Background: SodA is an important virulence factor in Borrelia burgdorferi.
Results: This SodA requires extraordinarily high intracellular manganese for activity and accumulates as either manganese or apoprotein, but not iron-bound.
Conclusion: B. burgdorferi SodA is a unique Mn-SOD based on metal requirements and predicted structure.
Significance: B. burgdorferi pathogenicity may be controlled by exploiting the unusual properties of SodA.
Keywords: Manganese, Metalloenzymes, Metalloproteins, Metals, Microbial pathogenesis, Superoxide Dismutase (SOD), Superoxide Ion, Yeast Metabolism, Yeast Physiology
Abstract
The Lyme disease pathogen Borrelia burgdorferi represents a novel organism in which to study metalloprotein biology in that this spirochete has uniquely evolved with no requirement for iron. Not only is iron low, but we show here that B. burgdorferi has the capacity to accumulate remarkably high levels of manganese. This high manganese is necessary to activate the SodA superoxide dismutase (SOD) essential for virulence. Using a metalloproteomic approach, we demonstrate that a bulk of B. burgdorferi SodA directly associates with manganese, and a smaller pool of inactive enzyme accumulates as apoprotein. Other metalloproteins may have similarly adapted to using manganese as co-factor, including the BB0366 aminopeptidase. Whereas B. burgdorferi SodA has evolved in a manganese-rich, iron-poor environment, the opposite is true for Mn-SODs of organisms such as Escherichia coli and bakers' yeast. These Mn-SODs still capture manganese in an iron-rich cell, and we tested whether the same is true for Borrelia SodA. When expressed in the iron-rich mitochondria of Saccharomyces cerevisiae, B. burgdorferi SodA was inactive. Activity was only possible when cells accumulated extremely high levels of manganese that exceeded cellular iron. Moreover, there was no evidence for iron inactivation of the SOD. B. burgdorferi SodA shows strong overall homology with other members of the Mn-SOD family, but computer-assisted modeling revealed some unusual features of the hydrogen bonding network near the enzyme's active site. The unique properties of B. burgdorferi SodA may represent adaptation to expression in the manganese-rich and iron-poor environment of the spirochete.
Introduction
Superoxide dismutases (SODs)3 represent families of metal-containing enzymes that catalyze the disproportionation of superoxide to hydrogen peroxide and oxygen. One family includes the Mn- and Fe-SOD enzymes that are well conserved from archaea to humans (1, 2). The manganese versus iron binding forms of this family are highly homologous to one another and can bind either metal with similar geometries and metal binding affinities (3–7), yet Mn-SODs are only active with manganese bound, and substitution with iron in the active site will destroy catalytic activity, largely due to disruption of redox potential. The converse is true with Fe-SODs; manganese binding inactivates the enzyme (8, 9). It is therefore critical that these SODs only capture their correct co-factor.
Most organisms are “iron-philic” and accumulate high micromolar to nearly millimolar levels of iron to catalyze a variety biochemical processes (10–12). Iron accumulation is typically 1–2 orders of magnitude higher than manganese and, based on the Irving-Williams series, is predicted to bind preferentially to cellular ligands over manganese, placing manganese at an apparent disadvantage for co-factor selection in SODs. Nevertheless, Mn-SOD enzymes have evolved methods for avoiding iron and inserting manganese into the active site, a classic example being the mitochondrial manganese Sod2p of Saccharomyces cerevisiae. Despite the 50-fold abundance of mitochondrial iron over manganese, Sod2p captures manganese and is virtually impervious to iron inactivation except under rare cases of manganese starvation or with certain yeast mutants of mitochondrial iron overload (1, 13, 14). Such exclusion of cellular iron appears conserved because the Mn-SodA from E. coli targeted to yeast mitochondria also acquires manganese over the more abundant metal, iron (14).
The need to avoid iron may be obviated with SOD enzymes from the Lyme disease pathogen, Borrelia burgdorferi. Elegant studies by Posey and Gherardini (15) have shown that this spirochete fails to accumulate any appreciable iron and does not express any known iron-specific enzymes. The total lack of an iron requirement is advantageous to B. burgdorferi during infection when the host attempts to starve pathogens of iron (15–17). B. burgdorferi expresses a single SOD of the Fe/Mn family that is essential for virulence (18). Based on the apparent lack of cellular iron, B. burgdorferi SodA is proposed to bind manganese (18), yet direct binding of manganese to B. burgdorferi SodA has not been demonstrated. Two independent studies have investigated the co-factor specificity of B. burgdorferi SodA based on differential H2O2 resistance (Mn-SOD enzymes should be resistant to peroxide), but the findings have been conflicting; one report (19) concludes the SOD binds iron, whereas a more recent study by Troxell et al. (20) concludes that B. burgdorferi SodA is a Mn-SOD. Furthermore, the implications for a SOD enzyme evolving in an iron-depleted cell have not been examined. Can a SOD enzyme that has only seen manganese still capture its co-factor in an iron-rich cellular environment?
Here we investigate the activity and metal requirement for B. burgdorferi SodA expressed in its native host versus a heterologous iron-philic host, namely the bakers' yeast S. cerevisiae. We find that B. burgdorferi can accumulate remarkably high levels of manganese that are needed to support activity of its SodA. Using a metalloproteomic approach, we demonstrate that B. burgdorferi SodA exists as active Mn-SOD enzyme as well as inactive apoprotein but does not bind other metals. When expressed heterologously in the iron-philic host S. cerevisiae, B. burgdorferi SodA is only active when the yeast accumulates vast quantities of manganese that exceed total cellular iron, a condition analogous to the natural B. burgdorferi host. Unlike the homologous Mn-Sod enzymes from yeast and E. coli, B. burgdorferi SodA does not appear to have evolved with the capacity for capturing manganese in an iron-rich environment.
EXPERIMENTAL PROCEDURES
Strains, Growth Media, and Plasmids
The B. burgdorferi WT strains ML23 and 297 and the bmtA mutant were described previously (18, 21). All yeast strains were derived from BY4741 and include the isogenic sod1Δ::kanMX4, sod2Δ::kanMX4 and the sod1Δ sod2Δ mutant AR142 (22). Escherichia coli strain DH5α was used.
B. burgdorferi was typically grown in BSK medium (pH 7.6) supplemented with 6% (v/v) rabbit serum (Sigma) also containing 0.05 mg/ml rifampicin, 0.1 mg/ml phosphomycin, and 5 μg/ml amphotericin b (18). BSK medium supplemented with synthetic Ex-cyte (Millipore) rather than rabbit serum was prepared precisely as described (15). B. burgdorferi cultures were typically inoculated from frozen stocks at a density of 104 and grown at 34 °C (unless indicated otherwise) to a density of 107 to 108 cells/ml. Yeast strains were grown in an enriched YPD (yeast extract, peptone, dextrose) at 30 °C, and E. coli was grown in BSK medium without antibiotics and at 37 °C.
The pAN002 plasmid for expressing E. coli SodA in the mitochondria of yeast and under the yeast SOD2 promoter and terminator was described previously (14). Plasmid pDA002 is a derivative of pAN002 in which the SodA coding region of E. coli was replaced with B. burgdorferi SodA. A DNA cassette was synthesized (Celtek Genes) consisting of the open reading frame of B. burgdorferi SodA that was codon-optimized for expression in yeast and engineered to contain flanking NdeI and BglII restriction sites at the start and stop codons, respectively. The cassette was inserted into the pGH vector (Celtek Genes), and following digestion with NdeI and BglII, the mobilized cassette was introduced into plasmid pAN002 digested with these same enzymes, replacing the E. coli SodA coding region with B. burgdorferi SodA. In the resultant plasmid, pDA002, B. burgdorferi SodA was fused in-frame to the mitochondrial leader sequence (MLS) of S. cerevisiae Sod2p and under the SOD2 gene promoter. Plasmid pSP002 for expressing B. burgdorferi SodA in the yeast cytosol was constructed by removing the MLS in plasmid pDA002. A NdeI site was introduced by oligonucleotide-directed mutagenesis at the yeast SOD2 start site for translation. Digestion with NdeI and religation resulted in removal of the MLS. All plasmids were verified by DNA sequencing.
