96 |
| >0.5 | 0.629 | 1.09 |
| CACGCG (+/−) | ||
| >0 | 0.001 | 1.38 |
| >0.1 | 0.003 | 1.36 |
| >0.5 | 0.070 | 1.32 |
The Non-canonical BS2 E Box Sequence Mediates Negative Regulation of the CLU Promoter by MYCN
To understand whether binding of MYCN to the CLU promoter was functionally relevant, we subcloned a section of the CLU promoter, containing BS2, in front of the luciferase gene. We also generated a luciferase vector with the CLU promoter containing point mutations that ablate the binding of MYCN (as shown in the gel shift assay in Fig. 1B). We then transfected the different constructs into SH-SY5Y neuroblastoma cells with or without a plasmid expressing MYCN. We observed that luciferase activity decreased in a statistically significant manner when the CLU promoter was co-transfected with the MYCN vector compared with the control vector. Importantly, mutation of the E box completely abrogated the repressive effect of MYCN, suggesting that binding to the E box is essential for negative regulation of the CLU promoter (Fig. 1D).
MYCN Causes a Bivalent Modification of the Chromatin Surrounding the E Box Binding Site in the CLU Promoter and Recruitment of Histone Deacetylases (HDACs) and Polycomb Proteins
To understand how mechanistically MYCN could cause transcriptional repression of the CLU promoter, we performed chromatin IP assays and quantitative PCR to quantitatively assess the binding of chromatin proteins, scanning ∼5 kb of the CLU promoter surrounding the BS2 MYCN-binding site. For these experiments, we used a neuroblastoma cell line, SHEP 21N, which contains an expression vector (MYCN Tet-Off) that allows the conditional expression of MYCN. As expected, the MYCN signal was enriched in correspondence of the E box BS2 in the absence of tetracycline. MAX was also found bound to the E box, but its presence was not dependent on MYCN expression. Notably, although binding of MYCN was associated with active chromatin marks, such as acetylated histone H3 and dimethylated lysine 4 on histone H3, negative marks such as trimethylated lysine 9 and 27 of histone H3 were observed immediately downstream of the E box. In agreement with this observation, several chromatin remodeling factors associated with transcriptional repression such as HDAC 1/2 and Polycomb complex factors (BMI1, EZH2, and SUZ12) were detected around the BS2 E box or 1000 bp downstream of the E box sequence in the presence of MYCN, respectively (Fig. 2). This “bivalent” configuration is typical of repressed, developmentally regulated genes, which are poised to be activated by physiological stimuli. Interestingly, in the absence of MYCN we observed a very strong increase of histone H3 pan-acetylation and dimethylated lysine 4, suggestive of a rearrangement of the chromatin compatible with transcriptional activation. Indeed, repressive proteins and negative epigenetic marks, such as the Polycomb factors and trimethylated lysine 9 and 27 on histone H3, were reduced or absent (Fig. 2).
FIGURE 2.
Binding of MYCN on the CLU promoter is associated with recruitment of chromatin remodeling proteins and bivalent epigenetic marks. Chromatin immunoprecipitation analysis of the CLU promoter region, surrounding the BS2 E box, was carried out in the presence or absence of MYCN, as indicated, using a Tet-Off system. Chromatin was immunoprecipitated using antibodies against the indicated proteins. The number of base pairs upstream (−) or downstream (+) the E box are indicated in the X axis. In the Y axis is indicated the fold of enrichment over the control IgG antibody. Error bars indicate S.D. ***, p < 0.001; **, p < 0.01 (n = 3).
