Background: ERK5 have distinct signaling roles compared with ERK2.
Results: New crystal structures of ERK5 reveal the structural basis for ERK5 versus ERK2 signaling.
Conclusion: Assembly of the ERK5 signaling cascade requires a unique cooperation between a linear binding motif and a modular protein-protein interaction domain.
Significance: Molecular principles underlying ERK-specific activation suggest how to interfere with their signaling pathways.
Keywords: MAP Kinases (MAPKs), Protein Complexes, Protein Evolution, Protein Structure, Signal Transduction, Linear Binding Motif
Abstract
Mitogen-activated protein kinase (MAPK) activation depends on a linear binding motif found in all MAPK kinases (MKK). In addition, the PB1 (Phox and Bem1) domain of MKK5 is required for extracellular signal regulated kinase 5 (ERK5) activation. We present the crystal structure of ERK5 in complex with an MKK5 construct comprised of the PB1 domain and the linear binding motif. We show that ERK5 has distinct protein-protein interaction surfaces compared with ERK2, which is the closest ERK5 paralog. The two MAPKs have characteristically different physiological functions and their distinct protein-protein interaction surface topography enables them to bind different sets of activators and substrates. Structural and biochemical characterization revealed that the MKK5 PB1 domain cooperates with the MAPK binding linear motif to achieve substrate specific binding, and it also enables co-recruitment of the upstream activating enzyme and the downstream substrate into one signaling competent complex. Studies on present day MAPKs and MKKs hint on the way protein kinase networks may evolve. In particular, they suggest how paralogous enzymes with similar catalytic properties could acquire novel signaling roles by merely changing the way they make physical links to other proteins.
Introduction
MAPKs are activated through phosphorylation by MAPK kinases (MKK).2 MKK5 is the specific activator of extracellular regulated kinase 5 (ERK5), which fulfills non-redundant physiological roles compared with its ERK1/2 MAPK paralogs (1–3). Mammals possess seven MKKs, where MKK1/2, MKK3/6, MKK7 activate ERK, p38, and JNK, respectively, MKK4 phosphorylates both p38 and JNK, and MKK5 activates ERK5 (4–6). ERK1/2 is generally involved in mediating mitogenic signals, but ERK5 controls distinct and more specific physiological responses in neuronal survival and differentiation or in cardiovascular development (7–10). Furthermore, ERK5 is the only critical MAPK required for the maintenance of blood vessel integrity and vascular homeostasis in the adult (11). ERK2 and ERK5 are expressed ubiquitously, and their activation patterns in different tissues may determine physiological outcomes in a synergistic or combinatorial manner (12, 13).
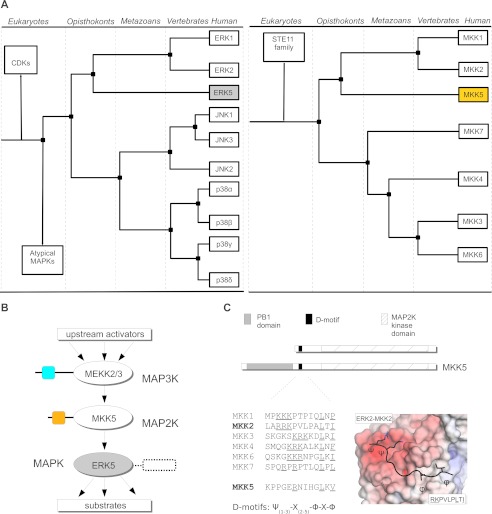
ERK5 is comprised of an N-terminal MAPK domain homologous to ERK1/2 and a long non-catalytic C-terminal tail that is involved in the regulation of its activity and cellular localization (14, 15). Present day MAPKs have emerged through whole genome or by individual gene duplication events (16, 17). The MAPK protein family arose at the dawn of eukaryotic evolution and separation of ERK-like and p38-like groups predates the divergence of animals and fungi (Opisthokonts). The four animal-specific MAPK subgroups (ERK1/2, ERK5, JNK1/2/3, p38α/β/γ/δ) came into existence early in Metazoan evolution. Interestingly, the ERK5 pathway is intact in all deuterostomes, but protostomes secondarily lost all components of this signaling pathway. MKKs belong to a separate branch of the kinome compared with MAPKs (STE7 protein kinase group), and MKK5 appears to be the closest homolog of ERK1/2 activating MKKs. Therefore, MKK5, with its unique PB1 (Phox and Bem1) domain among MKK enzymes, appeared after the separation of MKKs involved in ERK1/2 or p38/JNK MAPK activation (MKK1/2 versus MKK7/4/3/6, respectively). With the emergence of vertebrates, full genome duplications created many similar MAPK and MKK paralogs, but the cladogram of MAPKs and their MKK activators, in agreement with their biochemical specificity, concurs well (see Fig. 1A).
FIGURE 1.
A PB1 domain in a MAPK kinase is unique to the ERK5 signaling module. A, the cladogram for MAPKs and their specific MKK activators. Cladograms were calculated based on the sequences of kinase domains for MAPKs or MKKs found in KinBase (57). CDK, cyclin-dependent kinase. (Atypical MAPKs are ERK3/4/7 or NLK (58), STE11 family is comprised of MAPK kinase kinases.). B, schematic organization of the MEKK2/3-MKK5-ERK5 MAPK module. PB1 domains from MEKK3 and MKK5 are shown with colored squares, and a box next to the ERK5 kinase domain indicates the long non-catalytic C-terminal tail (407–816) of ERK5. C, all MKKs contain a linear D-motif: their sequences are shown for the seven human MKKs. In the MKK D-motif consensus Ψ, Φ, and X denotes basic, hydrophobic, and any amino acids, respectively. In addition, MKK5 also contains an evolutionary conserved PB1 globular domain. The panel on the right shows the MAPK docking surface colored according to its electrostatic potential (positive in blue and negative in red) from the ERK2 crystal structure complexed with a peptide containing the MKK2 linear D-motif (shown in black with the side chains of the consensus motif forming amino acids indicated) (26).
Although the three conventional MAPK pathways (ERK1/2, p38, and JNK) are highly studied, a lot less is known about the ERK5 pathway. The upstream activators of the three-tiered MEKK3-MKK5-ERK5 kinase module and its downstream targets are under intense investigation, particularly because this pathway is involved in a number of more aggressive, metastatic varieties of cancer due to its role in cell survival and proliferation (3). Growth factor-induced cellular stimulation and oxidative stress activates MEKK2/3, which in turn, activates MKK5 (see Fig. 1B). MEF2 transcription factors are common MAPK substrates and MEF2A, MEF2C were identified as physiological substrates for ERK5. Thus, in addition to their major regulatory role in skeletal and cardiac muscle development, these factors also play an important role in neuronal differentiation and survival through activation by the ERK5 pathway (18–20). MAPK substrates, similarly to other MAPK interacting partners such as MKKs, phosphatases, and protein scaffolds, frequently contain a (D)ocking motif consensus sequence in their unstructured region (21–23) These D-motifs generally possess a loose consensus sequence (Ψ1–3-X2–5-Φ-X-Φ, where Ψ, Φ, and X denote basic, hydrophobic, and any amino acids, respectively), and they bind to the MAPK docking groove (Fig. 1C) (23–26). In contrast to other MKK-MAPK systems depending only on linear D-motifs, efficient activation of ERK5 also requires a PB1 domain located in the N-terminal region of MKK5 (27–29). In addition, this domain was also shown to play an important role in MKK5 activation by its MAPK kinase kinase activators, MEKK2/3 (29, 30).
Here, we investigated the molecular mechanisms determining the signaling specificity of ERK5 versus ERK1/2 MAPK modules. These two systems are evolutionarily clearly related; however, they play distinct physiological roles in mammals. ERK5, in contrast to ERK2, has a long, non-catalytic C-terminal tail that is only known to be involved in downstream signaling events following ERK5 activation (1). Therefore, we focused on the differences between the ERK2 and ERK5 kinase domains. In this study, we examined the contribution of the MKK5 PB1 domain and the MKK5 linear docking motif toward ERK5-specific binding and activation. We determined the crystal structure of the ERK5 kinase domain alone and in complex with an MKK5 construct comprised of the MKK5 PB1 domain and the putative MAPK binding linear motif. These revealed that ERK5 and ERK2 docking surfaces are topographically different, suggesting that this is what sets these two MAPKs functionally apart. We also reconstituted the three-tiered ERK5 MAPK module (MEKK3-MKK5-ERK5) in vitro and show how the MKK5 PB1 domain can work as a bivalent adaptor between the upstream activator kinase and a downstream substrate. These mechanisms described here for the human ERK5 MAPK module could have collectively contributed to the emergence of a distinct ERK-based signaling pathway in most organisms during evolution, at least in relation to the better studied and more canonical MKK1/2-ERK1/2 system.