Biochemical Analyses
For preparation of B. burgdorferi cell lysates, cultures of B. burgdorferi were inoculated at a density of 104 cells/ml and grown to 3–8 × 107 cells/ml. Cells were harvested by centrifugation at 3200 × g at 4 °C and washed twice in PBS and twice in metal-free deionized water prior to resuspension in lysis buffer containing 10 mm sodium phosphate, pH 7.8, 5 mm EDTA, 5 mm EGTA, 50 mm NaCl, 0.45% (v/v) Nonidet P-40. Cells were lysed in a TissueLyser using 0.7-mm zirconium oxide beads (three cycles at 50 Hz for 3 min interspersed with 3 min on ice). Lysates were then clarified by centrifugation at 20,000 × g for 10 min at 4 °C. To prepare lysates for native and denaturing gel analyses, 45-ml cultures were used and cells were lysed in 150 μl of lysis buffer also containing 10% (v/v) glycerol. For large scale lysate preparations as required for multidimensional chromatography (see below), 600-ml cultures were used, and cells were lysed in 2.0 ml of buffer lacking glycerol. E. coli lysates for metal analysis were prepared as described above for B. burgdorferi, using E. coli grown in BSK medium at 37 °C to A600 ∼2.0. S. cerevisiae lysates were prepared from strains grown non-shaking for 20 h in YPD medium to a final A600 of ∼1.0–5.0. Cells were lysed by glass bead homogenization as described (23), except the lysis buffer also contained 10% (v/v) glycerol. In all cases, protein concentration was determined by the Bradford method.
For measurements of SOD protein and activity, lysates from S. cerevisiae or B. burgdorferi were partially enriched for SODs by heating at 42 °C for 20 min followed by centrifugation at 20,000 × g. This treatment removes ∼30% of total cellular protein with no loss in activity or protein levels of Cu/Zn-SODs or the Mn-SodAs of B. burgdorferi or E. coli. SOD activity was carried by the native gel assay (14, 24). Lysates from B. burgdorferi (2.5–25 μg of cellular protein) or from S. cerevisiae (50–75 μg) were subjected to native gel electrophoresis using 12% precast gels (Invitrogen) and staining with nitro blue tetrazolium as described (14, 24). For in-gel inactivation of SODs by peroxide, gels were soaked in 50 mm phosphate buffer, pH 8.1, containing the designated concentrations of H2O2 for 1 h prior to rinsing in H2O and incubating in nitro blue tetrazolium staining solution. To specifically inactivate yeast Cu/Zn-Sod1p, 5 mm H2O2 was used. For immunoblot analyses, 0.5–10.0 μg of B. burgdorferi or 50–75 μg of S. cerevisiae lysate protein was subject to denaturing gel electrophoresis on 10% polyacrylamide SDS gels, followed by transfer to membranes and hybridization to a mouse anti-SodA antibody (18) at 1:1500–2000 dilution and a secondary donkey anti-mouse antibody at 1:5000. Where indicated, S. cerevisiae blots were also probed with an anti-yeast Sod2p (14) and Pgk1p (23) antibodies as described.
For whole cell manganese analysis of B. burgdorferi by atomic absorption spectroscopy (AAS), ∼109 cells grown and harvested as described above were washed twice in either PBS or TE (10 mm Tris, 1 mm EDTA, pH 7.6) (results were identical with either PBS or TE), followed by dual washes in metal-free milliQ water. Cells were resuspended in 1 ml of 65–70% (v/v) nitric acid (Ultrex, high purity) and heated at 80 °C for 1 h. Cell debris was removed by centrifugation for 5 min at 20,000 × g. Samples were diluted 1:14 (WT) or 1:2 (bmtA mutant) in metal-free milliQ H2O prior to analysis by graphite furnace AAS (Analyst 600, PerkinElmer Life Sciences). AAS measurements of manganese in soluble protein lysates used lysates from B. burgdorferi, S. cerevisiae, and E. coli prepared as described above.
For iron and manganese analysis by Inductively coupled plasma mass spectrometry (ICP-MS), 109 to 1010 B. burgdorferi cells grown and harvested as above were washed twice in TE and once in metal-free milliQ water. As a blank control, the same volume of BSK medium incubated in parallel but with no cells was subjected to the identical centrifugation and washing treatments. The no cell control and B. burgdorferi pellet were heated in nitric acid and clarified by centrifugation as described above for AAS. Samples were diluted 1:30 in metal-free milliQ H2O and analyzed by ICP-MS (Agilent 7500ce; Johns Hopkins NIEHS Center Core Facility). Any elements detected in the blank control were subtracted from the B. burgdorferi sample. Under these conditions, there was no iron that could be detected above background in B. burgdorferi. ICP-MS analyses of whole yeast cells and E. coli were carried out in the same manner using 108 S. cerevisiae cells grown in YPD or 108 E. coli cells grown in BSK.
Multidimensional Chromatography for Metal Analysis of B. burgdorferi SodA
Soluble B. burgdorferi lysates were diluted in Tris buffer (50 mm, pH 8.8) and loaded onto an anion exchange column (1-ml HP HiTrap Q, GE Healthcare) at 0.5 ml/min. Proteins were eluted with 0.1, 0.2, 0.3, 0.4, 0.5, and 1 m sodium chloride Tris buffer (50 mm, pH 8.8) solutions. The 0.3 and 0.4 m NaCl elutions were concentrated using 3000 molecular weight cut-off spin columns (VIVASPIN 500, Sartorius Stedim Biotech) and then injected onto a size exclusion column (0.5 ml/min, 10 mm Tris buffer, 50 mm NaCl, pH 7.5, TSKgel G3000SWXL, TOSOH Bioscience) with fractions collected each minute. Aliquots of each eluted fraction were prepared for proteomic and ICP-MS mass spectrometry analyses. Proteomic samples were digested with trypsin (Trypsin Gold, Promega Corp.). For elemental analysis by ICP-MS, each fraction aliquot was diluted 1:4 into 5% (v/v) nitric acid containing 1 ppb Indium as an internal standard. ICP-MS analysis was performed on a Thermo Element 2 with an Aridus spray chamber (CETAC Technologies) with external calibration by plasma standards (SPEX CertiPrep Ltd.) and correction for matrix effects by Indium normalization.
LC/MS samples were concentrated onto a peptide cap trap and rinsed with 150 μl of 0.1% formic acid and 5% acetonitrile (v/v) in water, before gradient elution through a reversed phase Magic C18 AQ column (0.1 × 150 mm, 3-μm particle size, 200-Å pore size, Michrom Bioresources Inc.) on an Advance HPLC system (Michrom Bioresources Inc.) at a flow rate of 500 nl/min. The chromatography consisted of a gradient from 5% buffer A to 95% buffer B for 80 min, where A was 0.1% formic acid in water and B was 0.1% formic acid in acetonitrile. An LTQ linear ion trap mass spectrometer (Thermo Scientific Inc.) was used with an Advance CaptiveSpray source (Michrom Bioresources Inc.). The LTQ spectrometer was set to perform MS/MS on the top five ions using data-dependent settings, and ions were monitored over a range of 400–2000 m/z. Protein identifications were conducted using SEQUEST (Bioworks version 3.3, Thermo Inc.) using filters of ΔCN >0.1, >30% ions, Xcorr versus charge state of 1.9, 2.4, 2.9 for +1, +2, and +3 charges, respectively, and peptide probability of <1e−3. Protein identifications and relative protein abundances in each fraction (as normalized spectral counts) were also determined using Scaffold using protein and peptide probability settings of 99.9 and 95% and two tryptic peptides, respectively (Proteome Software version 3.5.1).
RESULTS
Activity of SodA in Its Native B. burgdorferi Host
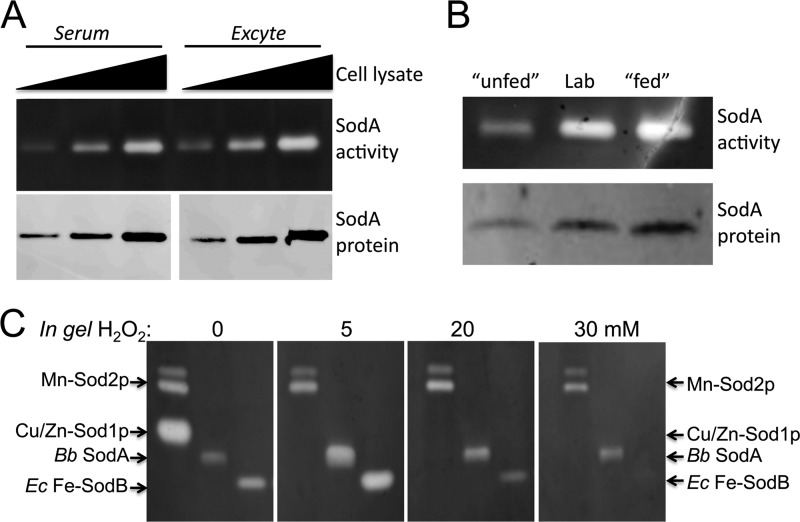
B. burgdorferi can be cultured outside the host in the laboratory using a serum-rich “BSK” growth medium. In these conditions, B. burgdorferi is seen to express a single SodA superoxide dismutase whose activity can be detected by a native gel assay (Fig. 1A) (18–20). To test whether SodA expression was dependent on serum factors, we replaced the 6% rabbit serum with Ex-cyte, a synthetic substitute (15). As seen in Fig. 1A, there is no change in SodA protein levels or enzymatic activity in cells cultured with synthetic Ex-cyte supplements, indicating that activity is not dependent on serum protein factors. We also examined effects of variations in growth conditions that have been reported to mimic the environment of the tick host, namely growth at pH 7.6 and 23 °C and at pH 6.7 and 37 °C, to simulate the unfed and post-blood meal conditions, respectively (25). As seen in Fig. 1B, B. burgdorferi SodA protein levels and activity are maximal under the laboratory conditions thought to best simulate the post-blood meal state.
