Background: The anti-tumor lipid edelfosine alters lipid raft integrity. How this signals inhibition of growth is not understood.
Results: Disruption of plasma membrane domain organization triggers the selective removal of lipid raft-associated transporters altering pH homeostasis.
Conclusion: Raft integrity controls pH homeostasis and growth.
Significance: Choline-containing lysolipid analogues induce biophysical modifications of microdomains leading to inhibition of cell growth through alteration of pH homeostasis.
Keywords: Lipid Raft, Lysophospholipid, pH Regulation, Sterol, Yeast, Edelfosine, Ether Lipid, Lysophosphatidylcholine Analogue, Nutrient H+ Symporter, pH Homeostasis
Abstract
The lysophosphatidylcholine analogue edelfosine is a potent antitumor lipid that targets cellular membranes. The underlying mechanisms leading to cell death remain controversial, although two cellular membranes have emerged as primary targets of edelfosine, the plasma membrane (PM) and the endoplasmic reticulum. In an effort to identify conditions that enhance or prevent the cytotoxic effect of edelfosine, we have conducted genome-wide surveys of edelfosine sensitivity and resistance in Saccharomyces cerevisiae presented in this work and the accompanying paper (Cuesta-Marbán, Á., Botet, J., Czyz, O., Cacharro, L. M., Gajate, C., Hornillos, V., Delgado, J., Zhang, H., Amat-Guerri, F., Acuña, A. U., McMaster, C. R., Revuelta, J. L., Zaremberg, V., and Mollinedo, F. (January 23, 2013) J. Biol. Chem. 288,), respectively. Our results point to maintenance of pH homeostasis as a major player in modulating susceptibility to edelfosine with the PM proton pump Pma1p playing a main role. We demonstrate that edelfosine alters PM organization and induces intracellular acidification. Significantly, we show that edelfosine selectively reduces lateral segregation of PM proteins like Pma1p and nutrient H+-symporters inducing their ubiquitination and internalization. The biology associated to the mode of action of edelfosine we have unveiled includes selective modification of lipid raft integrity altering pH homeostasis, which in turn regulates cell growth.
Introduction
The plasma membrane (PM)4 of eukaryotes represents one of the most complex biomembranes, featuring an asymmetric lipid distribution between the two leaflets of the bilayer as well as lateral domain organization within leaflets. Lipid rafts are dynamic areas of the membrane rich in sterols and sphingolipids that serve as platforms for the association of membrane proteins. At the heart of the lipid raft concept lies the idea that the lipid environment is not passive but is critical for regulation of protein function (1). In fact, it has been well documented that lipid rafts play important roles in membrane trafficking and signaling (1–3). Therefore, alteration of these lipid domains is expected to interfere with these pathways, eventually leading to a detrimental impact on cell fitness and survival. This is in fact a proposed mode of action for the anticancer lipid drug edelfosine (4–8).
The synthetic lipid edelfosine (1-O-octadecyl-2-O-methyl-rac-glycero-3-phosphocholine or ET-18-OCH3) is a prototypical member of a class of cancer chemotherapeutic drugs collectively known as alkyl-lysophospholipid analogues or antitumor lipids. Edelfosine is similar to lysophosphatidylcholine but has greater metabolic stability due to the presence of ether-linked groups to its glycerol backbone. The mode of action of this drug is still controversial. Two cellular compartments have emerged as main targets of edelfosine, the plasma membrane and the endoplasmic reticulum (ER). At the plasma membrane, edelfosine interacts with lipid rafts and induces changes in lipid and protein composition in these microdomains (6, 9, 10). At the ER, edelfosine can induce ER stress (11, 12) and inhibits phosphatidylcholine synthesis (13, 14).
The plasma membrane represents the first point of contact of edelfosine with cells. A major aspect of the anticancer activity of edelfosine lies in its selectivity to kill tumor cells, whereas normal cells are spared (9, 15, 16). Preferred uptake by cancer cells is thought to be crucial for its selective cytotoxicity (15, 17, 18).
Much of what we know on how edelfosine enters cells comes from studies in budding yeast. Uptake of the drug in yeast is mediated by a flippase and controlled by the Lem3p subunit (6, 19–21). Stimulated by the fact that edelfosine is cytotoxic to yeast at similar concentrations as those used to kill cancer cells, we initiated investigations to gain insight into its mode of action by performing unbiased genetic screens in this organism. A first screen provided evidence that edelfosine-mediated cytotoxicity is through modification of the biophysical structure of lipid rafts by inducing internalization of sterols and the essential proton pump Pma1p from the PM (6). In this study and the accompanying paper (69), we expanded our genetic screen approach to survey the yeast deletion collection for sensitivity and resistance to edelfosine, respectively. The results of these screens further support a key role for lipid rafts and the essential proton pump Pma1p in mediating edelfosine cytotoxicity in yeast. In addition, our results unveiled an important role for trafficking pathways from and to the PM in modulating mechanisms of sensitivity and resistance to edelfosine. We also found that failure to maintain pH homeostasis may be the signal that regulates communication between the PM and intracellular membranes.
EXPERIMENTAL PROCEDURES
Yeast Strains, Plasmids, and Growth Conditions
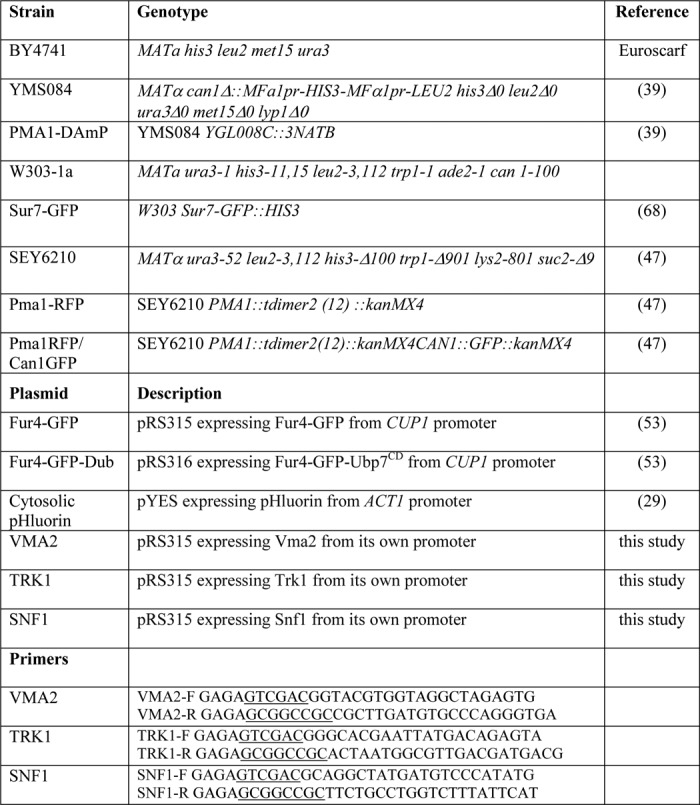
Detailed information on yeast strains and plasmids used in this study is provided in Table 1. Yeast strains were grown on standard rich medium YPD (1% yeast extract, 2% Bacto-peptone, 2% glucose) or synthetic minimal medium (SD: 0.67% yeast nitrogen base without amino acids, 2% glucose). Amino acids were supplemented to SD medium according to the requirements of the strains used. Agar (2%) was added for solid plates. All yeast cultures were incubated at 30 °C. Growth of cells treated with edelfosine was monitored by absorbance at a wavelength of 600 nm (A600).
TABLE 1.
Strains, plasmids, and primers used
Growth medium pH was adjusted to values in the range of 3–7.5 as indicated, by addition of HCl or NaOH. Glucose (2%) was added from a concentrated sterile stock after the medium was autoclaved.
The VMA2, SNF1, and TRK1 genes, including their own promoter and termination sequences, were PCR-amplified from genomic DNA obtained from the wild-type strain BY4741. SalI and NotI sites were engineered in the forward and reverse primer, respectively, to allow directional cloning into the centromeric plasmid pRS315 (LEU2). Specific primers used are listed in Table 1.
Identification of Edelfosine-hypersensitive Mutants in S. cerevisiae
A total of 4672 different yeast deletion mutants, generated by the international deletion consortium (22, 23), were obtained from Euroscarf. This collection of deletion mutants represents the total number of viable single mutants from a total of ∼6200 potential genes. All strains are derivatives of BY4741. The specific genes were disrupted with a kanamycin-resistant (kanR) cassette. Strains from the deletion collection were screened for hypersensitivity to edelfosine in solid medium. For this purpose, strains were arrayed individually on a series of rectangular OmniTray agar plates (Nalgene Nunc International) at 384 strains per plate and manipulated robotically using a Virtek Colony Arrayer (Bio-Rad). Edelfosine (the kind gift of Medmark Pharma GmbH and Inkeysa) was added to autoclaved synthetic defined (SD: 0.67% yeast nitrogen base without amino acids, 2% glucose and 2% agar and required supplements) medium cooled to ∼50 °C.
We and others have observed that the effect of lipids on cell growth in solid media can be affected by cell density (6, 24). The robot pins a large number of cells in one spot so the concentration of edelfosine had to be increased from those normally used in serial dilution assays. For the chemical-genetic screen, a final concentration of 190 μm edelfosine was used. This concentration did not affect growth of wild-type (BY4741) colonies, although it inhibited growth of a sensitive strain lacking Spo14p (25). Plates were incubated for 2 days at 30 °C and imaged using a Versa Doc (Bio-Rad) apparatus. Colony size comparison on yeast array plates was determined using a scoring system as described previously (26). The screen was run four independent times. Genes identified at least three times out of the four runs are reported herein (Table 2).
TABLE 2.
