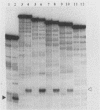
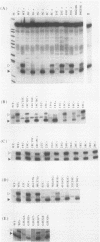
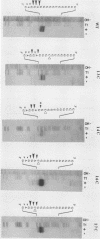
Abstract
RNase MRP is a site-specific endonuclease that processes primer mitochondrial RNA from the leading-strand origin of mitochondrial DNA replication. Using deletional analysis and saturation mutagenesis, we have determined the substrate requirements for cleavage by mouse mitochondrial RNase MRP. Two regions of sequence homology among vertebrate mitochondrial RNA primers, conserved sequence blocks II and III, were found to be critical for both efficient and accurate cleavage; a third region of sequence homology, conserved sequence block I, was dispensable. Analysis of insertion and deletion mutations within conserved sequence block II demonstrated that the specificity of RNase MRP accommodates the natural sequence heterogeneity of conserved sequence block II in vivo. Heterologous assays with human RNase MRP and mutated mouse mitochondrial RNA substrates indicated that sequences essential for substrate recognition are conserved between mammalian species.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981 Apr 9;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Baer M. F., Reilly R. M., McCorkle G. M., Hai T. Y., Altman S., RajBhandary U. L. The recognition by RNase P of precursor tRNAs. J Biol Chem. 1988 Feb 15;263(5):2344–2351. [PubMed] [Google Scholar]
- Bartkiewicz M., Gold H., Altman S. Identification and characterization of an RNA molecule that copurifies with RNase P activity from HeLa cells. Genes Dev. 1989 Apr;3(4):488–499. doi: 10.1101/gad.3.4.488. [DOI] [PubMed] [Google Scholar]
- Been M. D., Cech T. R. One binding site determines sequence specificity of Tetrahymena pre-rRNA self-splicing, trans-splicing, and RNA enzyme activity. Cell. 1986 Oct 24;47(2):207–216. doi: 10.1016/0092-8674(86)90443-5. [DOI] [PubMed] [Google Scholar]
- Burkard U., Willis I., Söll D. Processing of histidine transfer RNA precursors. Abnormal cleavage site for RNase P. J Biol Chem. 1988 Feb 15;263(5):2447–2451. [PubMed] [Google Scholar]
- Cech T. R., Bass B. L. Biological catalysis by RNA. Annu Rev Biochem. 1986;55:599–629. doi: 10.1146/annurev.bi.55.070186.003123. [DOI] [PubMed] [Google Scholar]
- Chang D. D., Clayton D. A. A mammalian mitochondrial RNA processing activity contains nucleus-encoded RNA. Science. 1987 Mar 6;235(4793):1178–1184. doi: 10.1126/science.2434997. [DOI] [PubMed] [Google Scholar]
- Chang D. D., Clayton D. A. A novel endoribonuclease cleaves at a priming site of mouse mitochondrial DNA replication. EMBO J. 1987 Feb;6(2):409–417. doi: 10.1002/j.1460-2075.1987.tb04770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D. D., Clayton D. A. Mouse RNAase MRP RNA is encoded by a nuclear gene and contains a decamer sequence complementary to a conserved region of mitochondrial RNA substrate. Cell. 1989 Jan 13;56(1):131–139. doi: 10.1016/0092-8674(89)90991-4. [DOI] [PubMed] [Google Scholar]
- Chang D. D., Clayton D. A. Precise assignment of the light-strand promoter of mouse mitochondrial DNA: a functional promoter consists of multiple upstream domains. Mol Cell Biol. 1986 Sep;6(9):3253–3261. doi: 10.1128/mcb.6.9.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D. D., Hauswirth W. W., Clayton D. A. Replication priming and transcription initiate from precisely the same site in mouse mitochondrial DNA. EMBO J. 1985 Jun;4(6):1559–1567. doi: 10.1002/j.1460-2075.1985.tb03817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster A. C., Jeffries A. C., Sheldon C. C., Symons R. H. Structural and ionic requirements for self-cleavage of virusoid RNAs and trans self-cleavage of viroid RNA. Cold Spring Harb Symp Quant Biol. 1987;52:249–259. doi: 10.1101/sqb.1987.052.01.030. [DOI] [PubMed] [Google Scholar]
- Forster A. C., Symons R. H. Self-cleavage of virusoid RNA is performed by the proposed 55-nucleotide active site. Cell. 1987 Jul 3;50(1):9–16. doi: 10.1016/0092-8674(87)90657-x. [DOI] [PubMed] [Google Scholar]
- Gold H. A., Topper J. N., Clayton D. A., Craft J. The RNA processing enzyme RNase MRP is identical to the Th RNP and related to RNase P. Science. 1989 Sep 22;245(4924):1377–1380. doi: 10.1126/science.2476849. [DOI] [PubMed] [Google Scholar]
- Green C. J., Vold B. S. Structural requirements for processing of synthetic tRNAHis precursors by the catalytic RNA component of RNase P. J Biol Chem. 1988 Jan 15;263(2):652–657. [PubMed] [Google Scholar]