FIGURE 1.
Activity of B. burgdorferi SodA in its native host and its sensitivity toward peroxide. A and B, whole cell B. burgdorferi lysates were prepared from strain ML23 and were analyzed for SOD activity by native gel electrophoresis and nitro blue tetrazolium staining (SodA activity) and for SodA protein levels by immunoblot (SodA protein), as described under “Experimental Procedures.” A, cells were grown in BSK medium supplemented with either 6% (v/v) rabbit serum or with the synthetic serum substitute Ex-cyte. Wedge represents increasing levels of lysate protein analyzed: 2.5, 5, and 10 μg for SodA activity and 0.5, 1, and 2.5 μg for immunoblot. B, cells were grown in serum containing BSK under the following conditions: unfed, pH 7.6, 23 °C to simulate unfed tick host (25); lab, pH 7.6, 34 °C standard laboratory culture conditions; fed, pH 6.7, 37 °C to simulate post-blood meal conditions in the tick host (25). C, samples containing the indicated SOD enzymes were subjected to native gel electrophoresis, and prior to staining with nitro blue tetrazolium for SOD activity, the gel was incubated with the indicated concentrations of H2O2 as described under “Experimental Procedures.” Mn-Sod2p and Cu/Zn-Sod1p, S. cerevisiae SOD enzymes present in 50 μg of total yeast lysate protein. Bb SodA, 5.0 μg of B. burgdorferi lysate protein; Ec Fe-SodB, 0.06 units of purified SodB enzyme from E. coli (Sigma).
Direct Association of Manganese with B. burgdorferi SodA
Based on the virtual absence of iron in B. burgdorferi, SodA is most likely a manganese enzyme, and this notion has been supported by recent studies examining peroxide resistance of the SOD (20). As seen in Fig. 1C, B. burgdorferi SodA follows the H2O2 resistance of the mitochondrial Mn-Sod2p of bakers' yeast (Fig. 1C, far right panel). However, peroxide resistance is not definitive proof of manganese binding because Fe-SOD enzymes can also be characterized as somewhat peroxide-resistant. As seen in Fig. 1C, both Fe- and Mn-SODs retain activity with millimolar levels of H2O2 that inactivate Cu/Zn-Sod1p. Hence, more direct methods of metal analysis are required. To this end, we carried out a metalloproteomic approach (26–30) to identifying the co-factor associated with B. burgdorferi SodA.
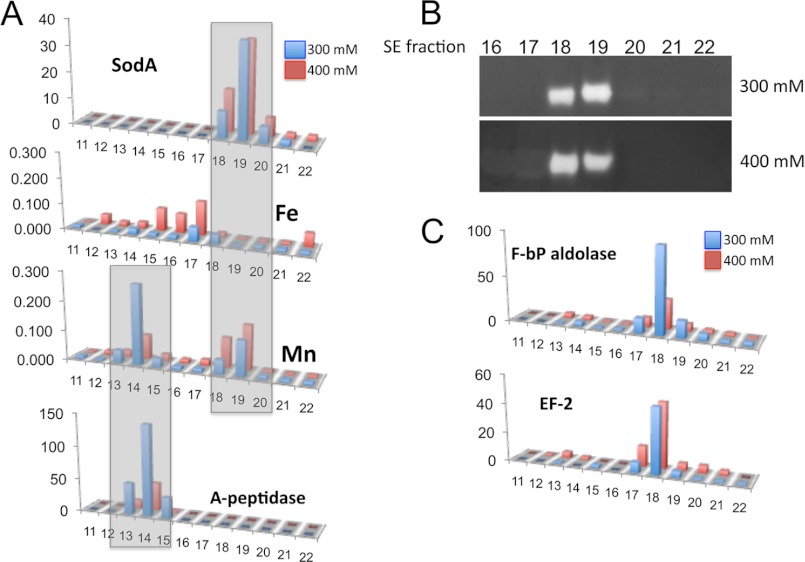
Soluble B. burgdorferi lysates were resolved by two-dimensional (strong anion exchange and size exclusion) chromatography under non-denaturing conditions (31), and fractionated proteins were digested with trypsin and identified by reversed-phase LC/MS and linear trap MS. Fig. 2 shows results from resolution of the 300 and 400 mm ion exchange fractions found to contain the SodA polypeptide. The peak of SodA protein identified by mass spectrometry in fractions 18 and 19 (Fig. 2A, top) retained full enzymatic activity (Fig. 2B); hence, the SOD retained its metal co-factor during fractionation. A shoulder of SodA protein eluting in fraction 20 (Fig. 2A, top) was devoid of enzymatic activity (Fig. 2B) and may be missing metal co-factor. Indeed, by ICP-MS, there is a well defined manganese peak that co-eluted with active SodA in fractions 18 and 19 but not with inactive SodA in fraction 20; moreover, there was no clear association between SodA and iron (Fig. 2, A and B). Likewise, zinc and copper failed to associate with the SodA protein (not shown). SodA was the only protein that closely overlapped this manganese peak. Although there was a partial overlap with a fructose bis-aldolase and EF-2 translation initiation factor in fraction 18, this could not account for the manganese signal in fraction 19 (Fig. 2, A and C). Together, these results demonstrating a tight association between active SodA enzyme and manganese (but not iron) establish SodA as a manganese-dependent SOD.
FIGURE 2.
Multidimensional chromatography of B. burgdorferi lysates. Soluble B. burgdorferi lysates were resolved by anion exchange, and the 300 and 400 mm NaCl elutions were resolved by size exclusion (SE) chromatography; shown are results from fractions 11–22 of increasing retention time on size exclusion. A, fractions were subjected to either metal analysis by ICP-MS (Fe and Mn), where y axis units represent relative abundance, or proteomic analysis by trypsin digestion, LC/MS, and MS/MS (SodA and A-peptidase), where y axis units represent spectral counts. Shaded boxes indicate manganese peak overlaps with SodA and with amino peptidase-1 (gene BB0366). B, fractions were analyzed for SodA activity by the native gel assay. C, proteomic analysis of fractions to illustrate that the manganese in fraction 19 shows poor correlation with a fructose bisphosphate aldolase (F-bP aldolase gene BB0445) and elongation factor EF-2 (EF-2 gene BB0540).
It is noteworthy that in addition to the SodA-containing peak in manganese, there was a second manganese peak eluting earlier by size exclusion (Fig. 2A). This second peak completely overlaps with a B. burgdorferi aminopeptidase (BB0366) that, in various organisms, uses iron, zinc, or manganese as a co-factor (32–35). This study underscores the power of multidimensional chromatography coupled to quantitative proteomics in identification of metal-protein partnerships.