Complete list of genes identified in our genetic screen
The genes discussed in this work are in boldface italic.
| Sensitivity | Gene | Cellular role | Localization |
|---|---|---|---|
| +++ | AFG3 | Protein phosphorylation | Mitochondria |
| +++ | BEM1 | Cell organization and biogenesis | Bud tip-bud neck-cell periphery |
| +++ | DBF2 | Protein phosphorylation | Bud neck-cytoplasm |
| +++ | ERG3 | Lipid metabolism | ER |
| +++ | GAL11 | Transcription | Nucleus |
| +++ | HTA3 (HTZ1) | Chromatin remodeling | Nucleus |
| +++ | PGD1 | Transcription | Nucleus |
| +++ | REI1 | Translation | Cytoplasm-ribosome |
| +++ | SIN3 | Chromatin remodeling | Mitochondria-nucleus |
| +++ | SIP3 | Energy homeostasis | Nucleus |
| +++ | SNF1 | Energy homeostasis | Nucleus-mitochondria-vacuole-cytoplasm |
| +++ | SNF2 | Chromatin remodeling | Nucleus |
| +++ | SNF6 | Chromatin remodeling | Nucleus |
| +++ | SPT7 | Transcription | Nucleus-mitochondria |
| +++ | VMA16 (PPA1) | Vacuolar ATPase | Vacuole |
| +++ | VMA2 | Vacuolar ATPase | Vacuole |
| +++ | VMA4 | Vacuolar ATPase | Vacuole |
| +++ | VMA5 | Vacuolar ATPase | Vacuole |
| +++ | VMA9 | Vacuolar ATPase | Vacuole |
| +++ | VPH2 (VMA12) | Vacuolar ATPase assembly | ER |
| +++ | YJL175W | Chromatin remodeling | Probably SWI3 (nucleus) |
| ++ | AKR1 | Protein palmitoylation | Golgi |
| ++ | ASC1 | Translation | Cytosol-ribosome |
| ++ | BIM1 | Cell organization and biogenesis | Microtubule |
| ++ | COX17 | Cytochrome c assembly | Mitochondria-nucleus? |
| ++ | CYC2 | Cytochrome c regulation | Mitochondria |
| ++ | DEF1 | Protein degradation | Nucleus-cytoplasm |
| ++ | ELO2 | Lipid metabolism | ER |
| ++ | ETR1 | Lipid metabolism | Mitochondria-nucleus |
| ++ | LIP5 | Lipid metabolism | Mitochondria |
| ++ | NAP1 | Chromatin remodeling | Nucleus-cytoplasm |
| ++ | PAA1 | Chromatin remodeling | Cytoplasm |
| ++ | PDA1 | Acetyl-CoA production (impact on lipid metabolism) | mitochondria |
| ++ | RPB4 | Transcription | Nucleus |
| ++ | SEC22 | Vesicular transport | Golgi-ER |
| ++ | SER1 | Serine metabolism (impact on lipid metabolism) | Cytoplasm |
| ++ | SHY1 | Cytochrome c assembly | Mitochondria |
| ++ | SNF4 | Energy homeostasis | Nucleus-PM-cytoplasm |
| ++ | SNF5 | Chromatin remodeling | Nucleus |
| ++ | SPF1 | Ion transport (P-type ATPase) | ER-mitochondria |
| ++ | SPO14 | Lipid metabolism | Endosomes? |
| ++ | SRB2 | Transcription | Nucleus-cytoplasm |
| ++ | SUR4 | Lipid metabolism | ER |
| ++ | SWI6 | Transcription | Nucleus-cytoplasm |
| ++ | TRK1 | pH homeostasis and K+ transport | PM |
| ++ | UBP6 | Protein degradation | Nucleus-cytoplasm |
| ++ | VMA10 | Vacuolar ATPase | Vacuole |
| ++ | VMA11 (TFP3) | Vacuolar ATPase | Vacuole-ER |
| ++ | VMA21 | Vacuolar ATPase assembly | Vacuole |
| ++ | VMA7 | Vacuolar ATPase | Vacuole |
| ++ | YBR178W | Lipid metabolism | Probably EHT1 (mitochondria-lipid particle) |
| ++ | YDR521W | Transcription | Probably YDR520C (nucleus) |
| + | PDX3 | Lipid metabolism | ? |
| + | VMA6 | Vacuolar ATPase | Vacuole |
Data Analysis and Functional Group Classification
Enrichment of data sets for Gene Ontology terms was performed using the Gene Ontology Term Finder at the Saccharomyces Genome Database and the MIPS database. Interaction networks were visualized with Osprey using data from several databases (27).
Funspec (28) was used to identify functional clusters of genes and statistical evaluation. Gene classification was done subjectively, supported by Saccharomyces Genome Database, MIPS, as well as the literature.
Cytosolic pH Measurements
pYES-ACT-pHluorin plasmid (kindly provided by Gertien Smits from University of Amsterdam, The Netherlands) alongside an empty pYES vector (Invitrogen), used as control, were transformed into wild-type or mutant yeast strains as indicated. pH determination was done according to the ratiometric method outlined by Orij et al. (29). Briefly, yeast transformants were grown to an A600 of 0.5 at 30 °C in modified synthetic media, lacking fluorescent compounds, p-aminobenzoic acid, riboflavin, and folic acid, as well as uracil for plasmid selection. For pHluorin calibration, cells in PBS were treated with 100 μg/ml digitonin (Sigma) and resuspended in buffers with pH values ranging from 5.95 to 7.80. Fluorescence emission of pHluorin was measured at 512 nm in a Cary Eclipse fluorescence Spectrophotometer (Varian) using excitation bands of 5 nm centered around 390 and 470 nm. The emission values were obtained using Cary Eclipse Ratio Application software; data and statistical analyses were done using Microsoft Excel and GraphPad Prism software. The ratio between emission intensities resulting from excitation at 390 and 470 nm (R390/470) was plotted against the corresponding buffer pH to obtain standard curves for cytosolic pHluorin. In all experiments, the emission intensity values (background fluorescence) obtained from empty vector transformants were subtracted. pHluorin as well as empty plasmid transformants were treated with 19 μm edelfosine, and emission intensity ratios were calculated as described above. Fluorescence measurements were performed for treated and untreated cultures every 15 min during the 1st h and at 90 min following initial addition of the drug. After the 90-min time interval, cells were treated with 100 μg/ml digitonin for 15 min to equilibrate them to the external pH. All pH determination experiments were repeated at least three times, in quadruplicates for each sample. Figures show one representative experiment with error bars indicating the standard deviation of the four replicates within the experiment.
Detergent-resistant Membrane (DRM) Isolation
Yeast DRMs were isolated as described previously (6). Briefly, log phase growing cultures at A600 of 0.1–0.2 in defined medium were incubated with or without 19 μm edelfosine for 2 h at 30 °C. Around 20 A600 cell equivalents were collected, washed, and resuspended in 500 μl of TNE buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 5 mm EDTA, protease inhibitor mixture (Roche Applied Science), 2.5 μg/ml pepstatin, 1 mm phenylmethanesulfonyl fluoride). Cells were broken in the presence of glass beads with a mini bead beater (Biospec) at maximum speed for 1 min at 4 °C. Unbroken cell and debris were removed by a 500 × g spin for 5 min at 4 °C. Protein concentration was determined (30), and 300 μg of protein in 500 μl of TNE buffer was incubated with Triton X-100 (1% final concentration) for 30 min on ice. The lysate was subsequently adjusted to 40% Optiprep (Nycomed) by adding 1 ml of 60% Optiprep solution and overlaid with 2.4 ml of 30% Optiprep in TXNE (TNE with 0.1% Triton X-100) and then with 400 μl of TXNE. The samples were centrifuged at 166,000 × g for 2 h in a swinging bucket TLS55 rotor (Beckman). Ten fractions of equal volume were collected from the top of the gradient. The interface between the 0 and 30% Optiprep contained the DRMs and was easily identified optically and collected as fraction 2. An aliquot of each fraction was analyzed by 8% SDS-PAGE followed by silver staining or Western blotting as indicated. Proteins were transferred to polyvinylidene difluoride (PVDF) membranes, and blots were incubated with antibodies to Pma1p, Gas1p (the kind gifts of Ramón Serrano, Universidad Politécnica de Valencia, and Howard Riezman, University of Geneva, respectively), or Pgk1p (Molecular Probes) and subsequently with horseradish peroxidase-conjugated secondary antibodies followed by detection using enhanced chemiluminescence. DRM isolations were performed ∼20 times in different experimental conditions.
Immunoprecipitation and Protein Analysis
Cultures in 200 ml of YPD were grown to an A600 of 1.0, and edelfosine was added at 19 μm. Cells were treated for the indicated times and collected by centrifugation, and pellets were kept frozen until used. Cell lysis was achieved by resuspending pellets in 400 μl of RIPA buffer (SDS 0.1%, Triton X-100 1%, sodium deoxycholate 1%) plus inhibitors (10 μg/ml aprotinin and leupeptin, 2 mm PMSF, 250 nm bortezomib, 10 mm iodoacetamide) and a similar volume of glass beads (0.5 mm diameter). The tubes were given three 15-s pulses in a Fastprep, with 30 s of cooling in between. Tubes were briefly spun down to pellet the beads; the supernatants were transferred to new tubes and centrifuged for 5 min at 500 × g to pellet unbroken cells and debris. Protein concentration of the supernatants was determined in triplicate by using the Bradford assay. Pma1p was immunoprecipitated from 160 μg of total protein with a rabbit polyclonal antibody raised against Pma1p, and 25 μl of CL-4B beads coated with protein A slurry (GE Healthcare) in 2 ml of PBS overnight at 4 °C. Alternatively, 500 μg of protein were incubated in the same conditions with 20 μl of beads covalently bound to a ubiquitin-interacting domain (ubiquitinated protein enrichment kit, Calbiochem). Beads were washed with PBS three times, resuspended in 30 μl of sample buffer, and heated at 50 °C for 10 min to avoid Pma1p aggregation. Samples were directly loaded in discontinuous 7% polyacrylamide gels, which were then electroblotted. Membranes were probed with the anti Pma1p polyclonal antibody and a goat secondary antibody linked to Alexa680 (Invitrogen) and imaged in a LiCor scanner.
Alternatively, proteins were transferred to PVDF membranes (GE Healthcare), and blots were incubated with antibodies to Pma1p or anti-ubiquitin (Invitrogen) and subsequently with horseradish peroxidase-conjugated secondary antibodies, followed by detection using enhanced chemiluminescence.
Microscopy
Filipin (Sigma) was used to examine sterol distribution as described previously (6). Filipin was freshly prepared as a 1 mg/ml stock in ethanol. Cells grown to log phase in defined medium were treated with 19 μm edelfosine, and aliquots of cells from the same culture were fixed at the indicated times with 3.7% EM-grade formaldehyde for 2 h at room temperature. Fixed cells were centrifuged and washed twice with phosphate-buffered saline, and spheroplasts were prepared by treatment with zymolyase. Cells were then incubated with 10 μg/ml filipin in the dark for 15 min at room temperature. Filipin fluorescence was observed with UV optics channel of a Zeiss Axiovert 200 M microscope fitted with a plan-neofluor ×100 oil immersion objective lens. Pma1-RFP was visualized using the rhodamine channel. Images were captured using a Zeiss Axio Cam HR and using Zeiss Axiovision Rel. 4.8 software.