MYCN Interacts with the PRC2 Complex via EZH2
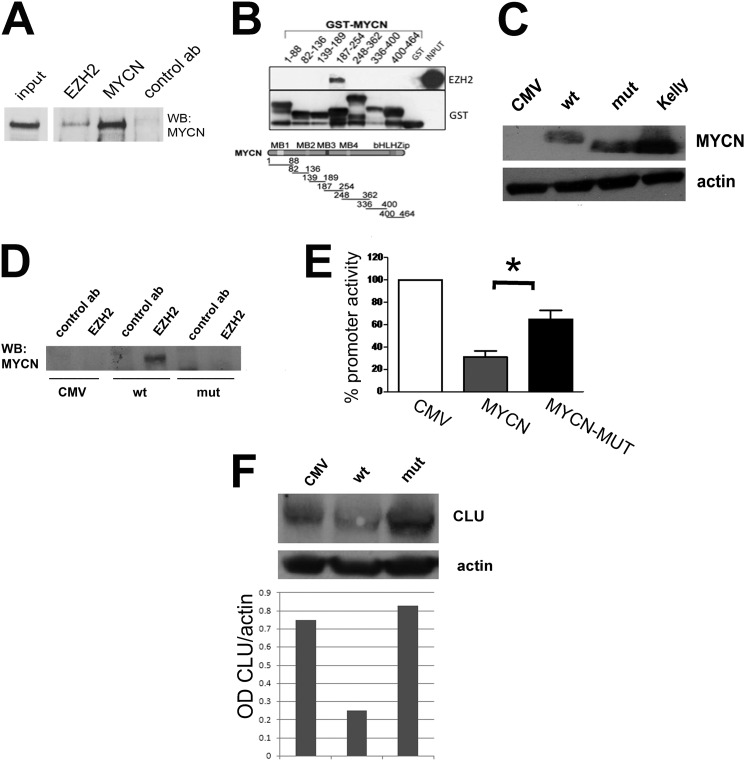
Enrichment of polycomb members in the proximity of the BS2 binding site prompted us to investigate whether MYCN could physically recruit inhibitory Polycomb proteins at the CLU promoter. Co-immunoprecipitation experiments indicated that endogenous EZH2 is physically associated with MYCN in MYCN-amplified IMR32 cells (Fig. 3A and supplemental Fig. S2). The histone methyltransferase EZH2 and SUZ12 are part of a complex called Polycomb repressive complex 2 that methylates lysine 27 on histone H3 (21). As expected, EZH2 and SUZ12 are tightly associated in IMR32 cells (supplemental Fig. S2), suggesting that MYCN could bind to the EZH2-SUZ12 complex. MYCN interacts with EZH2 via a short segment in the central part of the protein containing the MB3 (Fig. 3B). Notably, a mutant version of MYCN lacking the MB3 domain and incapable to bind to EZH2 in vitro and in vivo (Fig. 3, C and D, and supplemental Fig. S3) was significantly less efficient as a transcriptional repressor than the wild type MYCN protein in luciferase assays, although it bound to the non-canonical E box CACGCG in vitro with an efficiency similar to that of the wild type protein (Fig. 3E and supplemental Fig. S4). Furthermore, the MYCN mutant was unable to inhibit endogenous CLU expression in SH-SY5Y neuroblastoma cells (Fig. 3F and supplemental Fig. S5). The direct role of EZH2 in repressing CLU transcription in neuroblastoma cells has been recently demonstrated in a study from Thiele and co-workers (22). Overall, these results indicate that MYCN recruits EZH2 to the CLU promoter to induce transcriptional silencing.
FIGURE 3.