SodA Is Active in a Manganese-rich, Iron-free Host
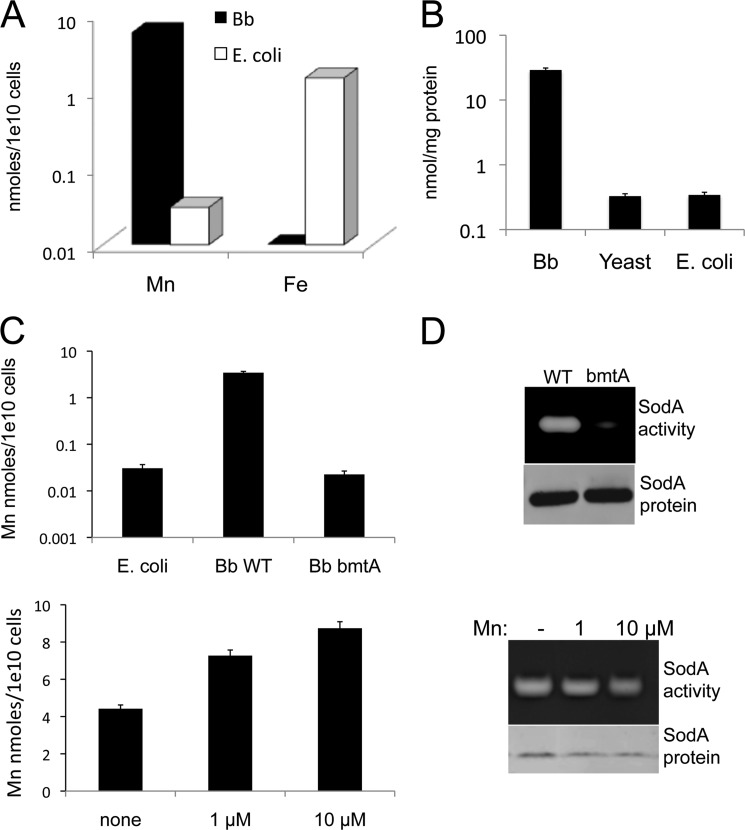
Posey and Gheridini (15) have reported that B. burgdorferi is virtually free of cellular iron, and we have confirmed these findings using ICP-MS (Fig. 3A). In the course of these metal analyses, we noted the spirochete accumulates unusually high levels of manganese. As seen in Fig. 3A, B. burgdorferi accumulated 2 orders of magnitude higher levels of manganese per cell than E. coli grown in BSK medium in parallel. This high level of manganese was seen with both the ML23 and the 297 strain backgrounds and by metal analysis with both AAS and ICP-MS (Fig. 3, A and C). Because cell volumes for the spirochete are difficult to estimate, we normalized manganese on the basis of soluble cellular protein and compared values in B. burgdorferi, E. coli, and the eukaryote, bakers' yeast. Yeast and E. coli are reported to accumulate similar micromolar concentrations of manganese (10, 11), and we also find similar manganese levels in these organisms when analyzed per mg of protein (Fig. 3B). By comparison, the level of manganese that accumulated in B. burgdorferi was 2 orders of magnitude higher (Fig. 3B).
FIGURE 3.
Metal analysis of B. burgdorferi versus iron-philic organisms and effects on SodA activity. A, ICP MS analysis of manganese and iron was carried out with whole cell B. burgdorferi strain ML23 versus E. coli cells grown in BSK medium, as described under “Experimental Procedures.” B, AAS measurements of manganese in soluble protein lysates from B. burgdorferi (Bb) strain ML23, E. coli, and S. cerevisiae as described under “Experimental Procedures.” C, AAS analysis of whole cell manganese in B. burgdorferi strain 297 and the corresponding bmtA mutant compared with E. coli (top) and B. burgdorferi strain ML23 grown in BSK supplemented with the indicated concentrations of MnCl2 (bottom). D, SodA activity and protein levels were examined as in Fig. 1A in B. burgdorferi strain 297 and the corresponding bmtA mutant (top) and B. burgdorferi strain ML23 grown with the indicated concentrations of MnCl2 (bottom).
The exceptionally high level of manganese in B. burgdorferi is required for maximal SodA activity. When the manganese transporter BmtA is deleted in B. burgdorferi, there is a 1–2-order of magnitude drop in cellular manganese (21) to levels within the range of WT E. coli (Fig. 3C, top). This drop in manganese results in inactive SodA (Fig. 3D, top). Unlike previous findings by Troxell et al. (20), we observed no change in B. burgdorferi SodA protein levels with this loss in enzymatic activity. Hence, high manganese is required to activate the enzyme and not regulate expression of SodA. We also addressed the effects of raising intracellular manganese on B. burgdorferi SodA activity. Manganese levels can double by growing B. burgdorferi in the presence of 10 μm MnCl2 (severe toxicity ensues above this) (Fig. 3C, bottom), but this rise in manganese does not increase the enzymatic activity nor protein levels of B. burgdorferi SodA (Fig. 3D, bottom). The enzyme appears maximally activated in the manganese-rich environment of B. burgdorferi without additional metal supplements.
Expression of B. burgdorferi SodA in an Iron-philic Host, S. cerevisiae
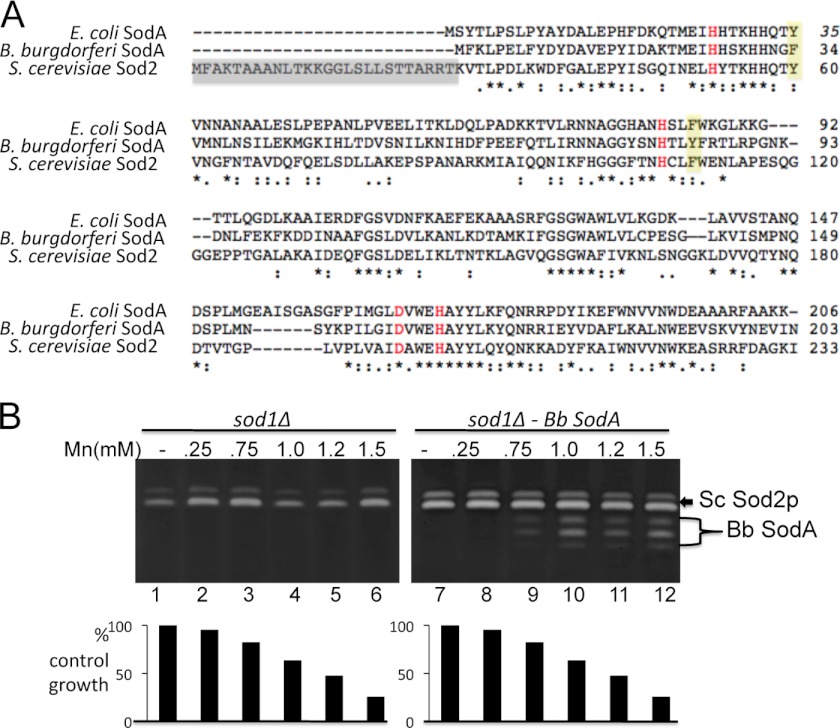
Manganese-containing SOD enzymes are fairly well conserved in evolution (2), as illustrated in the alignment of MnSODs from B. burgdorferi, E. coli, and yeast (Fig. 4A), yet unlike B. burgdorferi, the environment of E. coli and yeast would seem hostile to activation of a Mn-SOD enzyme. These organisms are iron-philic and accumulate intracellular levels of iron that far exceed manganese (10, 36). Can a SOD that evolved in a manganese-rich environment acquire its co-factor in an iron-rich host? To address this, we expressed SodA in the mitochondria of S. cerevisiae, where manganese activation of SOD enzymes has been well characterized (13, 14, 37, 38).
FIGURE 4.
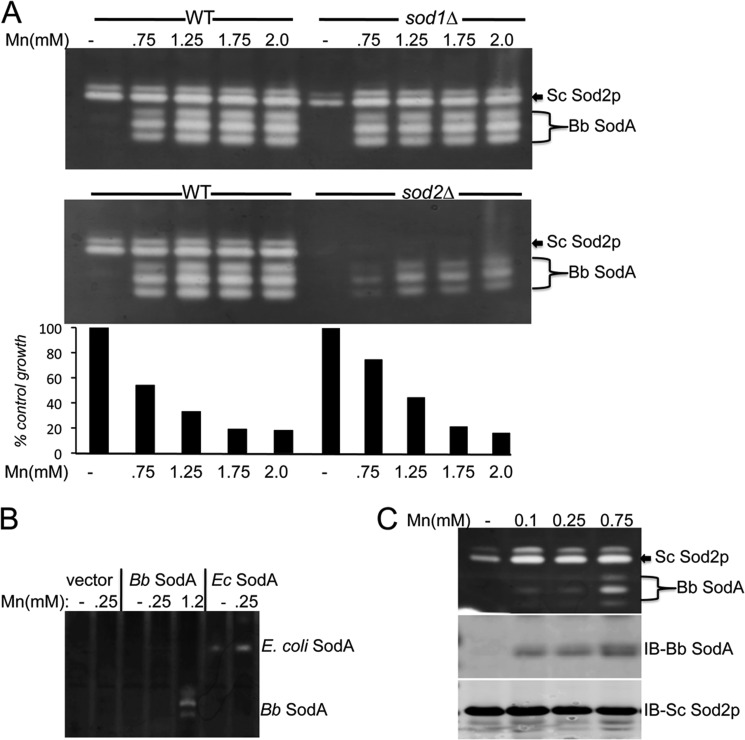
Targeting B. burgdorferi SodA to the mitochondria of S. cerevisiae. A, alignment of B. burgdorferi SodA with the Mn-Sod2p of S. cerevisiae and the Mn-SodA of E. coli using CLC sequencer viewer version 6.4 software. Asterisks mark identity, and dots represent similar residues; red marks, metal binding residues. The yellow shaded area marks the unique Phe-34 and Tyr-84 of B. burgdorferi SodA (see Fig. 8), and the gray shaded area shows the MLS of yeast Sod2p that was fused onto the bacterial SodA genes. B, a sod1Δ yeast strain was transformed where indicated with the pDA002 plasmid for expressing mitochondrial targeted B. burgdorferi SodA (lanes 7–12) and was grown with the indicated concentrations of MnCl2. Top, SOD activity was monitored by the native gel assay. The position of endogenous yeast Sod2p (Sc Sod2p) and heterologous B. burgdorferi SodA (Bb SodA) are indicated. Mitochondrial Sod2p runs as a doublet or triplet (14), and the same is true for B. burgdorferi SodA expressed in yeast mitochondria. Bottom, cell growth was monitored turbidimetrically at A600 nm and plotted as a percentage of control growth obtained in the absence of manganese.