In the case of transformants carrying plasmids containing FUR4-GFP or FUR4-GFP-DUb under the CUP promoter, expression was induced by addition of 100 μm CuCl2 into defined selective medium for 2 h. At this point cultures were split in two, and edelfosine was added to half of them at a final concentration of 19 μm. Control and edelfosine cultures were then incubated for 1 h at 30 °C before preparation of the slide for microscopy. Cells were then concentrated and placed on slabs of solid medium made from low fluorescent medium (as described above) and 2% agar. Coverslips were sealed, and digital images were obtained using a Leica SP5 confocal laser scanning system (Leica, Germany). Fluorescence signals of red fluorescent protein (excitation 543 nm, HeNe laser) were detected at emission range 565–635 nm, and fluorescence signals of GFP (excitation 488 nm, HeNe laser) were detected at emission range 499–561 nm. Images were then aligned using Adobe Photoshop (7.0) software. At least 100 cells per condition were analyzed for quantification of microscopy images as indicated. Experiments were conducted at least three times.
RESULTS
Genes That Control Intracellular pH Modulate Sensitivity to Edelfosine
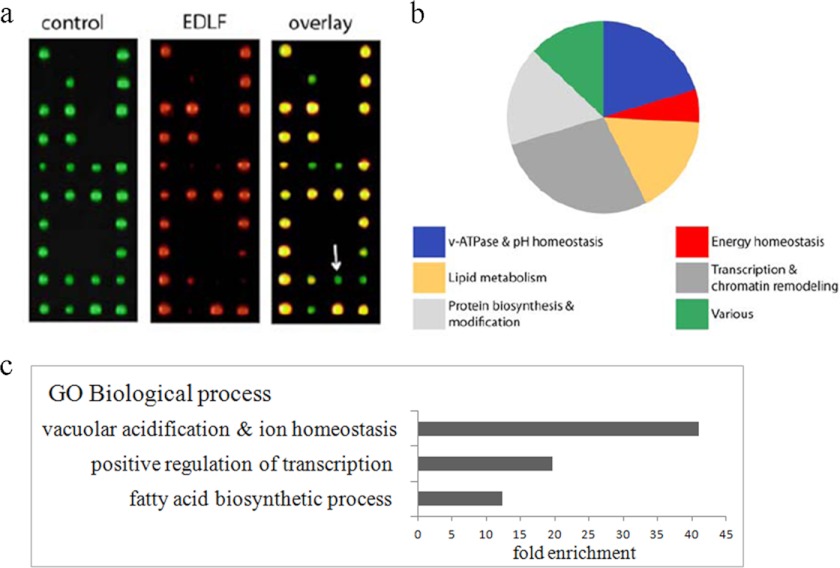
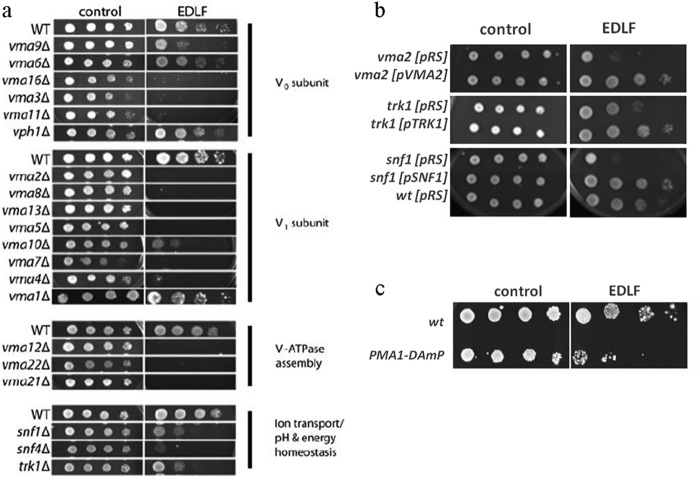
We performed a high throughput edelfosine sensitivity screen by robotically pinning an ordered array of 4672 haploid yeast gene deletion mutants onto defined medium in the absence or presence of edelfosine (Fig. 1). Fifty four genes whose inactivation reproducibly resulted in increased sensitivity to edelfosine compared with wild-type were identified (Table 2). This data set was enriched in genes known to participate in pH homeostasis, regulation of transcription, and lipid biosynthesis (Figs. 1 and 2). A cluster comprising genes coding for 9 of the 14 subunits of the vacuolar proton-translocating ATPase (V-ATPase), as well as VPH2- and VMA21-encoding proteins that participate in V-ATPase assembly in the ER (31), were identified (Table 2 and Figs. 2 and 3a). Serial dilution analysis expanded this result to include 12 V-ATPase subunits and one other gene coding for an assembly protein (VMA22) (Table 2 and Fig. 3a). In addition, TRK1, coding for the high affinity potassium transporter of the PM was among the genes identified (Table 2 and Figs. 2 and 3a). It is well documented that V-ATPase in the vacuole collaborates with Pma1p at the PM to maintain pH homeostasis in yeast (32). Trk1 also participates in regulation of intracellular pH by positively regulating Pma1p activity (32–34).
FIGURE 1.
High throughput edelfosine S. cerevisiae-sensitive screen. a, complete set of haploid S. cerevisiae yeast deletion mutants (∼4800 strains) was arrayed onto 20 plates and robotically pinned onto SD media (computerized colored green colonies) or SD + edelfosine (computerized colored red colonies). Putative edelfosine-sensitive mutants led to the formation of smaller colonies (or no colony) when grown on edelfosine-containing media (green on overlay, arrow). b, functional distribution of the 54 genes found to cause sensitivity to edelfosine when deleted. The functional categories corresponding to vacuolar acidification and ion transport, grouped as V-ATPase and pH homeostasis in the figure, had the lowest p values (2.784e-12 and 3.099e-08, respectively, Funspec analysis) indicative of a significant enrichment. c, enrichment of genes in our dataset clustering according to gene ontology (GO). Enrichment is calculated relative to the frequency of that cluster in the whole genome.
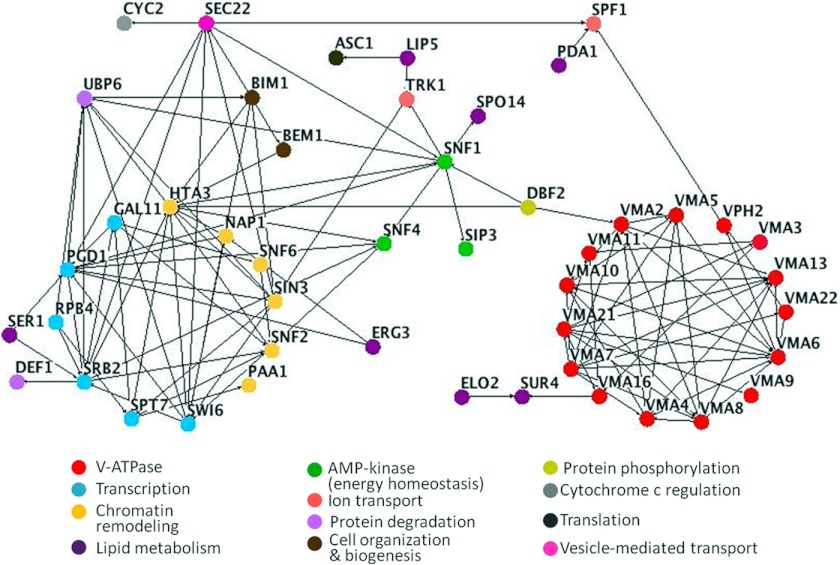
FIGURE 2.
Interactome map of genes identified in edelfosine-sensitive screen. Nodes represent genes identified as hypersensitive to edelfosine in our screen. Genes belonging to the same pathway or complex are colored according to the categories shown at the bottom of the figure. Edges indicate experimentally determined genetic or physical interactions. Nodes with a minimum of one connection are shown.
FIGURE 3.
Mutants with compromised pH homeostasis are hypersensitive to edelfosine. Strains scored as sensitive at least three times (from four screens completed) were verified by spotting serial dilutions of the cells onto media containing 19 μm edelfosine. Thus, a total of 54 deletion strains were confirmed to be edelfosine-sensitive. a, analysis was extended to all known V-ATPase-related genes by spotting serial dilutions of their corresponding deletion strains onto rich (YPD) medium control or plates containing 19 μm edelfosine (EDLF). Plates were incubated at 30 °C for 3 days. Data shown are representative of three independent experiments. b, deletion strains vma2, trk1, and snf1 were transformed with empty pRS315 (CEN) plasmids or the same plasmids containing the corresponding wild-type gene under their own promoters. Sensitivity to edelfosine was reverted to that displayed by the isogenic wild-type strain (BY4741) transformed with the empty plasmid. c, yeast strain with the PMA1-DAmP (decreased abundance by mRNA perturbation) allele displays increased sensitivity to edelfosine.
The second largest category of enrichment in our screen contained genes that positively regulate transcription. Genes coding for the yeast AMP-dependent kinase SNF1 and its γ subunit SNF4 were among these genes (Table 2, and Figs. 2 and 3a). AMP-dependent kinase/Snf1p plays a central role in controlling energy homeostasis in eukaryotes (35). Snf1 responds to glucose depletion and is a modulator of Trk1 activity (36, 37). Furthermore, cells deleted for SNF1 or SNF4 display pH-dependent phenotypes (34, 38). Deletion mutants vma2, trk1, and snf1 transformed with the corresponding wild-type genes displayed wild-type sensitivity to edelfosine (Fig. 3b). Therefore, the hypersensitive phenotype is not due to secondary mutations present in these strains.
The enrichment of genes related to maintenance of pH homeostasis in our sensitivity screen is in line with our previous findings involving Pma1p as the main target of edelfosine interaction with the yeast PM. Because Pma1p is an essential protein, it was not expected to be found in the screen for edelfosine-sensitive mutants using the S. cerevisiae nonessential gene deletion collection. We tested the sensitivity of cells carrying a hypomorphic allele of PMA1, PMA1-DAmP (Decreased Abundance by mRNA Perturbation) that results in substantially reduced levels of the protein (39). Consistent with our above results, PMA1-DAmP cells displayed increased sensitivity toward edelfosine (Fig. 3c), We hypothesized that edelfosine induced intracellular acidification, which in turn triggers a cellular response aimed at restoring pH homeostasis. Consequently, cells impaired in maintenance of pH homeostasis would display increased susceptibility to edelfosine.