- Greenberg B. D., Newbold J. E., Sugino A. Intraspecific nucleotide sequence variability surrounding the origin of replication in human mitochondrial DNA. Gene. 1983 Jan-Feb;21(1-2):33–49. doi: 10.1016/0378-1119(83)90145-2. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., van Belkum A., Pleij C. W., Altman S. Novel reactions of RNAase P with a tRNA-like structure in turnip yellow mosaic virus RNA. Cell. 1988 Apr 22;53(2):267–272. doi: 10.1016/0092-8674(88)90388-1. [DOI] [PubMed] [Google Scholar]
- Haseloff J., Gerlach W. L. Simple RNA enzymes with new and highly specific endoribonuclease activities. Nature. 1988 Aug 18;334(6183):585–591. doi: 10.1038/334585a0. [DOI] [PubMed] [Google Scholar]
- Hashimoto C., Steitz J. A. Sequential association of nucleolar 7-2 RNA with two different autoantigens. J Biol Chem. 1983 Feb 10;258(3):1379–1382. [PubMed] [Google Scholar]
- Hauswirth W. W., Clayton D. A. Length heterogeneity of a conserved displacement-loop sequence in human mitochondrial DNA. Nucleic Acids Res. 1985 Nov 25;13(22):8093–8104. doi: 10.1093/nar/13.22.8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauswirth W. W., Van de Walle M. J., Laipis P. J., Olivo P. D. Heterogeneous mitochondrial DNA D-loop sequences in bovine tissue. Cell. 1984 Jul;37(3):1001–1007. doi: 10.1016/0092-8674(84)90434-3. [DOI] [PubMed] [Google Scholar]
- Inoue T., Sullivan F. X., Cech T. R. Intermolecular exon ligation of the rRNA precursor of Tetrahymena: oligonucleotides can function as 5' exons. Cell. 1985 Dec;43(2 Pt 1):431–437. doi: 10.1016/0092-8674(85)90173-4. [DOI] [PubMed] [Google Scholar]
- Jacquier A., Michel F. Multiple exon-binding sites in class II self-splicing introns. Cell. 1987 Jul 3;50(1):17–29. doi: 10.1016/0092-8674(87)90658-1. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- McClain W. H., Guerrier-Takada C., Altman S. Model substrates for an RNA enzyme. Science. 1987 Oct 23;238(4826):527–530. doi: 10.1126/science.2443980. [DOI] [PubMed] [Google Scholar]
- Michel F., Jacquier A. Long-range intron-exon and intron-intron pairings involved in self-splicing of class II catalytic introns. Cold Spring Harb Symp Quant Biol. 1987;52:201–212. doi: 10.1101/sqb.1987.052.01.025. [DOI] [PubMed] [Google Scholar]
- Mount S. M., Pettersson I., Hinterberger M., Karmas A., Steitz J. A. The U1 small nuclear RNA-protein complex selectively binds a 5' splice site in vitro. Cell. 1983 Jun;33(2):509–518. doi: 10.1016/0092-8674(83)90432-4. [DOI] [PubMed] [Google Scholar]
- Reddy R., Tan E. M., Henning D., Nohga K., Busch H. Detection of a nucleolar 7-2 ribonucleoprotein and a cytoplasmic 8-2 ribonucleoprotein with autoantibodies from patients with scleroderma. J Biol Chem. 1983 Feb 10;258(3):1383–1386. [PubMed] [Google Scholar]
- Reimer G., Raska I., Scheer U., Tan E. M. Immunolocalization of 7-2-ribonucleoprotein in the granular component of the nucleolus. Exp Cell Res. 1988 May;176(1):117–128. doi: 10.1016/0014-4827(88)90126-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlenbeck O. C. A small catalytic oligoribonucleotide. Nature. 1987 Aug 13;328(6131):596–600. doi: 10.1038/328596a0. [DOI] [PubMed] [Google Scholar]
- Walberg M. W., Clayton D. A. Sequence and properties of the human KB cell and mouse L cell D-loop regions of mitochondrial DNA. Nucleic Acids Res. 1981 Oct 24;9(20):5411–5421. doi: 10.1093/nar/9.20.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood W. I., Gitschier J., Lasky L. A., Lawn R. M. Base composition-independent hybridization in tetramethylammonium chloride: a method for oligonucleotide screening of highly complex gene libraries. Proc Natl Acad Sci U S A. 1985 Mar;82(6):1585–1588. doi: 10.1073/pnas.82.6.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaug A. J., Been M. D., Cech T. R. The Tetrahymena ribozyme acts like an RNA restriction endonuclease. Nature. 1986 Dec 4;324(6096):429–433. doi: 10.1038/324429a0. [DOI] [PubMed] [Google Scholar]
- Zaug A. J., Cech T. R. The intervening sequence RNA of Tetrahymena is an enzyme. Science. 1986 Jan 31;231(4737):470–475. doi: 10.1126/science.3941911. [DOI] [PubMed] [Google Scholar]
- Zaug A. J., Grosshans C. A., Cech T. R. Sequence-specific endoribonuclease activity of the Tetrahymena ribozyme: enhanced cleavage of certain oligonucleotide substrates that form mismatched ribozyme-substrate complexes. Biochemistry. 1988 Dec 13;27(25):8924–8931. doi: 10.1021/bi00425a008. [DOI] [PubMed] [Google Scholar]