B. burgdorferi SodA codon-optimized for expression in yeast was fused to the MLS of yeast Sod2p (indicated in Fig. 4A) and placed under control of the yeast SOD2 promoter. Expression of SodA was first analyzed in the background of a sod1Δ yeast to avoid interference from yeast Cu/Zn-Sod1p that migrates to similar positions on the native gel for SOD activity (see Fig. 1C). As seen in Fig. 4B, sod1Δ cells co-expressing the endogenous yeast Sod2p and B. burgdorferi SodA in the mitochondria only exhibited activity of the endogenous yeast Mn-Sod2p (lane 7). However, B. burgdorferi SodA activity was gained with high levels of manganese (lanes 9–12) that were toxic to yeast, as indicated by growth inhibition (Fig. 4B, bottom). This dependence on high manganese for SodA activity was not due to oxidative damage from expression in the sod1 null strain because similar results were obtained in WT yeast, where Cu/Zn-SOD activity on the native gel was eliminated by peroxide treatment (Fig. 5A). Moreover, the yeast mitochondrial Sod2p does not compete with B. burgdorferi SodA for manganese because B. burgdorferi SodA activity was not increased in sod2Δ null mutants (Fig. 5A, middle). B. burgdorferi SodA exhibits 46% identity with the SodA from E. coli (Fig. 4A), yet E. coli SodA driven by the same yeast SOD2 promoter and MLS is not similarly dependent on toxic manganese for activity (Fig. 5B), as was previously published (14). Thus, the requirement for high manganese is not a general feature of bacterial Mn-SOD enzymes. In the case of B. burgdorferi SodA, high manganese is also needed for protein expression (Fig. 5C, middle). This is not a transcriptional effect because yeast Sod2p driven by the same SOD2 promoter remains constant with manganese (Fig. 5C, bottom). Instead, manganese activation of B. burgdorferi SodA seems to stabilize the protein expressed in yeast mitochondria.
FIGURE 5.
B. burgdorferi SodA requires high manganese for activity and expression in yeast mitochondria. A, the indicated WT or sod mutants of S. cerevisiae expressing mitochondrial targeted B. burgdorferi SodA were tested for manganese activation of SOD activity and for manganese effects on cell growth (for WT and sod2Δ, bottom) as described in the legend to Fig. 4, except yeast Cu/Zn-Sod1p was inactivated by in-gel treatment with 5 mm H2O2. The positions of endogenous yeast Sod2p (Sc Sod2p) and heterologous B. burgdorferi SodA (Bb SodA) on the native activity gels are indicated. B, the sod1Δ sod2Δ expressing either empty vector pRS315 (58) or the mitochondrial targeted SodA from either B. burgdorferi or E. coli on plasmids pDA002 and pAN002 (14) was grown with the indicated concentrations of MnCl2 and analyzed for SOD activity by the native gel assay. The positions of mitochondrial targeted E. coli and B. burgdorferi SodA are indicated. C, WT yeast strains expressing mitochondrial B. burgdorferi SodA were grown in the presence of the indicated concentrations of MnCl2 and were subjected to native gel assays for SOD activity as in A (top) or to immunoblot (IB) analysis of B. burgdorferi SodA and yeast Sod2p protein levels (middle and bottom).
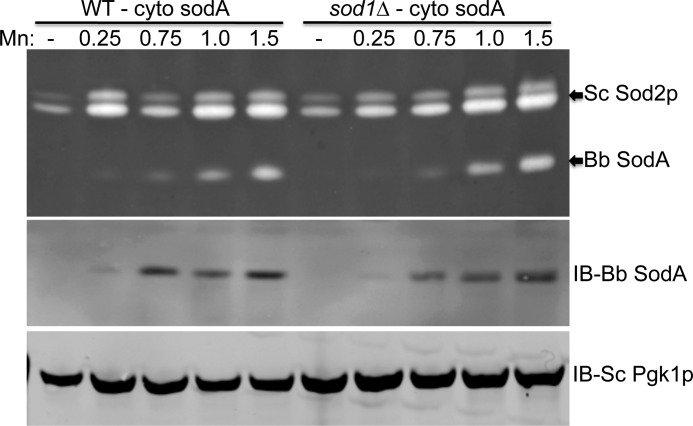
We also tested the effects of expressing B. burgdorferi SodA in the cytosol of S. cerevisiae by deleting the MLS for targeting to the mitochondria. As seen in seen Fig. 6, B. burgdorferi SodA in the cytosol exhibited the same dependence on high manganese for protein expression and enzymatic activity as was seen with mitochondrially expressed SodA. Identical results were obtained with expression in a WT strain and in a yeast sod1Δ mutant, where B. burgdorferi SodA represents the sole SOD enzyme of yeast cytosol (Fig. 6).
FIGURE 6.
Expression of B. burgdorferi SodA in yeast cytosol. The WT yeast strain or sod1Δ mutant was transformed with plasmid pSA002 for expressing B. burgdorferi SodA in yeast cytosol, and cells were cultured with the indicated millimolar concentrations of MnCl2. Top, SOD activity was assayed as in Fig. 5C. Middle and bottom, immunoblot (IB) analysis was conducted as in Fig. 5C using antibodies directed against B. burgdorferi (Bb) SodA and yeast cytosolic (Sc) Pgk1p.
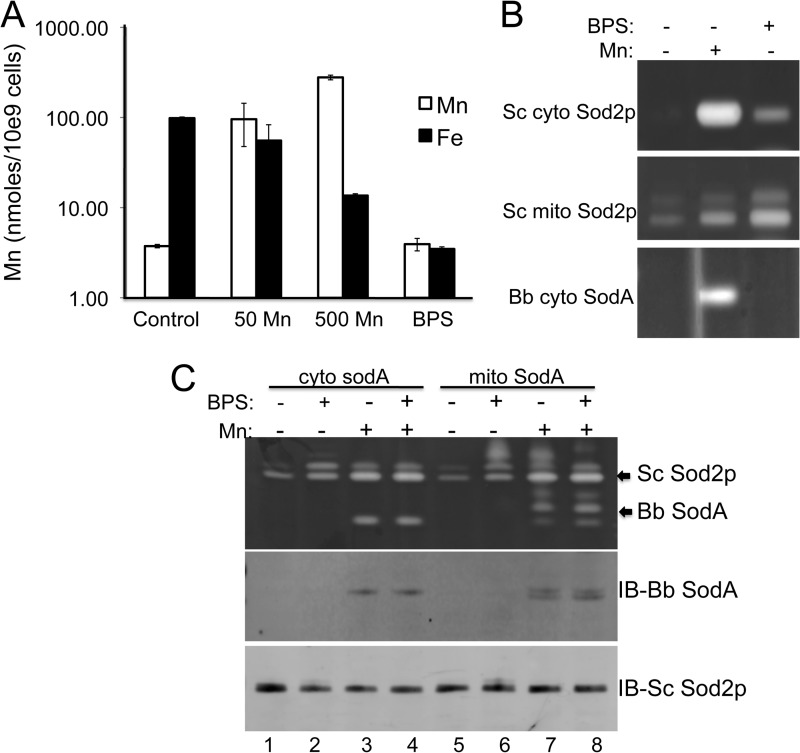
We sought to determine the level of intracellular manganese required to activate the heterologous B. burgdorferi SodA in yeast. Both mitochondrion- and cytosol- expressed B. burgdorferi SodA require roughly 500 μm extracellular manganese to detect any activity, and this reflects a 50–100-fold increase in intracellular accumulation of manganese (Fig. 7A). Interestingly, treatment with high manganese also results in a drastic reduction in cellular iron levels (Fig. 7A), perhaps due to competing effects of manganese on iron uptake.