Edelfosine Induces Intracellular Acidification in Yeast
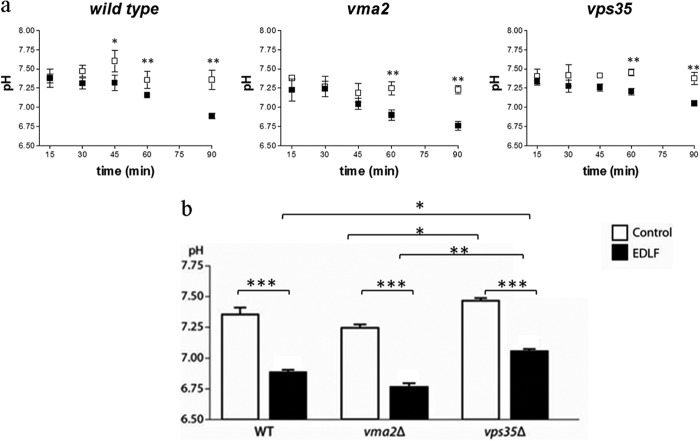
To assess the effect of edelfosine on intracellular pH, we used a method based on the expression of ratiometric pHluorin (29). This protein is a pH-sensitive green fluorescent protein that has been successfully used to measure pH in yeast (29, 32, 40, 41). To monitor the intracellular pH (pHi) of cells during treatment with edelfosine, cytosol-targeted pHluorin was expressed in wild-type cells as well as hypersensitive or resistant mutants vma2Δ and vps35Δ, respectively. Vma2p is a subunit of the vacuolar ATPase, and its deletion resulted in strong sensitivity to edelfosine (Fig. 3a). Moreover, Vps35p is a component of the yeast retromer, and cells with an inactivated VPS35 gene are resistant to edelfosine, although uptake of the drug is normal (69). A time frame of 90 min was chosen as we have previously determined it precedes cell death (6). The cytosolic pH in wild-type cells reached 7.35 ± 0.12 during the 90-min period but dropped to 6.89 ± 0.04 in the presence of edelfosine (Fig. 4). A steeper decline in pHi was observed in vma2Δ, dropping an extra 0.12–0.14 pH units after 1 h of treatment compared with wild type or vps35Δ in the same conditions (p < 0.001 WT or vps35Δ versus vma2Δ at 60 min, Fig. 4). Conversely, the resistant mutant vps35Δ displayed a less pronounced drop in pHi during the same period of treatment (69) (Fig. 4). These results indicate that the buffering capacity of the cell influences its ability to handle the disturbance in pH homeostasis induced by edelfosine, further supporting an important role for intracellular acidification in mediating the cytotoxic activity of edelfosine in S. cerevisiae.
FIGURE 4.
Edelfosine treatment leads to intracellular acidification. Cytosolic pH measurements were performed using pHluorin, as described under “Experimental Procedures.” a, intracellular pH was monitored during a 90-min period in control (open squares) or edelfosine (19 μm)-treated cells (closed squares). Time course is shown for wild type (BY4741), the resistant retromer mutant vps35Δ, and the hypersensitive V-ATPase mutant vma2Δ. b, comparison of the values obtained after a 90-min treatment. Data shown are mean values ± S.D. representative of three independent experiments. Asterisks indicate values that are significantly different from untreated control cells as follows: ***, p < 0.001; **, p < 0.01; *, p < 0.05.
We considered that if edelfosine compromises pH homeostasis, an aggravated phenotype should be displayed when cells are grown under stress conditions where control of cytosolic pH is part of the adaptation response, like in tolerance to extreme external pH (pHext). The lower and upper pHext tolerance limits for yeast growth are 2.5 and 8, respectively (42). Because of a tight control of intracellular pH, the kinetics of growth and fermentation are not affected in pHext between 3.5 and 6.0 (42). We therefore decided to test sensitivity to edelfosine at pHext 3.0 and 7.5, which allow for significant cell growth despite causing considerable stress (41). The hypomorphic strain PMA1-DAmP was included in the analysis to monitor the impact that the chosen pH conditions have on a strain that should (partially) mimic the effect of edelfosine. A decrease in Pma1p activity and expression has been previously described in yeast grown at pH 2.5 and 3 (41, 42). As expected, growth of the Pma1-DAmP strain was delayed when grown at pH 3 (Fig. 5 control plates 2 days). Interestingly, a similar delay was also observed in control plates at pHext 7.5, although no major differences were detected at pHext 5. As expected, edelfosine effect was stronger at pHext 3, but surprisingly, it was even more potent at alkaline pHext 7.5 (Fig. 5, 3 days). These results suggest that additional compensatory pathway(s) uniquely triggered by alkaline media are either impaired by treatment with edelfosine or result in enhanced sensitivity to the drug.
FIGURE 5.
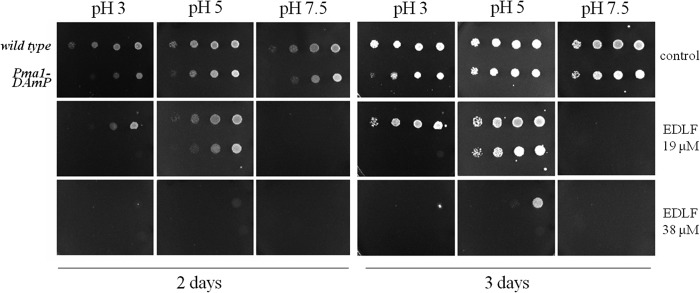
Edelfosine sensitivity at various external pH conditions. The yeast strain PMA1-DAmP and its isogenic wild type were grown to log phase in defined medium, pH 5.2, and then serially diluted in plates containing the indicated concentrations of edelfosine at different external pH values of 3, 5, or 7.5. Plates were incubated at 30 °C for 3 days. Data shown are representative of three independent experiments. EDLF, edelfosine.
Edelfosine Induces Ubiquitination of Pma1p
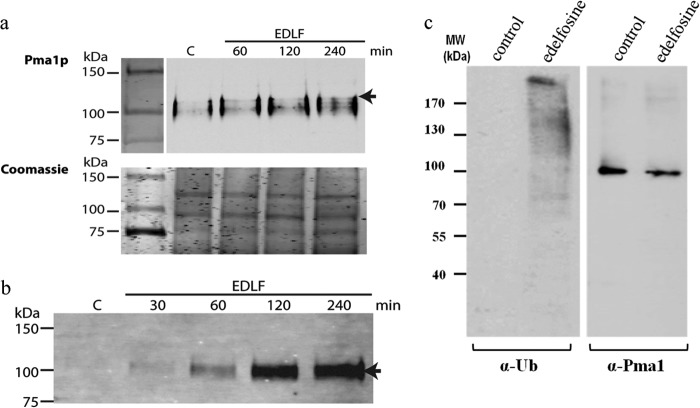
We have previously shown that edelfosine induces sterol and Pma1p internalization in yeast, with Pma1p being internalized via endocytosis and then degraded in the vacuole (6). In this study, co-localization studies using Pma1-RFP and filipin to detect sterol localization were performed (Fig. 6, a and b). Sterols and Pma1p were initially localized to the PM. Intracellular accumulation of sterols was evident after a 30-min incubation with edelfosine. No intracellular co-localization with Pma1p was observed at this time point (Fig. 6b). The filipin signal became very weak at the PM by 50–60 min of treatment, although the majority of the cells (77%) still displayed Pma1-RFP at the PM. Therefore, the kinetics of internalization of sterols and Pma1p as well as their intracellular localization differed. These results suggest that sterol internalization may precede Pma1p exit from the PM and that lipid and protein raft components are internalized through different routes (Fig. 6). Partitioning of Pma1p into detergent-resistant membranes (DRMs), rich in sterols and sphingolipids, is significantly reduced after treatment with edelfosine (Fig. 6, c and d).
FIGURE 6.
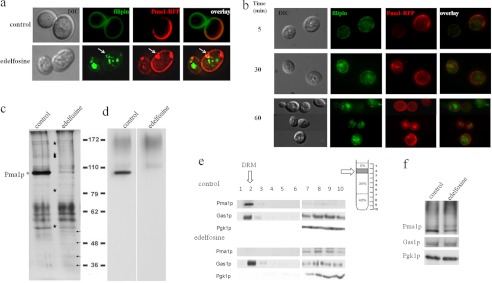
Edelfosine induces multiple changes in lipid raft organization. a, cells expressing Pma1p-RFP were treated with edelfosine (19 μm) or just vehicle (control) for 50 min at 30 °C followed by a 5-min staining with filipin to visualize sterols. Arrows point to an area of the PM enriched in Pma1p-RFP that seems to be partially surrounded by sterols on the intracellular side. Note: lack of Pma1-RFP signal in growing areas of the plasma membrane of budding yeast (areas containing newly synthesized Pma1-RFP) is due to the slow maturation of the red fluorescent protein fluorophore, although the protein is present in those areas (47). b, time course of sterols and Pma1 internalization during treatment with edelfosine (19 μm) in fixed cells. DIC, differential interference contrast. c–f, wild-type (BY4741) cells were treated with edelfosine (19 μm) for 2 h in defined medium at 30 °C. DRMs were prepared as indicated under “Experimental Procedures.” Silver staining (c) shows whole protein profile and Western blot for Pma1p (d). The most prominent change induced by edelfosine is the decrease in the abundance of Pma1p from DRMs. Several other proteins are also affected by treatment with edelfosine. Stars denote proteins that decrease, and arrows indicate protein bands that increase their presence. Several other bands do not change. The protein profile associated with DRMs changes according to growth conditions, incubation times, and edelfosine concentration. This is a representative experiment from at least 20 performed in similar or different conditions (see also protein profile from three different genetic backgrounds from Ref. 6). e, Western blot analysis of Pma1p, Gas1p glycosylphosphatidylinositol-anchored protein that partitions into DRMs, and Pgk1p (cytosol) showing the distribution of all three proteins in the entire gradient. f, Western blot analysis of the same proteins as in e in whole cell lysate samples (loading control).