MYCN physically interacts with Polycomb repressive complex 2 via EZH2. A, co-immunoprecipitation assay showing the interaction of endogenous EZH2 and MYCN in IMR-32 cells. Cell lysates were immunoprecipitated with the antibodies indicated on the top of the gel and subjected to Western blot analysis (WB) with the MYCN antibody. B, immobilized GST-MYCN polypeptides were incubated with equal amounts of the in vitro translated EZH2 protein, separated by SDS-PAGE, and probed with an anti-EZH2 antibody. The diagram in the bottom shows full-length MYCN protein in which the functional domains are indicated by different shades of gray. The MYCN segments cloned in the GST expression vector are in black, and numbers indicate amino acid positions. C, Western blot analysis showing equal expression of the wild type and mutant (mut; lacking amino acids 187–241) MYCN proteins in SH-SH5Y cells stably transfected with the different MYCN constructs. The MYCN-amplified cell line Kelly is used as a positive control for MYCN expression. D, co-immunoprecipitation analysis in SH-SY5Y cells transfected with the wild type or mutant MYCN constructs showing interaction of EZH2 wild type, but not mutant, MYCN protein. E, luciferase assay of SH-SY5Y cells expressing the wild type or mutant MYCN constructs, transiently transfected with the CLU promoter. Error bars indicate S.D., and the asterisk indicates statistical significance (Student's t test; p = 0.01, n = 4). F, Western blot analysis showing expression of CLU in SH-SY5Y cells stably transfected with the wild type or mutant MYCN plasmids. The bottom panel depicts the densitometric analysis of MYCN expression relative to actin.
Therapeutic Potential of CLU Reactivation in MYCN-amplified Neuroblastomas
In the next set of experiments, we assessed whether CLU reactivation could have clinical relevance in the context of MYCN-amplified tumors. HDAC inhibitors are compounds that inhibit, in addition to their specific HDAC targets, Polycomb group proteins (23). Exposure of the MYCN-amplified cell lines LAN-1 and Kelly to valproic acid and trichostatin A but not the demethylating drug azacytidine strongly induced CLU expression after 24 h (Fig. 4A). Notably, treatment of neuroblastoma cells with trichostatin A or valproic acid alone or in combinations caused a marked increase of pan-acetylated histone H3 of LAN-1 cells, strengthening the hypothesis that the reversal of epigenetic silencing at the CLU locus is responsible for the reactivation of gene expression (supplemental Fig. S6). Next, we investigated whether the epigenetic drugs caused modifications of the biological behavior of the neuroblastoma cell lines. We first evaluated the proliferation rates of LAN-1 and Kelly in the presence or absence of drugs. Trichostatin A and valproic acid strongly inhibited the proliferation of neuroblastoma cells (Fig. 4B). The inhibitory effect was caused by apoptosis, indicated by the presence of a large increase of cells with fragmented sub-G1 DNA, and reduced cell cycle activity, demonstrated by decreased number of cells in the S phase (Fig. 4C).
FIGURE 4.
HDAC inhibitors reactivate CLU expression in MYCN amplified cell lines. A, MYCN amplified cell lines Kelly and LAN-1 were exposed to azacytidine (A), trichostatin A (T), valproic acid (V), or their combinations as indicated. C indicates control untreated cells. Expression of CLU or actin used as a reference control was assessed by Western blot analysis. B, proliferation curves of Kelly and LAN-1 cells in the presence or absence of TSA or valproic acid (VPA). The error bars indicate S.D. of values from triplicate wells. The experiment was repeated twice with similar results; a representative experiment is shown. C, cell cycle profiles of LAN-1 cells were evaluated after 48 h in the presence or absence of trichostatin A or valproic acid. The experiment was repeated twice with similar results; a representative experiment is shown. PI, phosphatidylinositol.
Short term cultures of cells dissected from fresh tumors deriving from a MYCN transgenic mouse (TH-MYCN) or a child with metastatic neuroblastoma with amplification of MYCN were cultured in the presence or absence of TSA. We observed that, in the presence of the drug, CLU expression was reactivated in parallel with changes in cell cycle profiles and induction of sub-G1 fragmented DNA, diagnostic of apoptosis, validating the effect of HDAC inhibitors in primary tumor cells (Fig. 5). The induction of apoptosis in human neuroblastoma cells was confirmed by annexin V staining (supplemental Fig. S7).
FIGURE 5.