FIGURE 7.
Iron chelation does not help activate B. burgdorferi SodA expressed in yeast. A, ICP-MS analysis of iron and manganese in whole cells of S. cerevisiae (Sc) grown in the presence of the indicated concentrations of MnCl2 or 100 μm BPS as described under “Experimental Procedures.” B, yeast strains transformed with pEL124 (37) for expressing yeast Sod2p in the cytosol (top) or with pDA002 for expressing mitochondrial B. burgdorferi (Bb) SodA (bottom) were grown in the presence of 1.0 mm MnCl2 (Mn: +) and/or 0.1 mm of the iron chelator BPS (BPS: +) and subjected to SOD activity analysis. C, yeast cells expressing either mitochondrial or cytosolic B. burgdorferi SodA were treated with manganese or BPS and tested for SOD activity as in B (top) and for levels of B. burgdorferi SodA and S. cerevisiae Sod2p by immunoblot as in Fig. 5C (middle and bottom). Error bars, standard deviation.
We tested whether the loss in cellular iron seen with high manganese contributes to the activation of B. burgdorferi SodA in yeast. Iron levels in yeast cells can be lowered by treatment with the iron chelator, bathophenanthroline disulfonate (BPS) (14), and as seen in Fig. 7A, BPS effectively lowered cellular iron levels 25-fold without changes in intracellular manganese. This lowering of iron was sufficient to induce activity of a yeast Mn-SOD expressed in yeast cytosol and also increased the activity of mitochondrial yeast Sod2p (Fig. 7B), consistent with the notion that a certain pool of yeast Sod2p is iron-bound inactive enzyme (13, 14). It is noteworthy that the cytosolic version of S. cerevisiae Sod2p is more strongly activated by manganese than by BPS compared with endogenous mitochondrial Sod2p (Fig. 7B). Apparently, in the cytosol, where manganese is limiting, iron depletion on its own cannot maximally activate S. cerevisiae Sod2p. Although BPS was effective in increasing activity of yeast Sod2p, it failed to activate the B. burgdorferi SodA enzyme or stabilize the SodA polypeptide expressed in either the cytosol (Fig. 7, B (bottom) and C (lane 2)) or in the mitochondria of yeast (Fig. 7C, compare lanes 5 and 6). BPS also did not enhance the effects of high manganese in activating B. burgdorferi SodA (Fig. 7C, compare lanes 3 and 4 and lanes 7 and 8). Therefore, lowering cellular iron is by itself insufficient to activate B. burgdorferi SodA, and high levels of manganese are essential. It is noteworthy that the high manganese/iron ratio required to activate B. burgdorferi SodA in yeast is not unlike the situation in the native B. burgdorferi host, where manganese levels are exceedingly high and iron is virtually absent.
DISCUSSION
B. burgdorferi has uniquely evolved without a cellular requirement for iron, and we show here that the organism accumulates high levels of manganese compared with other more iron-philic organisms, such as E. coli and S. cerevisiae. This environment of high manganese and a virtual absence of iron is well suited for activation of a manganese SodA. Through a metalloproteomic approach, we firmly establish SodA as a manganese enzyme and show that in Borrelia, the active enzyme is bound to manganese, whereas a smaller pool of inactive enzyme is apo, not bound to any other metal. By comparison, the Mn-SODs from other organisms, such as yeast, E. coli, and humans can bind intracellular iron (5, 6, 39–41). We have no evidence for iron binding to B. burgdorferi SodA in either its native spirochete host or in the iron-rich environment of yeast mitochondria. In addition, B. burgdorferi SodA activity requires exceedingly high levels of intracellular manganese. When expressed in S. cerevisiae, the enzyme is only active when manganese levels exceed mitochondrial iron, conditions that simulate the native B. burgdorferi host.
The accumulation of unusually high manganese in B. burgdorferi that we report here has not been previously documented, although our values are very similar to those published by Ouyang et al. (21). In studies by Posey and Gherardini (15), the manganese in B. burgdorferi cell lysates was reported to be only 2–3-fold higher than that of E. coli and might reflect differential growth conditions used because our cells were grown to near stationary phase. In any case, our findings clearly demonstrate a tremendous capacity for manganese uptake without toxicity in this spirochete. In fact, in our preliminary studies comparing manganese across various species (not shown), the levels of the metal in whole cell B. burgdorferi are comparable with Lactobacillus plantarum, notoriously known for hyperaccumulating manganese without a SOD enzyme (42).
The high manganese in B. burgdorferi may serve dual purposes in the adaptation of this pathogen. First, in the absence of iron-requiring enzymes, manganese may be more widely used as a co-factor. Consistent with this, we observe a close association with B. burgdorferi manganese and an aminopeptidase (Fig. 2A), a metalloenzyme that employs iron in other organisms (35). Moreover, the ability of B. burgdorferi to accumulate high manganese may represent yet another fascinating adaptation of the organism to the metal starvation response of innate immunity. When infected, the host not only systemically starves pathogens of iron (16, 17), but macrophages and neutrophils attempt to limit manganese bioavailability for the invading species (43–45). High manganese is essential for virulence in B. burgdorferi (21), and SodA may only be part of the story. Non-proteinaceous complexes of manganese to small metabolites (so-called Mn-antioxidants) are receiving increasing attention as critical factors in microbial oxidative stress resistance and pathogenesis (1, 46–51).
Very recently, Wang et al. (52) have reported iron accumulation in B. burgdorferi. This report appears in direct conflict with the previous findings of Posey and Gherardini (15) and with our ICP-MS analysis of iron. The basis for the iron reported by Wang et al. (52) cannot be reconciled at this time but might reflect the differential extraction methods used for metal analysis. Alternatively, under certain non-standard laboratory conditions, the bacteria may be capable of iron uptake.
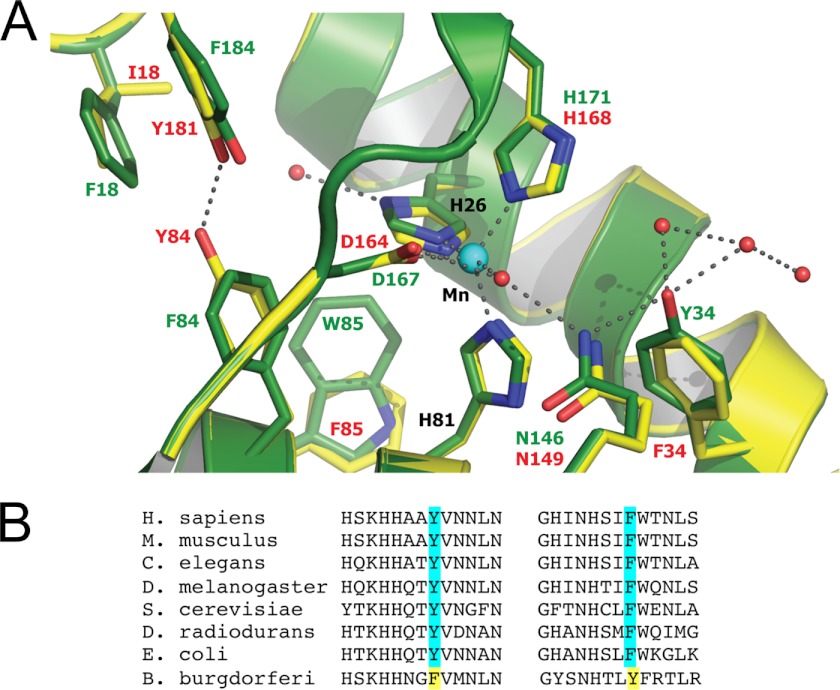
Last, why does B. burgdorferi SodA require such high levels of cellular manganese for activity? Currently, there are no structural data available on B. burgdorferi SodA; however, we were able to generate a computer-assisted model of B. burgdorferi SodA using MODELLER (53) based on known structures of E. coli SodA and S. cerevisiae Sod2p. A comparison of the active site regions of E. coli and B. burgdorferi SodA molecules is shown in Fig. 8A. The manganese coordination site is identical between the two SODs; however, several interesting features emerge. For example, the second sphere residue Phe-34 in B. burgdorferi SodA is a tyrosine in Mn-SOD molecules ranging from bacteria (e.g. E. coli and Deinococcus radiodurans) to fungi (S. cerevisiae), invertebrates (C. elegans and Drosophila melanogaster), and mammals (Fig. 8B). Tyr-34 is well-known to participate in a hydrogen bonding network at the active site (simulated in Fig. 8A), and in fact the Y34F derivatives of human and yeast Mn-SOD have been analyzed in detail and shown to dramatically alter the kinetics of the SOD reaction, disrupting the “prompt protonation pathway” (54–56). However, there were no reports of Y34F affecting manganese binding in human and yeast Mn-SOD. Thus, the unique Phe-34 in B. burgdorferi SodA may well account for some enzyme catalysis effects but not the requirement for high manganese. A second noteworthy residue in B. burgdorferi SodA is Tyr-84, which is a phenylalanine in other Mn-SOD enzymes (Fig. 8B). As seen in Fig. 8A, the model predicts that Tyr-84 forms nearly an ideal hydrogen bond with Tyr-181, which could potentially occlude access of manganese to the active site. Such an occlusion would be consistent with the conformationally gated metal uptake mechanism proposed for Mn-SOD molecules (57). However, we observed that the single Y84F substitution in B. burgdorferi SodA did not alter the requirement for high manganese (data not shown), indicating that other residues of B. burgdorferi SodA must be involved. Our structural model will provide a useful guide in unraveling the unique properties of the enzyme that force its requirement for high manganese in vivo.