A Pma1p mutant (pma1-7) that fails to associate with sphingolipid and ergosterol-rich membrane microdomains en route to the PM is targeted to the endosomal/vacuolar system (43) in a ubiquitin-dependent process (44). Because the effect of edelfosine resembles that of the pma1-7 mutant, we then investigated if edelfosine would also induce Pma1p ubiquitination. In fact, when cell extracts were immunoprecipitated with protein A-Sepharose beads coupled to an antibody against Pma1p, an extra band with a molecular mass ∼8 kDa higher than the Pma1p band from untreated cells was observed at early time points (1–2 h, Fig. 7a). This is compatible with a monoubiquitination signal and was further supported by the immunoprecipitation of ubiquitinated proteins followed by Western blot using an anti-Pma1p antibody (Fig. 7b). Ubiquitinated Pma1p was readily detected after drug treatment, although it was absent in untreated cells (Fig. 7b). Analysis of total cell lysates suggests Pma1p is not the only protein being ubiquitinated upon treatment with edelfosine (Fig. 6c). Furthermore, the unbiased results from our chemogenomic screen testing for enhanced resistance to edelfosine (69) identified genes involved in maintenance of the ubiquitin pool, E3-ubiquitin ligase adaptor proteins, as well as mutants of pathways that recognize ubiquitinated cargo like the ESCRT and retromer systems. Together, these results indicate that edelfosine induces exit of sterols and Pma1p from the PM followed by Pma1p internalization and degradation in a ubiquitin-mediated process.
FIGURE 7.
Edelfosine induces Pma1p ubiquitination. a, Pma1p immunoprecipitation allows for detection of a discrete band ∼8 kDa larger than Pma1p (arrow) upon edelfosine (EDLF) treatment, which could account for ubiquitinated species (upper panel). Coomassie-stained gels (lower panel) are shown as loading control. b, immunoprecipitation of total ubiquitinated proteins leads to detection of Pma1p (arrow) by Western blot only in edelfosine-treated samples. c, wild-type (BY4741) cells were grown in the absence or presence of edelfosine (19 μm) in defined medium for 2 h at 30 °C. Cells were collected and lysates prepared. Fifteen μg of total protein from each sample were separated by SDS-PAGE and blotted against ubiquitin (left panel) or Pma1p (right panel).
Edelfosine Alters Plasma Membrane Domain Organization, Inducing Ubiquitination and Internalization of Nutrient H+ Symporters
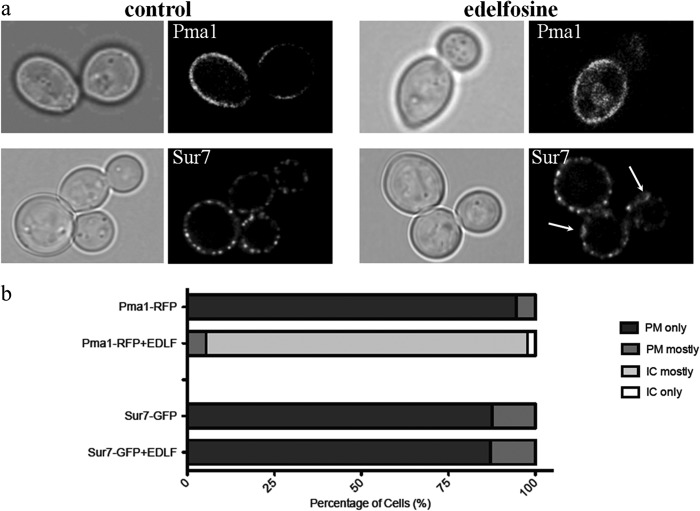
The emerging picture of the yeast PM indicates it is highly organized displaying an intricate array of domains with most membrane-associated proteins segregating into distinct areas (45, 46). Different distribution patterns have been observed for PM proteins by fluorescence microscopy, ranging from “patch-like” to “network”-shaped as described for the eisosome protein Sur7p and the proton pump Pma1p, respectively (45–47). These differences observed in vivo are lost when crude fractionation approaches, like isolation of DRMs, are used. Notoriously, all yeast PM proteins analyzed to date associate with DRMs (47–50). Therefore, the profile of PM proteins should be contained in that of DRMs. A comparison of the protein profile from control versus treated cells suggested that Pma1p was not the only protein disappearing from DRMs upon treatment with edelfosine (Fig. 6c) (6). We noticed that although the band corresponding to Pma1p was clearly affected, other bands also became fainter after edelfosine treatment (Fig. 6c, stars). This prompted us to analyze the effect of edelfosine on several well studied proteins that partition in a compartment that has been named MCC, for membrane compartment occupied by Can1 as opposed to MCP, the membrane compartment occupied by Pma1p (50, 51). The first MCC protein analyzed was Sur7p, as it forms distinct, nonoverlapping domains with MCP (45).
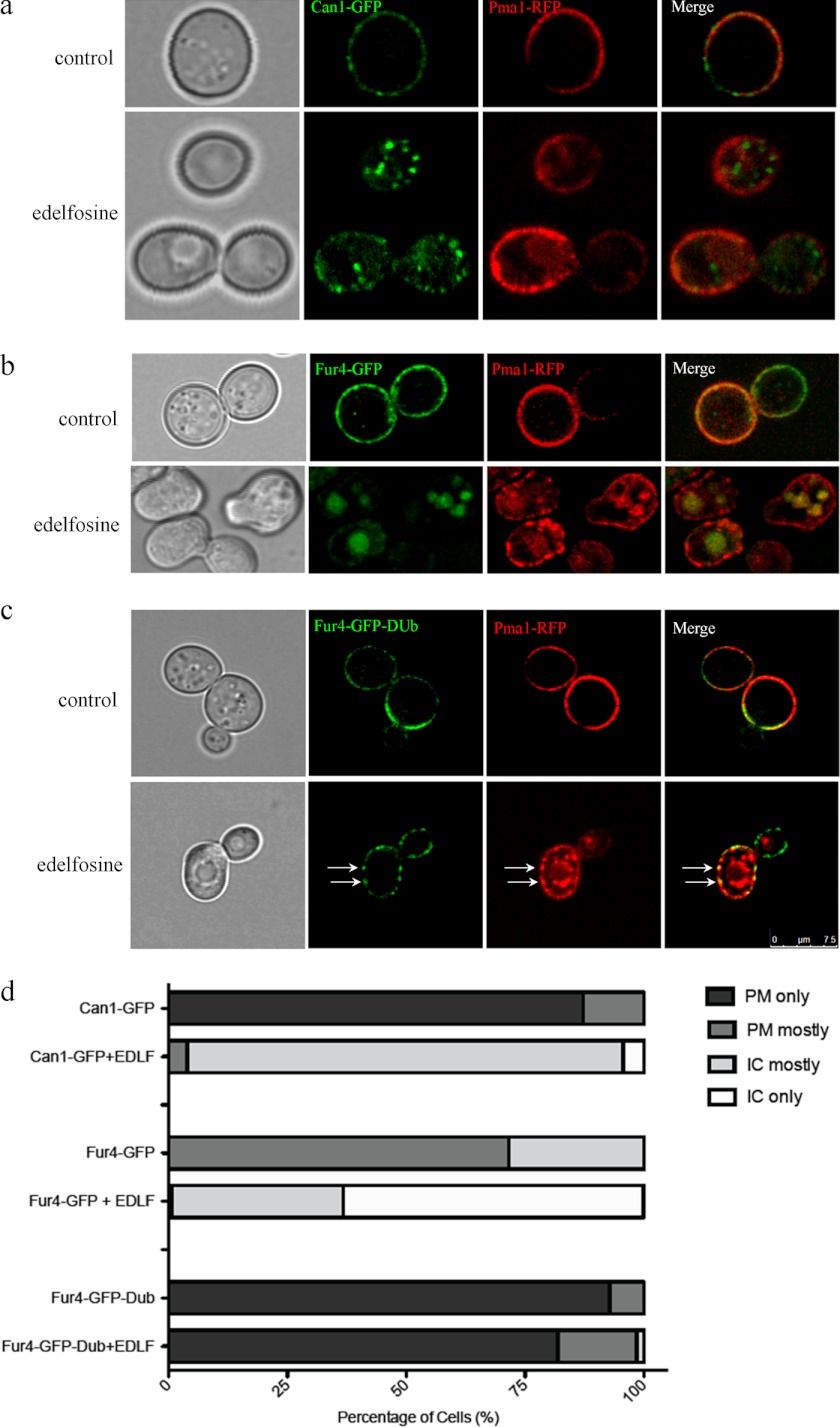
The effect of edelfosine treatment was studied by confocal microscopy using cells expressing Pma1p-RFP or Sur7p-GFP. Although edelfosine induced internalization of Pma1p and its accumulation in the vacuole, Sur7p remained associated with the PM in a patch-like distribution (Fig. 8). Closer inspection of Sur7p in edelfosine-treated cells revealed the formation of larger patches in the form of rings not observed in control cells (Fig. 8). The nutrient H+-symporters Can1p (arginine transporter) and Fur4p (uracil transporter) co-localize with Sur7p at MCC domains. Interestingly, in contrast to Sur7p and similar to Pma1p, edelfosine efficiently induced internalization of Can1p and Fur4p (Fig. 9, a and b). These results indicate that edelfosine alters the organization of the yeast PM by selectively inducing the internalization of Pma1p as well as the MCC resident nutrient H+-symporters Can1p and Fur4p, but not the structural eisosome protein Sur7p.
FIGURE 8.
Edelfosine does not induce internalization of the MCC resident protein Sur7p. a, cells with endogenously expressed Pma1p-RFP (top panel) or Sur7p-GFP (bottom panel) were grown to log phase in the absence (control) or presence of edelfosine (19 μm) for 1 h at 30 °C. Arrows indicate changes in the distribution pattern of Sur7p after treatment with edelfosine. b, quantitation of at least 100 cells from each condition. IC, intracellular; EDLF, edelfosine.
FIGURE 9.