EXPERIMENTAL PROCEDURES
DNA Constructs, Protein Expression, and Purification
All expression constructs were human proteins. Complementary DNAs (cDNAs) encoding human proteins were produced from mRNA derived from HEK293 cells. Reverse transcription was followed by polymerase chain reaction (PCR) with protein-specific PCR primer pairs. All sequences were verified by DNA sequencing after subcloning into pET expression vectors. All experiments were carried out using the ERK5 MAPK construct containing the N-terminal region of ERK5-(1–431) (Uniprot ID Q13164). This and different MKK5 (Uniprot ID Q13163) constructs were cloned into a modified pET vector allowing the expression of recombinant proteins with an N-terminal maltose binding protein (MBP) fusion tag and a C-terminal hexahistidine tag. (MKK5 constructs contained the following regions: A, 1–448; B, 113–448; C, 127–448; D, 166–448; E, 16–130; F:1–112; and G, 113–129). Proteins were expressed in Esherichia coli Rosetta(DE3) pLysS (Novagen) cells using standard techniques. Recombinant proteins were purified with a procedure involving a nickel and/or MBP affinity chromatography step that, depending on further use, was followed by an ion exchange step using a Resource Q 1 ml column (GE Healthcare). The MBP tag was removed by tobacco etch virus protease cleavage except for MBP pulldown experiments. Final protein samples were dialyzed into storage buffer containing 20 mm Tris, pH 8.0, 100 mm NaCl, 10% glycerol, and 2 mm tris(2-carboxyethyl)phosphine. All peptides were synthesized on an ABI 431A peptide synthesizer using Fmoc strategy.
Expression and purification of ERK2 was done as earlier described in Ref. 23. MKK1 and MKK2 constructs were expressed in E. coli with N-terminal GST and C-terminal hexahistidine tags and double affinity purified with standard procedures. The MEKK3 PB1 domain (residues 42–126) was expressed as GST fusion protein using a modified pET vector enabling fusion protein production in bacteria. MEKK3 constructs containing the kinase domain were expressed as MBP fusion proteins in SF9 cells with the Bac-to-Bac baculoviral expression system (Invitrogen). Proteins were purified from SF9 cell lysates similarly as described previously for proteins expressed in E. coli.
Protein-protein Binding Assays
For MBP pulldown experiments, the amylose resin (New England BioLabs) was first equilibrated with binding buffer (20 mm Tris, 100 mm NaCl, 0.1% octylphenoxypolyethoxyethanol, 2 mm β-mercaptoethanol) and 10 μg of immobilized MBP fusion protein (and MBP protein as negative control) was incubated in the presence of 10 μm prey in 200 μl of binding buffer for 30 min at room temperature. Binding reactions typically contained 10–20 μl of resin saturated with baits. Amylose beads were pelleted with centrifugation and washed three times. Retained proteins were eluted from the resin with SDS loading buffer. Samples were subjected to SDS-PAGE and stained with Coomassie protein dye. GST pulldowns were done similarly, but glutathione beads were used (GE Healthcare).
For fluorescence polarization (FP)-based binding affinity measurements reporter peptides, pepMKK5 (ASKPPGERNIHGLKVNTRA) or pepMEF2 (SRKPDLRVVIPPS) were N-terminally labeled with carboxyfluorescein or tetramethylrhodamin fluorescent dyes, respectively. PB1-D was labeled with 5-iodoacetamidofluorescein (Sigma, I9271) according to the following procedure: dimethylformamide dissolved dye was added in ∼5-fold molar excess to purified PB1-D sample that was formerly dialyzed in PBS (pH ∼ 7.0) and then incubated in the dark for 2 h at room temperature. Unreacted dye was removed on a PD-10 buffer exchange column (GE Healthcare). Labeling efficiency was determined to be ∼75%.
Change in the FP signal in direct binding affinity measurements was monitored as a function of increasing concentration of purified MAPKs with an Analyst GT (Molecular Devices) or with a Synergy H4 (BioTek Instruments) plate reader in 384-well plates. The labeled peptides or FP labeled PB1-D were at 10 nm in 20 mm Tris, pH 8.0, 100 mm NaCl, 0.05% Brij35P, 2 mm DTT. The resulting binding isotherms were fit to a quadratic binding equation with the OriginPro software (version 7, OriginLab Corp.). The affinity of the unlabeled peptides or different MKK5 constructs to MAPKs was measured in steady-state competition experiments: 10 nm labeled reporter was mixed with MAPK samples in a concentration to achieve ∼60–80% complex formation. Subsequently, increasing amounts of unlabeled peptide or protein was added and the FP signal was measured as described earlier for direct titration experiments. The dissociation constant (Kd) for each MAPK-unlabeled peptide or protein interaction was determined by fitting the data to a competition binding equation. Titration experiments were carried out in triplicates and the average FP signal was used for fitting the data with OriginPro (version 7).
In Vitro Protein Kinase Assays
Reactions were carried out in 50 mm HEPES, pH 7.5, 100 mm NaCl, 5 mm MgCl2, 0.05% octylphenoxypolyethoxyethanol, 5% glycerol, 2 mm DTT using recombinantly expressed and purified proteins in the presence of 400 μm ATP and ∼5 μCi of [γ-32P]ATP. Reactions were stopped with protein loading sample buffer complemented with 20 mm EDTA, boiled, and then subjected to SDS-PAGE. Gels were dried before analysis by phosphorimaging or by Western blots on a Typhoon Trio+ scanner (GE Healthcare). Western blots were done by using anti-phospho-ERK5 antibody (E7153, Sigma). The constitutive active form of MKK1 (MKK1_EE, S218E/S222E), MKK2 (MKK2_EE, S222E/S226E), MKK5 (MKK5DD, S311D/T315D) and the kinase inactive version of ERK5 (ERK5_KI: D182A), which was used as substrate in kinase assays, were generated by QuikChange site-directed mutagenesis or by PCR, and proteins were expressed and purified as described above. Phosphorylation reporters (MEF2A, region 269–329; RSK1c, region 411–735, MNK1, full-length, 1–465) were expressed in bacteria as GST fusion proteins with an N-terminal GST and C-terminal hexahistidine tag and then double-affinity purified.
Structure Solution of Apo ERK5 and the ERK5·PB1-D Complex
The MAP kinase domain of ERK5-(1–431) and the MKK5 PB1-D construct (16–130) were expressed as N-terminal MBP fusion proteins with a C-terminal non-cleavable hexahistidine tag in E. coli. After double-affinity purification, the MBP fusion tag was cleaved off by the tobacco etch virus protease, and samples were further purified by ion-exchange on a Resource Q column. Purified ERK5 and PB1-D samples were mixed so that the PB1-D would be in excess, and the sample was gel-filtrated on a Superdex 75 column (GE Healthcare). Fractions corresponding to the ERK5·PB1-D complex were pooled, and the sample was concentrated to 10 mg/ml.
The stock solution of the final protein sample was supplemented with 2 mm adenylyl-imidodiphosphate (AMP-PNP) and 2 mm MgCl2. Crystallization conditions were found by using a custom in-house PEG crystallization screen in standard sitting drop vapor diffusion set-up at 23 °C. ERK5 apo and ERK5·PB1-D complex crystals grew in 35% PEG1000 buffered with 100 mm sodium citrate (pH 5.0) or in 48% PEG200 buffered with 100 mm MIB (composite buffer of malonate, imidazol, and boric acid) (pH 6.5), respectively. Crystals were directly flash cooled in liquid nitrogen. Data were collected on the PXIII beam line of the Swiss Light Source (Paul Scherrer Institute, Villigen, Switzerland).
Data were processed with XDS (31). The structure was solved by molecular replacement in PHASER using a starting model of the MKK5 PB1 domain (Protein Data Bank code 2O2V) and the kinase domain from ERK2 (Protein Data Bank code 2Y9Q) (23, 32, 33). The molecular replacement search at 3 Å resolution identified two ERK5 molecules for the apo crystal structure and two ERK5·PB1 complexes for the ERK5·PB1-D crystal structure per asymmetric unit. Structure refinement was carried out in PHENIX (F > 1.35 σF), and structure remodeling/building was done in Coot (34, 35). The final refined models had good geometry, as assessed by Molprobity, with 90.5 and 0.3% of the amino acid residues in the favored and disallowed regions of the Ramachandran plot, respectively (36). X-ray data collection and structure refinement statistics are given in Table 1. Crystal structures of apo ERK5 and the ERK5·PB1-D complex are deposited in the Protein Data Bank with identifiers 4IC8 and 4IC7, respectively.