FIGURE 8.
The predicted active site of B. burgdorferi SodA. A, a model of B. burgdorferi SodA was generated with the program MODELLER (53) using the 0.9 Å structure of E. coli SodA (Protein Data Bank accession number 1IX9) as the structural template. Residues of B. burgdorferi SodA are shown in yellow with numbering in red, and the equivalent positions in E. coli SodA are marked in green. Red balls indicate water molecules, and dotted lines represent hydrogen bonds or the coordination of the manganese ion (aqua ball) to its four amino acid ligands and a single water molecule. B, a comparison of Tyr-34 and Phe-84 in B. burgdorferi SodA with the equivalent positions in Mn-SOD molecules from the indicated organisms.
Acknowledgments
We thank Dr. M. Norgard for the bmtA mutant and Jana Mihalic for ICP-MS.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 ES08996 and GM50016 (to V. C. C.) and SC1 AI078559 (to J. S.). This work was also supported in part by the Johns Hopkins University NIEHS, National Institutes of Health, center, by Robert A. Welch Foundation Grant AQ-1399, and in part by Veterans Affairs Department Grant I01BX000506, South Texas Veterans Health Care System (to P. J. H.). Metalloproteomic studies were funded by the Gordon and Betty Moore Foundation Marine Microbiology Program and National Science Foundation Chemical Oceanography Grant OCE-1031271 (to M. A. S.).
- SOD
- superoxide dismutase
- YPD
- yeast extract, peptone, dextrose medium
- AAS
- atomic absorption spectroscopy
- ICP-MS
- inductively coupled plasma mass spectrometry
- MLS
- mitochondrial leader sequence
- BPS
- bathophenanthroline disulfonate.
REFERENCES
- 1. Aguirre J. D., Culotta V. C. (2012) Battles with iron. Manganese in oxidative stress protection. J. Biol. Chem. 287, 13541–13548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wintjens R., Noël C., May A. C., Gerbod D., Dufernez F., Capron M., Viscogliosi E., Rooman M. (2004) Specificity and phenetic relationships of iron- and manganese-containing superoxide dismutases on the basis of structure and sequence comparisons. J. Biol. Chem. 279, 9248–9254 [DOI] [PubMed] [Google Scholar]
- 3. Mizuno K., Whittaker M. M., Bächinger H. P., Whittaker J. W. (2004) Calorimetric studies on the tight-binding metal interactions of Escherichia coli manganese superoxide dismutase. J. Biol. Chem. 279, 27339–27344 [DOI] [PubMed] [Google Scholar]
- 4. Iranzo O. (2011) Manganese complexes displaying superoxide dismutase activity. A balance between different factors. Bioorg. Chem. 39, 73–87 [DOI] [PubMed] [Google Scholar]
- 5. Kang Y., He Y. X., Zhao M. X., Li W. F. (2011) Structures of native and Fe-substituted SOD2 from Saccharomyces cerevisiae. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 67, 1173–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yamakura F., Kawasaki H. (2010) Post-translational modifications of superoxide dismutase. Biochim. Biophys. Acta 1804, 318–325 [DOI] [PubMed] [Google Scholar]
- 7. Yamakura F., Kobayashi K., Furukawa S., Suzuki Y. (2007) In vitro preparation of iron-substituted human manganese superoxide dismutase. Possible toxic properties for mitochondria. Free Radic. Biol. Med. 43, 423–430 [DOI] [PubMed] [Google Scholar]
- 8. Vance C. K., Miller A. F. (1998) A simple proposal that can explain the inactivity of metal-substituted superoxide dismutases. J. Am. Chem. Soc. 120, 461–467 [Google Scholar]
- 9. Jackson T. A., Brunold T. C. (2004) Combined spectroscopic/computational studies on Fe- and Mn-dependent superoxide dismutases. Insights into second-sphere tuning of active site properties. Acc. Chem. Res. 37, 461–470 [DOI] [PubMed] [Google Scholar]
- 10. Outten C. E., O'Halloran T. V. (2001) Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292, 2488–2492 [DOI] [PubMed] [Google Scholar]
- 11. Rosenfeld L., Reddi A. R., Leung E., Aranda K., Jensen L. T., Culotta V. C. (2010) The effect of phosphate accumulation on metal ion homeostasis in Saccharomyces cerevisiae. J. Biol. Inorg. Chem. 15, 1051–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eide D. J., Clark S., Nair T. M., Gehl M., Gribskov M., Guerinot M. L., Harper J. F. (2005) Characterization of the yeast ionome. A genome-wide analysis of nutrient mineral and trace element homeostasis in Saccharomyces cerevisiae. Genome Biol. 6, R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang M., Cobine P. A., Molik S., Naranuntarat A., Lill R., Winge D. R., Culotta V. C. (2006) The effects of mitochondrial iron homeostasis on cofactor specificity of superoxide dismutase 2. EMBO J. 25, 1775–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Naranuntarat A., Jensen L. T., Pazicni S., Penner-Hahn J. E., Culotta V. C. (2009) The interaction of mitochondrial iron with manganese superoxide dismutase. J. Biol. Chem. 284, 22633–22640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Posey J. E., Gherardini F. C. (2000) Lack of a role for iron in the Lyme disease pathogen. Science 288, 1651–1653 [DOI] [PubMed] [Google Scholar]
- 16. Correnti C., Strong R. K. (2012) Mammalian siderophores, siderophore-binding lipocalins, and the labile iron pool. J. Biol. Chem. 287, 13524–13531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Johnson E. E., Wessling-Resnick M. (2012) Iron metabolism and the innate immune response to infection. Microbes Infect. 14, 207–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Esteve-Gassent M. D., Elliott N. L., Seshu J. (2009) sodA is essential for virulence of Borrelia burgdorferi in the murine model of Lyme disease. Mol. Microbiol. 71, 594–612 [DOI] [PubMed] [Google Scholar]
- 19. Whitehouse C. A., Williams L. R., Austin F. E. (1997) Identification of superoxide dismutase activity in Borrelia burgdorferi. Infect. Immun. 65, 4865–4868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Troxell B., Xu H., Yang X. F. (2012) Borrelia burgdorferi, a pathogen that lacks iron, encodes manganese-dependent superoxide dismutase essential for resistance to streptonigrin. J. Biol. Chem. 287, 19284–19293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ouyang Z., He M., Oman T., Yang X. F., Norgard M. V. (2009) A manganese transporter, BB0219 (BmtA), is required for virulence by the Lyme disease spirochete, Borrelia burgdorferi. Proc. Natl. Acad. Sci. U.S.A. 106, 3449–3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Reddi A. R., Culotta V. C. (2011) Regulation of manganese antioxidants by nutrient sensing pathways in Saccharomyces cerevisiae. Genetics 189, 1261–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gleason J. E., Corrigan D. J., Cox J. E., Reddi A. R., McGinnis L. A., Culotta V. C. (2011) Analysis of hypoxia and hypoxia-like states through metabolite profiling. PLoS One 6, e24741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Flohé L., Otting F. (1984) Superoxide dismutase assays. Methods Enzymol. 105, 93–104 [DOI] [PubMed] [Google Scholar]
- 25. Karna S. L., Sanjuan E., Esteve-Gassent M. D., Miller C. L., Maruskova M., Seshu J. (2011) CsrA modulates levels of lipoproteins and key regulators of gene expression critical for pathogenic mechanisms of Borrelia burgdorferi. Infect. Immun. 79, 732–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yannone S. M., Hartung S., Menon A. L., Adams M. W., Tainer J. A. (2012) Metals in biology. Defining metalloproteomes. Curr. Opin. Biotechnol. 23, 89–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lancaster W. A., Praissman J. L., Poole F. L., 2nd, Cvetkovic A., Menon A. L., Scott J. W., Jenney F. E., Jr., Thorgersen M. P., Kalisiak E., Apon J. V., Trauger S. A., Siuzdak G., Tainer J. A., Adams M. W. (2011) A computational framework for proteome-wide pursuit and prediction of metalloproteins using ICP-MS and MS/MS data. BMC Bioinformatics 12, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cvetkovic A., Menon A. L., Thorgersen M. P., Scott J. W., Poole F. L., 2nd, Jenney F. E., Jr., Lancaster W. A., Praissman J. L., Shanmukh S., Vaccaro B. J., Trauger S. A., Kalisiak E., Apon J. V., Siuzdak G., Yannone S. M., Tainer J. A., Adams M. W. (2010) Microbial metalloproteomes are largely uncharacterized. Nature 466, 779–782 [DOI] [PubMed] [Google Scholar]