Edelfosine induces internalization of nutrient H+-symporters Can1p and Fur4p of the MCC, ubiquitination of Fur4p by edelfosine. Cells co-expressing Pma1p-RFP and either Can1p-GFP, Fur4p-GFP, or Fur4p-GFP-Dub were grown to log phase in the presence or absence of edelfosine (19 μm) for 1 h at 30 °C before imaging using confocal microscopy. a, Pma1p (MCP) and Can1p (MCC) are internalized upon treatment with edelfosine. BF, bright field. b, Pma1p (MCP) and Fur4p (MCC) are internalized upon treatment with edelfosine. c, fusion of the catalytic domain of Ubp7p (DUb) to the carboxyl-terminal end of Fur4p-GFP (53) prevents internalization of Fur4p, whereas Pma1p still gets internalized upon treatment with edelfosine. Arrows point to the co-localization of Pma1p and Fur4p-GFP-Dub in large patches at the PM induced by edelfosine. Such distribution is absent from control cells. d, quantitation of at least 100 cells from each condition. IC, intracellular; EDLF, edelfosine.
Because ubiquitination of Fur4p is a known cell surface event that is required prior to internalization (52), investigations were conducted to test if this was also true in the case of treatment with edelfosine. We expressed Fur4p-GFP fused to the catalytic domain of the deubiquitinating peptidase Ubp7p (DUb), which has been shown to be a constitutive nonubiquitinatable form of Fur4p (53). These cells were also expressing Pma1p-RFP to allow co-localization studies. Cells were challenged with edelfosine and live cells imaged by confocal microscopy (Fig. 9c). In contrast to Pma1p-RFP and Fur4p-GFP, the presence of the DUb catalytic domain abolished Fur4p internalization induced by edelfosine. It is worth noting that although Fur4p-GFP-DUb remained at the PM, its distribution did indeed change, becoming patchier. A similar pattern was observed for Pma1p-RFP on the way to being internalized. Altogether these results indicate that internalization of nutrient H+-symporters like Fur4p depends on ubiquitination induced by edelfosine. They also suggest edelfosine induces lateral movement of PM proteins prior to ubiquitination.
DISCUSSION
Our findings clearly show that edelfosine alters the domain organization of the yeast plasma membrane by inducing changes in the distribution of lipids and proteins within domains. This in turn affects pH homeostasis, which emerged from this study as a major contributor to sensitivity toward edelfosine. Pma1p at the PM is a key regulator of pH homeostasis in yeast, and the displacement of Pma1p from lipid rafts appears to be a critical event that mediates the edelfosine effect. Indeed, treatment with edelfosine induced intracellular acidification in wild-type yeast. In this context, cells defective in V-ATPase activity were more susceptible to edelfosine as this vacuolar H+-pump is known to collaborate with Pma1p in preserving a proper cytosolic pH (32). In fact, Pma1p is missorted to the vacuole in vma mutants (32), and it has been recently shown that aberrant cytosolic pH in these mutants is responsible for missorting of Pma1p and other cargo proteins from the Golgi (54). Thus, intracellular pH controls sorting of PM proteins, and in the case of edelfosine mediates communication from the PM to internal compartments. Interestingly, the retromer vps35Δ mutant that is resistant to edelfosine displayed a higher buffering capacity (Fig. 4). This probably reflects recycling of Pma1p back to the PM upon treatment with edelfosine (69).
The functional categories of transporters and trafficking machinery were significantly enriched in a recent genome-wide survey of vacuolar pH under different stress conditions (55). These categories were also enriched in our resistance genetic screen presented in the accompanying paper (69). Of particular interest is the fact that the same phospholipid flippases mediating edelfosine uptake emerged as pH regulators, as did retromer and ESCRT mutants (55). This further highlights the role that intracellular pH plays in the communication between membranes in the cell. Our findings strongly support a proposed signaling role for cytosolic pH in the control of growth in yeast (41).
Our current model of action of edelfosine in yeast starts with edelfosine inducing the internalization of sterols from the PM. Indeed, reduced accessibility of sterols to edelfosine alleviates its cytotoxic effect (7). We hypothesize that edelfosine induces changes in the sterol retention capacity of the PM (56) by interfering with the interaction between sterols and sphingolipids (57). Perturbation of lipid composition of the PM and reduction in lipid complexity have in fact been associated with reduced protein segregation (45). We show in this study that segregation of several transporters, including Pma1p, Can1p, and Fur4p, is affected by edelfosine. These proteins act in consortium with Pma1p producing the protonmotive force that is consumed by nutrient H+-symporters like Can1p and Fur4p (47). Displacement of transport proteins leads to their ubiquitination, internalization, and vacuolar degradation. In contrast, Sur7p, a structural protein of eisosomes, remained at the PM after treatment with edelfosine. A similar scenario to the one depicted by edelfosine was observed upon induction of PM depolarization. MCC resident H+-symporters, including Can1p and Fur4p, moved out of MCC patches together with ergosterol resulting in growth inhibition (58, 59). Under the same conditions, Sur7p localization remained unaffected (58, 59).
Edelfosine triggers ubiquitination of proteins, including Pma1p (Fig. 7). Furthermore, we show herein that internalization of Fur4p induced by edelfosine is also mediated by ubiquitination, as the nonubiquitinatable form of Fur4p remains at the PM (Fig. 9). Fur4p has been extensively studied, and it is known that its sorting to the endosomal system is controlled by Rsp5p-dependent ubiquitination, which also involves the adaptor proteins Bul2p and Bsd2p (60–63). Rsp5p is an E3 ubiquitin ligase, and the null mutant is absent from the yeast deletion collection used in our chemogenomic screens. Interestingly, the genes coding for the Rsp5p protein adaptors BUL2 and BSD2 were identified in our resistance screen (69) suggesting they could mediate ubiquitination of Fur4p and other proteins in response to the changes in the PM introduced by edelfosine.
It is not clear at this point if changes in intracellular pH could be sensed by the ubiquitination machinery. In this regard, it is interesting to note that mutations in the PMA1 gene have been found to suppress the temperature sensitivity phenotype of rsp5 mutants (64). It is worth highlighting that the changes in lateral segregation of proteins targeted for internalization seem to precede ubiquitination, as Fur4-GFP-DUb alters its PM distribution upon treatment with edelfosine, coinciding with that of Pma1p en route to being internalized (Fig. 9).
Snf1p, the AMP-dependent kinase yeast orthologue, sits at the intersection of distinct organelles, including the vacuole, mitochondria, and nucleus (65). The interactome map for genes identified in our screen shows that SNF1 and SNF4 intersect many of them (Fig. 2), providing a link between the different functional categories enriched in our screen.
Snf1p senses ATP availability and controls cellular metabolism regulating the switch from fermentation to respiration, a condition known to induce a rapid drop in intracellular pH (32). Our results indicate that signaling through Snf1p modulates sensitivity of yeast to edelfosine. It is worth noting that fermentation is characteristic of cancer cells, and therefore the role of AMP-dependent kinase in the cellular response to edelfosine may be similar in cancer cells as in yeast (35). In this regard, we observed that susceptibility of wild-type yeast to edelfosine increases in the presence of nonfermentable or semi-fermentative carbon sources, like glycerol and galactose, respectively (data not shown). Interestingly, growth on galactose or glycerol induces cytosolic acidification (29), and we suggest that this lower pH threshold, analogous to vma2, trk1, and pma1 mutants, causes increased sensitivity to edelfosine. Furthermore, it has been reported Snf1p is a possible modulator of Trk1p. Cells lacking Snf1p and Snf4p are sensitive to acidic pH, and this depends on lipid metabolism (34). Snf1 also plays a critical role in response to alkaline pH stress (38).
Edelfosine sensitivity was exacerbated at acidic pHext and, interestingly, was also aggravated at a slightly alkaline pHext. We speculate that edelfosine may also affect proper partitioning of high affinity glucose transporters (i.e. Hxt2p) as well as iron and copper transporters into their unique co-existing microdomains at the PM (45). Uptake of glucose, iron, and copper has been shown to be critical to improve fitness of yeast at alkaline pH (38, 66). In addition, a critical role for Snf1 in tolerance to alkalinization due to the function of this kinase in regulating the switch from fermentative to respiration metabolism has been proposed (38, 65). This switch to respiration may also be responsible for the stronger effect of edelfosine in alkaline pH. In line with this rationale, a novel role for mitochondria in the maintenance of pH homeostasis has been recently unveiled (41). Furthermore, our resistance genetic screen was enriched in mitochondrial functional categories (69), suggesting impaired respiration alleviates edelfosine sensitivity. Another possibility is that edelfosine, or its effect on intracellular ergosterol accumulation, may affect proper V-ATPase assembly/function that is expected to confer susceptibility to high pHext (41, 55). In support of this hypothesis, a requirement of ergosterol for proper V-ATPase function has been established (67). Finally, uptake of edelfosine may be enhanced at alkaline pH. It is worth noting that expression of Drs2p, one of the yeast flippases, is augmented in response to high pHext (66). Experiments are underway to test these possibilities.
All the changes induced by edelfosine at the PM occur within the 1st h of treatment and up to this point are reversible, as cell viability is not compromised (6, 7). We have noticed that uptake of choline, cysteine, and methionine decreases within 30 min of incubation with edelfosine (6). This probably suggests a more general effect of edelfosine on nutrient uptake leading to a quiescent phase of growth. Therefore, we propose that during the 1st h, the effect of edelfosine is mostly cytostatic due to lack of nutrient uptake. After 1 h, edelfosine uptake reaches a maximum and accumulates in the endoplasmic reticulum (6, 69). Therefore, cells that can correct the changes induced by edelfosine at the PM would be able to resume growth despite the intracellular accumulation of edelfosine. This is the behavior we have noticed for strains that were able to recycle Pma1p back to the PM due to slower endocytosis and decreased protein degradation (6).
Our work clearly identifies the plasma membrane as the first target of edelfosine action. Uptake of edelfosine into the plasma membrane of cells alters sterol organization that in turn affects the organization of specific proteins that reside in sterol-enriched domains inducing a quiescent growth phase. Many of the PM transporters affected pump protons into and out of the cell with the end result being a reduction in cytoplasmic pH and eventually cell death. The buffering capacity of cells and their ability to restore pH homeostasis is critical for edelfosine efficacy.
Acknowledgments
We thank Drs, Smits, Walther, Tanner, Piper, Riezman, and Serrano for the kind gifts of strains, plasmids, and antibodies. We also thank J. Kearley for technical assistance.