TABLE 1.
Crystal structure solution and refinement of apo ERK5 and the ERK5·PB1-D complex
| Apo ERK5 | ERK5·PB1-D | |
|---|---|---|
| Data collection | ||
| Space group | P21 | P43 |
| Cell dimensions | a = 59.9, b = 93.2, and c = 69.3 Å; α = 90.0, β = 89.9, and γ = 90.0° | a = 69.4, b = 69.4, and c = 271.2 Å; α = 90.0, β = 90.0, and γ = 90.0° |
| Resolution (Å)a | 45.29–2.8 (2.87–2.8) | 48.27–2.6 (2.67–2.6) |
| Rmergea | 0.057 (0.459) | 0.051 (0.719) |
| I/σ(I)a | 17.46 (2.69) | 19.03 (2.52) |
| Completeness (%)a | 98.1 (100.0) | 99.8 (98.6) |
| Redundancya | 3.5 (3.3) | 4.5 (4.5) |
| Refinement | ||
| No. of reflections | 18,523 | 39,029 |
| Rwork/Rfree | 0.262/0.289 | 0.211/0.258 |
| No. of atoms | ||
| Macromolecules | 4807 | 7250 |
| Ligands | 62 | |
| Water | 83 | |
| B factors | ||
| Wilson B-factor | 58.6 | 61.2 |
| Average | 91.0 | 90.8 |
| Macromolecules | 91.0 | 90.8 |
| Solvent | - | 62.5 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.006 | 0.008 |
| Bond angles | 1.52° | 1.59° |
a Values in parentheses are for the highest resolution shell.
RESULTS
Characterization of the MKK5-ERK5 Interaction
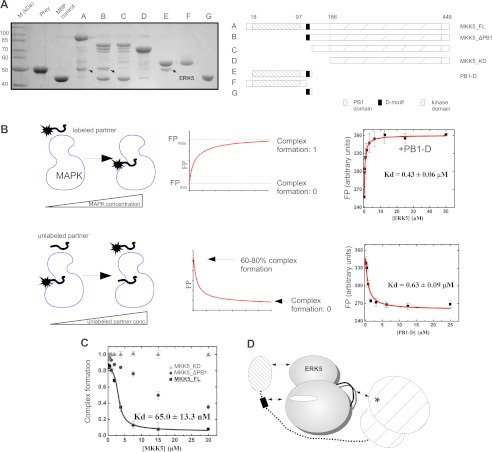
MKK5 contains a C-terminal kinase domain, an N-terminal PB1 domain, and a short linear D-motif located between the former two domains. We tested the binding of different MKK5 constructs to the kinase domain of ERK5-(1–431) in MBP pull-down experiments. Full-length MKK5 and several deletion constructs, missing one or two of the regions mentioned above, were expressed as MBP fusion proteins in bacteria and then bound to amylose resin that was incubated with recombinantly expressed and purified ERK5 (Fig. 2A). These binding studies showed that neither the PB1 domain nor the MKK5 kinase domain (MKK5_KD) alone was sufficient to mediate detectable binding to ERK5. Interestingly, all constructs that bound ERK5 contained the D-motif in combination with at least one structured MKK5 domain. These results suggest that multiple regions in MKK5 synergistically contribute to ERK5 binding.
FIGURE 2.
Characterization of the MKK5-ERK5 interaction. A, three distinct MKK5 regions contribute to its interaction with ERK5. MBP pulldown results with the ERK5 kinase domain (residues 1–431) as prey and different deletion constructs (A, A–G panels) of MKK5 as baits. Interaction of bait and prey was detected on an SDS-PAGE gel stained with Coomassie protein dye. Arrows indicate the position of ERK5 in the pulldown experiments. MBP control was used as negative control to assess unspecific binding of the prey. Bands appearing in addition to baits or preys are degradation products of MBP-fusion proteins. The panel shows the results of a representative MBP pulldown assay from two independent experiments. B, schematic diagram of direct (top panels) and competitive (bottom panels) FP-based titration experiments for determining protein-peptide or protein-protein binding affinities (Kd). Panels on the right show binding isotherms for direct (on the top) and for competitive (on the bottom) ERK5·PB1-D binding experiments. Errors indicate uncertainty in the fit, and error bars on the binding isotherms show S.D. based on three independent measurements. C, competitive binding experiments to monitor binding of MKK5_KD, MKK5_ΔPB1, and MKK5_FL to ERK5. The panel shows the competitive binding curve using unlabeled PB1-D as the competitor and labeled ERK5·PB1-D complex as the reporter (PB1-D labeled with 5-iodoacetamidofluorescein, IAF). The y axis shows the degree of complex formation that was determined based on arbitrary FPmin and FPmax units from numerical fits to competitive binding equations as shown on B. Data were fit to a competition binding equation. Errors indicate uncertainty in the fit, and error bars on the binding isotherms show S.D. based on three independent measurements. D, interactions contributing to the formation of the MKK5·ERK5 binary complex: (i) the MKK5 PB1 domain binds to the ERK5 kinase domain, (ii) MKK5 D-motif presumably binds in the ERK5 docking groove, and (iii) the MKK5 kinase domain binds to the ERK5 kinase domain on an area distinct from the PB1 and D-motif-interacting surfaces. This latter interaction might speculatively form between the MKK5 active site and the ERK5 activation loop (shown with an asterisk or with a black line, respectively). conc., concentration.
To assess the contribution of different MKK5 domains toward MKK5-ERK5 binding quantitatively, we set up a fluorescence polarization based protein-protein interaction assay for the determination of steady-state binding affinities (Fig. 2B). The PB1-D MKK5 construct was labeled by 5-iodoacetamidofluorescein on a cysteine residue and its complex formation with ERK5 was monitored in a FP-based binding assay (Fig. 2B). This construct bound with ∼0.5 μm affinity to ERK5, which was also confirmed in a competitive titration experiment using unlabeled PB1-D (Fig. 2B). Next, we measured the affinity of the MKK5-ERK5 interaction using the 5-iodoacetamidofluorescein-labeled PB1-D as the reporter and three different MKK5 constructs as the competitor in a competition titration experiment (Fig. 2C). Full-length MKK5 (MKK5_FL) competed with ERK5·PB1-D binding and its binding affinity to ERK5 was determined to be <0.1 μm. MKK5_KD, however, did not compete with PB1-D, suggesting that the MKK5 kinase domain contacts ERK5 on a surface that is not used in the ERK5·PB1-D complex. In contrast, MKK5_ΔPB1 did compete; however, its titration curve could not be fit to a simple competition binding equation. This shows that competition with the ERK5-PB1-D interaction by the D-motif in MKK5_ΔPB1 does not confer to a simple one-site binding model: the binding of the PB1 domain and the D-motif to ERK5 may be synergistic for example. Results of these quantitative measurements are consistent with the results of MBP pulldown experiments. They suggest that all three regions of MKK5, the PB1 domain, the D-motif, and the kinase domain, contact ERK5 on complementary surfaces (Fig. 2D). All contacts seem to contribute to the high affinity (<100 nm) of the MKK5·ERK5 complex.
Crystal Structure of the ERK5·PB1-D Complex
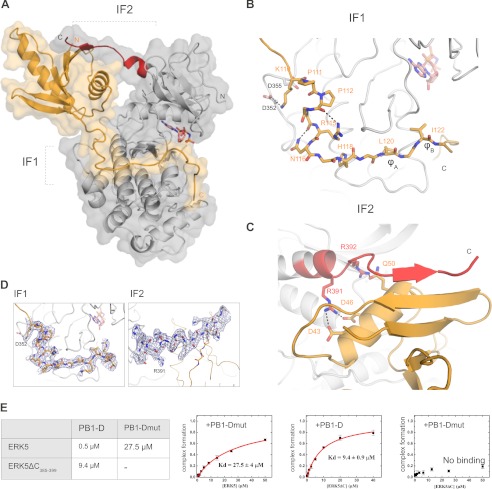
To gain insight into the structural basis of MKK5-ERK5 binding, we determined the crystal structure of the ERK5 kinase domain bound to an MKK5 construct that mediated submicromolar binding and contained both the PB1 domain as well as the linear D-motif (PB1-D) (Fig. 3A). The PB1-D construct contacts the MAPK on two distinct surfaces: the linear motif binds in the MAPK docking groove (IF1), whereas the PB1 domain interacts with ERK5 on the N-terminal kinase lobe (IF2). Overall, this complex displays 1600 Å2 buried surface area. The final crystallographic model of the ERK5·PB1-D complex contains two complexes in the asymmetric unit related by non-crystallographic symmetry. The two complexes are greatly similar (r.m.s.d. is 1.0 Å for 451 Cα atoms), and the only difference between the two complexes appears to be that the C-terminal region of the PB1-D construct is less structured in one of the complexes. In addition, we also determined the crystal structure of the ERK5 kinase domain without its interaction partner (apo ERK5). This crystallographic model also contains two ERK5 molecules related by non-crystallographic symmetry (Table 1). The two ERK5 molecules are also highly similar (r.m.s.d. of 0.6 Å for 324 Cα atoms); however, a short region (150–155) corresponding to an important part of the MAPK docking groove is disordered in one of the ERK5 molecules (see below).