Trichostatin A induces apoptosis and CLU reactivation in primary neuroblastoma cells. Mouse neuroblastoma cells from MYCN transgenic mice (A) or a human neuroblastoma metastasis (B) were cultured in vitro in the presence or absence of TSA for 24 h. Cells were stained with propidium iodide, and cell cycle profiles were assessed by FACS analysis. Western blot analysis with a CLU antibody confirmed that CLU is reactivated after incubation of mouse (C) and human (D) neuroblastoma cells with TSA.
Valproic acid is an orally available drug currently used in the clinic as an antiepileptic; thus, it was of interest to investigate whether it showed anticancer activity in xenotransplantation experiments. We injected the flanks of immunodeficient mice with LAN-1 cells and randomize the mice in valproic acid and control groups. There was a significant suppression of tumor growth in mice consuming valproic acid, suggesting that the drug could potentially be useful for the treatment of this childhood neoplasia (supplemental Fig. S8).
CLU Reactivation Is Key for the Anti-invasive effects of Epigenetic Drugs on MYCN-expressing Tumor Cells
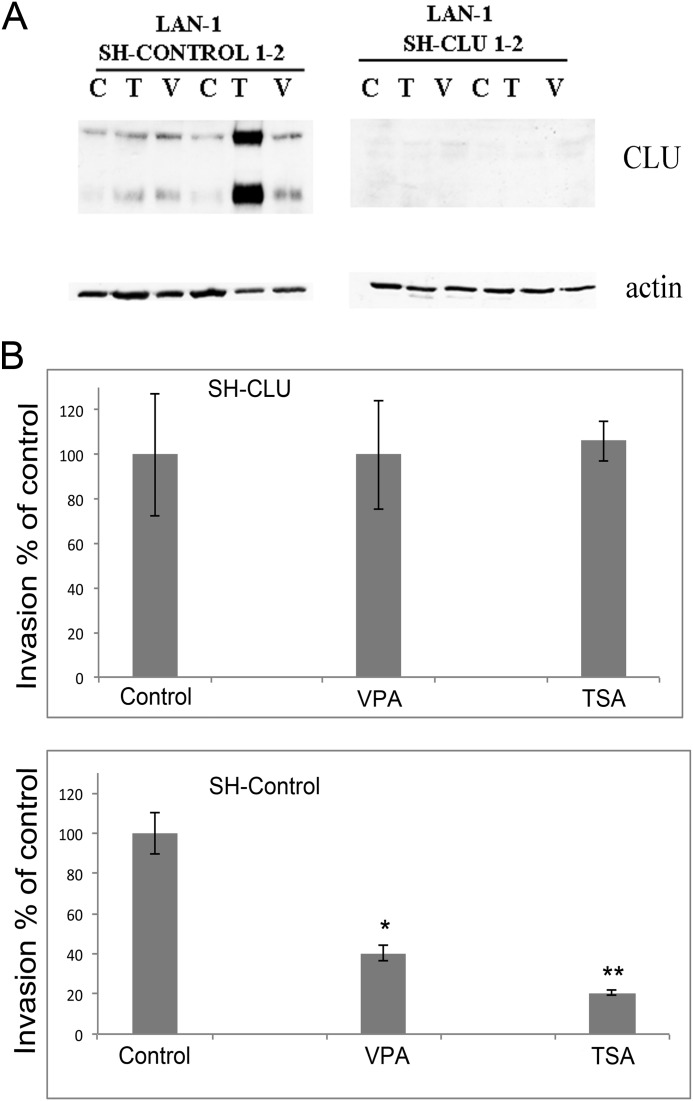
To understand whether reactivation of CLU is required to mediate the biological effects of trichostatin A and valproic acid, we transfected the LAN-1 (MYCN-amplified) neuroblastoma cell line with a CLU-specific shRNA construct validated in a previous study (13). After selection with G418, we chose two clones that showed a reduced expression of CLU in the presence or absence of epigenetic drugs (Fig. 6A). We showed previously that CLU inhibits the metastatic potential of neuroblastoma cells (13, 24), we thus investigated whether the epigenetic drugs acted as inhibitors of neuroblastoma metastasis using an in vitro invasion assay. Invasion of LAN-1 cells was significantly inhibited in the presence of the epigenetic drugs. This effect was not a consequence of cell death because neuroblastoma cells were exposed to the drugs for 24 h, and only viable cells were seeded for the assay, which was carried out in the absence of the drugs. Importantly, ablation of CLU by the shRNA construct completely abolished the antimetastatic effects of the epigenetic drugs (Fig. 6B and supplemental Fig. S9).