This work was supported in part by a Natural Sciences and Engineering Research Council of Canada discovery grant, a seed grant from the University of Calgary, a Natural Sciences and Engineering Research Council of Canada University Faculty award (to V. Z.), Canadian Institutes of Health Research Grant 14124 (to C. R. M.), Spanish Ministerio de Economia y Competitividad Grants SAF2008-02251 and SAF2011-30518, Red Temática de Investigación Cooperativa en Cáncer, Instituto de Salud Carlos III, co-funded by the Fondo Europeo de Desarrollo Regional of the European Union Grants RD06/0020/1037 and RD12/0036/0065, European Community's Seventh Framework Programme FP7-2007-2013 Grant HEALTH-F2-2011-256986, (PANACREAS), and Junta de Castilla y León Grants CSI052A11-2 and CSI221A12-2 (to F. M.).
- PM
- plasma membrane
- ER
- endoplasmic reticulum
- DRM
- detergent-resistant membrane
- V-ATPase
- vacuolar proton-translocating ATPase.
REFERENCES
- 1. Simons K., Gerl M. J. (2010) Revitalizing membrane rafts: new tools and insights. Nat. Rev. Mol. Cell Biol. 11, 688–699 [DOI] [PubMed] [Google Scholar]
- 2. Simons K., Toomre D. (2000) Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 1, 31–39 [DOI] [PubMed] [Google Scholar]
- 3. Simons K., Sampaio J. L. (2011) Cold Spring Harbor Perspect. Biol. 3, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mollinedo F., Fernández M., Hornillos V., Delgado J., Amat-Guerri F., Acuña A. U., Nieto-Miguel T., Villa-Pulgarín J. A., González-García C., Ceña V., Gajate C. (2011) Involvement of lipid rafts in the localization and dysfunction effect of the antitumor ether phospholipid edelfosine in mitochondria. Cell Death Dis. 2, e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wright M. M., Howe A. G., Zaremberg V. (2004) Cell membranes and apoptosis: role of cardiolipin, phosphatidylcholine, and anticancer lipid analogues. Biochem. Cell Biol. 82, 18–26 [DOI] [PubMed] [Google Scholar]
- 6. Zaremberg V., Gajate C., Cacharro L. M., Mollinedo F., McMaster C. R. (2005) Cytotoxicity of an anti-cancer lysophospholipid through selective modification of lipid raft composition. J. Biol. Chem. 280, 38047–38058 [DOI] [PubMed] [Google Scholar]
- 7. Bitew T., Sveen C. E., Heyne B., Zaremberg V. (2010) Vitamin E prevents lipid raft modifications induced by an anti-cancer lysophospholipid and abolishes a Yap1-mediated stress response in yeast. J. Biol. Chem. 285, 25731–25742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gajate C., Mollinedo F. (2001) The antitumor ether lipid ET-18-OCH(3) induces apoptosis through translocation and capping of Fas/CD95 into membrane rafts in human leukemic cells. Blood 98, 3860–3863 [DOI] [PubMed] [Google Scholar]
- 9. Gajate C., Del Canto-Jañez E., Acuña A. U., Amat-Guerri F., Geijo E., Santos-Beneit A. M., Veldman R. J., Mollinedo F. (2004) Intracellular triggering of Fas aggregation and recruitment of apoptotic molecules into Fas-enriched rafts in selective tumor cell apoptosis. J. Exp. Med. 200, 353–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. van der Luit A. H., Budde M., Ruurs P., Verheij M., van Blitterswijk W. J. (2002) Alkyl-lysophospholipid accumulates in lipid rafts and induces apoptosis via raft-dependent endocytosis and inhibition of phosphatidylcholine synthesis. J. Biol. Chem. 277, 39541–39547 [DOI] [PubMed] [Google Scholar]
- 11. Nieto-Miguel T., Fonteriz R. I., Vay L., Gajate C., López-Hernández S., Mollinedo F. (2007) Endoplasmic reticulum stress in the proapoptotic action of edelfosine in solid tumor cells. Cancer Res. 67, 10368–10378 [DOI] [PubMed] [Google Scholar]
- 12. Gajate C., Matos-da-Silva M., Dakir el-H., Fonteriz R. I., Alvarez J., Mollinedo F. (2012) Antitumor alkyl-lysophospholipid analog edelfosine induces apoptosis in pancreatic cancer by targeting endoplasmic reticulum. Oncogene 31, 2627–2639 [DOI] [PubMed] [Google Scholar]
- 13. Boggs K. P., Rock C. O., Jackowski S. (1995) Lysophosphatidylcholine and 1-O-octadecyl-2-O-methyl-rac-glycero-3-phosphocholine inhibit the CDP-choline pathway of phosphatidylcholine synthesis at the CTP:phosphocholine cytidylyltransferase step. J. Biol. Chem. 270, 7757–7764 [DOI] [PubMed] [Google Scholar]
- 14. Boggs K. P., Rock C. O., Jackowski S. (1995) Lysophosphatidylcholine attenuates the cytotoxic effects of the antineoplastic phospholipid 1-O-octadecyl-2-O-methyl-rac-glycero-3-phosphocholine. J. Biol. Chem. 270, 11612–11618 [DOI] [PubMed] [Google Scholar]
- 15. Mollinedo F., Fernández-Luna J. L., Gajate C., Martín-Martín B., Benito A., Martínez-Dalmau R., Modolell M. (1997) Selective induction of apoptosis in cancer cells by the ether lipid ET-18-OCH3 (Edelfosine): molecular structure requirements, cellular uptake, and protection by Bcl-2 and Bcl-X(L). Cancer Res. 57, 1320–1328 [PubMed] [Google Scholar]
- 16. Gajate C., Mollinedo F. (2007) Edelfosine and perifosine induce selective apoptosis in multiple myeloma by recruitment of death receptors and downstream signaling molecules into lipid rafts. Blood 109, 711–719 [DOI] [PubMed] [Google Scholar]
- 17. Gajate C., Mollinedo F. (2002) Biological activities, mechanisms of action, and biomedical prospect of the antitumor ether phospholipid ET-18-OCH(3) (edelfosine), a proapoptotic agent in tumor cells. Curr. Drug Metab. 3, 491–525 [DOI] [PubMed] [Google Scholar]
- 18. Gajate C., Fonteriz R. I., Cabaner C., Alvarez-Noves G., Alvarez-Rodriguez Y., Modolell M., Mollinedo F. (2000) Intracellular triggering of Fas, independently of FasL, as a new mechanism of antitumor ether lipid-induced apoptosis. Int. J. Cancer 85, 674–682 [DOI] [PubMed] [Google Scholar]
- 19. Hanson P. K., Malone L., Birchmore J. L., Nichols J. W. (2003) Lem3p is essential for the uptake and potency of alkylphosphocholine drugs, edelfosine and miltefosine. J. Biol. Chem. 278, 36041–36050 [DOI] [PubMed] [Google Scholar]
- 20. Pomorski T., Holthuis J. C., Herrmann A., van Meer G. (2004) Tracking down lipid flippases and their biological functions. J. Cell Sci. 117, 805–813 [DOI] [PubMed] [Google Scholar]
- 21. Chen R., Brady E., McIntyre T. M. (2011) Human TMEM30a promotes uptake of antitumor and bioactive choline phospholipids into mammalian cells. J. Immunol. 186, 3215–3225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Winzeler E. A., Shoemaker D. D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J. D., Bussey H., Chu A. M., Connelly C., Davis K., Dietrich F., Dow S. W., El Bakkoury M., Foury F., Friend S. H., Gentalen E., Giaever G., Hegemann J. H., Jones T., Laub M., Liao H., Liebundguth N., Lockhart D. J., Lucau-Danila A., Lussier M., M'Rabet N., Menard P., Mittmann M., Pai C., Rebischung C., Revuelta J. L., Riles L., Roberts C. J., Ross-MacDonald P., Scherens B., Snyder M., Sookhai-Mahadeo S., Storms R. K., Véronneau S., Voet M., Volckaert G., Ward T. R., Wysocki R., Yen G. S., Yu K., Zimmermann K., Philippsen P., Johnston M., Davis R. W. (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906 [DOI] [PubMed] [Google Scholar]
- 23. Giaever G., Chu A. M., Ni L., Connelly C., Riles L., Véronneau S., Dow S., Lucau-Danila A., Anderson K., André B., Arkin A. P., Astromoff A., El-Bakkoury M., Bangham R., Benito R., Brachat S., Campanaro S., Curtiss M., Davis K., Deutschbauer A., Entian K. D., Flaherty P., Foury F., Garfinkel D. J., Gerstein M., Gotte D., Güldener U., Hegemann J. H., Hempel S., Herman Z., Jaramillo D. F., Kelly D. E., Kelly S. L., Kötter P., LaBonte D., Lamb D. C., Lan N., Liang H., Liao H., Liu L., Luo C., Lussier M., Mao R., Menard P., Ooi S. L., Revuelta J. L., Roberts C. J., Rose M., Ross-Macdonald P., Scherens B., Schimmack G., Shafer B., Shoemaker D. D., Sookhai-Mahadeo S., Storms R. K., Strathern J. N., Valle G., Voet M., Volckaert G., Wang C. Y., Ward T. R., Wilhelmy J., Winzeler E. A., Yang Y. H., Yen G., Youngman E., Yu K. X., Bussey H., Boeke J. D., Snyder M., Philippsen P., Davis R. W., Johnston M. (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387–391 [DOI] [PubMed] [Google Scholar]
- 24. Fyrst H., Oskouian B., Kuypers F. A., Saba J. D. (1999) The PLB2 gene of Saccharomyces cerevisiae confers resistance to lysophosphatidylcholine and encodes a phospholipase B/lysophospholipase. Biochemistry 38, 5864–5871 [DOI] [PubMed] [Google Scholar]
- 25. Zaremberg V., McMaster C. R. (2002) Differential partitioning of lipids metabolized by separate yeast glycerol-3-phosphate acyltransferases reveals that phospholipase D generation of phosphatidic acid mediates sensitivity to choline-containing lysolipids and drugs. J. Biol. Chem. 277, 39035–39044 [DOI] [PubMed] [Google Scholar]
- 26. Fairn G. D., McMaster C. R. (2005) Studying phospholipid metabolism using yeast systematic and chemical genetics. Methods 36, 102–108 [DOI] [PubMed] [Google Scholar]
- 27. Breitkreutz B. J., Stark C., Tyers M. (2003) Osprey: a network visualization system. Genome Biol. 4, R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Robinson M. D., Grigull J., Mohammad N., Hughes T. R. (2002) FunSpec: a web-based cluster interpreter for yeast. BMC Bioinformatics 3, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Orij R., Postmus J., Ter Beek A., Brul S., Smits G. J. (2009) In vivo measurement of cytosolic and mitochondrial pH using a pH-sensitive GFP derivative in Saccharomyces cerevisiae reveals a relation between intracellular pH and growth. Microbiology 155, 268–278 [DOI] [PubMed] [Google Scholar]