FIGURE 3.
Crystal structure of the ERK5·PB1-D complex. A, transparent surface representation of the ERK5·PB1-D complex. The ERK5 kinase domain is shown in gray, and its C-terminal region (from 385 to 399) is shown in red. The MKK5 PB1-D construct is shown in orange. B, zoomed-in view of the protein-protein interface in the MAPK docking groove (IF1). C, zoomed-in view of the protein-protein interface on the PB1 domain (IF2). Only amino acid side chains appearing to play an important role at the interfaces are indicated. Specific hydrogen bonds are shown with dashed lines, and the non-hydrolyzable ATP analog, AMP-PNP, is shown in pink. D, representative examples of electron density maps shown around the MKK5 D-motif at IF1 and the ERK5 C-terminal extension at IF2. 2Fo − Fc maps were calculated with the ERK5·PB1-D final model and contoured at 1σ. E, results of binding affinity measurements on ERK5·PB1-D complex formation. The table displays the binding affinities between wild-type (see also Fig. 2B) or modified versions of MKK5 PB1-D and ERK5 kinase domain constructs. ERK5ΔC385–399 lacks the C-terminal ERK5 kinase domain region shown in red on A and C. PB1-Dmut is an MKK5 PB1-D construct in which Arg-115, Leu-120, and Ile-122 are mutated to alanines. (− indicates no detectable binding; Kd > 100 μm). Errors indicate uncertainty in the fit, and error bars on the binding isotherms show S.D. based on three independent measurements. mut, mutant.
The ERK5·PB1-D complex crystal structure demonstrates how the MKK5 PB1 domain and the D-motif contacts ERK5. The N-terminal part of the MKK5 D-motif unexpectedly does not engage Arg-115 in binding the aspartate residues of the common docking site (Asp-352 and Asp-355) (Fig. 1C) (28). It is Lys-110 that contacts Asp-352 from ERK5, and the intervening region between this basic amino acid and the hydrophobic motif (Leu-120 and Leu-122) adopts an L-shaped turn stabilized by side chain-specific hydrogen bonds (Arg-115 and Asn-116) and by a short polyproline type II helix (Pro-111, Pro-112, and Gly-113) (Fig. 3, B and D). Leu-120 and Leu-122 from PB1-D bind into the classical ϕxϕ MAPK docking groove similarly to other MAPK binding linear motifs (23). At the other ERK5·PB1-D interface, the C-terminal MAPK region (from residue 385 to 399) makes several main chain atom mediated hydrogen bonds to residues of a β-strand (between residue 26 and 32) from the PB1 domain (Fig. 3, C and D). These side chain-independent interactions are complemented by side chain-mediated hydrogen bonds formed by Arg-391 and Arg-392 from ERK5. Importance of contacts observed by the ERK5 MAPK docking groove (IF1) or by the novel interface on the MKK5 PB1 domain (IF2) was confirmed by fluorescence polarization-based protein-protein binding assays. Although the ERK5 MAPK construct bound PB1-D with ∼0.5 μm affinity, there was no interaction detected between two modified constructs where the D-motif and the C-terminal ERK5 kinase domain extension both were mutated or deleted, respectively (Fig. 3E). The PB1-Dmut construct contained alanine replacements at Arg-115, Leu-120, and Leu-122; ERK5ΔC385–399 lacked its PB1 domain interacting region. These also bound their corresponding wild-type partner with greatly reduced (50-fold or 20-fold) binding affinities (Fig. 3E). These results demonstrate how the MKK5 PB1 domain and an MKK docking motif forms synergistic contacts and how MKK5 engages canonical as well as novel protein-protein interaction surfaces to mediate high affinity binding to ERK5.
Distinct Protein Interactions of ERK5 and ERK2
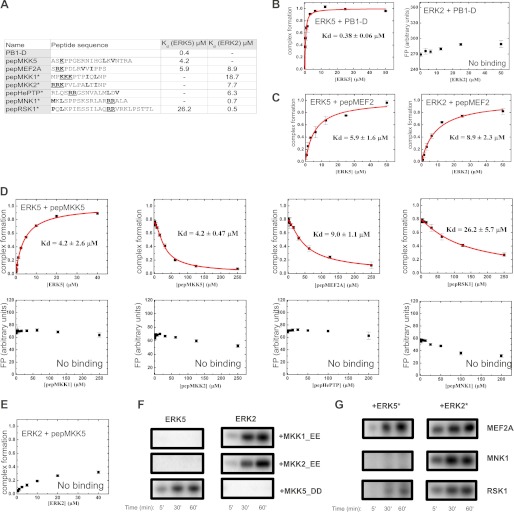
Binding affinity measurements showed that although PB1-D bound ERK5 with submicromolar affinity, ERK2 did not bind to the same MKK5 construct (Fig. 4, A and B). Similarly, the linear D-motif from MKK5 bound only to ERK5 but not to ERK2 (Fig. 4, A, D, and E). These findings are in line with the observation that MKK5 could not phosphorylate ERK2 in cell-based assays (28). ERK5 stimulates the activity of MEF2 transcription factors; it binds to the D-motif in MEF2A/C, and it promotes phosphorylation of critical MAPK regulatory target sites (37). Moreover, it was shown earlier that a D-motif containing peptide from MEF2A binds to ERK2 and p38α (23, 24). We tested the binding of a labeled MEF2A peptide (pepMEF2) to ERK5 and found that this linear motif containing peptide indeed bound to ERK5 as expected (Fig. 4C). We also tested the binding specificity of ERK5 to other ERK2 binding D-motifs derived from known ERK2 interaction partners. Of five additional linear motifs, it was only the RSK1 peptide that mediated modest binding to ERK5 (Fig. 4A,D) (38). These suggest that ERK5 can discriminate against canonical ERK2 partners via its docking groove. Conversely, short linear motifs can contribute to biologically relevant MAPK-partner protein binding specificity. They may be ERK2- or ERK5-specific, or nondiscriminatory.
FIGURE 4.
Specificity of the ERK5 MAPK docking groove compared with ERK2. A, ERK5 and ERK2 binding affinities of chemically synthesized peptides compared with the binding affinities of the MKK5 PB1-D construct. Asterisks indicate peptides for which ERK2 binding affinities are listed from Ref. 23. A minus sign indicates no detectable binding; Kd > 100 μm. Amino acids corresponding to the ϕxϕ motif are shown in boldface type, and basic amino acids binding to the negatively charged common docking (CD) groove are underlined and shown in boldface type. Note that linear motifs may bind into the MAPK docking groove in two different N-to-C-terminal orientations. In addition to the classical D-motif, reverse D-motifs also exist (e.g. pepMNK1 or pepRSK1) (23). Shown are the results of PB1-D (B) and MEF2A (C) docking peptide binding experiments with ERK5 and ERK2, ERK5 competitive titration experiments with various chemically synthesized peptides (D), ERK2 binding experiment with pepMKK5 (E). Errors indicate uncertainty in the fit, and error bars on the binding isotherms show S.D. based on three independent measurements. F, MKK1, MKK2, and MKK5 phosphorylate only their cognate MAPK in vitro. Recombinant ERK5 and ERK2 (5 μm) were incubated as substrates with constitutively activated forms of MKK enzymes (MKK1_EE, MKK2_EE, MKK5_DD; 0.5 μm) in the presence of radioactive [γ-32P]ATP. G, phosphorylation of different MAPK substrates by ERK5 and ERK2. Recombinantly expressed wild-type MAPKs (1 μm) were preincubated with ATP and constitutively active MKK5 or MKK1 enzymes (1–1 μm) creating activated MAPKs (*), in turn phosphorylation of MEF2A, MNK1, and RSK1 phosphorylation reporters was monitored by adding 5-fold excess of substrates (5 μm) in the presence of radioactive [γ-32P]ATP. F and G show typical phosphorimaging results of in vitro kinase assays from two independent experiments.