- 29. Waldron K. J., Tottey S., Yanagisawa S., Dennison C., Robinson N. J. (2007) A periplasmic iron-binding protein contributes toward inward copper supply. J. Biol. Chem. 282, 3837–3846 [DOI] [PubMed] [Google Scholar]
- 30. Robinson N., Waldron K., Tottey S., Bessant C. (2008) Metalloprotein metal pools. Identification and quantification by coupling native and non-native separations through principal component analysis. Protocol Exchange, doi: 10.1038/nprot.2008.236 [DOI] [Google Scholar]
- 31. Tottey S., Patterson C. J., Banci L., Bertini I., Felli I. C., Pavelkova A., Dainty S. J., Pernil R., Waldron K. J., Foster A. W., Robinson N. J. (2012) Cyanobacterial metallochaperone inhibits deleterious side reactions of copper. Proc. Natl. Acad. Sci. U.S.A. 109, 95–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gu Y. Q., Holzer F. M., Walling L. L. (1999) Overexpression, purification and biochemical characterization of the wound-induced leucine aminopeptidase of tomato. Eur. J. Biochem. 263, 726–735 [DOI] [PubMed] [Google Scholar]
- 33. Hafkenscheid J. C., Kohler B. E. (1985) A continuous method for the determination of leucine aminopeptidase in human serum with l-leucinamide as substrate. J. Clin. Chem. Clin. Biochem. 23, 393–398 [DOI] [PubMed] [Google Scholar]
- 34. Kale A., Pijning T., Sonke T., Dijkstra B. W., Thunnissen A. M. (2010) Crystal structure of the leucine aminopeptidase from Pseudomonas putida reveals the molecular basis for its enantioselectivity and broad substrate specificity. J. Mol. Biol. 398, 703–714 [DOI] [PubMed] [Google Scholar]
- 35. Sule N., Singh R. K., Zhao P., Srivastava D. K. (2012) Probing the metal ion selectivity in methionine aminopeptidase via changes in the luminescence properties of the enzyme bound europium ion. J. Inorg. Biochem. 106, 84–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rosenfeld L., Culotta V. C. (2012) Phosphate disruption and metal toxicity in Saccharomyces cerevisiae. Effects of RAD23 and the histone chaperone HPC2. Biochem. Biophys. Res. Commun. 418, 414–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Luk E., Carroll M., Baker M., Culotta V. C. (2003) Manganese activation of superoxide dismutase 2 in Saccharomyces cerevisiae requires MTM1, a member of the mitochondrial carrier family. Proc. Natl. Acad. Sci. U.S.A. 100, 10353–10357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Luk E. E., Culotta V. C. (2001) Manganese superoxide dismutase in S. cerevisiae acquires its metal co-factor through a pathway involving the Nramp metal transproter, Smf2p. J. Biol. Chem. 276, 47556–47562 [DOI] [PubMed] [Google Scholar]
- 39. Whittaker M. M., Whittaker J. W. (2012) Metallation state of human manganese superoxide dismutase expressed in Saccharomyces cerevisiae. Arch. Biochem. Biophys. 523, 191–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Privalle C. T., Fridovich I. (1992) Transcriptional and maturation effects of manganese and iron on the biosynthesis of manganese-superoxide dismutase in Escherichia coli. J. Biol. Chem. 267, 9140–9145 [PubMed] [Google Scholar]
- 41. Beyer W. F., Jr., Fridovich I. (1991) In vivo competition between iron and manganese for occupancy of the active site region of the manganese-superoxide dismutase of Escherichia coli. J. Biol. Chem. 266, 303–308 [PubMed] [Google Scholar]
- 42. Archibald F. S., Fridovich I. (1981) Manganese and defenses against oxygen toxicity in Lactobacillus plantarum. J. Bacteriol. 145, 442–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cassat J. E., Skaar E. P. (2012) Metal ion acquisition in Staphylococcus aureus. Overcoming nutritional immunity. Semin. Immunopathol. 34, 215–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kehl-Fie T. E., Skaar E. P. (2010) Nutritional immunity beyond iron: a role for manganese and zinc. Curr. Opin. Chem. Biol. 14, 218–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Govoni G., Gros P. (1998) Microphage NRAMP1 and its role in resistance to microbial infections. Inflamm. Res. 47, 277–284 [DOI] [PubMed] [Google Scholar]
- 46. Horsburgh M. J., Wharton S. J., Cox A. G., Ingham E., Peacock S., Foster S. J. (2002) MntR modulates expression of the PerR regulon and superoxide resistance in Staphylococcus aureus through control of manganese uptake. Mol. Microbiol. 44, 1269–1286 [DOI] [PubMed] [Google Scholar]
- 47. Horsburgh M. J., Wharton S. J., Karavolos M., Foster S. J. (2002) Manganese. Elemental defence for a life with oxygen. Trends Microbiol. 10, 496–501 [DOI] [PubMed] [Google Scholar]
- 48. Seib K. L., Tseng H. J., McEwan A. G., Apicella M. A., Jennings M. P. (2004) Defenses against oxidative stress in Neisseria gonorrhoeae and Neisseria meningitidis. distinctive systems for different lifestyles. J. Infect. Dis. 190, 136–147 [DOI] [PubMed] [Google Scholar]
- 49. Tseng H. J., Srikhanta Y., McEwan A. G., Jennings M. P. (2001) Accumulation of manganese in Neisseria gonorrhoeae correlates with resistance to oxidative killing by superoxide anion and is independent of superoxide dismutase activity. Mol. Microbiol. 40, 1175–1186 [DOI] [PubMed] [Google Scholar]
- 50. Kehl-Fie T. E., Chitayat S., Hood M. I., Damo S., Restrepo N., Garcia C., Munro K. A., Chazin W. J., Skaar E. P. (2011) Nutrient metal sequestration by calprotectin inhibits bacterial superoxide defense, enhancing neutrophil killing of Staphylococcus aureus. Cell Host Microbe 10, 158–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Daly M. J., Gaidamakova E. K., Matrosova V. Y., Kiang J. G., Fukumoto R., Lee D. Y., Wehr N. B., Viteri G. A., Berlett B. S., Levine R. L. (2010) Small-molecule antioxidant proteome-shields in Deinococcus radiodurans. PLoS One 5, e12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang P., Lutton A., Olesik J., Vali H., Li X. (2012) A novel iron- and copper-binding protein in the Lyme disease spirochaete. Mol. Microbiol. 86, 1441–1451 [DOI] [PubMed] [Google Scholar]
- 53. Sali A., Blundell T. L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 54. Sheng Y., Butler Gralla E., Schumacher M., Cascio D., Cabelli D. E., Selverstone Valentine J. (2012) Six-coordinate manganese(3+) in catalysis by yeast manganese superoxide dismutase. Proc. Natl. Acad. Sci. U.S.A. 109, 14314–14319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Guan Y., Hickey M. J., Borgstahl G. E., Hallewell R. A., Lepock J. R., O'Connor D., Hsieh Y., Nick H. S., Silverman D. N., Tainer J. A. (1998) Crystal structure of Y34F mutant human mitochondrial manganese superoxide dismutase and the functional role of tyrosine 34. Biochemistry 37, 4722–4730 [DOI] [PubMed] [Google Scholar]
- 56. Perry J. J., Hearn A. S., Cabelli D. E., Nick H. S., Tainer J. A., Silverman D. N. (2009) Contribution of human manganese superoxide dismutase tyrosine 34 to structure and catalysis. Biochemistry 48, 3417–3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Whittaker J. W. (2010) Metal uptake by manganese superoxide dismutase. Biochim. Biophys. Acta 1804, 298–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sikorski R. S., Hieter P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces ceresiviae. Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]