FIGURE 6.
CLU mediates the antimetastatic activity of HDAC inhibitors in in vitro invasion assays. A, Western blot assays showing the expression of CLU or actin used as a loading control in LAN-1 cell lines transfected with a shRNA control (the shRNA control 1 cell line is shown in lanes 1–3, and the shRNA control 2 in lanes 4–6) or an shRNA vector targeting CLU (the shRNA CLU 1 and 2 cell lines are shown in lanes 1–3 and 4–6, respectively). T, trichostatin A; V, valproic acid; C, vehicle control. B, LAN-1 cells with or without CLU expression (clone 1) were plated in the top wells of in vitro invasion chambers after 24 h of exposure to the HDAC inhibitors TSA and valproic acid (VPA). Only live cells (as assessed by trypan blue dye exclusion assay) were plated in the invasion chamber. The number of vehicle-treated cells seeded in the top well divided by the number of cells passing the Matrigel membrane and recovered in the bottom chamber was set as 100% invasion index. The error bars indicate S.D. from triplicate wells. *, p = 0.001; **, p = 0.0006 (n = 3).
DISCUSSION
CLU is a multifunctional protein that we have recently shown to act as a haploinsufficient tumor suppressor gene in neuroblastoma. MYCN inhibits the expression of CLU mRNA and protein in neuroblastoma cell lines and primary tumors in part by inducing oncogenic microRNAs belonging to the miR17–92 cluster (13). However, the miR17–92 cluster inhibits CLU indirectly, through the TGF-β pathway (25). The presence of MYC binding motifs in the CLU promoter prompted us to ask whether MYCN could regulate CLU expression directly.
We first observed that MYCN binds to and transcriptionally regulates the CLU gene in a negative manner, through a non-canonical E box element in the 5′-flanking sequence. MYC proteins have been shown to bind to different versions of the classical E box motif “CACGTG,” but the E box characterized in this study “CACGCG” is peculiar because it mediates negative instead of positive regulation. In previous studies, it was reported that MYCN represses other tumor suppressor genes indirectly, by interacting with the transcription factors SP1 and MIZ1 (12, 19, 26). To our knowledge, our study is the first to report that a MYC member causes transcriptional repression by recruiting histone-modifying enzymes such as Polycomb proteins via an E box. MYCN interacts with the Polycomb member EZH2 through the MYC box domain 3, MB3. This is a notable finding because it was shown that this element is crucially required for MYC transcriptional repression, and deletion of MB3 impairs the transforming activity of MYC (27). An intriguing hypothesis stemming from these observations is that the interaction between the MB3 element and EZH2 critically links MYC transforming activity to the recruitment of Polycomb transcriptional repressors.
Another notable finding of our study is that the binding of MYCN to the CLU promoter causes a chromatin rearrangement that is compatible with the bivalent state, typical of developmentally regulated genes in stem cells or genes silenced in cancer (28–30). Thus, MYC oncoproteins could confer stem-like features and aggressive phenotype to cancer cells by physically recruiting Polycomb group proteins and HDACs at tumor suppressor genes such as CLU and inducing the bivalent epigenetic state. This notion is corroborated by a recent study from the Einsenman group in which c-MYC was shown to impose widespread changes in histone methylation patterns in ES cells. Furthermore, it was noted that a subset of Polycomb-bound genes with bivalent histone methylation patterns are bound and regulated in response to altered c-MYC levels (31). It will be of interest to assess whether the ability of c-MYC to induce cell reprogramming and/or transformation also involves the formation of bivalent epigenetic marks around E boxes at the promoters of other tumor suppressor genes.