Moreover, binding specificity of MKK linear motifs showed agreement with the in vitro phosphorylation pattern of MKK-MAPK pairs when the MAPK phosphorylation capacity of constitutively activated forms of MKK1, MKK2, and MKK5 were tested: MKK1/2 phosphorylated only ERK2 and MKK5 phosphorylated only ERK5 (Fig. 4F). We also tested the substrate phosphorylation profile of preactivated MAPKs using three different substrate proteins (MEF2A, RSK1, and MNK1) (23). Similarly to the ERK2 and ERK5 phosphorylation experiments by MKKs described above, the goal of these experiments was to test how linear docking motifs mediate binding and activation of longer protein constructs or full-length proteins. The MEF2 phosphorylation reporter contained a ∼50-amino acid-long region of MEF2A containing the region spanning from the D-motif to the MAPK target sites (Thr-312 and Thr-319) (23), the RSK1 construct contained the C-terminal RSK1 kinase domain in addition to the RSK1 docking motif (RSK1c) (23), and the MNK1 construct was full-length. Preactivated wild-type ERK2 and ERK5 kinase domain with constitutively activated forms of MKK1 (MKK1_EE) and MKK5 (MKK5_DD), respectively, phosphorylated these substrates with a specificity profile that showed agreement with their linear motif binding profile determined with short peptides (Fig. 4G). These results suggest that short linear motifs contribute to biologically relevant MAPK activation or MAPK substrate phosphorylation profiles also in the context of their full protein sequence.
Different Sequence and Surface Features on ERK5 and ERK2
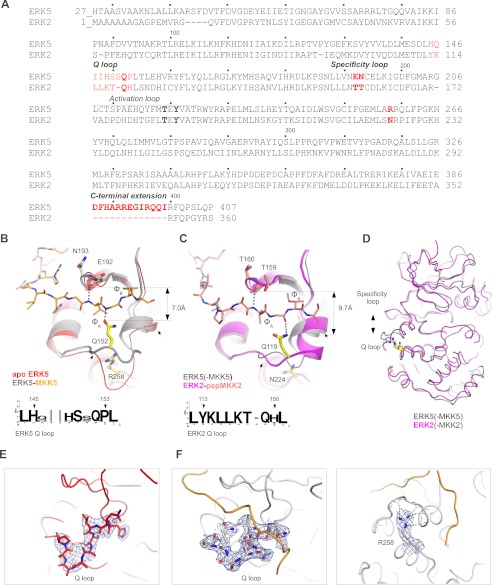
ERK2 and the ERK5 kinase domains are ∼50% identical and ∼80% similar in sequence (Fig. 5A). Moreover, the D-motif-bound kinase domains of these two MAPKs superimpose well (with average r.m.s.d. of 1.0 Å for 240 Cα atoms). What is the structural basis for the distinct interaction partner selection for ERK5 compared with ERK2? Interestingly, the ϕxϕ groove of the MAPK docking groove greatly differs in the apo ERK5 and in the ERK5·PB1-D complex crystal structures (Fig. 5, B–D). A loop region forming the base of this hydrophobic MAPK groove changes its conformation upon binding the MKK5 D-motif (Fig. 5, E and F). This region lies between residues 145 and 153 and is directly involved in mediating protein-protein interactions as observed in the ERK5·PB1-D complex (IF1). Interestingly, the corresponding loop region is well ordered in all apo ERK2 structures and, in contrast to ERK5, it does not show any conformational change upon D-motif peptide binding (23, 39–42). A conserved glutamine in all classical MAPKs (Gln-152 in ERK5 and Gln-119 in ERK2) plays an important role in D-motif binding at this region (referred to as the Q loop henceforth). This conserved residue mediates hydrogen bonding interaction with the backbone amide or carbonyl group of the amino acid located between the ϕA and ϕB positions of the D-motif consensus. As shown on Fig. 5, B–D, the width of this pivotal D-motif binding region, and ultimately its overall topography, are determined by the position of the conserved glutamine residue and amino acids located in the so-called specificity loop (Glu-192 and Asn-193 in ERK5) (23). The Q loop appears to be supported and held in place for H-bonding with the D-motif by a network of hydrogen bonds around Arg-258 (Fig. 5F).
FIGURE 5.
Different sequence and surface features set ERK5 apart from ERK2. A, sequence alignment of the ERK5 kinase domain and full-length human ERK2. Sequence regions that are important and responsible for different ERK5 versus ERK2 protein-protein binding surface topography are highlighted in red. The threonine and tyrosine phosphorylation target sites of the upstream kinase, which are located in the MAPK activation loop, are shown in boldface type. B, comparison of the ϕxϕ MAPK docking groove region from apo ERK5 and the ERK5·MKK5(PB1-D) complex. Note that the conformation of the Q-loop (shown between two small arrows) differs between the apo (red) and the complexed (gray) crystal structures. (This panel shows the conformation of the ERK5 Q loop from one of the non-crystallographic symmetry-related ERK5 molecules, whereas this region is disordered in the other molecule.) C, comparison of the ϕxϕ MAPK docking groove region from the ERK5·MKK5(PB1-D) and the ERK2-pepMKK2 complex crystal structures (26). Note the difference in the width of the ϕxϕ binding groove. The width is defined by the distance between two H-bond forming amino acids from the MAPK specificity loop on top and from the so-called Q loop from below. These two H-bonds are observed in all known MAPK-docking peptide complex structures, and they are shown with dashed lines. Sequence logos on B and C show the evolutionarily conserved but distinct ERK2 and ERK5 Q-loop sequences (from various annotated vertebrate, primitive metazoan, and opisthokont MAPKs.) D, superposition of the ERK5·MKK5(PB1-D) and the ERK2-pepMKK2 complexes. The figure shows the Cα trace of the two MAPKs when they bind to MKK5 or MKK2 constructs. Note the different width of the MAPK docking groove, which is set by the amino acid supporting the Q loop (Arg-258 in ERK5 and Asn-224 in ERK2). E, 2Fo − Fc electron density map contoured at 1σ and shown around the Q loop from apo ERK5. F, 2Fo − Fc maps contoured at 1σ and shown around the Q loop (left) or around Arg-258 (right) from the ERK5·PB1-D complex. Dashed lines show H-bonds.
Furthermore, the Q loop, the specificity loop, as well as the residue corresponding to Arg-258 in ERK5 or to Asn-224 in ERK2 are evolutionarily conserved within members of a certain MAPK group (ERK1/2, ERK5, p38, and JNKs); however, they are distinct across members belonging to different MAPK groups (Fig. 5, B and C). For example, the Q loop is longer by one amino acid in all known and annotated ERK5 orthologs compared with ERK2 orthologs. Furthermore, the Q-loop has different amino acid composition in the two MAPKs, resulting in different flexibility and protein-protein interaction surface topography. Moreover, ERK5 has a short C-terminal kinase domain extension, which is involved in MKK5 PB1 domain binding and is unique among all MAPKs. These differences between ERK2 and ERK5 in turn form the basis for their differential protein-protein interaction capacity.