- 30. Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951) Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 31. Graham L. A., Hill K. J., Stevens T. H. (1998) Assembly of the yeast vacuolar H+-ATPase occurs in the endoplasmic reticulum and requires a Vma12p/Vma22p assembly complex. J. Cell Biol. 142, 39–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Martínez-Muñoz G. A., Kane P. (2008) Vacuolar and plasma membrane proton pumps collaborate to achieve cytosolic pH homeostasis in yeast. J. Biol. Chem. 283, 20309–20319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yenush L., Merchan S., Holmes J., Serrano R. (2005) pH-responsive, posttranslational regulation of the Trk1 potassium transporter by the type 1-related Ppz1 phosphatase. Mol. Cell. Biol. 25, 8683–8692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Young B. P., Shin J. J., Orij R., Chao J. T., Li S. C., Guan X. L., Khong A., Jan E., Wenk M. R., Prinz W. A., Smits G. J., Loewen C. J. (2010) Phosphatidic acid is a pH biosensor that links membrane biogenesis to metabolism. Science 329, 1085–1088 [DOI] [PubMed] [Google Scholar]
- 35. Zhang J., Vemuri G., Nielsen J. (2010) Systems biology of energy homeostasis in yeast. Curr. Opin. Microbiol. 13, 382–388 [DOI] [PubMed] [Google Scholar]
- 36. Portillo F., Mulet J. M., Serrano R. (2005) A role for the non-phosphorylated form of yeast Snf1: tolerance to toxic cations and activation of potassium transport. FEBS Lett. 579, 512–516 [DOI] [PubMed] [Google Scholar]
- 37. Ariño J., Ramos J., Sychrová H. (2010) Alkali metal cation transport and homeostasis in yeasts. Microbiol. Mol. Biol. Rev. 74, 95–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Casamayor A., Serrano R., Platara M., Casado C., Ruiz A., Ariño J. (2012) The role of the Snf1 kinase in the adaptive response of Saccharomyces cerevisiae to alkaline pH stress. Biochem. J. 444, 39–49 [DOI] [PubMed] [Google Scholar]
- 39. Schuldiner M., Collins S. R., Thompson N. J., Denic V., Bhamidipati A., Punna T., Ihmels J., Andrews B., Boone C., Greenblatt J. F., Weissman J. S., Krogan N. J. (2005) Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell 123, 507–519 [DOI] [PubMed] [Google Scholar]
- 40. Orij R., Brul S., Smits G. J. (2011) Intracellular pH is a tightly controlled signal in yeast. Biochim. Biophys. Acta 1810, 933–944 [DOI] [PubMed] [Google Scholar]
- 41. Orij R., Urbanus M. L., Vizeacoumar F. J., Giaever G., Boone C., Nislow C., Brul S., Smits G. J. (2012) Genome-wide analysis of intracellular pH reveals quantitative control of cell division rate by pH(c) in Saccharomyces cerevisiae. Genome Biol. 13, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Carmelo V., Bogaerts P., Sá-Correia I. (1996) Activity of plasma membrane H+-ATPase and expression of PMA1 and PMA2 genes in Saccharomyces cerevisiae cells grown at optimal and low pH. Arch. Microbiol. 166, 315–320 [DOI] [PubMed] [Google Scholar]
- 43. Bagnat M., Chang A., Simons K. (2001) Plasma membrane proton ATPase Pma1p requires raft association for surface delivery in yeast. Mol. Biol. Cell 12, 4129–4138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pizzirusso M., Chang A. (2004) Ubiquitin-mediated targeting of a mutant plasma membrane ATPase, Pma1–7, to the endosomal/vacuolar system in yeast. Mol. Biol. Cell 15, 2401–2409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Spira F., Mueller N. S., Beck G., von Olshausen P., Beig J., Wedlich-Söldner R. (2012) Patchwork organization of the yeast plasma membrane into numerous coexisting domains. Nat. Cell Biol. 14, 640–648 [DOI] [PubMed] [Google Scholar]
- 46. Ziółkowska N. E., Christiano R., Walther T. C. (2012) Organized living: formation mechanisms and functions of plasma membrane domains in yeast. Trends Cell Biol. 22, 151–158 [DOI] [PubMed] [Google Scholar]
- 47. Malínská K., Malínský J., Opekarová M., Tanner W. (2003) Visualization of protein compartmentation within the plasma membrane of living yeast cells. Mol. Biol. Cell 14, 4427–4436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Olivera-Couto A., Aguilar P. S. (2012) Eisosomes and plasma membrane organization. Mol. Genet. Genomics 287, 607–620 [DOI] [PubMed] [Google Scholar]
- 49. Bagnat M., Keränen S., Shevchenko A., Shevchenko A., Simons K. (2000) Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc. Natl. Acad. Sci. U.S.A. 97, 3254–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lauwers E., André B. (2006) Association of yeast transporters with detergent-resistant membranes correlates with their cell-surface location. Traffic 7, 1045–1059 [DOI] [PubMed] [Google Scholar]
- 51. Surma M. A., Klose C., Simons K. (2012) Lipid-dependent protein sorting at the trans-Golgi network. Biochim. Biophys. Acta 1821, 1059–1067 [DOI] [PubMed] [Google Scholar]
- 52. Marchal C., Haguenauer-Tsapis R., Urban-Grimal D. (1998) A PEST-like sequence mediates phosphorylation and efficient ubiquitination of yeast uracil permease. Mol. Cell. Biol. 18, 314–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stringer D. K., Piper R. C. (2011) A single ubiquitin is sufficient for cargo protein entry into MVBs in the absence of ESCRT ubiquitination. J. Cell Biol. 192, 229–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Huang C., Chang A. (2011) pH-dependent cargo sorting from the Golgi. J. Biol. Chem. 286, 10058–10065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Brett C. L., Kallay L., Hua Z., Green R., Chyou A., Zhang Y., Graham T. R., Donowitz M., Rao R. (2011) Genome-wide analysis reveals the vacuolar pH-stat of Saccharomyces cerevisiae. PLoS One 6, e17619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Baumann N. A., Sullivan D. P., Ohvo-Rekilä H., Simonot C., Pottekat A., Klaassen Z., Beh C. T., Menon A. K. (2005) Transport of newly synthesized sterol to the sterol-enriched plasma membrane occurs via nonvesicular equilibration. Biochemistry 44, 5816–5826 [DOI] [PubMed] [Google Scholar]
- 57. Hąc-Wydro K., Dynarowicz-L̸atka P., Wydro P., Bąk K. (2011) Edelfosine disturbs the sphingomyelin-cholesterol model membrane system in a cholesterol-dependent way–the Langmuir monolayer study. Colloids Surf. B Biointerfaces 88, 635–640 [DOI] [PubMed] [Google Scholar]
- 58. Grossmann G., Opekarová M., Malinsky J., Weig-Meckl I., Tanner W. (2007) Membrane potential governs lateral segregation of plasma membrane proteins and lipids in yeast. EMBO J. 26, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Grossmann G., Malinsky J., Stahlschmidt W., Loibl M., Weig-Meckl I., Frommer W. B., Opekarová M., Tanner W. (2008) Plasma membrane microdomains regulate turnover of transport proteins in yeast. J. Cell Biol. 183, 1075–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Blondel M. O., Morvan J., Dupré S., Urban-Grimal D., Haguenauer-Tsapis R., Volland C. (2004) Direct sorting of the yeast uracil permease to the endosomal system is controlled by uracil binding and Rsp5p-dependent ubiquitylation. Mol. Biol. Cell 15, 883–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Nikko E., Pelham H. R. (2009) Arrestin-mediated endocytosis of yeast plasma membrane transporters. Traffic 10, 1856–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Novoselova T. V., Zahira K., Rose R. S., Sullivan J. A. (2012) Bul proteins, a nonredundant, antagonistic family of ubiquitin ligase regulatory proteins. Eukaryot. Cell 11, 463–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lauwers E., Erpapazoglou Z., Haguenauer-Tsapis R., André B. (2010) The ubiquitin code of yeast permease trafficking. Trends Cell Biol. 20, 196–204 [DOI] [PubMed] [Google Scholar]
- 64. Kamińska J., Tobiasz A., Gniewosz M., Zol̸adek T. (2000) The growth of mdp1/rsp5 mutants of Saccharomyces cerevisiae is affected by mutations in the ATP-binding domain of the plasma membrane H+-ATPase. Gene 242, 133–140 [DOI] [PubMed] [Google Scholar]
- 65. Vincent O., Townley R., Kuchin S., Carlson M. (2001) Subcellular localization of the Snf1 kinase is regulated by specific β subunits and a novel glucose signaling mechanism. Genes Dev. 15, 1104–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Serrano R., Bernal D., Simón E., Ariño J. (2004) Copper and iron are the limiting factors for growth of the yeast Saccharomyces cerevisiae in an alkaline environment. J. Biol. Chem. 279, 19698–19704 [DOI] [PubMed] [Google Scholar]
- 67. Zhang Y. Q., Gamarra S., Garcia-Effron G., Park S., Perlin D. S., Rao R. (2010) Requirement for ergosterol in V-ATPase function underlies antifungal activity of azole drugs. Plos Pathog. 6, e1000939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Walther T. C., Brickner J. H., Aguilar P. S., Bernales S., Pantoja C., Walter P. (2006) Eisosomes mark static sites of endocytosis. Nature 439, 998–1003 [DOI] [PubMed] [Google Scholar]
- 69. Cuesta-Marbán Á., Botet J., Czyz O., Cacharro L. M., Gajate C., Hornillos V., Delgado J., Zhang H., Amat-Guerri F., Acuña A. U., McMaster C. R., Revuelta J. L., Zaremberg V., Mollinedo F. (2013) Drug uptake, lipid rafts, and vesicle trafficking modulate resistance to an anticancer lysophosphatidylcholine analogue in yeast. J. Biol. Chem. 288, 8405–8418 [DOI] [PMC free article] [PubMed] [Google Scholar]