Infants and children with cancer are particularly sensitive to the toxic effects of chemotherapeutic drugs, which cause a number of adverse sequelae, including cognitive impairments, retarded development, and secondary neoplasms (4, 32). In neuroblastoma, amplification of MYCN marks a class of patients with metastatic disease whom need to be treated aggressively. Despite therapy, many children die with relapsing disease. It is therefore essential to explore other therapeutic avenues to find a treatment approach that is more effective and safer for children with aggressive forms of neuroblastoma.
There is a growing interest in histone deacetylase inhibitors for cancer therapy because they are relatively safe and have shown promising results in clinical trials (33, 34). An important advance in the understanding of the mechanisms of action and the clinical potential of these compounds would be to determine the critical biomarkers that predict a positive response to treatment. For example, in a cohort of patients with cutaneous T cell lymphoma, oral administration of the HDAC inhibitor lbh589 caused the up-regulation of 23 genes in biopsies taken after the treatments (20). We have shown here that MYC-mediated repression of CLU can be reversed by HDAC inhibitors, resulting in anticancer activity. In the clinic, the presence of CLU, a secreted protein, in the sera of cancer patients could serve as a biomarker to predict a favorable response after treatments with epigenetic drugs.
This work was supported by grants from SPARKS, The Neuroblastoma Society, a Wellcome Trust grant (to A. S.), and the Italian Association for Cancer Research (to G. P.).

This article contains supplemental “Experimental Procedures,” Table S1, and Figs. S1–S9.
- Tet
- tetracycline
- HDAC
- histone deacetylase
- MB3
- MYC box domain 3
- TSA
- trichostatin A.
REFERENCES
- 1. Garinis G. A., Patrinos G. P., Spanakis N. E., Menounos P. G. (2002) DNA hypermethylation: when tumour suppressor genes go silent. Hum. Genet. 111, 115–127 [DOI] [PubMed] [Google Scholar]
- 2. Marchion D., Münster P. (2007) Development of histone deacetylase inhibitors for cancer treatment. Expert Rev. Anticancer Ther. 7, 583–598 [DOI] [PubMed] [Google Scholar]
- 3. Momparler R. L. (2003) Cancer epigenetics. Oncogene 22, 6479–6483 [DOI] [PubMed] [Google Scholar]
- 4. Brodeur G. M. (2003) Neuroblastoma: biological insights into a clinical enigma. Nat. Rev. Cancer 3, 203–216 [DOI] [PubMed] [Google Scholar]
- 5. Weiss W. A., Aldape K., Mohapatra G., Feuerstein B. G., Bishop J. M. (1997) Targeted expression of MYCN causes neuroblastoma in transgenic mice. EMBO J. 16, 2985–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bell E., Premkumar R., Carr J., Lu X., Lovat P. E., Kees U. R., Lunec J., Tweddle D. A. (2006) The role of MYCN in the failure of MYCN amplified neuroblastoma cell lines to G1 arrest after DNA damage. Cell Cycle 5, 2639–2647 [DOI] [PubMed] [Google Scholar]
- 7. Burkhart C. A., Cheng A. J., Madafiglio J., Kavallaris M., Mili M., Marshall G. M., Weiss W. A., Khachigian L. M., Norris M. D., Haber M. (2003) Effects of MYCN antisense oligonucleotide administration on tumorigenesis in a murine model of neuroblastoma. J. Natl. Cancer Inst. 95, 1394–1403 [DOI] [PubMed] [Google Scholar]