The MKK5 PB1 Domain Organizes MEKK3, MKK5, and ERK5 into One Signaling Competent Complex
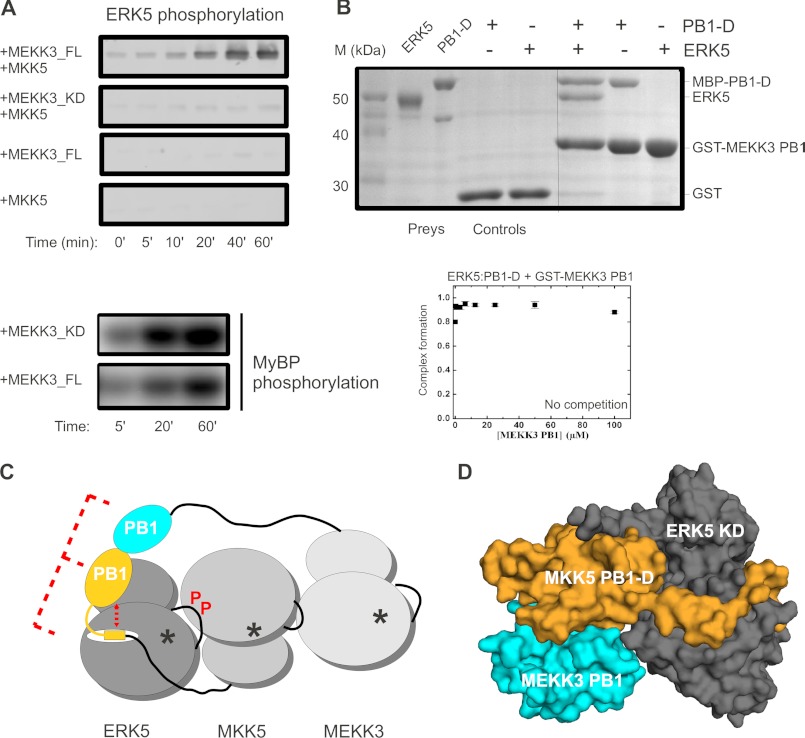
Formerly, the MKK5 PB1 domain was shown to mediate MEKK3 binding via PB1 domain-mediated heterodimerization (27, 30). Here, we set up an in vitro kinase assay to monitor MEKK3 → MKK5 → ERK5 activation with recombinantly expressed and purified ERK5 MAPK module components. The ERK5 MAPK module was reconstituted in vitro, and ERK5 phosphorylation was monitored by anti-phospho-ERK5 Western blots upon addition of full-length MKK5, full-length MEKK3, or a MEKK3 construct lacking the MEKK3 PB1 domain but containing an intact kinase domain (MEKK3_KD, 354–626) (Fig. 6A). We detected efficient ERK5 phosphorylation only with full-length MEKK3 but not with the MEKK3_KD construct that lacked the MEKK3 PB1 domain. The inability of MEKK3_KD to phosphorylate ERK5, however, was not due to its lack of kinase activity because this MEKK3 construct phosphorylated a general protein kinase substrate (myelein basic protein) as efficiently as full-length MEKK3 (Fig. 6A).
FIGURE 6.
The MKK5 PB1 domain serves as an adaptor between MEKK3 and ERK5. A, results of in vitro reconstituted kinase assays with MEKK3-MKK5-ERK5 kinase module components (0.1 μm MEKK3, 0.2 μm MKK5, and 2 μm ERK5). ERK5 phosphorylation was detected by anti-phospho-ERK5 Western blots. Panels show the phosphorimaging results of kinase activity experiments on a general kinase substrate. MEKK3_FL, full-length; MEKK3_KD, kinase domain; MyBP, myelin basic protein. Panels show a representative set of results from two independent experiments. B, ERK5 kinase domain, PB1-D from MKK5, and the PB1 domain from MEKK3 form a ternary complex. Results of a GST pulldown experiment, with GST-MEKK3(PB1) as the bait and the ERK5 MAPK domain and a MBP fusion of MKK5(PB1-D) as prey, are shown in the SDS-PAGE gel stained with Coomassie protein dye (top). Lanes 1 and 2 corresponding to prey show the position of ERK5 and MBP-PB1-D proteins. Control lanes (lanes 3 and 4) indicate the lack of unspecific binding to GST protein-loaded glutathione beads. The panel shows the results of a representative GST pulldown assay from two independent experiments. The panel below shows the results of a fluorescence polarization-based binding assay where complex formation between ERK5 and a labeled MKK5 PB1-D construct was attempted to be competed by the addition of increasing amounts of GST-MEKK3(PB1). Error bars on the binding isotherms show S.D. based on three independent measurements. Results of this experiment are also consistent with the existence of a ternary ERK5·PB1-D·MEKK3(PB1) complex. C, role of PB1 domains in ERK5 activation: (i) synergism between the linear MKK5 D-motif and the MKK5 PB1 domain contributes to high affinity MAPK specific binding (ii) adapter/scaffold function of the MKK5 PB1 domain ensures efficient phosphorylation of ERK5 initiated by MEKK3. An asterisk indicates kinase active sites. P in red highlights ERK5 activation through activation loop phosphorylation by MKK5. D, model of the ERK5·PB1-D·MEKK3(PB1) ternary complex (shown in surface representation). The binary complex crystal structure of the MKK5 and MKK3 PB1 domains (Protein Data Bank code 2O2V) was superimposed on the ERK5·PB1-D complex via the MKK5 PB1 domain.
Next, we tested whether the MKK5 PB1 domain may simultaneously bind to its upstream activator, MEKK3, as well as to its downstream substrate, ERK5. Results of GST pulldown experiments showed that ERK5 could be recruited to the MEKK3 PB1 domain (used as baits in this experiment) only via the MKK5 PB1-D construct (Fig. 6B). In addition, fluorescence polarization-based protein-protein binding assay with labeled PB1-D and ERK5 showed that binding between the MKK5 PB1 domain and ERK5 cannot be competed off by a MEKK3 PB1 domain construct (Fig. 6B). These suggest that MKK5 PB1 domain can serve as an adaptor between MEKK3 and ERK5.
As a summary, Fig. 6C schematically depicts a signaling competent complex of the MEKK3-MKK5-ERK5 MAPK module. It highlights the different molecular mechanisms through which the unique PB1 domain in MKK5, which is not found in other MKKs, contributes to ERK5-specific activation. These include synergism with a linear D-motif to mediate ERK5-specific binding and adapter/scaffold function for the recruitment of the MAPK kinase kinase activator and the MAPK substrate.
DISCUSSION
D-motifs are simple protein-protein interactions tools that MAPKs use to make connections to their partner proteins. These short linear motifs (7 to 17 amino acids) were shown to be able to efficiently discriminate JNK MAPKs from ERK1/2 and p38 MAPKs because JNK has an evolutionary remodeled docking groove compared with more ancient ERK/p38 kinases. (Namely, JNK has a narrowed docking groove that can be bound only by D-motifs with a shorter intervening region between Ψ and ϕ positions) (23).) Most D-motifs cannot specifically target the topographically more similar ERK2 and p38 docking grooves, and additional factors contribute in determining the signaling specificity of ERK1/2- and p38 MAPK-based systems. For example, the MKK1/2 and the MKK6 D-motif can bind to cognate and non-cognate MAPKs as well. However, these MKK enzymes can phosphorylate their cognate MAPK only. This is because the MAPK activation loop sequence, which serves as the substrate for MKK enzymes, is highly divergent for ERK1/2 and p38 kinases. For the ERK1/2 and ERK5 system, a similar mechanism is not likely to play an important role. First, the activation loop sequence of these MAPKs are very similar, and second, it was shown earlier that if the MKK1 catalytic domain is artificially recruited to ERK5, it can activate this noncognate MAPK (29). In this study, we examined the specificity of protein-protein interactions underpinning ERK5 versus ERK1/2 signaling.
Results of binding affinity measurements with some chemically synthesized peptides containing linear D-motif sequences showed that these bound with biologically relevant specificity to ERK5 or ERK2. For ERK5, however, a longer MKK5 construct containing the PB1 domain in addition to its D-motif bound its cognate MAPK with an order of magnitude stronger (Kd = 4.2 μm versus Kd = 0.4 μm). Interestingly, similar examples for synergism between a MAPK docking groove binding D-motif and other linear motifs, microdomains, or domains have already been described (43–47). In these examples, MAPK protein partners may use both docking motifs and structured domains simultaneously, where these function together to enable high binding affinity or novel MAPK interaction specificity. In the MAPK-responsive ATF2 transcription factor, for example, both its D-motif as well as the Zn finger directly preceding it in its regulatory region needs to be intact, and they appear to be synergistic in mediating binding between JNK and ATF2 (48, 49). The topography of the linear D-motif binding docking groove of most MAPKs is similar. Thus, for signaling networks relying on the use of linear binding motifs to govern their protein-protein interaction specificity, it is vital to use mechanisms based on positive and/or negative selection (50, 51). We showed that the linear D-motifs from MKK1 and MKK2 could not bind to the evolutionarily modified ERK5 docking groove as an example for negative selection. Conversely, the MKK5 PB1 domain increased the binding affinity of the MKK5 D-motif as an example for positive selection. The sequence of ERK1/2 and ERK5 activation loops are identical around the MKK target sites. Furthermore, it was previously demonstrated that an MKK5(PB1-D)-MKK1 chimera, where the first 131 amino acids of MKK5 was fused to full-length MKK1, can efficiently phosphorylate and activate ERK5 in cell-based assays (29). These suggest that signaling specificity in these evolutionarily related two MKK-MAPK systems is not based on different catalytic properties but is rather determined by the distinct protein-protein recruitment capacity of their components.
PB1 domains are compact modular protein-protein interaction domains comprised of less than 100 amino acids: they mediate homo- and heterodimerization between diverse proteins (30, 52). The MKK5 PB1 domain did indeed bind to ERK5 in addition to binding to the MEKK3 PB1 domain. Recently, the crystal structure of the MKK5 PB1/MEKK3 PB1 domain heterodimer was determined by x-ray crystallography (Protein Data Bank code 2O2V) and the MKK5 PB1 domain binding surface on the MEKK3 PB1 domain was studied by NMR (30). When the heterodimer of the MEKK3 and MKK5 PB1 domains is superimposed on the binary ERK5·PB1-D complex through their common MKK5 PB1 domains (489 atoms, r.m.s.d. of 0.7 Å), the MEKK3 PB1 domain shows no sterical clash with ERK5 in the MEKK3 PB1·MKK5 PB1-D·ERK5 ternary complex model (Fig. 6D). This is in agreement with the results of our protein-protein association studies on this ternary complex. Thus, the MKK5 PB1 domain seems to function as an adaptor: it engages two distinct surfaces to bind MEKK3 and ERK5, and it assembles the MEKK2/3-MKK5-ERK5 module into one signaling competent complex (29, 53).
MKK5 is the specific activator of ERK5, which fulfills non-redundant physiological roles compared with its ERK1/2 paralogs. Present day MAPKs have emerged through whole genome or by individual gene duplication events during evolution (16). After gene duplication events, a new paralog may distinguish itself by forming new connections with proteins from other signaling circuits (54). For example, JNK3 sets itself apart from the other two JNK paralogs by possessing a unique ∼40-amino acid-long N-terminal extension to its kinase domain. This enables JNK3 to be specifically recruited to and to be activated by the β-arrestin2 scaffold complex (55). Interestingly, two cladograms drawn based on available present day MAPK and MKK sequences concur well (Fig. 1A). MKK-MAPK signaling thus serve as an excellent paradigm how protein-protein interaction elements, structurally and functionally unrelated, may come together to determine signaling specificity during the evolution of paralogous signaling enzymes. Our results on the MKK5-ERK5 system particularly demonstrates how a C-terminal extension to the MAPK domain and the acquisition of a dedicated protein-protein interaction tool in the MKK enzyme (i.e. a PB1 domain) may have enabled a new MKK-MAPK pair to distinguish itself from its MKK1/2-ERK1/2 homologs (56). In addition, we also demonstrated that the topography of the MAPK docking groove is different among related present-day MAPKs. Naturally, this also contributes to the differential partner protein preferences of ERK2 and ERK5. The characterized signaling relevant protein-protein interactions of MKK5·ERK5 and MKK1/2·ERK1/2 complexes show how signaling enzymes with common origin could acquire novel protein-protein interaction profiles. This is a pivotal step that ultimately could lead to the emergence of distinct signaling pathways.
Finally, our results provide structural insight into how protein-protein interaction specificity is determined for closely related but functionally distinct MAPK-based systems. This knowledge will be a great asset for interfering with ERK1/2- or ERK5-based pathways in a targeted fashion.
Acknowledgment
We thank András Patthy for excellent quality peptide synthesis.
This work was supported by the Hungarian Scientific Research Fund (OTKA NK81950 and NKTH-OTKA H07A 74216).
The atomic coordinates and structure factors (codes 4IC7 and 4IC8) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- MKK
- MAPK kinase(s)
- MBP
- maltose binding protein
- FP
- fluorescence polarization
- AMP-PNP
- adenylyl-imidodiphosphate
- KD
- kinase domain
- r.m.s.d.
- root mean square deviation
- FL
- full length.
REFERENCES
- 1. Nishimoto S., Nishida E. (2006) MAPK signalling: ERK5 versus ERK1/2. EMBO Rep. 7, 782–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang X., Tournier C. (2006) Regulation of cellular functions by the ERK5 signalling pathway. Cell Signal. 18, 753–760 [DOI] [PubMed] [Google Scholar]
- 3. Drew B. A., Burow M. E., Beckman B. S. (2012) MEK5/ERK5 pathway: the first fifteen years. Biochim. Biophys. Acta 1825, 37–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhou G., Bao Z. Q., Dixon J. E. (1995) Components of a new human protein kinase signal transduction pathway. J. Biol. Chem. 270, 12665–12669 [DOI] [PubMed] [Google Scholar]
- 5. Kato Y., Tapping R. I., Huang S., Watson M. H., Ulevitch R. J., Lee J. D. (1998) Bmk1/Erk5 is required for cell proliferation induced by epidermal growth factor. Nature 395, 713–716 [DOI] [PubMed] [Google Scholar]
- 6. Raman M., Chen W., Cobb M. H. (2007) Differential regulation and properties of MAPKs. Oncogene 26, 3100–3112 [DOI] [PubMed] [Google Scholar]
- 7. Cavanaugh J. E., Ham J., Hetman M., Poser S., Yan C., Xia Z. (2001) Differential regulation of mitogen-activated protein kinases ERK1/2 and ERK5 by neurotrophins, neuronal activity, and cAMP in neurons. J. Neurosci. 21, 434–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu L., Cundiff P., Abel G., Wang Y., Faigle R., Sakagami H., Xu M., Xia Z. (2006) Extracellular signal-regulated kinase (ERK) 5 is necessary and sufficient to specify cortical neuronal fate. Proc. Natl. Acad. Sci. U.S.A. 103, 9697–9702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Regan C. P., Li W., Boucher D. M., Spatz S., Su M. S., Kuida K. (2002) Erk5 null mice display multiple extraembryonic vascular and embryonic cardiovascular defects. Proc. Natl. Acad. Sci. U.S.A. 99, 9248–9253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sohn S. J., Sarvis B. K., Cado D., Winoto A. (2002) ERK5 MAPK regulates embryonic angiogenesis and acts as a hypoxia-sensitive repressor of vascular endothelial growth factor expression. J. Biol. Chem. 277, 43344–43351 [DOI] [PubMed] [Google Scholar]
- 11. Hayashi M., Kim S. W., Imanaka-Yoshida K., Yoshida T., Abel E. D., Eliceiri B., Yang Y., Ulevitch R. J., Lee J. D. (2004) Targeted deletion of BMK1/ERK5 in adult mice perturbs vascular integrity and leads to endothelial failure. J. Clin. Invest. 113, 1138–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pearson G., English J. M., White M. A., Cobb M. H. (2001) ERK5 and ERK2 cooperate to regulate NF-κB and cell transformation. J. Biol. Chem. 276, 7927–7931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Watson F. L., Heerssen H. M., Bhattacharyya A., Klesse L., Lin M. Z., Segal R. A. (2001) Neurotrophins use the Erk5 pathway to mediate a retrograde survival response. Nat. Neurosci. 4, 981–988 [DOI] [PubMed] [Google Scholar]
- 14. Buschbeck M., Ullrich A. (2005) The unique C-terminal tail of the mitogen-activated protein kinase ERK5 regulates its activation and nuclear shuttling. J. Biol. Chem. 280, 2659–2667 [DOI] [PubMed] [Google Scholar]
- 15. Kondoh K., Terasawa K., Morimoto H., Nishida E. (2006) Regulation of nuclear translocation of extracellular signal-regulated kinase 5 by active nuclear import and export mechanisms. Mol. Cell. Biol. 26, 1679–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Caffrey D. R., O'Neill L. A., Shields D. C. (1999) The evolution of the MAP kinase pathways: coduplication of interacting proteins leads to new signaling cascades. J. Mol. Evol. 49, 567–582 [DOI] [PubMed] [Google Scholar]
- 17. Li M., Liu J., Zhang C. (2011) Evolutionary history of the vertebrate mitogen activated protein kinases family. PloS One 6, e26999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kato Y., Kravchenko V. V., Tapping R. I., Han J., Ulevitch R. J., Lee J. D. (1997) BMK1/ERK5 regulates serum-induced early gene expression through transcription factor MEF2C. EMBO J. 16, 7054–7066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu L., Cavanaugh J. E., Wang Y., Sakagami H., Mao Z., Xia Z. (2003) ERK5 activation of MEF2-mediated gene expression plays a critical role in BDNF-promoted survival of developing but not mature cortical neurons. Proc. Natl. Acad. Sci. U.S.A. 100, 8532–8537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Potthoff M. J., Olson E. N. (2007) MEF2: a central regulator of diverse developmental programs. Development 134, 4131–4140 [DOI] [PubMed] [Google Scholar]