- 8. Kang J. H., Rychahou P. G., Ishola T. A., Qiao J., Evers B. M., Chung D. H. (2006) MYCN silencing induces differentiation and apoptosis in human neuroblastoma cells. Biochem. Biophys. Res. Commun. 351, 192–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hogarty M. D., Norris M. D., Davis K., Liu X., Evageliou N. F., Hayes C. S., Pawel B., Guo R., Zhao H., Sekyere E., Keating J., Thomas W., Cheng N. C., Murray J., Smith J., Sutton R., Venn N., London W. B., Buxton A., Gilmour S. K., Marshall G. M., Haber M. (2008) ODC1 is a critical determinant of MYCN oncogenesis and a therapeutic target in neuroblastoma. Cancer Res. 68, 9735–9745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brenner C., Deplus R., Didelot C., Loriot A., Viré E., De Smet C., Gutierrez A., Danovi D., Bernard D., Boon T., Pelicci P. G., Amati B., Kouzarides T., de Launoit Y., Di Croce L., Fuks F. (2005) Myc represses transcription through recruitment of DNA methyltransferase corepressor. EMBO J. 24, 336–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Eilers M., Eisenman R. N. (2008) Myc's broad reach. Genes Dev. 22, 2755–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu T., Tee A. E., Porro A., Smith S. A., Dwarte T., Liu P. Y., Iraci N., Sekyere E., Haber M., Norris M. D., Diolaiti D., Della Valle G., Perini G., Marshall G. M. (2007) Activation of tissue transglutaminase transcription by histone deacetylase inhibition as a therapeutic approach for Myc oncogenesis. Proc. Natl. Acad. Sci. U.S.A. 104, 18682–18687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chayka O., Corvetta D., Dews M., Caccamo A. E., Piotrowska I., Santilli G., Gibson S., Sebire N. J., Himoudi N., Hogarty M. D., Anderson J., Bettuzzi S., Thomas-Tikhonenko A., Sala A. (2009) J. Natl. Cancer Inst. 101, 663–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fontana L., Fiori M. E., Albini S., Cifaldi L., Giovinazzi S., Forloni M., Boldrini R., Donfrancesco A., Federici V., Giacomini P., Peschle C., Fruci D. (2008) Antagomir-17–5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PLoS One 3, e2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schulte J. H., Horn S., Otto T., Samans B., Heukamp L. C., Eilers U. C., Krause M., Astrahantseff K., Klein-Hitpass L., Buettner R., Schramm A., Christiansen H., Eilers M., Eggert A., Berwanger B. (2008) MYCN regulates oncogenic MicroRNAs in neuroblastoma. Int. J. Cancer 122, 699–704 [DOI] [PubMed] [Google Scholar]
- 16. Lutz W., Stöhr M., Schürmann J., Wenzel A., Löhr A., Schwab M. (1996) Conditional expression of N-myc in human neuroblastoma cells increases expression of α-prothymosin and ornithine decarboxylase and accelerates progression into S-phase early after mitogenic stimulation of quiescent cells. Oncogene 13, 803–812 [PubMed] [Google Scholar]
- 17. Seeger R. C., Rayner S. A., Banerjee A., Chung H., Laug W. E., Neustein H. B., Benedict W. F. (1977) Morphology, growth, chromosomal pattern and fibrinolytic activity of two new human neuroblastoma cell lines. Cancer Res. 37, 1364–1371 [PubMed] [Google Scholar]
- 18. Schwab M., Alitalo K., Klempnauer K. H., Varmus H. E., Bishop J. M., Gilbert F., Brodeur G., Goldstein M., Trent J. (1983) Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumour. Nature 305, 245–248 [DOI] [PubMed] [Google Scholar]
- 19. Iraci N., Diolaiti D., Papa A., Porro A., Valli E., Gherardi S., Herold S., Eilers M., Bernardoni R., Della Valle G., Perini G. (2011) A SP1/MIZ1/MYCN repression complex recruits HDAC1 at the TRKA and p75NTR promoters and affects neuroblastoma malignancy by inhibiting the cell response to NGF. Cancer Res. 71, 404–412 [DOI] [PubMed] [Google Scholar]