- 21. Tanoue T., Adachi M., Moriguchi T., Nishida E. (2000) A conserved docking motif in MAP kinases common to substrates, activators, and regulators. Nat. Cell Biol. 2, 110–116 [DOI] [PubMed] [Google Scholar]
- 22. Bardwell A. J., Frankson E., Bardwell L. (2009) Selectivity of docking sites in MAPK kinases. J. Biol. Chem. 284, 13165–13173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Garai Á., Zeke A., Gógl G., Töriő I., Fördős F., Blankenburg H., Bárkai T., Varga J., Alexa A., Emig D., Albrecht M., Reményi A. (2012) Specificity of linear motifs that bind to a common mitogen-activated protein kinase docking groove. Sci. Signal. 5, ra74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chang C. I., Xu B. E., Akella R., Cobb M. H., Goldsmith E. J. (2002) Crystal structures of MAP kinase p38 complexed to the docking sites on its nuclear substrate MEF2A and activator MKK3b. Mol. Cell 9, 1241–1249 [DOI] [PubMed] [Google Scholar]
- 25. Reményi A., Good M. C., Bhattacharyya R. P., Lim W. A. (2005) The role of docking interactions in mediating signaling input, output, and discrimination in the yeast MAPK network. Mol. Cell 20, 951–962 [DOI] [PubMed] [Google Scholar]
- 26. Gógl G., Törő I., Reményi A. (2012) Protein-peptide complex crystallization: a case study on the ERK2 mitogen-activated protein kinase. Acta Crystallogr. D Biol. Crystallogr., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nakamura K., Johnson G. L. (2003) PB1 domains of MEKK2 and MEKK3 interact with the MEK5 PB1 domain for activation of the ERK5 pathway. J. Biol. Chem. 278, 36989–36992 [DOI] [PubMed] [Google Scholar]
- 28. Nakamura K., Johnson G. L. (2007) Noncanonical function of MEKK2 and MEK5 PB1 domains for coordinated extracellular signal-regulated kinase 5 and c-Jun N-terminal kinase signaling. Mol. Cell. Biol. 27, 4566–4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nakamura K., Uhlik M. T., Johnson N. L., Hahn K. M., Johnson G. L. (2006) PB1 domain-dependent signaling complex is required for extracellular signal-regulated kinase 5 activation. Mol. Cell. Biol. 26, 2065–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hu Q., Shen W., Huang H., Liu J., Zhang J., Huang X., Wu J., Shi Y. (2007) Insight into the binding properties of MEKK3 PB1 to MEK5 PB1 from its solution structure. Biochemistry 46, 13478–13489 [DOI] [PubMed] [Google Scholar]
- 31. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Barsyte-Lovejoy D., Galanis A., Clancy A., Sharrocks A. D. (2004) ERK5 is targeted to myocyte enhancer factor 2A (MEF2A) through a MAPK docking motif. Biochem. J. 381, 693–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ranganathan A., Pearson G. W., Chrestensen C. A., Sturgill T. W., Cobb M. H. (2006) The MAP kinase ERK5 binds to and phosphorylates p90 RSK. Arch Biochem. Biophys. 449, 8–16 [DOI] [PubMed] [Google Scholar]
- 39. Zhang F., Strand A., Robbins D., Cobb M. H., Goldsmith E. J. (1994) Atomic structure of the MAP kinase ERK2 at 2.3 A resolution. Nature 367, 704–711 [DOI] [PubMed] [Google Scholar]
- 40. Zhou T., Sun L., Humphreys J., Goldsmith E. J. (2006) Docking interactions induce exposure of activation loop in the MAP kinase ERK2. Structure 14, 1011–1019 [DOI] [PubMed] [Google Scholar]
- 41. Liu S., Sun J. P., Zhou B., Zhang Z. Y. (2006) Structural basis of docking interactions between ERK2 and MAP kinase phosphatase 3. Proc. Natl. Acad. Sci. U.S.A. 103, 5326–5331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ma W., Shang Y., Wei Z., Wen W., Wang W., Zhang M. (2010) Phosphorylation of DCC by ERK2 is facilitated by direct docking of the receptor P1 domain to the kinase. Structure 18, 1502–1511 [DOI] [PubMed] [Google Scholar]
- 43. Jacobs D., Glossip D., Xing H., Muslin A. J., Kornfeld K. (1999) Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes Dev. 13, 163–175 [PMC free article] [PubMed] [Google Scholar]
- 44. Bhattacharyya R. P., Reményi A., Good M. C., Bashor C. J., Falick A. M., Lim W. A. (2006) The Ste5 scaffold allosterically modulates signaling output of the yeast mating pathway. Science 311, 822–826 [DOI] [PubMed] [Google Scholar]
- 45. Murakami Y., Tatebayashi K., Saito H. (2008) Two adjacent docking sites in the yeast Hog1 mitogen-activated protein (MAP) kinase differentially interact with the Pbs2 MAP kinase kinase and the Ptp2 protein tyrosine phosphatase. Mol. Cell. Biol. 28, 2481–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Aberg E., Torgersen K. M., Johansen B., Keyse S. M., Perander M., Seternes O. M. (2009) Docking of PRAK/MK5 to the atypical MAPKs ERK3 and ERK4 defines a novel MAPK interaction motif. J. Biol. Chem. 284, 19392–19401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Piserchio A., Warthaka M., Devkota A. K., Kaoud T. S., Lee S., Abramczyk O., Ren P., Dalby K. N., Ghose R. (2011) Solution NMR insights into docking interactions involving inactive ERK2. Biochemistry 50, 3660–3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gupta S., Campbell D., Dérijard B., Davis R. J. (1995) Transcription factor ATF2 regulation by the JNK signal transduction pathway. Science 267, 389–393 [DOI] [PubMed] [Google Scholar]
- 49. Sharrocks A. D., Yang S. H., Galanis A. (2000) Docking domains and substrate-specificity determination for MAP kinases. Trends Biochem. Sci. 25, 448–453 [DOI] [PubMed] [Google Scholar]
- 50. Zarrinpar A., Park S. H., Lim W. A. (2003) Optimization of specificity in a cellular protein interaction network by negative selection. Nature 426, 676–680 [DOI] [PubMed] [Google Scholar]
- 51. Alexander J., Lim D., Joughin B. A., Hegemann B., Hutchins J. R., Ehrenberger T., Ivins F., Sessa F., Hudecz O., Nigg E. A., Fry A. M., Musacchio A., Stukenberg P. T., Mechtler K., Peters J. M., Smerdon S. J., Yaffe M. B. (2011) Spatial exclusivity combined with positive and negative selection of phosphorylation motifs is the basis for context-dependent mitotic signaling. Sci. Signal. 4, ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Moscat J., Diaz-Meco M. T., Albert A., Campuzano S. (2006) Cell signaling and function organized by PB1 domain interactions. Mol. Cell 23, 631–640 [DOI] [PubMed] [Google Scholar]
- 53. Zeke A., Lukács M., Lim W. A., Reményi A. (2009) Scaffolds: interaction platforms for cellular signalling circuits. Trends Cell Biol. 19, 364–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bhattacharyya R. P., Reményi A., Yeh B. J., Lim W. A. (2006) Domains, motifs, and scaffolds: the role of modular interactions in the evolution and wiring of cell signaling circuits. Ann Rev. Biochem. 75, 655–680 [DOI] [PubMed] [Google Scholar]
- 55. Guo C., Whitmarsh A. J. (2008) The beta-arrestin-2 scaffold protein promotes c-Jun N-terminal kinase-3 activation by binding to its nonconserved N terminus. J. Biol. Chem. 283, 15903–15911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Won A. P., Garbarino J. E., Lim W. A. (2011) Recruitment interactions can override catalytic interactions in determining the functional identity of a protein kinase. Proc. Natl. Acad. Sci. U.S.A. 108, 9809–9814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Milanesi L., Petrillo M., Sepe L., Boccia A., D'Agostino N., Passamano M., Di Nardo S., Tasco G., Casadio R., Paolella G. (2005) Systematic analysis of human kinase genes: a large number of genes and alternative splicing events result in functional and structural diversity. BMC Bioinformatics 6, S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Coulombe P., Meloche S. (2007) Atypical mitogen-activated protein kinases: structure, regulation and functions. Biochim. Biophys. Acta 1773, 1376–1387 [DOI] [PubMed] [Google Scholar]