- 20. Ellis L., Pan Y., Smyth G. K., George D. J., McCormack C., Williams-Truax R., Mita M., Beck J., Burris H., Ryan G., Atadja P., Butterfoss D., Dugan M., Culver K., Johnstone R. W., Prince H. M. (2008) Histone deacetylase inhibitor panobinostat induces clinical responses with associated alterations in gene expression profiles in cutaneous T-cell lymphoma. Clin. Cancer Res. 14, 4500–4510 [DOI] [PubMed] [Google Scholar]
- 21. Pasini D., Bracken A. P., Jensen M. R., Lazzerini Denchi E., Helin K. (2004) Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 23, 4061–4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang C., Liu Z., Woo C. W., Li Z., Wang L., Wei J. S., Marquez V. E., Bates S. E., Jin Q., Khan J., Ge K., Thiele C. J. (2012) EZH2 Mediates epigenetic silencing of neuroblastoma suppressor genes CASZ1, CLU, RUNX3, and NGFR. Cancer Res. 72, 315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bommi P. V., Dimri M., Sahasrabuddhe A. A., Khandekar J., Dimri G. P. (2010) The polycomb group protein BMI1 is a transcriptional target of HDAC inhibitors. Cell Cycle 9, 2663–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Santilli G., Aronow B. J., Sala A. (2003) Essential requirement of apolipoprotein J (clusterin) signaling for IκB expression and regulation of NF-κB activity. J. Biol. Chem. 278, 38214–38219 [DOI] [PubMed] [Google Scholar]
- 25. Dews M., Fox J. L., Hultine S., Sundaram P., Wang W., Liu Y. Y., Furth E., Enders G. H., El-Deiry W., Schelter J. M., Cleary M. A., Thomas-Tikhonenko A. (2010) The myc-miR-17∼92 axis blunts TGFβ signaling and production of multiple TGFβ-dependent antiangiogenic factors. Cancer Res. 70, 8233–8246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Seoane J., Le H. V., Massagué J. (2002) Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature 419, 729–734 [DOI] [PubMed] [Google Scholar]
- 27. Herbst A., Hemann M. T., Tworkowski K. A., Salghetti S. E., Lowe S. W., Tansey W. P. (2005) A conserved element in Myc that negatively regulates its proapoptotic activity. EMBO Rep. 6, 177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rodriguez J., Muñoz M., Vives L., Frangou C. G., Groudine M., Peinado M. A. (2008) Bivalent domains enforce transcriptional memory of DNA methylated genes in cancer cells. Proc. Natl. Acad. Sci. U.S.A. 105, 19809–19814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ohm J. E., McGarvey K. M., Yu X., Cheng L., Schuebel K. E., Cope L., Mohammad H. P., Chen W., Daniel V. C., Yu W., Berman D. M., Jenuwein T., Pruitt K., Sharkis S. J., Watkins D. N., Herman J. G., Baylin S. B. (2007) A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat. Genet. 39, 237–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Balch C., Nephew K. P., Huang T. H., Bapat S. A. (2007) Epigenetic “bivalently marked” process of cancer stem cell-driven tumorigenesis. Bioessays 29, 842–845 [DOI] [PubMed] [Google Scholar]
- 31. Lin C. H., Lin C., Tanaka H., Fero M. L., Eisenman R. N. (2009) Gene regulation and epigenetic remodeling in murine embryonic stem cells by c-Myc. PLoS One 4, e7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fox E., Citrin D., Balis F. M. (2009) The legacy of cancer therapy in children. J, Natl, Cancer Inst. 101, 1105–1107 [DOI] [PubMed] [Google Scholar]
- 33. Glaser K. B. (2007) HDAC inhibitors: clinical update and mechanism-based potential. Biochem. Pharmacol. 74, 659–671 [DOI] [PubMed] [Google Scholar]
- 34. Prince H. M., Bishton M. J., Harrison S. J. (2009) Clinical studies of histone deacetylase inhibitors. Clin. Cancer Res. 15, 3958–3969 [DOI] [PubMed] [Google Scholar]